Abstract
Background:
Over the past five years, interest in and use of DNA array technology has increased dramatically, and there has been a surge in demand for different types of arrays. Although manufacturers offer a number of pre-made arrays, these are generally of utilitarian design and often cannot accommodate the specific requirements of focused research, such as a particular set of genes from a particular tissue. We found that suppliers did not provide an array to suit our particular interest in testicular toxicology, and therefore elected to design and produce our own.
Results:
We describe the procedures used by members of the US Environmental Protection Agency MicroArray Consortium (EPAMAC) to produce a mouse testis expression array on both filter and glass-slide formats. The approaches used in the selection and assembly of a pertinent, nonredundant list of testis-expressed genes are detailed. Hybridization of the filter arrays with normal and bromochloroacetic acid-treated mouse testicular RNAs demonstrated that all the selected genes on the array were expressed in mouse testes.
Conclusion:
We have assembled two lists of mouse (950) and human (960) genes expressed in the mouse and/or human adult testis, essentially all of which are available as sequence-verified clones from public sources. Of these, 764 are homologous and will therefore enable close comparison of gene expression between murine models and human clinical testicular samples.
Background
DNA arrays, variously called microarrays, complementary DNA (cDNA) arrays, gene arrays and gene expression arrays, have been widely heralded and are becoming increasingly integrated into the current research and future plans of many laboratories [1]. The main utility of DNA arrays lies in their ability to report the expression level of thousands of genes simultaneously, although other uses are being continually introduced. There are an increasing number of different commercially available DNA array formats. These include: the glass-slide based oligonucleotide array system [2,3,4] developed by Affymetrix; the glass-slide-based cDNA clone Gene Expression Microarray (GEM) line from Incyte Genomics (IGI); the filter and glass-slide-based 'Atlas' arrays developed by Clontech Laboratories, composed of selected regions of PCR-amplified gene sequences; filter-based GeneFilters from Research Genetics (RGI) which, like IGI's GEMs, utilize partial or full-length cDNA clones. Many research institutions are also investing in core facilities that, after the fashion of the Brown laboratory [5,6], are producing their own filter and/or glass arrays from PCR products of clones from the Integrated Molecular Analysis of Genomes and their Expression (IMAGE) consortium, or other clones. A detailed explanation of the differences between these various array formats and other aspects of current DNA array technology can be found in the review by Rockett and Dix [7].
The efficiency, sensitivity and reproducibility of commercially available arrays is good and improving, but they are limited in three main areas: their cost is still somewhat prohibitive for some labs; the number of species for which arrays are available is small; the genes available are limited in scope and number. Although many companies offer a custom service for producing arrays, the process is expensive and cumbersome, and does not lend itself well to the commonly changing needs of the researcher. Most commercially available off-the-shelf arrays contain a cosmopolitan series of well characterized genes expressed across many cell types. As most researchers focus on one or two tissues only, this global approach to array production is somewhat wasteful. In many cases, it would be far more useful to report on the expression of only those genes that are part of the transcriptome of the tissue of interest. Thus, a group interested in the liver may be better served by a chip containing only those genes expressed in the liver, and a group, such as our own, interested in the testis, should find more value in a testis expression chip. We are interested in the effects of environmentally induced male infertility, with a focus on using components of the stress response as a means to indicate, qualitate or even quantitate the degree of harmful exposure. It may also be possible to use this gene expression data to elucidate the mechanisms of action of toxic agents and identify the causative agent, thus facilitating better preventive or palliative action. The development of an in-house array is a complex process, however, which must overcome a number of logistical problems.
We describe here our approach to developing a testis expression array, including the gene selection and printing procedures. We have assembled two lists of genes (one of 950 mouse genes, and one of 960 human genes) that are expressed in either mouse testis or human testis or both. These two distinct sets of testis-expressed genes, containing 764 homologs, have enabled us to focus our resources more efficiently and begin to develop a picture of gene expression patterns in environmentally and genetically challenged mouse models. It is anticipated that the production of human testis expression arrays will follow, thus permitting the direct comparison of gene expression networks in mouse models and human clinical samples. This may aid in the elucidation of the molecular mechanisms underlying genetically and environmentally induced male infertility.
Results and discussion
The continuing development of miniaturization technologies and the completion of genome sequencing projects will soon make it possible to fit an entire mammalian genome onto one array. In most cases, however, researchers do not require such a vast amount of information, and arrays containing select tissue transcriptomes will be more cost-effective to produce and the data from them easier to manipulate. Commercial availability of such arrays is currently very limited, so there is often a need for in-house development and production where they are required. Our work is focused on analyzing the effects of potentially hazardous environmental agents, such as arsenic [8] and water disinfectant by-products [9], on embryonic development and the male reproductive tract. One approach to fulfilling this mission is to use genomic technology, particularly DNA arrays, to examine gene expression in model animal systems following treatment with selected agents. As a means to this end, we developed and produced an in-house mouse cDNA array, in both filter (TestisFilter) and glass-slide (TestisSlide) format, for the specific analysis of testis-expressed genes. This was a time-consuming and complex process requiring the overcoming of a number of logistical problems. We thus describe here the decision processes and procedures used to develop and produce the TestisFilter and TestisSlide arrays (see Figure 1 for overview of process).
Figure 1.
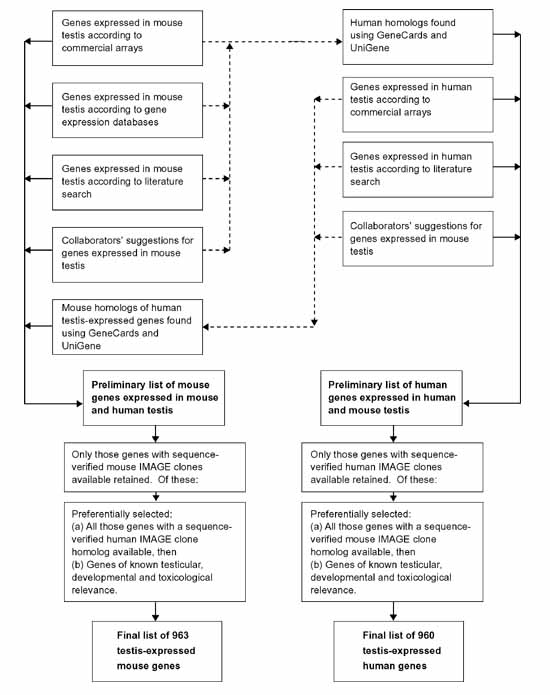
Flow chart of the procedure used to compile mouse and human testis expression array gene lists.
The first step was to assemble two lists of 950 genes that represent part of the adult C57BL/6 mouse and adult human testis transcriptomes. The first decision in planning a custom array is whether or not to include clones of expressed sequence tags (ESTs) in addition to known genes. Where resources are limited, it is prudent to use only named genes that have been previously characterized. It may be possible to obtain several thousand such clones, depending on the species of interest. ESTs from tissue-specific cDNA libraries can be included if resources or circumstances dictate, or if the researcher is more interested in finding new genes that have not been previously associated with a particular model. In most instances we did not use ESTs, as there are a large number of clones of named mouse genes available from various suppliers. In some cases, however, highly relevant genes were only available as EST sequences. Where the choice was available, mouse ESTs described as 'highly similar' to the gene of another species were used (26 in the mouse list, 21 in the human list). 'Moderately similar' EST sequences were used as sparingly as possible (seven in the mouse list, once in the human list), and 'weakly similar' EST sequences were included only as a last resort (zero in mouse, two in human).
Our approach to developing a list of testis-expressed genes was to gather and integrate gene expression data from a large number of sources, including several different arrays. To identify some of the many genes expressed in the normal adult 'mammalian' testis, we hybridized adult mouse and human testicular RNA to a variety of different commercially available DNA arrays that, in total, contained several thousands of different named genes. In the case of the mouse, we also used RNA from heat-shocked testes in order to identify genes induced by the stress response. Pooling of mouse RNA for this experimental step is important, as previous experience has shown that there are differences in gene expression even between individuals of the same strain. The larger the number of individuals used to derive the pool, the more representative the final list of expressed genes is likely to be. Public databases and the literature were also used to identify genes expressed in adult mouse and human testes. In this case, genes expressed in all stages of testicular development and diseases were used, with a particular emphasis on toxicological and stress responses.
A combined species approach was used in all cases, whereby genes present on the arrays of, or literature for, one species, but not the other, were used to search for gene homologs. The identification of homologs is, predictably, time-consuming, but a database such as GeneCards [10,11] can facilitate the process. Of course, gene homologs are not always known or available. Furthermore, the expression of a gene in one species of mammal does not necessarily mean it will be expressed in another. Since the developmental and physiological characteristics of the male reproductive tract are highly conserved across mammalian species, however, this was deemed an acceptable procedure for determining testis-expressed genes. Indeed, in many instances, we were able to select genes that have known homologs in humans and mice. This will, in future, allow us to make direct comparisons between the gene expression patterns seen in mouse models and human clinical samples.
One of the most difficult and time-consuming aspects of assembling multiple lists of expressed genes in this manner is the efficient elimination of duplicate genes from the master list. This procedure is complicated by the fact that most genes have more than one name or designation both within and between species (Table 1, available with the complete version of this article online). In consolidating data from numerous disparate sources, as in this case, a careful search of a database such as GeneCards is a pertinent step for identifying and removing redundant gene names. A second confounding factor is the fact that each selected gene must be individually assessed for the following factors.
Availability of a publicly accessible clone
Although suppliers such as Clontech, IGI and the IMAGE consortium [12] provide public access to a vast number of full-length and partial clones from a number of species, not all genes are represented by a publicly accessible clone of the gene or gene fragment. This problem is most often encountered when newly discovered or characterized genes have been selected from the literature.
Availability of a sequence-verified clone
It is important to select sequence-verified clones where possible to ensure that correct sequences are obtained. For example, clones provided by IMAGE (by far the largest selection of publicly available clones) can be mislabeled (between 7 and 13% [13]).
3'-end clones
When the choice is available (with UniGene database [14] verification), selection of a 3'-end clone is desirable, as the inclusion of the 3' UTR will, in most cases, enhance the specificity of the gene fragment for its gene in the hybridization process.
Length of clone
Selected clones should be checked to confirm that they are not so small that they allow a high degree of non-specific hybridization, and not so long as to introduce concerns about being able to amplify the insert by PCR for printing on the array. Gene fragments in the range of 200-1,000 base pairs (bp) are optimal.
It should also be recognized that IMAGE and other clones are frequently reassigned to different gene clusters in the UniGene database, and that UniGene numbers and the genes/clones they represent can be removed at any point as new information becomes available. Consequently, the tracking and naming of clones is of central importance in projects such as this, and gene lists should be constructed to provide as much detail as possible. Thus, it is prudent to include for each gene such details as: the UniGene number (and note the build from which it was taken); the IMAGE or other clone number; the GenBank accession number [15]; the SwissProt number [16]; the source of selection, which may be of value if questions subsequently arise concerning specific clones.
Selected details (UniGene number, IMAGE number and gene name) of the final gene lists for our mouse and human DNA arrays are shown in Tables 2 and 3, respectively, available with the complete version of this article online. The 764 homologous clones are shown in Table 1, available with the complete version of this article, online. More detailed lists have been posted on the US Environmental Protection Agency MicroArray Consortium (EPAMAC) website [17], where they are freely accessible as a public resource.
Assembling the list of genes to be included in a tissue-specific array is perhaps the most challenging part of the process. The subsequent acquisition of clones can be as simple as ordering the entire set from a sole supplier. In this case, the assembly of the gene list will be limited by the supplier's library. However, distributors of IMAGE clones clearly have a very large selection and are continuously sequence-verifying and purging this vast resource of phage contamination. Proprietary clones are also available from companies such as Clontech and IGI. Although these tend to be a little more costly than IMAGE clones, they include genes that are not available through IMAGE.
To prepare DNA arrays, clones of the genes to be printed are grown, the plasmids containing the gene fragments isolated, and the gene fragments amplified using PCR (see [18] for detailed protocols). This can be done in-house or, as in this case, provided as a contract service by the clone supplier. In preparing PCR products of a clone set for printing, it is prudent to note that clone growth may not occur in some cases. Six of our 963 requested clones (0.6%) were scored as 'no grows'. However, spots representing two of these clones produced positive results in our TestisFilter hybridizations. This suggests that, in these cases, limited bacterial growth did actually occur, permitting the isolation of small amounts of plasmid and subsequent production of small amounts of PCR product, whose levels were below the detection limit of the gels used to check them. Hybridization signals from such spots should be treated with caution, as it is not known whether enough PCR product was deposited onto the membrane to be in excess of the target cDNA in the test sample. In addition, PCR amplification of IMAGE (and other) clones can be expected to fail at a small rate. Of the 963 original amplifications (carried out by RGI), 45 (4.7%) did fail and had to be repeated under more carefully controlled conditions to obtain product. Furthermore, a small number of PCR products can be expected to produce multiple bands or smearing. Such results are most likely the result of mispriming events in the PCR reaction, although it is possible that the original stocks may be contaminated with one or more additional clones. In fact we observed that 5.5% (53/963) of our PCR products contained a double PCR band, 3.0% (29/963) contained a triple band, 0.3% (3/963) contained a quadruple band and 0.5% (5/963) produced a smear. Despite these anomalous amplifications, we printed from all wells without re-racking to eliminate the anomalous PCR products. Although this saved time and resources, the caveat is that significant results from all subsequent array hybridizations must be checked against this list of potentially spurious clones. If hybridizations with the spots containing multiple-band PCR products repeatedly give statistically significant changes, it should be possible to isolate and sequence the different PCR products, and use this information for confirmatory analysis, perhaps using reverse transcriptase-polymerase chain reaction (RT-PCR).
Genes from the mouse testis-expressed gene list were procured as both clones and PCR products from a single supplier (RGI). Five additional reactions were added from PCR amplifications of heat-shock protein clones available in our laboratory. This final set of PCR products was prepared for printing. PCR products were then printed as filter arrays by a contractor (Radius Biosciences). Relatively small amounts of DNA were required to produce multiple arrays. We obtained 84 arrays, with each spot printed in duplicate, from approximately 3 μg of PCR product resuspended in 12 μl 3x SSC.
Preliminary hybridizations were carried out on the arrays with adult mouse testicular RNA from C57BL/6 mice (Figure 2a,b). Bromochloroacetic acid (BCA, 216 mg/kg/day) or water was administered daily by gavage for 14 days. RNA extracted from these testes and hybridized against the TestisFilters produced measurable signal in the duplicate spots of virtually all the arrayed genes, confirming the expression of the genes we selected. At the time of writing, there were a small number of genes (14) that did not appear to be expressed in the testis of either this BCA model or a second, metallothionein knockout (129SvPCJ background [19]), model which was also investigated. Four of these genes were 'no grows'. The other ten may have represented genes that were not expressed in the two models we tested, although they could also represent clones that had been UniGene-clustered with the wrong gene. As described earlier, the selection of some of the genes on this mouse testis array was based on results with human expression arrays, showing that our cross-species approach to gene selection is a viable means of identifying candidate genes for such tissue-specific arrays.
Figure 2.
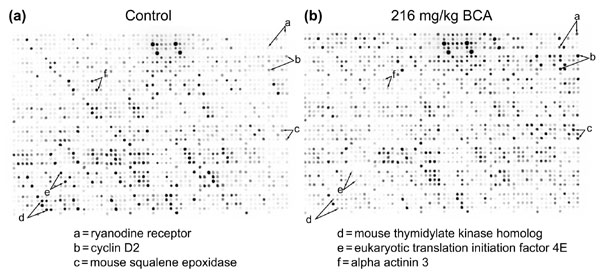
Hybridization of mouse TestisFilter against mouse testicular RNA. Eight-week-old male mice were dosed for 14 days via gavage with (a) water (control) or (b) 216 mg/kg acid BCA. Four hours after the final dose, total RNA was extracted from half a testis from each animal, and 1 μg was used to make [33P]CTP-labeled cDNA using reverse transcription. The labeled cDNA population was hybridized against the filter overnight at 42°C. After washing, the image was captured on a Kodak phosphorimaging screen using three days of exposure and visualized using a BioRad FX phosphorimager. Selected genes with demonstrably altered expression in the BCA-treated versus control testis are highlighted (a-f).
We also produced slide-based DNA arrays of our testis-expressed clone set at the Center for Molecular Medicine and Genetics (Wayne State University, Detroit, MI). A picture of one such array, stained with POPO-3 iodide (Molecular Probes), is shown in Figure 3. Experimental samples had not been applied to this array at the time of writing, but the POPO-3 stain clearly shows the quality of printing and the configuration of the sub-grid of spots within the array. The TestisSlides were printed on bar-coded slides, the numbers being visible to both the naked eye and in the fluorescence-scanned image. The use of such slides is highly recommended, as it ensures that hybridizations with different RNA populations can be identified quickly and linked unambiguously to subsequent image and data analysis.
Figure 3.
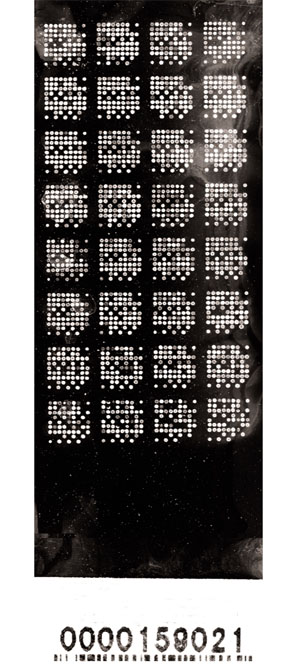
POPO-3 iodide staining of mouse TestisSlide array. POPO-3 iodide was diluted 1:10,000 in water. The TestisSlide was wetted with water and incubated in the POPO-3 solution with gentle agitation for 30 min at room temperature. Excess stain was washed away for 10 min under a gentle flow of reverse osmosis-distilled water, and the image captured using a ScanArray 4000. The number at the bottom of the TestisSlide is a unique identifier, in numeric and barcode form, for this particular slide and can be used to quickly link the image to the specific experimental conditions used to generate it.
Data acquisition from both the TestisFilter and TestisSlide requires that the identity of the cDNA spots be related to their respective positions on filter or glass slide. This information can be embedded in appropriate Excel spreadsheets. For the TestisFilters, we are currently testing two approaches for acquiring intensify values from digital phosphorimages of the hybridized arrays. In the first approach, we designed templates matching the arrangement of cDNA spots on the filters using ImageQuant for Macintosh (Molecular Dynamics). These templates were used for volume integration as a measure of signal intensify. Our second approach utilizes Phoretix Array v2.0 software (Nonlinear USA) to automatically generate an acquisition template and perform volume integrations for the cDNA spots. Both these software packages accommodate background determination and subtraction, and the Nonlinear software also supports data normalization. TestisSlides are being imaged using a ScanArray 4000 confocal laser scanner (Packard Biochip Technologies) and data acquisition accomplished using QuantArray software from the same vendor.
Although it is widely acknowledged that DNA arrays are most powerful when they contain as many different gene probes as possible, the cost of purchasing or producing such large arrays is prohibitive for most researchers. Thus, the primary advantage of developing a relatively small tissue-specific array is its economy, whereby the researcher is not obliged to obtain prefabricated arrays or libraries containing genes that are uninformative for their favorite tissue and/or genomic interests. Another advantage is that the clone set is also retained by the researcher and can be reused at will to prepare nucleic acid for further array printing. New clones can also be added to the set as they become available. Such a set also allows the researcher to be more or less focused in their choice of genes for expression analysis, permitting use of the full set or subsets. This approach enables the researcher to select genes that are of specific, probable or possible interest in a particular context, and thus to tailor the final array exclusively to the model(s) of interest. For example, the researcher may wish to examine the effects of a chemical treatment on a particular gene pathway (for example, glycolysis) or family of genes (for example, heat-shock proteins, cytochrome P450s) in a particular tissue. Relatively small targeted arrays can be used in this context to provide a focused platform to generate leads for the new kind of data-driven research that is gradually supplanting hypothesis-driven experimentation in genomics.
Despite these advantages, it should nevertheless be recognized that, like commercially available arrays, the final list of genes selected for this array is arbitrary in that they were selected on the basis of the somewhat restrictive combination of individual judgment and the limited availability of sequence-verified clones, and therefore may not reflect a priori the most versatile or optimal set of genes for such an array. For example, at the time of purchase we were unable to acquire sequence-verified IMAGE clones of Prm1, Dmc1, Msh4 and a number of important cyclins, all of which have key roles in testicular function. Fortunately, the number of available sequence-verified IMAGE clones is increasing daily, as is the understanding of molecular mechanisms underlying cell function. Thus, arrays developed in future will benefit from this increased resource and knowledge base, and will therefore be less arbitrary than current versions, permitting an even more efficient use of resources.
In an ideal situation, one would perhaps like to array whole tissue transcriptomes so that all possible transcription profiles can be accounted for. It should be noted, however, that a complete 'normal adult mouse testis' transcriptome (or that of any other tissue or organ) will be extremely difficult to assemble, given that expression differences undoubtedly exist between strains and even between individuals of the same inbred strain. Migrating cells such as immune cells can also complicate the issue. Completion of the sequencing of mouse and other laboratory animal genomes will facilitate the production of more directed arrays, but until the full transcriptional and post-transcriptional (that is, alternative splicing) repertoire of every cell type in every condition in every strain at every age is mapped out, developing a fully representative array is unrealistic. Nevertheless, we have described the first steps, and the caveats to consider, for those wishing to begin developing the kind of tissue- or cell-specific arrays that will, we believe, be used with increasing frequency by the array community.
Materials and methods
Obtaining a mouse testis transcriptome
Animals
Male C57BL/6 mice were obtained from Charles River Breeding Laboratories at 8 weeks old, and maintained in a temperature- and humidity-controlled room on a 12 h light/dark cycle. The animals were housed singly in polycarbonate cages with pine shavings bedding and free access to food and water.
Heat-shock treatment
At 10 weeks of age, two animals were sedated by an intraperitoneal injection of 100 μl per 10 g body weight of 10% ketamine (Fort Dodge Laboratories) in phosphate-buffered saline (pH 7.4). The lower half of the torso of each animal was submerged in a 43°C water bath for 20 min, after which the animals were dried off and returned to their cages. The testes from these animals were harvested 4 h later. The testes of two untreated, adult male C57BL/6 mice were harvested concurrently.
Arrays
Total RNA was extracted individually from the testes of the four mice using two extractions through Tri Reagent (Sigma). The control and heat-shocked total RNA samples were then pooled and passed twice through the Oligotex™ mRNA Purification System (Qiagen) to produce poly(A) RNA. One microgram each of normal and heat-shocked testicular poly(A) RNA was submitted to Genome Systems (now part of IGI) for contract hybridization against their mouse Gene Expression Microarray I (GEM 1 [20]). Hybridization of the samples and analysis of the gene expression changes was carried out by GSI staff and the data returned electronically. All named expressed genes were added to the mouse testis transcriptome gene list. The same RNA samples were used to probe Atlas mouse cDNA expression array (588 genes) and Stress/Tox array (149 genes) (Clontech Laboratories), 32P-labeled cDNAs were produced from 2 μg of each pooled RNA, using the reagents supplied with the membrane array kits. Hybridization to the membrane arrays was carried out according to the manufacturer's instructions. The control and heat-shock RNA pools were each hybridized against two different membranes of each membrane type. During exposures, the arrays were kept moist with 2x SSC saturated filter paper wrapped in plastic wrap. Images were developed using a Phosphorlmager (Molecular Dynamics). Images were analyzed using Atlas Image v1.01 (Clontech). Genes whose adjusted signal intensity (average intensity minus background) were not at least twice their respective background value on both membranes were not considered genuine signals and discarded, as were those which were expressed on only one membrane. The remaining 'expressed' genes were added to the mouse testis transcriptome gene list.
Homolog search
In addition to the mouse array expression data, a search for mouse homologs of genes expressed in human testes (according to the Clontech and RGI arrays), which were not represented in the mouse arrays, was carried out using UniGene and/or GeneCards. Genes from successful searches were added to the mouse testis transcriptome gene list.
Database search for testicular transcriptome genes
The NCBI UniGene, Jackson Laboratories Mouse Gene Expression (GXD), and National Library of Medicine (MEDLINE) databases were searched to identify genes expressed in mouse testis [21,22]. Genes that were not already present in the mouse testis transcriptome gene list were added.
Literature search for testicular transcriptome genes
An extensive literature survey was conducted to find murine testis-expressed genes. Genes that were not already present in the mouse testis transcriptome gene list were added.
Collaborator contributions for testicular transcriptome genes
EPAMAC collaborators contributed the names of murine testis-expressed genes not present on the master list.
Obtaining a human testis transcriptome
Genes expressed in human testis
Adult human testicular total RNA was purchased from Clontech. This pooled RNA originated from samples taken from 19 Caucasian males (trauma victims) ranging from 19 to 54 years of age. Two types of Atlas human filter arrays were used: the 588-gene cDNA Expression v1.1 array and the 234-gene Stress/Toxicology array. Two micrograms of total RNA was reverse transcribed with 32P-dATP, and the labeled fractions collected using ChromaSpin (human) columns. One microgram of RNA was also hybridized against 5,184-element human GeneFilter membrane arrays (RGI; GeneFilters I to IV, and Named-Genes) following purification of the [33P]dCTP-labeled probe with BioSpin6 columns (BioRad). Hybridization to all the membrane arrays was carried out according to the manufacturer's instructions. After washing, the arrays were exposed to phosphorimaging screens for 1-7 days and images developed using a Phosphorlmager (Molecular Dynamics) or a Molecular Imager FX (BioRad). Clontech array images were analyzed using Atlas Image v1.01. RGI array images were analyzed using Pathways v.2.01 software, and all expressed genes added to the human testis transcriptome gene list.
Homolog search
In addition to the human array expression data, a search for human homologs of genes expressed in mouse testes (according to the Clontech and IGI arrays and the database search), which were not represented in the human arrays, was carried out using UniGene and/or GeneCards. Genes from successful searches were added to the human testis transcriptome gene list.
Literature search for testicular transcriptome genes
An extensive literature survey was conducted to find human testis-expressed genes. Genes that were not already present in the human testis transcriptome gene list were added.
Collaborator contributions for testicular transcriptome genes
EPAMAC collaborators contributed the names of human testis-expressed genes not present on the master list.
Assembly of a master mouse-human testis transcriptome gene list
The mouse and human testis transcriptome gene lists were brought together for comparison. Paring of the lists down to the final 960 genes was carried out sequentially according to the following criteria: all genes which did not have a sequence-verified mouse IMAGE clone available were removed (assessed by checking against RGI inventory of svIMAGE clones);
All genes present in both lists were retained; all genes of specific interest to ourselves and our collaborators were retained, as were those with proven or suspected toxicological relevance; genes shown to be expressed in the array experiments were selectively retained over those obtained from the literature, which were in turn selectively retained over those obtained from database searching, which were in turn selectively retained over those obtained from homolog searches.
In this way we were able to produce two final master lists of 960 testis-expressed genes for both human and mouse, all of which, except five, were obtainable from a single source (RGI).
Procurement of mouse clones and PCR products
Sequence-verified mouse IMAGE clones of the 963 genes on the final testicular transcriptome gene list were obtained from RGI, along with PCR products of the same genes containing 3-5 μg DNA per gene. Owing to undetected IMAGE or Unigene number duplications on the submitted list of 955 genes, only 945 unique clones were received. PCR products of five heat-shock proteins (HSP70-1, HSP70-2, HSP70-3, Hsc70 and HSC70t) were produced in-house and added to the library, to produce a final total of 950 unique gene PCR products.
Array printing
The PCR products were precipitated, washed, dried and resuspended in 12 μl 3x SSC in V-bottomed 96-well microtiter plates. These samples were used by Radius BioSciences to print 84 12 cm × 8 cm filters. Each gene was printed in duplicate on each filter. Slide-based arrays were prepared by the array facility at the Center for Molecular Medicine and Genetics (Wayne State University) directly from the clone set. Arrays were printed using a Flexys robot (Genomic Solutions) on a 9 × 9 double-spotted grid with 400 μm spacing between spots. Bar-coded CMT-GAPS (gamma amino propyl silane) coated slides (Corning) were used.
Hybridization of TestisFilter with adult mouse RNA
Eight-week-old male C57BL/6 mice were dosed via gavage with either water or 216 mg/kg BCA for 14 days. Four hours after the final dose, animals were sacrificed and testes harvested. Total RNA was extracted from half a testis by homogenization in Tri Reagent (Sigma) and treated with two units of DNase (Ambion). One microgram of the total RNA from a single animal was primed with oligo dT and incubated at 70°C for 10 min followed by a brief chill on ice. Six microliters of 5x first-strand buffer (RGI) (1 μl 100 mM DTT, 1.5 μl dNTPs (20 mM), 1.5 μl reverse transcriptase (Superscript II, Gibco BRL), 10 μl [33P]dCTP (ICN)) were then added to the primed RNA. Elongation was carried out at 37°C for 90 min, after which 70 μl water was added. Unincorporated isotope was separated using BioSpin6 columns (BioRad). Filters were prehybridized in a solution containing 5 ml Microhyb (RGI), 5 μl mouse Cot-1 DNA and 5 μl poly(A) for 2 h at 42°C. The probe was added and incubated overnight at 42°C. Filters were washed twice with 2x SSC, 1% SDS at 50°C for 30 min, and once with 0.5x SSC, 1% SDS at room temperature for 30 min. The filters were then exposed to a Kodak phosphorimaging screen for three days and visualized with a Molecular Imager FX (BioRad).
Data acquisition from TestisFilter and TestisSlide
An ImageQuant for Macintosh (Molecular Dynamics) template matching the arrangement of cDNA spots on the TestisFilter was used for volume integration as a measure of signal intensity. Alternatively, Phoretix Array v2.0 automatically generated an acquisition template and performed volume integrations. TestisSlides are imaged using a ScanArray 4000 confocal laser scanner (Packard Biochip Technologies) and data acquired with QuantArray software from the same vendor.
Additional data files
The following additional data files are included with the online version of this article: Table 1, The 764 homologous mouse and human genes present in testis-expression gene lists; Table 2, 950 testis-expressed genes included in mouse TestisFilter and TestisSlide arrays; and Table 3, 960 human testis-expressed genes selected for producing a human TestisFilter and TestisSlide.
Supplementary Material
The 764 homologous mouse and human genes present in testis-expression gene lists.
The 950 testis-expressed genes included in mouse TestisFilter and TestisSlide arrays.
The 960 human testis-expressed genes selected for producing a human TestisFilter and TestisSlide.
Acknowledgments
Acknowledgements
The following members of EPAMAC suggested genes for inclusion in the testis DNA arrays: Sally Darney (US EPA), Sue Fenton (US EPA), Norman Hecht (University of Pennsylvania), Jeff Welch (US EPA) and Tim Zacharewski (Michigan State University). We also thank Sue Fenton (US EPA) and Jie Liu (NIEHS) for scientific review of this manuscript before submission. The information in this document has been funded in part by the US Environmental Protection Agency. It has been subjected to review by the National Health and Environmental Effects Research Laboratory and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
References
- Rockett JC, Dix DJ. Application of DNA arrays to toxicology. Environ Health Perspect. 1999;107:681–686. doi: 10.1289/ehp.99107681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor SPA, Read JL, Pirrung MC, Stryer L, Tsai Lu A, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Morris MS, Chee M, Hubbell E, Kozal MJ, Shah N, Shen N, Yang R, Fodor SPA. Using oligonucleotide probe arrays to access genetic diversity. BioTechniques. 1995;19:442–447. [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- The Brown Lab http://cmgm.Stanford.EDU/pbrown/
- Rockett JC, Dix DJ. DNA arrays: technology, options and toxicological applications. Xenobiotica. 2000;30:155–177. doi: 10.1080/004982500237758. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Garges JB, Hong RL. Inhibition of hsp70-1 and hsp70-3 expression disrupts preimplantation embryogenesis and heightens embryo sensitivity to arsenic. Mol Reprod Dev. 1998;51:373–380. doi: 10.1002/(SICI)1098-2795(199812)51:4<373::AID-MRD3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Luft JC, Garges JB, Rockett JC, Dix DJ. Male reproductive toxicity of bromochloroacetic acid in mice. Biol Reprod. 2000;62 (Suppl 1):1–246. [Google Scholar]
- GeneCards: human genes, maps, proteins and diseases http://www.dkfz-heidelberg.de/GeneCards/
- Nelson M. A one-step source for information about human genes. Genome Biol. 2000;1:reports2049. [Google Scholar]
- The I.M.A.G.E. consortium http://image.llnl.gov/
- I.M.A.G.E. quality control - process and results http://image.llnl.gov/image/qc/bin/display_error_rates#Results
- UniGene Resources http://www.ncbi.nlm.nih.gov/UniGene/
- GenBank Database http://www.psc.edu/general/software/packages/genbank/genbank.html
- Swiss-Prot http://www.ebi.ac.uk/swissprot/
- Environmental Protection Agency MicroArray Consortium http://www.epa.gov/nheerl/epamac/
- Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Earle-Hughes J, Snesrud E, Lee N, Quackenbush J. A concise guide to cDNA microarray analysis. BioTechniques. 2000;29:548–562. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- Masters BA, Kelly EJ, Quaife CJ, Brinster RL, Palmiter RD. Targeted disruption of metallothionein I and II genes increases sensitivity to cadmium. Proc Natl Acad Sci USA. 1994;91:584–588. doi: 10.1073/pnas.91.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse GEM 1 http://www.incyte.com/reagents/gem/products.shtml
- Halgren RG, Fielden MR, Zacharewski TR. Gene expression in the mouse testis: development of a murine testis transcriptome by mining public databases. The Toxicologist. 2000;54:386. [Google Scholar]
- dbTEST - Database of Transcripts Expressed in Spermatogenesis and Testis http://35.8.60.132/dbtest.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 764 homologous mouse and human genes present in testis-expression gene lists.
The 950 testis-expressed genes included in mouse TestisFilter and TestisSlide arrays.
The 960 human testis-expressed genes selected for producing a human TestisFilter and TestisSlide.


