Abstract
Erwin Schrödinger pointed out in his 1944 book “What is Life” that one defining attribute of biological systems seems to be their tendency to generate order from disorder defying the second law of thermodynamics. Almost parallel to his findings, the science of complex systems was founded based on observations on physical and chemical systems showing that inanimate matter can exhibit complex structures although their interacting parts follow simple rules. This is explained by a process known as self-organization and it is now widely accepted that multi-cellular biological organisms are themselves self-organizing complex systems in which the relations among their parts are dynamic, contextual and interdependent. In order to fully understand such systems, we are required to computationally and mathematically model their interactions as promulgated in systems biology. The preponderance of network models in the practice of systems biology inspired by a reductionist, bottom-up view, seems to neglect, however, the importance of bidirectional interactions across spatial scales and domains. This approach introduces a shortcoming that may hinder research on emergent phenomena such as those of tissue morphogenesis and related diseases, such as cancer. Another hindrance of current modeling attempts is that those systems operate in a parameter space that seems far removed from biological reality. This misperception calls for more tightly coupled mathematical and computational models to biological experiments by creating and designing biological model systems that are accessible to a wide range of experimental manipulations. In this way, a comprehensive understanding of fundamental processes in normal development or of aberrations, like cancer, will be generated.
Keywords: reductionism, emergentism, systems biology, self-organization, agent-based modeling, tissue morphogenesis, early carcinogenesis
Introduction
Fifty years ago at the dawn of the molecular biology revolution, unprecedented enthusiasm was generated by the idea that biology was finally reduced to chemistry and consequently, the proposed way to understand organisms was to study them from the bottom up. Central to this view was genetic determinism, i.e. the perception that the organism was determined by a genetic program. The origin of systems biology, in contrast, attributed to Ludwig von Bertalanffy, a biologist and philosopher, and Paul Alfred Weiss, a biologist, emphasized an organicist view where both bottom-up and top-down causation are considered. These two opposed views are represented by two discrete approaches in a new version of the systems biology discipline. O’Malley and Dupre call the genetic approach ‘pragmatic systems biology,’ which is centered around large-scale molecular interactions, such as gene networks, while the organicist approach, called ‘systems-theoretic biology’, is centered on system principles [OS05]. The differences between both approaches are not technical but rather philosophical, given that both are committed to mathematical modeling.
Philosophy is central to all scientific endeavors, including experimental and systems biology. Although many biologists ignore it, their research is guided by unstated ontological and epistemological stances. The inescapable fact is that, whether biologists like it or not, there are no theory-free data. As put by the philosopher D.C. Dennett: “There is no such thing as philosophy-free science; there is only science whose philosophical baggage is taken on board without examination” [D95, p. 21]. Hence, in this review we will address the philosophical underpinnings of systems biology and of the science of complex systems. The incorporation of network models in the practice of systems biology over the theoretical framework of an interacting bottom-up and top-down system suggest a reductionist slant that hinders research on emergent phenomena. In addition, we are proposing a systems biology approach beyond networks.
Philosophical underpinnings
Reductionism
There are three types of reductionisms, namely, ontological, methodological, and epistemic [S92]. Ontological reductionism, also called physicalism, claims that organisms are made up by molecules and their interactions. This form of reductionism represents the worldview of the practitioners of the other two kinds of reductionism. Epistemic reduction claims that higher order phenomena can be reduced to another more basic level. This line of thought entails a ‘hard-core’ view, whereby biology could be reduced to chemistry and physics and, hence, biology would not be an independent science. According to the Stanford Encyclopedia of Philosophy [SEP], “methodological reduction is the idea that biological systems are most fruitfully investigated at the lowest possible level, and that experimental studies should be aimed at uncovering molecular and biochemical causes.” This is another way of saying that molecular biology can, in principle, fully explain all biological facts. This type of reductionism is also pervasive in other fields of biology where causality is sought using a bottom-up approach. A great number of biologists insist that explanations should always be sought for at the gene and/or gene product level, regardless of the level of organization at which the phenomenon of interest is observed. Thus, genetic reductionism together with its twin, genetic determinism, predicates that everything in biology may be reduced to genes because the genome is the exclusive repository of transmissible information. It then follows that genes are the only units of selection [D76] and development is just the unfolding of a genetic program. In sum, genes would be the building units of the organism and have a privileged metaphysical status (for an extended analysis of this subject, see [GG04]).
A main obstacle to the success of reductionism is the historicity of the organism, i.e., evolution and ontogeny. As Francois Jacob noted, nature is not an engineer, but a tinker — a given molecule is put to different uses [J77]. Evolutionary history confronts us with the fact that these transformations were lost with the extinction of over 95% of the species that once existed. We are then forced to reconstruct this history from the organisms that exist today. This reconstruction is further hampered by evidence pointing to the fact that even in the same organism a protein may have different functions in different cells. For example, lactate dehydrogenase and crystalline are the same molecule; the former is an enzyme in muscle while the latter plays a structural role in the eye’s lens. Beta-catenin is both a transcription factor and a cell-adhesion protein [GS00]. Also, a signal pathway effector may lead to the induction of different gene products and therefore distinct differentiation programs in different cell lineages [BS02]. This lack of a unique correlation between a given protein and its function was addressed by Hull as the problem of “the many and the many” [H74]. In other words, one phenotype may result from several different molecular mechanisms, while a single molecule may be involved in different phenotypes. A clear example of this divergence is polyphenism, where a single genotype produces different phenotypes. These examples of diversity make reduction difficult, if not impossible.
Organicism and emergentism
Organicism is a philosophical stance that, contrary to reductionism, considers both bottom-up and top-down causation. It claims that “…Wholes are so related to their parts that not only does the existence of the whole depend on the orderly cooperation and interdependence of their parts, but the whole exercises a measure of determinative control over its parts” [RB28]. Implicit in this description is the concept of emergence, meaning that at each level of biological organization new properties manifest, which could not have been predicted from the analysis of the lower levels.
The existence of emergent properties is dismissed by physicalists because in their metaphysical stance, the belief on the causal closure of the physical word precludes the existence of emergents. However, as organisms are open systems, external constraints are always operating on them. The internal constraints defining a system are always disturbed by external ones; thus, in order to understand what is going on in a system, we must jump simultaneously to multiple levels on which this system is integrated [S97]. For instance, a cell is integrated in a more complex system, the tissue. Organisms and their cells are ontogenetically linked. For example, a zygote is a cell as well as an organism. It divides, producing more cells, which are organized in a three-dimensional pattern. When gastrulation takes place, cells dramatically change their positions relative to one another followed by the formation of germ layers and a new series of rearrangements, local cell proliferation, cell movement, cell migration and cell specialization resulting in the emergence of tissues and organs. Even in a simpler system, like a muscle cell in the heart, its components are proteins that channel calcium and potassium ions, and they carry currents that change the cell voltage, which in turn changes the ion channels [N06]. Thus, the components alter the behavior of the heart and the heart alters the behavior of the components, yet both components and the heart are integrated in a higher multi-cellular structure, the organism. This means that the working of such systems is never defined by initial internal constraints. When dealing with open systems, new systemic properties emerge as time elapses which can modify the initial properties. Thus what is described at an early time point (T1) is not the essence of the system. In other words, when one states that the biological facts at T1 cause physical facts at a later time point T2, and that they compete with the explanation of these facts as purely physical ones, we are making a mere idealization. At T2, the system is not the same as the one at T1, because it has acquired new properties that were absent at T1. Therefore, a system’s description of natural events is not a complete description of what this system does. Diachronic emergence then means that in specific natural or formal systems the initial relations and properties of elements cannot teach us how they would be applied as the system evolves. Thus, the historical way by which a system of natural events operates is not a consequence of its description. It acts and it produces novelty in the real world (novel qualities and novel structures). In conclusion, emergence has an ontological meaning [B04] and is not a simple epistemic property [SS08].
Complex systems
The last half of the 20th century and the first decade of the current one were characterized by the dominance of reductionist approaches to biology which were mainly driven by molecular biology. This type of reductionism was inspired by the influential 1944 book “What is life” by Erwin Schrödinger [S44] who postulated that the chromosome formed an “aperiodic crystal” that is durable, an important prerequisite for hereditary matter. Schrödinger called it the “material carrier of life”. Parts of the chromosomes are formed by genes, which themselves are large, durable and responsible for the observed inheritance mechanism, thus making animate matter unique. Schrödinger’s ideas were driven by quantum mechanical reasoning applied to biology and were seminal in triggering the molecular biology revolution and lead to an increasingly gene-centric view of nature, a view further extended by another influential book, “The selfish gene” by Richard Dawkins [D76]. However, now that the human genome has been decoded (see e.g. [HGS04]), one may ask whether (a) knowing all parts of the system, can we fix or repair it if something goes wrong, and (b) can we put the parts back together?
The first question has been addressed by Yuri Lazebnik in several entertaining public lectures at systems biology conferences (e.g. ICSB Conference in Heidelberg 2004) and is summarized in his paper “Can a biologist fix a radio?” [L04]. Lazebnik, an engineer, concludes that a more systematic and quantitative approach has to be adopted in modern biology while referring to general systems theory (GST) developed by Ludwig von Bertalanffy and others contemporaneous of Erwin Schrödinger [B68]. The latter already pointed out that living organisms must have developed ways that let them defy the second law of thermodynamics constructing ‘order from disorder’ and allowing them to decrease their entropy by adding ‘negative entropy’ to the environment [S44, p. 79ff]. von Bertalanffy took this idea further and argued that living systems are “open systems having a steady state” [B68, pp. 39–40] and opened the door to an organicist view of biology. But only in recent years, accrued evidence is telling us that by understanding the parts of a system we do not necessarily understand the overall systems behavior (see diverse examples on complexity in [G08]).
This realization brings us back to the second question about reassembling the system even when knowing all its parts. Staying with the radio metaphor of Lazebnik, one would conclude that if we identified and carefully disassembled all parts of the system and recorded all their connections and positions we should obtain a blueprint of the radio. Next, we should be able to reassemble the system. This, however, does not imply that from the knowledge gained we would be able to repair or even modify the radio such that we would e.g. improve its reception. To accomplish this, the engineer would have to find, first, functional units that could be subsequently analyzed in isolation and in concert with other components to which it is connected. He might then find out that by enlarging the antenna, the reception of the radio might be substantially improved. Biological systems, however, are much more intricate than a man-made and designed apparatus like a radio, where all of its component can be studied in isolation under equilibrium conditions. Biological systems operate in non-equilibrium conditions and “comprise many interacting parts with the ability to generate a new quality of macroscopic collective behavior the manifestations of which are the spontaneous formation of distinctive temporal, spatial or functional structures…” thus matching the most commonly found definitions of complex systems (as defined by the editors of the Springer series “Understanding Complex Systems”, see also [G08] for similar definitions). This has remarkable consequences and implications for the question “Can we put the parts back together?” which we will attempt to elucidate next.
Interpretations of ideas about complex systems have been discussed since the 1940s. Several new fields and theories carrying different names emerged from these discussions (e.g. Synergetics, Dynamic Systems Theory, Chaos Theory, Cybernetics, Tensegrity). A common denominator in all these areas is that even a system that consists of very simple parts that interact with each other in a non-linear fashion can exhibit complex systems-level or emergent properties, such as structure and organization. These properties are quite surprising and unexpected when one examines the properties of the individual parts alone. In other words, the system itself is more than just the sum of its parts [G08]. Denis Noble, who followed up on the second question “Can we put the parts back together?” in his book “The Music of Life,” relates a telling anecdote about his attempts to mathematically model the oscillatory behavior of the heart. He was asked: “Mr Noble, where is the oscillator in your equations? What is that you expect to drive the rhythm?” Only decades later, he found the answer to this question: “Indeed, it is an eminently necessary question, if we are talking about some man-made, mechanical systems. But we are not. Instead, we can have a system that operates rhythmically and yet contains no specific ‘oscillator’ component. There is no need for one. The reason is that the rhythm is an integrative activity that emerges as a result of the interactions of a number of protein (channel) mechanisms.” (see p. 60, [N06]).
This explanation implies that the key to emergent phenomena and system-level properties of complex systems must lie in the interaction between the elements comprising the system. It is therefore intrinsically difficult to predict the future behavior of such systems as the interactions between the system parts are shielding their specific individual features from the system-level properties. Due to the lack of derivable laws, computational and mathematical tools are indispensable for complex system scientists, in general, or the systems biologist, in particular.
Networks and graphs
The above definition of complex systems consisting of interacting parts leads naturally to the use of mathematical tools based on networks or graphs where the individual parts translate to nodes and the interactions translate to edges or links. In his book “Linked” Albert L. Barabási summarizes the most common properties found among numerous naturally formed networks ranging from the Internet to social and gene regulatory networks [B02]. When analyzing the network topology of these diverse complex systems, some important overarching rules emerge. It is not completely surprising that these networks deviate substantially from randomly built networks as studied by Paul Erdős and Alfréd Rényi [ER60]. We therefore do not observe a bell-shaped frequency distribution of the number of links per node as expected from randomly formed networks; instead, we observe a power-law distribution, which is characteristic of small world or scale-free networks [AS00]. This implies that a large majority of nodes have only a few links, whereas very few nodes have a large number of links. Those nodes are called hubs or connectors [B02] and play a vital role in our understanding of, for instance, how diseases spread and epidemics can be stopped by targeting hubs identified in the network (e.g. [LE01]).
A scale-free network topology can be reproduced when dynamically constructing a network by adding nodes iteratively and linking them preferentially to already well connected (or fit) nodes in the existing network. This concept was termed “Rich get richer” by Barabási [B02] and works analogous to increasing returns in economy, an idea hatched in the early 1980s by Belfast-born economist W. Brian Arthur to describe high technology. In such networks, two randomly picked nodes are usually connected by a quite short path (sequence of links to neighbors) which is another characteristic of small world networks (“Six degrees of separation”, [B02]). Although the underlying topology makes the network vulnerable to direct attacks to hubs, the resulting network is very robust against random perturbations [CN00]. Many naturally growing networks also exhibit various level of modularity where sub-clusters are more strongly connected with each other (e.g. cortical networks, see e.g. [KG07]) describing a hierarchy of scale-free networks where hubs connect the different modular layers thus conserving the overall scale-free network topology [RS02].
The above-referred observations contributed to the fast rise of systems biology. However, we are skeptic of the view voiced by Barabasi in [B02] that by establishing the “map of life” that describes the complete metabolic (biochemical), regulatory (gene or protein interaction) and cellular networks of an organism, we will hold the keys to an understanding of how an organism works despite the fact that scale-free networks are an emerging feature of various complex systems or networks. We share the concern of Yaneer Ban-Yam who says that “[t]he biggest current danger to the field [of complexity] is that it will be hijacked by people who don’t understand the essence of the field. Many are adopting the terminology without understanding what complex systems are really about. Systems biology, systems engineering and other systems related fields are often (but not always) just using the words but continuing a reductionist approach.” [B02, p. 15ff] It is reductionist to believe that by understanding the interactions between the molecules contained in a cell we will be able to understand how the cell works, a tissue is formed or cancer arises; these assumptions are only driven by upward causation in the “map of life”. The upward causation assumption completely neglects the contribution of the environment and of the emergent structure itself (by downward causation).
Although scale-freeness emerges in complex networks like the Internet, the World Wide Web, social and biological networks as well as larger parts of the modern, globalized economy, it is not a universal feature of complex systems [K05]. Other network topologies emerge also naturally not showing small world properties. One prominent example is a road network connecting cities. In this case, each node is not only a point but has a certain size, cannot freely move and roads themselves (or edges) are restricted by the topographic and geological settings. This implies that having spatial constraints limiting the dynamic construction process can yield different network topologies. Therefore, it seems important to include spatial localization information when building gene regulatory networks or protein-protein interaction maps given that a substance can only react with another substance when both reside in the same spatial compartment [D03].
So how do domain boundaries emerge in complex systems? Could these boundaries relate to the individual modules found in hierarchical networks? Is there a correlation between functional units and compartmentalization? Is there a form-function relationship to be found in living systems? Although we are yet unable to answer these questions, it is worth making a simple thought-experiment by revisiting Lazebnik’s radio example described above. We can ask ourselves whether it makes sense to decompose the whole radio down to its molecular constituents so to understand its workings. Going back to Schrödinger’s ‘order from disorder’ principle, we would certainly suspect that there might be a correlation between spatial domains and functional units. Next, one might consider looking at the apparent, spatial patterns visible in the radio and hope that these units can be studied in isolation. In the case of an individual cell, this would mean that the cell’s anatomy should be taken into account, looking at sub-cellular compartments [H05] and the protein interactions therein giving rise to protein clusters potentially describing functional regions (the toponome, see [SB06]). In the case of tissues or organs, we could first try to focus, for instance, on understanding how typically found patterns in glandular tissues are formed (e.g. acini and ducts, see [KM08]).
Self-organization
So, how are complex spatial patterns formed? Suppose that we can find an explanation for at least one complex system that is exclusively composed of simple, inanimate compounds such as atoms or molecules where, obviously, no overall blueprint exists nor can be executed. In this case, one will have to accept that the ‘order from disorder’ principle is also applicable to systems consisting of much more complex parts, like those found in biological systems ranging from bacteria to multicellular organisms, where interactions are not only governed by physical laws, but by more complex physiological and behavioral responses [CD01]. The simple answer to the above question is… through self-organization.
Although rather unknown and not well studied when Schrödinger wrote his book, several very simple self-organizing systems have been since discovered not only in physics and chemistry showing stunning emerging spatial patterns (see e.g. the rock formation of the Giant’s Causeway in Figure 1, soap bubbles that build when a flask of dish-washing detergent is shaken, the well-known Benard convection [CD01] or some more recent finding on the physics of Type-I superconductors [P07]) underpinning the fact that self-organization might also be present in more complex systems (as is shown in [CD01]). As physical laws rule the interactions between the parts in physical systems, we can exclude alternative explanations of pattern formation that require intervention from outside the system, such as (i) the presence of a leader, (ii) the existence of a blueprint, (iii) the execution of a recipe, or (iv) the use of a template [CD01]. Although (i)-(iv) are relevant to biological systems, self-organization is certainly an option when it comes to explaining biological pattern formation where “the rules in self-organizing systems can be quite economical in the physiological and behavioral machinery needed to implement them” [CD01, p. 63]. This simplicity might give self-organization an evolutionary advantage over the alternative solutions (i)-(iv), making it more prevalent in biological systems. Having said this, it is certainly possible that a mixture of these mechanisms is present in the same system.
Figure 1.

Example of self-organization of inanimate matter: left panel shows an overview of the rock formation found at the Giant’s Causeway in County Antrim, Northern Ireland. The right panel shows a close-up photographed downwards onto the rock formation showing a regular, polygonal structure that emerged from volcanic activity.
Let us consider the alternative explanations (i)–(iv) first and then, see if they are applicable when it comes to tissue (or organ) morphogenesis: The presence of a leader (i) can almost certainly be excluded as we are not aware of any molecular mechanisms that would enable a single entity to receive all the information signaled from all other cells and instructing them to perform certain actions as a result of processing the incoming information. The first mathematical model proving that an aggregation of single-celled units into larger cooperative entities can be explained without requiring a leader, such as a founder or pacemaker cell, was published by Evelyn Keller and Lee Segel in 1970 [KS70] for the slime mould (Dictyostelium discoideium). Furthermore, it seems unlikely that a template (iv) is used when cells aggregate to form tissues since tissues can be grown in vitro without the presence of any template structure. This brings us to the alternative explanation requiring the existence of a blueprint (ii) that describes the parts and the spatial layout of the tissue to be built. Such blueprint does not, however, describe how tissue is to be built and consequently requires each cell to have a global picture of the tissue being formed at any point in time. This seems very unlikely as there is no known molecular mechanism conveying such information to each individual cell. This then leaves the remaining option that each cell is following a strict recipe (iii) describing a set of instructions to be carried out. Although such set of instructions might explain how an individual like a spider builds a cocoon for its eggs [E70], it is unlikely that each cell can follow and execute each of the encoded rules independently of the crowded environment present in a tissue or organ. Since cells can only sense their local environment, the emergence of tissues can only be driven by rules governed by coordinated interactions with the local environment of each cell. This leads to the conclusion that the dynamic process of tissue formation must mainly be governed by self-organization.
How does self-organization work? First, the components need to be able to interact with or get feedback from other neighboring components, but also from the local environment or from the emerging structure itself (stigmergy, [CD01, p. 56]). In the case of tissues, this would correspond to interactions with other cells, nutrients and the extracellular matrix. This feedback can be either negative or positive. It turns out that positive feedback is prevalent in self-organizing systems as it leads to aggregation, but bears the risk of overamplification. In order to control and stabilize positive feedback mechanisms, negative feedback is needed. This feedback can either be built into the system (e.g. cells get quiescent) or be offered through physical constraints (e.g. cell-migration depends on forces exerted by the extracellular matrix). Components of such system can interact with each other using either cues that specifically convey information (e.g. like ants when leaving a trail of pheromones leading to their food source) or cues that convey information incidentally (e.g. like a deer leaving a trail when walking through the wood, see also [CD01]). In cellular systems, we observe biochemical (e.g. morphogen gradients generate diverse cell types in distinct spatial regions) as well as biomechanical cues (e.g. fibroblasts degrading collagen fibers giving way to epithelial cell-migration).
Self-organizing systems are usually very stable over a large range of parameters, but can exhibit sudden and abrupt changes in the emergent pattern due to minimal changes of one or more parameters thus moving from one stable state to another or showing criticality at the edge of chaos (see [NP08] for a biological example). If we now classify phenotypes for one species according to the emergent patterns observed, we can observe a change in phenotype close to the bifurcation by altering only the parameters governed by the environment (e.g. the raid patterns of army ants [DG89]). This implies that the same genotype can exhibit different phenotypes depending on the environment.
Environmental determination of the phenotype was first documented at the end of the 19th century in Lepidoptera. The European map butterfly exhibits strikingly different wing phenotypes depending on the season of eclosion of the butterflies: while the spring morph shows orange wings with black spots, the summer morph is black with a white band. This dimorphism misled Carl Linnaeus, the father of taxonomy, to classify the morphs as distinct species. In 1875, by incubating the caterpillars in different conditions, August Weissman found that the seasonal pattern of the wings of certain butterflies is temperature-induced. Indeed, the discipline of Ecological Developmental Biology deals with this phenomenon, called polyphenism, and other aspects of environmental determination of the phenotype [GE09]. These phenomena were mostly ignored by mainstream biologists under the spell of genetic determinism. However, the discoveries of hormonally-active man-made chemicals and that human adult diseases often have their origins during fetal life has greatly contributed to the revival of the eco-devo tradition [SS10]
Modeling tissue morphogenesis
The findings published in 1952 in Alan Turing’s seminal paper about the chemical basis of morphogenesis [T52] offered a possible mathematical explanation of patterns forming in developing biological systems which can be seen as yet another manifestation of self-organization. This theory of temporarily emerging stationary waves starting from homogeneously distributed reactants (or morphogens) was influential in developmental biology as explained by Claude Wilson Wardlaw [W53], who remarked “That diffusion-reaction systems are present in all growing regions, indeed in all living matter, is basic to studies of metabolism. What is novel in Turing’s theory is his demonstration that, under suitable conditions, many different diffusion-reaction systems will eventually give rise to stationary waves; in fact, to a patternized distribution of metabolites. Thus, in the present writer’s view, the theory would appear to afford an explanation of the inception of the symmetrical, radiate histological pattern that appears adjacent to the embryonic region of the root apex. Not all kinds of pattern, however, are referable to the development of stationary waves—the major feature of Turing’s theory as thus far developed—but all may eventually be related to some kind of diffusion-reaction system. The inception of polarity in an embryo, i.e. of axiate development, is probably due to a particular distribution of metabolites in an initially homogeneous system; this could be regarded as a very simple case of a stationary wave.”
This success might also explain the prevalence of partial differential equations being used as mathematical tools for the analysis of spatially distributed dynamic systems and for the exploration of self-organization mechanisms under the influence of positive and negative feedbacks that give rise to patterns in plant and animal morphology or to electrostatic waves in the heart [WQ03]. There are, however, several tools available to the modeler of biological systems, which have been summarized by Bassingthwaighte et al. under the following application areas [BHN09]:
Evolutionary biology and genetics: quantitative, model based mathematical or statistical analysis studying mutation, selection, genotype-phenotype mapping as well as morphogenesis using agent-based or individual based modeling,
Biophysics and electrophysiology: signal transduction across membranes through channels using combinations of ordinary differential (ODE), partial differential (PDE), algebraic and differential algebraic equations (DAE) as well as Markov state models for channel gating,
Mathematical biology: DAEs, ODEs and PDEs based on reaction-diffusion systems applied to cancer modeling, cell cycle and pattern formation in embryogenesis,
Computational physiology: biophysical models based on solid and fluid mechanics applied at the levels of cells, tissues, organs using conservation laws, continuum mechanics and finite element methods,
Computational chemistry: applying quantum mechanics and molecular dynamics to investigate protein-protein and protein-ligand interactions as well as protein-(un)folding at the atomic level,
Network systems biology: DAEs or ODEs are applied to gene regulatory, signal transduction and metabolic networks using mainly the biophysics encapsulated in mass balance equations of chemical species. Analytical tools include bifurcation theory, non-linear control theory, Bayesian statistics and linear algebra, and
Systems physiology: DAEs applied to study physiological function at the organ system level such as blood pressure control or exocrine signaling at the cell level.
Furthermore, Bassingthwaighte et al. state that “[o]ne of the central principles is that complex systems like the heart are inevitably multiscalar, composed of elements of diverse nature, constructed spatially in a hierarchical fashion” which “requires linking together different types of modeling at the various levels”, but noting that “[i]n multiscalar systems with feedback and feedforward loops between the scale levels, there may be no privileged level of causation” [BHN09]. This implies that it is important that a successful model of a complex system has to include relevant scales and only subsequent system analysis might reveal at which level biological function might be integrated [BHN09].
Emerging structures in glandular tissues
Before starting to model tissue morphogenesis it is therefore important to take a closer look at emergent structures and the potential scales involved. For this review, we would like to focus on prominent structures found in glandular tissues and on the question how these might be formed through self-organization. It is widely acknowledged that cells cultured in 2D have different patterns of gene expression from their 3D counterparts [PR07]. This is increasingly stimulating interest in 3D tissue models such as the one we have developed [KM08]. We summarize below some results concerning factors that may influence epithelial structure formation that concern the main structures found in glandular tissues, which are round, hollow acini and tubular branching structures called ducts (see Figure 2). These fundamental structures only comprise a few hundred cells and depend on the composition of the extracellular matrix (ECM).
Figure 2.
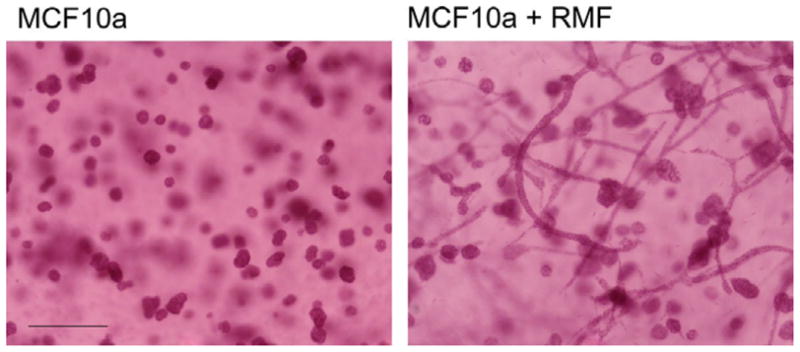
Whole mounts of MCF10A cells (left panel) and MCF10A cells and RMF grown for three weeks within a matrix made of 1 mg col-I-50% Matrigel. Note that ducts only formed in the presence of fibroblasts. Scale bar 200 m (taken form [KM08]).
ECM compliance and mechanical force
Human breast MCF10A cells formed branching ducts or rounded structures (acini) depending on the rigidity and isotropy of the extracellular matrix [PZ05].
Stromal cells
When cultured alone, the epithelial cells formed acini in an isotropic matrix; however, when co-cultured with normal breast fibroblasts (RMF, see Figure 2) or with pre-adipocytes [KM08], epithelial structures elongated forming branching ducts.
ECM fiber organization
In a cellular gels, collagen formed thin fibers without any defined organizational pattern. Both epithelial and stromal cells organized the collagen fibers, a phenomenon evident during the first 24 h of culture. As epithelial structures formed later on, small and short collagen fibers organized radially were found in the vicinity of the acini, while long fibers were found parallel to the long axis of the ducts. Branching appeared to occur through the formation of a projecting sprout from an existing duct and was usually associated with the development of a collagen bundle along the branch axis extending to a nearby epithelial structure (see Figure 3).
Figure 3.
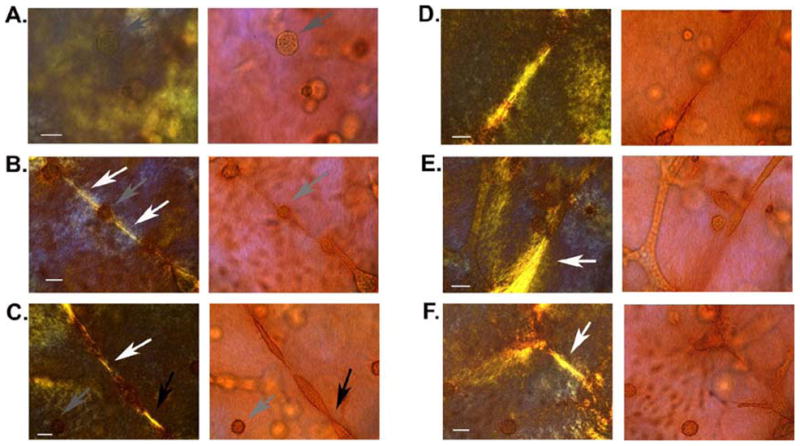
MCF10A + RMF co-culture in floating gel at the 10th day in culture. Whole mount picrosirius red staining; images were taken under polarized (left) and non-polarized light (right). (A) Acini in the lower layer of the gel (arrow). (B) Acini loosing the spherical symmetry (gray arrow) and interacting with neighboring structures through modified collagen fibers (white arrows). (C) Elongating structures interacting via modified collagen fibers (white arrow) and fusing with each other into tubular structures (black arrow). Notice the absence of modified collagen fibers nearby the non-elongating acini in lower left corner (gray arrow). (D) Bundle of thick collagen fibers formed between two structures along their elongation axis. (E) Collagen bundle (white arrow) formed along the elongation axis of a duct. (F) Tubular structure interacting with neighboring structure though collagen fibers (white arrow) and forming branching sprout. Scale bars 20 m (taken form [DM10]).
Culture dish topology and epithelial organization
In addition to the matrix composition the shape and rigidity of the culture dish also influence the organization of epithelial structures. In floating 1mg/ml Col-I gels, ducts appeared mainly in the upper layers, while the lower layers contained almost exclusively acini, suggesting that factors that promote tubulogenesis are heterogeneously localized along the Z axis (see Figure 4). Thus, an asymmetrical spatio-temporal distribution of biomechanical and/or biochemical factors appears to regulate tubulogenesis within the floating gel during matrix reorganization.
Figure 4.

Schematic representation of contracted floating gel; i: periphery zone, ii: intermediate area, iii: central area. Circular and rectangular shapes denote the distribution of acinar and ductal structures respectively (taken from [DM10]).
In summary, perhaps the most remarkable finding of our studies has been the plasticity of the 3D tissue model as revealed by the local and temporal changes observed both in the distribution of epithelial structures, their phenotype, and in collagen fiber organization. These dynamic changes became apparent by the systematic observation of the whole-mounted gels. This plasticity suggests a dynamic process initiated by the cell-mediated collagen organization that resulted in reciprocal interaction between the newly organized and biomechanically enabled fibers and the emerging epithelial structures. Once fibers start to form elongated bundles they, in turn, exert forces upon the epithelial structures. Hence, the heterogeneity resulting from the local interplay of fibers and cells generate forces that shape and remodel the epithelial structures [DM10].
Agent-based modeling of tissue morphogenesis
Discrete computational simulation methods are often used when complex spatial arrangements are less amenable to abstraction and a more mechanistic model is required. The agent-based model is currently being eagerly adopted in the life sciences, because it is well-suited for modeling tissue morphogenesis as they allow for intra-cellular decision processes [GHA09]. Agent-based or individual-based modeling is a computational method in which a complex system is decomposed into a number of discrete entities called agents. The agents’ movement in virtual space, their behavior and interactions with other agents and the temporal evolution of their internal state are determined by a single rule set, that is, repeatedly applied independently for each agent. Agents are either restricted to sites on a regular lattice (cellular automaton) or are lattice-free allowing more realistic, quantitative models [GHA09].
This rule-set can be seen as a formal description of the processes that underlie the emergent phenomena observed in simulation or, by analogy, in an experiment. Thus, derivation of the rule-set is the central task in modeling, and the rule-set itself typically represents a major piece of knowledge gained in the modeling process. This explains the fact that the empirical process used to identify the rule-set requires tight coupling with experimental observations [TB07]. Early models of tissue morphogenesis only considered cell proliferation rates and apoptosis to model cell population kinetics, but ignored the fact that cells are volumetric objects that interact with their environment, which in turn impacts on model parameters such as migration, proliferation, “differentiation” and apoptosis [HI99, I03]. Later individual modeling attempts included these feedback components as biomechanical properties.
In simple agent-based models, isolated cells are described as elastic spheres of variable volume [GHA09]. When cells get in contact with the substrate or other cells, they exhibit an adhesive energy that is proportional to the contact area formed, but they also get deformed and compressed all adding to a total internal energy. If no external stimulus is applied, cells migrate randomly in a friction dominated environment. The cell cycle is simplified into an interphase where cells stochastically increase in size up to twice their original volume and a mitotic phase where the cell divides into two daughter cells of equal size. Different proliferation, apoptotic and migratory behavior can be simulated depending on the cell type, and on cell-cell, cell-substrate and cell-matrix contact areas. Such models are mostly used to reproduce growth colony dynamics and morphology in 2D, but also 3D in vitro assays.
For reliably modeling the emergence of stable acinar structures from epithelial cells, a more realist model of the cellular shape had been introduced, as encapsulated in the immersed boundary framework used by Kasia A. Rejniak [R07]. This model “captures interactions between immersed elastic tumor cells and a viscous incompressible fluid, representing the cytoplasm inside the cells and the extracellular matrix outside the tumor tissue. The fluid flow is influenced by sources of fluid located inside the growing cells, as well as by forces generated by the immersed, elastic boundaries, while at the same time the elastic structures move at the local fluid velocity. The cell cycle and cell processes are related to the concentration of external factors, such as oxygen, sensed by the cell from its local environment” [R05]. The underlying temporal distributions of nutrients and other molecular constituents are usually modeled using PDEs and are combined with the agent-based system into a hybrid model (or hybrid discrete-continuum model (HDC), [AW06, CL10]). HDC systems make it possible to model collagen fiber orientation as a continuous vector field while other cell types such as fibroblast remain discrete entities that interact with the collagen matrix and alter the underlying PDEs [MD06].
The work by Rejniak [RA08] assumes that acinar structures originate from a single mother cell which self-organizes through subsequent proliferation, migration, polarization and apoptosis through nutrient starvation into a hollow structure, the acini, where the hollow core is formed. The ECM is assumed to be homogeneous which allows for a less computational intensive 2D model. If we envisage the cells being embedded in a heterogeneous 3D ECM that exhibits different mechanical properties along and perpendicular to local collagen fiber orientation, the Rejniak model [RA08] could possibly explain how hollow, tubular ductal structures emerge in such tissues. It remains to be shown experimentally, which properties of the microenvironment finally decide on the developmental faith of branching ducts or acini in the same medium and whether these structures always originate from one mother cell.
Understanding early carcinogenesis using mathematical modeling
Cancer is diagnosed by pathologists while examining the tissue level of biological organization. We have proposed that carcinogenesis is due to altered tissue organization akin to development gone awry [SS05]. Therefore, one would have hoped that by understanding normal tissue development we would have been able to define in our model those parameters in normal tissue development that reach a critical threshold beyond which regular structures found in healthy glandular tissues such as acini and ducts suddenly disappear (see also above notion of criticality and the edge of chaos). It seems therefore logical, that the modeling machinery used for tissue morphogenesis can also be used to model cancer development. Helen Byrne recently summarized the mathematical modeling approaches to carcinogenesis, avascular and vascular tumor growth and angiogenesis [B10] that span the complete range of multi-scale, hybrid and three-dimensional models. She states that it can therefore be difficult to choose the correct model for a particular question, in particular since different approaches can yield the same results. In such situation, she thinks it might be appropriate to appeal to Occam’s razor or the Law of Parsimony using “the philosophical principle that one should not look for multiple causes of any effect if a single cause can provide a suitable explanation” [RB02]. Although this approach may be useful as a first approximation, it negates the fact that in a complex system there is no privileged level of causality. More importantly, more organic models that facilitate the collaboration between modelers and experimentalists should be adopted [B10]. This might also explain the increasing popularity of agent or individual-based modeling approaches towards tumor growth in recent years [IC04, R05, AW06, ZA07, ZW09, DZ09]. All models produce comparable results to those presented by the immersed boundary framework presented by Rejniak [R05]. This model uses a similar parameter setup as for the computation of the emergence of acinar structures in healthy tissue [RA08], but with the main difference that tumor cells do not polarize and form stable structures at the growth boundary. Dependent on model parameters such as proliferation rates and structure of the ECM, more rounded or more perforated, finger-like structures, are being generated [AR09].
Limitations of current models
Although current models allow us to hypothesize on the effects of certain model conditions on cancer growth [AW06, AR09], they face very severe limitations. Besides the effect that an increased complexity of the computational model leads to a significant restriction on the tissue volume and time-frame that can be computationally modeled [DZ09], the most limiting factor is the lack of direct coupling of the mathematical or computational model to experimental data [B10]. This problem manifests itself in the fact that although numerous models on tumor progression and growth implicitly assume that “[m]ost tumors in vivo arise from a single cell that has escaped the growth-controlling mechanism” [R05], nobody has ever observed a tumor in statu nascendi. The experimental difficulty to do so was already identified by Theodor Boveri in 1914 as being the main hindrance to study the early events leading of carcinogenesis. Despite this fact and its incompatibility with nonmutagenic carcinogenesis, the prevailing paradigm for carcinogenesis underlying almost all mathematical models remains the somatic mutation theory [W07, BS09] and alternative explanations such as the tissue organization field theory only slowly are being introduced [BS09, BCP10]. Furthermore, without direct coupling between experiment and mathematical model, most parameters estimated throughout simulation and validation processes are difficult to be related to real, biologically relevant entities and quantities.
An example of a prominent parameter that causes quite controversial debates amongst biologists relates to cell proliferation. For a modeler, the default modus operandi of a cell can be both, proliferation or quiescence. To increase cell proliferation rates, a modeler either reduces the concentration of an inhibiting substance where proliferation is seen as the cell’s default or increases the concentration of a stimulating substance where quiescence is seen as default. Both models will yield the same qualitative behavior in their respective simulation, whereas the biological ‘truth’ is very likely to be reflected by only one of both scenarios. Unless such substance is directly found in the experiment itself, both modeling assumptions have to be seen as equivalent and undistinguishable as both models validate the same experimental observations (please note that indirect and intermediate processes might make it experimentally difficult to unequivocally discriminate between inhibitory and stimulating substances).
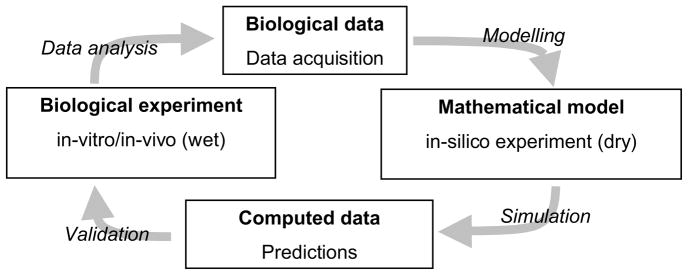
To overcome this problem, we not only should have to adopt an organicist or systems view of biology that makes us aware of the connectedness of living systems where interactions between molecules, genes, cells, species and the environment are regulating biological function, but more importantly, we should have to tightly couple biological experiments with an organic mathematical model of an inherently complex and adaptive system. Such model would allow us to derive quantitative measures and eventually make predictions about the biological system through simulation or execution of a computational implementation of the mathematical model. By relating predicted quantities back to the real biological system and conducting experiments to validate the predictions, we close the systems biology cycle for knowledge discovery (see Figure 5). If the predictions get validated it means that our mathematical model encapsulated our current knowledge of that particular system correctly. If not, we should have to go back to the drawing board and modify our model and rerun the cycle (see also the introductory section in [P03]).
Figure 5.
The systems biology cycle for knowledge discovery shows how biological experiments are being tightly coupled with mathematical models through data analysis, modeling, simulation and validation. Only by completing a full cycle, actual knowledge is generated and our understanding of a biological system is furthered.
Therefore, as long as mathematical models are not tightly coupled to biological experiments, most modeling attempts will remain descriptive in nature giving us only phenomenological insights into ongoing processes not allowing us to directly predict experimental outcomes. However, since predictions derived from a mathematical model that are subsequently validated through novel biological experiments are the ultimate Holy Grail [CD01] that would lead to an understanding of the biological system itself, we might fall short in our attempts to gain more profound insights into complex biological processes, such as tissue morphogenesis or early carcinogenesis, in the absence of a highly controllable, observable and flexible biological model system that can be directly coupled to a mathematical model.
In vivo vs. in vitro models
A long standing controversy relates to the choice of the correct biological model system, where, for obvious reasons, common sense prefers the in vivo situation. For ethical reasons, this choice becomes problematic when dealing with human subjects. This then makes it necessary to use alternative biological model systems. Are animal models in general more reflective of human physiology than in vitro assays? A low percentage of drugs (ranging from 5–20%) found to be effective in the animal model itself (Phase I) actually proved to be successful in the human trial phase and make it through registration [KL04]. This low success rate suggests that this thought process should be guided by the desire to make a biological model system tractable and accessible to mathematical modeling so that both can be tightly coupled through the systems biology cycle for knowledge discovery. This means that although simplified models such as the 3D tissue model of the mammary gland shown above might not be reflective in all details of the human physiology, they might allow us to generate deeper insights into microenvironment-dependent cell-cell interactions.
Conclusions and future directions
This review intended to highlight the mechanisms underlying self-organization and their importance for the life sciences in general and for tissue morphogenesis and cancer modeling in particular. Our journey following the more recent history of science and philosophy has revealed that although it is important to identify the constituents of a system and to study the working of its individual parts, this approach we may not necessarily provide understanding of the system-level properties. This view is certainly embraced by systems biology as promoted by Hiroaki Kitano [K02]. The success of network modeling tools applied to gene regulatory or metabolic networks of individual organisms seems to convey the message that instead of the reductionist view where one single gene regulates function, it is now a concert of genes responsible for the same task. This, however, is still a reductionist viewpoint as it does not consider the interplay between upward and downward causation and the role of biomechanics and topology as determinants of biological structure and function. The dominance of network related modeling in the young field of systems biology and its inherent focus on the ‘map of life’ as proposed by Barabasi [B02], further deviates our attention from the fact that there are other mechanisms prevalent in complex systems that can create order from disorder through self-organization, a process fundamental for living matter [S44]. The fact that spatial aggregation and compartmentalization are prevalent not only in biological systems does imply that there are form-function relationships waiting to be uncovered and that spatial organization is indeed an important parameter that needs to be considered in mathematical models.
As pointed out by Scott Camazine et al [CD01], mathematical modeling is central to understand complex systems. In particular, computational models need to be tightly coupled with in-vivo or in-vitro models not only to validate the mathematical model, but also to predict system properties yet unknown. In the modeling field, this activity currently mostly revolves around agent-based models [TB07], which can be computationally expensive, but best reflect the nature of tissues as self-organizing systems. Experimentally, highly controllable in vitro model systems are needed for systematic investigation of the association rules, parameters and processes that yield biological tissue formation. To this purpose, we have developed a novel 3D tissue organogenesis model of the mammary gland that contains both epithelium and stroma (cellular and ECM) [KM08]. This model is aimed at identifying the key physical processes that regulate epithelial organization into cylindrical structures (ducts and branching ducts) and spherical structures (acini), prevalent structures also found in other glandular tissues. By using time-lapse microscopy [PH07], we expect to uncover the local rules that govern cell proliferation, migration and aggregation depending on the microenvironment, such as collagen density and collagen fiber orientation [PE06, PI08], as well as the potential cues of which they are targets. We anticipate that this new methodology that operates in glandular tissue model systems will bring us closer to the goal of “putting the parts back together”.
Key points
Because the physical world is causally open, emergent phenomena are to be expected, where both bottom-up and top-down causation must be taken into consideration.
Spatial patterns can emerge through self-organization, which creates order from disorder.
Hybrid discrete-continuum based modeling approaches provide a natural way to computationally describe self-organizing phenomena at the cellular/tissue level.
Adaptive complex systems are inherently multiscalar and hierarchical with upward and downward causation across multiple scales.
The systems biology knowledge discovery cycle is not limited to ‘network systems biology’ and should embrace the wider framework adopted by the Physiome and the Virtual Human Project.
When studying tissue morphogenesis and early carcinogenesis, biological models that can be tightly linked to mechanistic models which in turn allow us to generate testable predictions and new insights are hard to come by.
Because animal models for testing cancer drugs have not proven to be very efficient in predicting outcomes in humans, the need for alternative models is urgently desirable.
Acknowledgments
We would like to thank Dr Hendrik Fuss for his suggestions. This work was funded in part by grants from the National Institutes of Health-National Institute of Environmental Health Sciences (ES08314 and ES018822) and a grant from The Parsemus Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AR09.Anderson ARA, Rejniak KA, Gerlee P, Quaranta V. Microenvironment driven invasion: a multiscale multimodel investigation. J Math Biol. 2009;58:579–624. doi: 10.1007/s00285-008-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AS00.Amaral LA, Scala A, Barthelemy M, et al. Classes of small-world networks. Proc Natl Acad Sci U S A. 2000;97:11149–11152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AW06.Anderson ARA, Weaver AM, Cummings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127:905–915. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- B02.Barabasi A-L. Linked: How Everything is Connected to Everything Else and What it Means for Business and Everyday Life. Plume Books; 2002. [Google Scholar]
- B04.Bunge M. Emergence and Convergence. Toronto: University of Toronto Press; 2004. pp. 13–14. [Google Scholar]
- B10.Byrne HM. Dissecting cancer through mathematics: from the cell to the animal model. Nat Rev Cancer. 2010;10:221–230. doi: 10.1038/nrc2808. [DOI] [PubMed] [Google Scholar]
- B68.von Bertalanffy L. General System Theory: Foundations, Development, Applications George Braziller. 1968. [Google Scholar]
- BHN09.Bassingthwaighte J, Hunter P, Noble D. The Cardiac Physiome: perspectives for the future. Exp Physiol. 2009;94:597–605. doi: 10.1113/expphysiol.2008.044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BPC10.Baker SG, Cappuccio A, Potter JD. Research on Early-Stage Carcinogenesis: Are We Approaching Paradigm Instability? J Clin Oncol. 2010 doi: 10.1200/JCO.2010.28.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BS02.Brisken C, Socolovsky M, Lodish HF, et al. The signaling domain of the erythropoietin receptor rescues prolactin receptor-mutant mammary epithelium. Proc Nat Acad Sci USA. 2002;99:14241–14245. doi: 10.1073/pnas.222549599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BS09.Baker SG, Soto AM, Sonnenschein C, Cappuccio A, Potter JD, Kramer BS. Plausibility of stromal initiation of epithelial cancers without a mutation in the epithelium: a computer simulation of morphostats. BMC Cancer. 2009;9:89. doi: 10.1186/1471-2407-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CD01.Camazine S, Deneubourg JL, Franks NR, et al., editors. Self-organization in biological systems. Princeton University Press; 2001. [Google Scholar]
- CL10.Cristini V, Lowengrub J. Multiscale modelling of cancer. Cambridge University Press; 2010. [Google Scholar]
- CN00.Callaway DS, Newman MEJ, Strogatz SH, et al. Network robustness and fragility: Percolation on random graphs. Physical Review Letters. 2000;85:5468. doi: 10.1103/PhysRevLett.85.5468. [DOI] [PubMed] [Google Scholar]
- D03.Davis T. Protein Localization in proteomics. Current Opinion in Chemical Biology. 2004;8:49–53. doi: 10.1016/j.cbpa.2003.11.003. [DOI] [PubMed] [Google Scholar]
- D76.Dawkins R. The Selfish Gene. Oxford: Oxford University Press; 1976. [Google Scholar]
- D95.Dennett DC. Darwin’s Dangerous Idea. New York, NY: Simon & Schuster; 1995. p. 21. [Google Scholar]
- DM10.Dhimolea E, Maffini MV, Soto AM, Sonnenschein C. The role of collagen reorganization on mammary epithelial morphogenesis in a 3D culture model. Biomaterials. 2010;31:3622–3630. doi: 10.1016/j.biomaterials.2010.01.077. [DOI] [PubMed] [Google Scholar]
- DG89.Deneubourg JL, Goss S, Franks N, et al. The blind leading the blind: Modeling chemically mediated army ant raid patterns. Journal of Insect Behavior. 1989;2:719–725. [Google Scholar]
- DZ09.Deisboeck TS, Zhang L, Yoon J, Costa J. In silico cancer modeling: is it ready for prime time? Nat Clin Pract Oncol. 2009;6:34–42. doi: 10.1038/ncponc1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E70.Eibl-Eibesfeldt I. Ethology: the biology of behavior Holt. Rinehart and Winston; 1970. [Google Scholar]
- ER60.Erdos P, Rényi A. On the evolution of random graphs. Publ Math Inst Hung Acad Sci. 1960;5:17–61. [Google Scholar]
- G08.Gershenson C, editor. Complexity: 5 Questions. Automatic Press; 2008. [Google Scholar]
- GE09.Gilbert SF, Epel D. Ecological Developmental Biology and Epigenesis: An Integrated Approach to Embryology, Evolution, and Medicine. Sinauer Associates; 2009. [Google Scholar]
- GG04.Griffiths PE, Gray RD. The Developmental Systems Perspective: Organism-environment systems as units of evolution. In: Pigliucci M, Preston K, editors. The Evolutionary Biology of Complex Phenotypes. Oxford and New York: Oxford University Press; 2004. [Google Scholar]
- GHA09.Galle J, Hoffmann M, Aust G. From single cells to tissue architecture - a bottom-up approach to modeling the spatio-temporal organization of complex multi-cellular systems. Journal of Mathematical Biology. 2009;58:261–283. doi: 10.1007/s00285-008-0172-4. [DOI] [PubMed] [Google Scholar]
- GS00.Gilbert SF, Sarkar S. Embracing complexity: Organicism for the 21st century. Developmental Dynamics. 2000;219:1–9. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1036>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- H05.Harold FM. Molecules into cells: specifying spatial architecture. Microbiol Mol Biol Rev. 2005;69:544–564. doi: 10.1128/MMBR.69.4.544-564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- H74.Hull D. The Philosophy of Biological Science. Englewood Clifts NJ: Prentice Hall; 1974. pp. 8–44. [Google Scholar]
- HGS04.Consortium International HGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- HI99.Huang S, Ingber DE. The structural and mechanical complexity of cell-growth control. Nat Cell Biol. 1999;1:E131–E138. doi: 10.1038/13043. [DOI] [PubMed] [Google Scholar]
- HM06.Hermann-Meyer M, Maini P, Iber D. An analysis of B cell selection mechanisms in germinal centers. Math Med Biol. 2006;23(3):255–277. doi: 10.1093/imammb/dql012. [DOI] [PubMed] [Google Scholar]
- I03.Ingber DE. Tensegrity I. Cell structure and hierarchical systems biology. J Cell Sci. 2003;116:1157–1173. doi: 10.1242/jcs.00359. [DOI] [PubMed] [Google Scholar]
- IC04.Izaguirre JA, Chaturvedi R, Huang C, Cickovski T, Coffland J, Thomas G, Forgacs G, Alber M, Hentschel G, Newman SA, Glazier JA. CompuCell, a multi-model framework for simulation of morphogenesis. Bioinformatics. 2004;20:1129–1137. doi: 10.1093/bioinformatics/bth050. [DOI] [PubMed] [Google Scholar]
- J77.Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- K02.Kitano H. Systems Biology A Brief Overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- K05.Keller EF. Revisiting “scale-free” networks. Bioessays. 2005;27:1060–1068. doi: 10.1002/bies.20294. [DOI] [PubMed] [Google Scholar]
- KG07.Kaiser M, Gorner M, Hilgetag CC. Criticality of spreading dynamics in hierarchical cluster networks without inhibition. New Journal of Physics. 2007;9:110. [Google Scholar]
- KL04.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- KM08.Krause S, Maffini MV, Soto AM, et al. A Novel 3D In Vitro Culture Model to Study stromal-Epithelial Interactions in the Mammary Gland. Tissue Eng Part C Methods. 2008;14:261–71. doi: 10.1089/ten.tec.2008.0030. [DOI] [PubMed] [Google Scholar]
- KS70.Keller EF, Segel LA. Initiation of slime mold aggregation viewed as an instability. J Theor Biol. 1970;26:399–415. doi: 10.1016/0022-5193(70)90092-5. [DOI] [PubMed] [Google Scholar]
- L04.Lazebnik Y. Can a Biologist Fix a Radio? - or, What I Learned while Studying Apoptosis. Biochemistry (Moscow) 2004;69:1403–1406. doi: 10.1007/s10541-005-0088-1. [DOI] [PubMed] [Google Scholar]
- LE01.Liljeros F, Edling CR, Amaral LA, et al. The web of human sexual contacts. Nature. 2001;411:907–908. doi: 10.1038/35082140. [DOI] [PubMed] [Google Scholar]
- MD06.McDougall S, Dallon J, Sherratt J, et al. Fibroblast migration and collagen deposition during dermal wound healing: mathematical modeling and clinical implications. Phil Trans R Soc A. 2006;364:1385–1405. doi: 10.1098/rsta.2006.1773. [DOI] [PubMed] [Google Scholar]
- N06.Noble D. The Music of Life: Biology beyond the Genome. Oxford: Oxford University Press; 2006. [Google Scholar]
- NP08.Nykter M, Price ND, Aldana M, et al. Gene expression dynamics in the macrophage exhibit criticality. Proc Natl Acad Sci U S A. 2008;105:1897–1900. doi: 10.1073/pnas.0711525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OS05.O’Malley MA, Dupre J. Fundamental issues in systems biology. BioEssays. 2005;27:1270–1276. doi: 10.1002/bies.20323. [DOI] [PubMed] [Google Scholar]
- P03.Preziosi L, editor. Cancer Modeling and Simulation. Chapman & Hall/CRC; 2003. [Google Scholar]
- P07.Prozorov R. Equilibrium Topology of the Intermediate State in Type-I Superconductors of Different Shapes Physical Review Letters. APS. 2007;98:257001. doi: 10.1103/PhysRevLett.98.257001. [DOI] [PubMed] [Google Scholar]
- PE06.Provenzano PP, Eliceiri KW, Campbell JM, et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PH07.Pearson GW, Hunter T. Real-time imaging reveals that noninvasive mammary epithelial acini can contain motile cells. J Cell Biol. 2007;179:1555–1567. doi: 10.1083/jcb.200706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PI08.Provenzano PP, Inman DR, Eliceiri KW, et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PR07.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- PZ05.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- R05.Rejniak KA. A single-cell approach in modeling the dynamics of tumor microregions. Math Biosci Eng. 2005;2:643–655. doi: 10.3934/mbe.2005.2.643. [DOI] [PubMed] [Google Scholar]
- R07.Rejniak KA. An immersed boundary framework for modeling the growth of individual cells: An application to the early tumor development. J Theor Biol. 2007;247:186–204. doi: 10.1016/j.jtbi.2007.02.019. [DOI] [PubMed] [Google Scholar]
- RA08.Rejniak KA, Anderson ARA. A computational study of the development of epithelial acini: I. Sufficient conditions for the formation of a hollow structure. Bull Math Biol. 2008;70:677–712. doi: 10.1007/s11538-007-9274-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RB02.Re VL, Bellini LM. William of Occam and Occam’s razor. Ann Intern Med. 2002;136:634–635. doi: 10.7326/0003-4819-136-8-200204160-00022. [DOI] [PubMed] [Google Scholar]
- RB28.Ritter WE, Bailey EW. The organismal conception: its place in science and its bearing on philosophy. Univ Calif Pub Zool. 1928;31:307–58. [Google Scholar]
- RS02.Ravasz E, Somera AL, Mongru DA, et al. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- S44.Schrödinger E. What is life. Cambridge University Press; 1944. [Google Scholar]
- S92.Sarkar S. Models of reduction and categories of reductionism. Synthese. 1992;91:167–194. [Google Scholar]
- S97.Stengers I. Cosmopolitiques. VI. La decouverte; Paris: 1997. [Google Scholar]
- SB06.Schubert W, Bonnekoh B, Pommer AJ, et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotechnol. 2006;24:1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- SEP.Stanford Encyclopedia of Philosophy. http://plato.stanford.edu/entries/reduction-biology/
- SS05.Soto AM, Sonnenschein C. Emergentism as a default: cancer as a problem of tissue organization. J Biosci. 2005;30:103–118. doi: 10.1007/BF02705155. [DOI] [PubMed] [Google Scholar]
- SS08.Soto A, Sonnenschein C, Miquel P. On physicalism and Downward Causation in Developmental and Cancer Biology. Acta Biotheoretica. 2008;56:257–274. doi: 10.1007/s10441-008-9052-y. [DOI] [PubMed] [Google Scholar]
- SS10.Soto AM, Sonnenschein C. Environmental causes of cancer: endocrine disruptors as carcinogens. Nat Rev Endocrinol. 2010;6:363–370. doi: 10.1038/nrendo.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- T52.Turing AM. The chemical basis of morphogenesis. Philosophical Transactions of the Royal Society. 1952;237:37–72. [Google Scholar]
- TB07.Thorne BC, Bailey AM, Peirce SM. Combining experiments with multi-cell agent-based modeling to study biological tissue patterning. Brief Bioinform. 2007;8:245–257. doi: 10.1093/bib/bbm024. [DOI] [PubMed] [Google Scholar]
- W07.Wunderlich V. Early references to the mutational origin of cancer. Int J Epidemiol. 2007;36:246–247. doi: 10.1093/ije/dyl272. [DOI] [PubMed] [Google Scholar]
- W53.Wardlaw CW. A commentary on turing’s diffusion-reaction theory of morphogenesis. New PhytologistWardlaw_1953_commentaryturingsdiffusion-reaction. 1953;52:40–47. [Google Scholar]
- WQ03.Weiss JN, Qu Z, Garfinkel A. Understanding biological complexity: lessons from the past. FASEB J. 2003;17(1):1–6. doi: 10.1096/fj.02-0408rev. [DOI] [PubMed] [Google Scholar]
- ZA07.Zhang L, Athale CA, Deisboeck TS. Development of a three-dimensional multiscale agent-based tumor model: simulating gene-protein interaction profiles, cell phenotypes and multicellular patterns in brain cancer. J Theor Biol. 2007;244:96–107. doi: 10.1016/j.jtbi.2006.06.034. [DOI] [PubMed] [Google Scholar]
- ZW09.Zhang L, Wang Z, Sagotsky JA, Deisboeck TS. Multiscale agent-based cancer modeling. J Math Biol. 2009;58:545–559. doi: 10.1007/s00285-008-0211-1. [DOI] [PubMed] [Google Scholar]