1. Introduction
Expression of breast cancer resistance protein (BCRP/ABCG2) limits the transport of xenobiotics across cellular membranes; hence BCRP expression on the apical surface of the human intestinal epithelium might limit transport across the gut lumen, thereby limiting oral bioavailability of pharmaceuticals [1–5]. The effect of BCRP on the absorption of xenobiotics may be antagonized by inhibiting BCRP function or by BCRP gene knockdown [6–9]. To date, several classes of functional BCRP inhibitors are available [10–13] but modulating intestinal BCRP expression to improve drug absorption remains largely unexplored.
Intestinal BCRP expression is regulated transcriptionally by alternative promoter usage. Previous work from our lab at the University of Maryland identified tissue-specific expression of BCRP mRNA isoforms (E1U, E1A, E1B and E1C) in humans. The BCRP mRNA isoforms identified shared the same coding region hence, they translated the same protein. However, their first exons were transcribed from distinct genomic regions upstream of BCRP Exon 2, indicating regulation of BCRP expression by alternative promoters. In the same study, analyzing the human small intestine for BCRP mRNA isoform expression identified only the E1B and E1C isoforms. The first exons of the E1B and E1C isoforms have unique 3′ splice acceptor sites but their sequences overlap towards the 5′ end [14]. This suggested transcription of intestinal BCRP mRNA from a single alternative promoter, which we designated as E1B/C. The E1B/C promoter is a functional promoter with confirmed cis regulatory elements [14, 15]. Interference with E1B/C promoter to enhance in vivo oral drug absorption requires the development of a suitable animal model.
Mouse Bcrp1 protein and human BCRP protein are structurally and functionally similar. They share 87% sequence homology and efflux identical substrates [10]. Murine in vivo models harboring Bcrp1 gene knockouts in FVB or C57BL/6J mouse strains are used to predict BCRP regulation of human small intestinal drug absorption [16, 17]. Mouse Bcrp1 mRNA and protein are expressed throughout the mouse intestine with highest expression in the mouse ileum [18, 19]. Humans too may have region-specific variation in expression of BCRP but the data available in the literature are controversial [20–22]. Similarities in BCRP and Bcrp1 regulation emerged when Zong et al identified three alternative Bcrp1 mRNA isoforms (E1a, E1b and E1c) with transcription initiation from alternative promoters. Using an unspecified region of the mouse intestine, they attributed mouse small intestinal Bcrp1 mRNA expression to the Bcrp1 E1b mRNA isoform [23].
Since intestinal region-specific differences exist for mouse Bcrp1 mRNA and protein expression [18, 19], we hypothesize that similar intestinal region-specific differences may also exist for Bcrp1 alternative promoter usage. In addition, we recently identified a novel Bcrp1 mRNA isoform designated E1u because of its location 5′ upstream of the other alternative first exons on mouse chromosome 6 (to be published separately). E1u expression in the mouse intestine is yet to be characterized. Hence, in this study, we first quantified the four Bcrp1 mRNA isoforms in the mouse duodenum, jejunum and ileum. We established the predominance of the E1b isoform in all regions of the mouse small intestine. Using reporter assays we established functional activity for the promoter upstream of E1b for the first time, narrowing the core promoter region to −231/−42 bps in relation to the transcription start site of E1b. Finally, using functional studies and ChiP assays we established phospho-CREB (p-CREB) as a positive regulatory factor of the mouse E1b Bcrp1 promoter region.
2. Materials and Methods
2.0 Materials
The cAMP analog, 8Br-cAMP and phosphatidylinositol-3-kinase (PI3K), inhibitor LY294002 were purchased from Sigma-Aldrich, Inc (St. Louis, MO). The protein kinase A (PKA) inhibitor, H89 and the extracellular signal-regulated kinase (ERK) inhibitor PD98059 were obtained from LKT Laboratories, Inc (St. Paul, MN) and Cell Signaling Technology (Danvers, MA) respectively. The rat anti-mouse Bcrp1 antibody (BXP-9) was purchased from Abcam (Cambridge, MA), the rabbit anti-mouse GAPDH and the horse radish peroxidase (HRP) labeled secondary antibodies were purchased from Cell Signaling Technology.
2.1. Collection of mouse and human intestinal tissues
The animal use protocol was approved by University of Maryland School of Medicine Institutional Animal Care and Use Committee. Male FVB/NCr mice (6 weeks old) purchased from the National Cancer Institute were allowed to acclimatize for a minimum of two weeks with food and water ad libitum. Mice were euthanized by cervical dislocation, the entire intestine removed and the intestinal contents flushed three times with 1 ml of ice cold saline. The duodenum (1 cm region distal to the stomach), jejunum (16-19 cm from duodenum) and ileum (3 cm from the tip of the cecum) were collected in individual cryovials and flash frozen in liquid nitrogen [24]. Human intestinal samples were collected under an IRB-approved protocol as described previously [14].
2.2. Cell Culture
The mtsA58 transgenic mice, harboring a thermo-labile mutant of SV40 Large T antigen (tsA58) inserted downstream of an interferon-γ inducible H-2K promoter element [25]. The MSIE cell line is grown at a permissive temperature of 33°C (T-antigen stable) in RPMI media (Biosource, Invitrogen, Carlsbad, CA) supplemented with 5% FBS (Biosource), 10% Penicillin-Streptomycin and 5 units of mouse interferon-γ/ml (Peprotech Inc, Rocky Hill, NJ). Interferon-γ was added every two to three days. Growing the cells at the permissive temperature (33°C) preserves the activity of the temperature sensitive T-antigen (T-Ag) mutant tsA58 and facilitates MSIE cell proliferation while the interferon-γ supplementation drives the tsA58 mutant T-Ag expression. MSIE cells grown in interferon-free conditions for at least a week were used for the experiments. MSIE cells remain viable for at least 10–15 days in culture after interferon-γ withdrawal. MSIE cells grown to 90% confluence in 100 mm dishes for at least a week under interferon-free conditions were used to prepare mRNA and protein. The NIH3T3 cells (a kind gift from Dr. Anne Hamburger, University of Maryland, Greenebaum Cancer Center) were grown in DMEM media (Biosource) supplemented with 10% FBS (Biosource) and 10% Penicillin-Streptomycin.
2.3. RNA and cDNA preparation
Mouse intestinal sections were individually homogenized with a mortar and pestle and the resulting homogenate was used to prepare both RNA and protein. RNA was extracted from each intestinal section and also from the MSIE cell line using Trizol (Invitrogen, Carlsbad, CA) and reverse transcribed with M-MULV reverse transcriptase (Roche Diagnostics, Basel, Switzerland). Human small intestinal RNA was prepared as described previously [14].
2.4. Quantitative RT-PCR
The alternative Bcrp1 mRNA isoforms from the different regions of the mouse intestine were absolutely quantified with standard curves generated by serial dilution of the respective full-length isoforms inserted in pcDNA3 vector. 1μg of the reverse-transcribed RNA was then PCR-amplified in real-time with IQ SYBR green mix (Bio-Rad, Hercules, CA) using the MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad). Each reaction was performed in duplicate. Total Bcrp1 mRNA was amplified with primers spanning Exon 6 and Exon 7 while the individual isoforms were amplified using isoform specific forward primers and a common reverse primer in Exon 2. The standard curves for the individual isoforms were prepared from the mouse Bcrp1 cDNA inserted into pBluescript I KS (−) vector (kind gift from Dr. Naoko Takebe MD, PhD, CTEP, NCI). To prepare the full-length Bcrp1 cDNA isoforms with alternative 5′ UTRs, the Bcrp1 cDNA from pBluescript I KS (−) vector was first restriction enzyme digested and inserted into the pcDNA3 expression vector. Next, the E1u, E1a, E1b and E1c 5′UTR sequences were PCR-amplified using 5′ UTR specific forward primers spanning the 5′ end of the individual 5′ UTR sequences and a common Bcrp1 specific reverse primer designated as Bcrp1 OP R in Exon 2 (Table 1) and cloned into pCR2.1 vector using the TOPO TA cloning kit (Invitrogen). Each of the four TOPO cloned Bcrp1 5′ UTRs were then restriction enzyme digested and inserted upstream of the translational start site in Exon 2 of the Bcrp1 cDNA in the pcDNA3 vector. The sequence of each promoter construct was confirmed by sequence analysis (Biopolymer/Genomics shared services). The data from three different mice were collected and analyzed for significant differences in mRNA isoform expression along the mouse small intestine. The data for the MSIE cell line is representative of at least three different passages. The sequences for 5′ UTR cloning and qRT-PCR primers are given in Table 1.
Table 1.
Primers
| Primer Name | Sequence | Function | Digestion site |
|---|---|---|---|
| Bcrp1 OP R | cgcctgtgggtcccagaata | 5′ RACE, 5′ UTR cloning | |
| Bcrp1 IP R | gcgacattggtactaacacg | 5′ RACE | |
| E1u 5′ Bcrp1 F | aggaatcacacgaatggaag | 5′ UTR cloning | |
| E1a 5′ Bcrp1 F | ctccttgccagataagag | 5′ UTR cloning | |
| E1b 5′ Bcrp1 F | agacgctcctggcgggcgag | 5′ UTR cloning | |
| E1c 5′ Bcrp1 F | aggttgaatatttggagac | 5′ UTR cloning | |
| Bcrp1 F | cgaatgctgtccttttgct | Real-time qRT-PCR | |
| Bcrp1 R | ataccgaggctgatgaatgg | Real-time qRT-PCR | |
| E1u Bcrp1 F | agacaccctgatgttacc | Real-time qRT-PCR | |
| E1a Bcrp1 F | gacacctcattacacatagc | Real-time qRT-PCR | |
| E1b Bcrp1 F | gcggctgctgctcc | Real-time qRT-PCR | |
| E1c Bcrp1 F | gaagaaccagaccaataagggaaa | Real-time qRT-PCR | |
| E2 Bcrp1 R | ctccttctgcgagcgtcct | Real-time qRT-PCR | |
| E1a −1875 F | cacctgggagctctgctacagga | Deletion construct | Sac1 |
| E1a +220 R | gcctcaagagacaggcaaaatcaag | Deletion construct | |
| E1u −1906 F | tgcagtggaaagagctcccttta | Deletion construct | Sac1 |
| E1u +64 R | gcagagaagctagcatcagggtgtc | Deletion construct | Nhe1 |
| E1b −1847 F | caggtaccgaggctgtggcttggcacttgt | Deletion construct | Kpn1 |
| E1b +60 R | cggctagccagccgcgccagaactcg | Deletion construct | Nhe1 |
| E1c −1904 F | gtctctggggaatgagctcaag | Deletion construct | Sac1 |
| E1c +83 R | tcccttattggacgcgttcttctca | Deletion construct | Mlu1 |
| E1b −1027 F | tttgcaagccatggtaccgc | Deletion construct | Kpn1 |
| E1b −305 F | cccagcctggtaccgttat | Deletion construct | Kpn1 |
| E1b −231 F | cacagggtgaaggtaccctga | Deletion construct | Kpn1 |
| E1b −75 F | cctggtaccggcttct | Deletion construct | Kpn1 |
| E1b PM1S | cggcgcgagctgaccacacggcggtc | Site directed mutagenesis | |
| E1b PM1AS | gaccgccgtgtggtcagctcgcgccg | Site directed mutagenesis | |
| E1b CRE F | tctcatcaaaaggccatcca | ChiP real-time qPCR | |
| E1b CRE R | aacctggaggacccacaaga | ChiP real-time qPCR |
The restriction enzyme digestion sites are underlined in the primers used for preparing the deletion constructs. For the primers used for site-directed mutagenesis, the CRE site is underlined; the sites of the point mutations are indicated in BOLD.
2.5. 5′ Rapid amplification of cDNA ends (RACE) PCR
RNA obtained from mouse jejunum epithelial cells and human small intestine was subjected to 5′ RACE PCR according to First Choice RLM-RACE kit instructions (Ambion Inc, Austin, TX) using forward primers supplied in the kit and Bcrp1 specific reverse primers designated as Bcrp1 OP R and Bcrp1 IP R in Table 1. The Bcrp1 5′ RACE products were inserted into pCR2.1-TOPO vector (Invitrogen), cloned and sequenced at the Biopolymer/Genomics shared services at University of Maryland Greenebaum Cancer Center (UMGCC). The mouse Bcrp1 5′ RACE sequences were compared with the 80kb genomic region upstream of mouse Bcrp1 Exon 2 and human BCRP 5′ RACE sequences were compared with 100kb genomic region upstream of human BCRP Exon 2 using the BLAST program at NCBI and mapped in relation to the translational start site in Exon 2. The Bcrp1 5′ RACE products sequences are available in the NIH genetic sequence database, GenBank (Accession No: 1316841). See supplemental table S1.
2.6. Western Blotting
Intestinal protein supernatants were prepared by sonicating each intestinal homogenate in RIPA buffer (50 mM Tris-HCl, pH 7.4; 1% NP-42; 0.25% sodium deoxycholate; 150 mM NaCl; 1 mM EDTA; 1 mM sodium orthovanadate; 1μg aprotinin/ml and 2 mM Pefabloc SC, Roche Diagnostics) and clearing the sample of cell debris by centrifugation at 10,000 × g for 10 min. MSIE cells were gently scraped off the culture surface and the supernatant from the cells was prepared as mentioned above for intestinal homogenate. Protein concentrations were then measured with Bio-Rad Protein assay (Bio-Rad). 20 μg protein from each intestinal sample and 80 μg protein from cell culture samples were separated on 10% SDS-PAGE gel. The electrophoretically separated proteins were transferred to PVDF membranes and probed with rat anti-mouse Bcrp1 (1:50) and anti-GAPDH (1:3000) primary antibodies followed by HRP labeled goat anti-rat and goat anti-rabbit secondary antibodies (1:3000 each). The immune complexes were visualized with a chemiluminescent substrate. The expression of Bcrp1 protein and GAPDH protein in each intestinal section was quantified by densitometric analysis (Visionworks LS image acquisition and analysis software, UVP, Upland, CA).
2. 7. Deletion constructs
The regions flanking the transcription start site of Bcrp1 alternative 5′ UTRs (E1u −1906/+64, E1a −1875/+10, E1b −1847/+60 and E1c −1904/+83) were PCR-amplified from BAC clones RP23-285A12 and RP24-314E24 (BACPAC Resources Center, Children’s Hospital Oakland Research Institute, Oakland, CA) using restriction digestion site-inserted forward and reverse primers (Table 1). The PCR-amplified fragments were restriction digested and inserted into the pGL3-Basic vector (Promega, Madison, WI). Nucleotide sequences of the selected clones were confirmed by sequencing. The 5′ end shortened deletion constructs for E1b −1847/+60 were prepared by shortening the 5′ end to −1027, −305, −231 and −75 bps while maintaining the 3′ end at +60 bps by using a common reverse primer and sequence specific forward primers (Table 1). On sequencing the clones from E1b −231/+60 deletion construct, some clones were found to have a deletion from bp −58 to bp −42, likely due to a PCR artifact. Clones with such deletions were hence designated as the Δ −58/−42 Bcrp1 E1b −231/+60 deletion construct. Two point mutations T-33A and G-32C were introduced on the CRE in the E1b −1847/+60 deletion construct using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies Inc, Santa Clara,CA) as per manufacturer instructions. The sense (E1b PM1S) and antisense (E1b PM1AS) primers for generating the two point mutant, designated as E1bPM −1847/+60, are given in Table 1. The CRE on the primers is given in bold and the point mutations are underlined.
2.8. Reporter assay
MSIE cells were seeded and grown in complete growth media in the absence of interferon-γ for at least a week before the experiments. On the day of the experiment, MSIE cells were plated at a density of 50,000 cells/well in 48 well plates and allowed to attach for 6 hrs. Each of the above-mentioned Bcrp1 deletion constructs (100 ng/mL) and the pGL3-Basic vector (100 ng/mL, negative control) were then transiently co-transfected with the internal control, pRL-TK vector (20 ng/mL, Promega) using FuGENE 6 transfection reagent (Roche Diagnostics). Following transfection, the cells were cultured for an additional 48 hours at 33°C, after which the dual luciferase assay (Promega) was performed using the 20/20n Single Tube Luminometer (Turner Biosystems, Sunnyvale, CA) in accordance with kit instructions. Each experiment was performed at least thrice with triplicate samples per experiment. The luminescence of each deletion construct and of the pGL3-Basic vector relative to that of pRL-TK was calculated. To compare the promoter activity between the four alternative promoters, the relative luminescence of each alternative promoter construct was normalized over pGL3-Basic vector. For deletion analysis of the E1b promoter region, the relative luminescence of each shorter E1b deletion construct was normalized over luminescence of the -1847/+60 construct. For determining the promoter activity of the E1bPM −1847/+60, NIH3T3 fibroblasts cultured in 24-well plates at a density of 500,000 cells/well were transfected with either Bcrp1 E1b −1847/+60 deletion construct, the E1bPM −1847/+60 deletion construct or pGL3-Basic vector. After 24 hrs of transfection, cells were serum starved overnight and then stimulated with 2 mM 8Br-cAMP (final concentration) in SFM for 6 hours. Following stimulation, cells were lysed in passive lysing buffer and dual luciferase assays were performed as detailed above. pRL-TK vector was used as internal control for transfection. For promoter assays in presence of analogs and inhibitors of cAMP, NIH3T3 fibroblasts were plated in 24-well plate with density of 500,000 cells/well. After 6 hr plating, cells were transfected with 1μg of pGL3-basic vector or Bcrp1 E1b/PGL3 reporter plasmid and co-transfected with 4 ng of the internal control, pRL-TK in 20 μl SFM plus 1.5 μl of FuGENE 6 (Roche diagnostics). After 24 hr of transfection, cells were starved in SFM overnight, then incubated with a inhibitor mix (20 μM of H89, 50 μM of LY294002 and 50 μM of PD98059, final concentration in SFM for 20 min. After washing, followed by stimulation with 2 mM 8Br-cAMP for 6 hrs, cells were lysed with passive lysing buffer and luciferase activity was determined with Dual-Luciferase reporter assay kit (Promega). The results are expressed as the ratio of the luciferase activity over the internal control.
2.9. Identification of cis and trans-elements on Bcrp1 E1b and BCRP E1B/C promoters
A 500 bp region upstream of E1b 5′ untranslated region (5′ UTR) was scanned for transcription factor binding sites using the PROSCAN [26] promoter scan program as well as the PAGEN [27] program and also searched against the Transcription Regulatory Regions Database [28] using BLAST. All three programs were used to identify additional transcription factor binding sites on the 500 bp region upstream of human BCRP E1C 5′ UTR.
2.10. ChiP assay
NIH3T3 fibroblasts, cultured in 100 mm dishes after serum starvation overnight, were stimulated with 2 mM 8Br-cAMP in SFM for 30 min and subjected to ChiP assay as per manufacturer instructions (Millipore, Temecula, CA). Briefly, cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Cells were then harvested in sodium dodecyl sulfate lysis buffer and the DNA sheared to 200–1000 bps by sonication. The sonicated cell lysates were precleared with protein A agarose beads pre-blocked in Bovine Serum Albumin (Millipore). Immunoprecipitation of protein-DNA complexes was then performed on the pre-cleared lysates with rabbit anti-mouse p-CREB antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Simultaneously, pre-cleared lysates were also incubated with anti- rabbit IgG (Cell Signaling Technology) to control for potential non-specific co-immunoprecipitations. DNA was quantified by real-time qPCR using promoter-specific primers for p-CREB site on E1b. The primers for qRT-PCR are given in Table 1.
2.11. Statistical analysis
Linear regression analysis was used to quantify the alternative Bcrp1 mRNA isoforms as well as the total Bcrp1 mRNA isoform expression from each region of the mouse intestine using MYIQ™ real-time PCR detection system software version 1.0. All other statistical analyses were performed with GraphPad Prism software version 4.03. Statistical significance for real-time qRT-PCR and the promoter activity of the alternative 5′ untranslated regions was determined using the Kruskal-Wallis non-parametric test with post hoc Dunn’s test. The statistical significance of the promoter activity for the shorter E1b deletion constructs was determined (p<0.005) using non-parametric t-test with Welch’s correction. The statistical significance of the promoter activity for the E1b −1847/+60 deletion construct and its point mutant, both in the presence and absence of analog/inhibitor mix of cAMP as well as the statistical significance of the ChiP assays was measured with non-parametric Student’s t-test.
3. Results
3.1. E1b is the major mouse intestinal Bcrp1 mRNA isoform
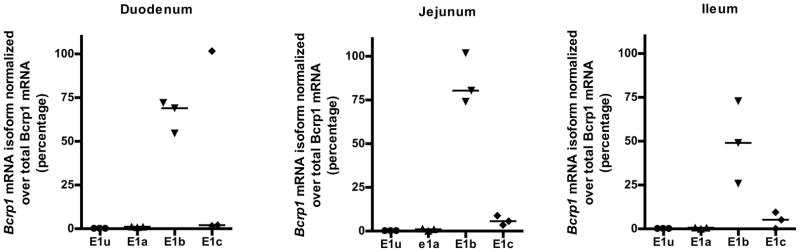
Alternative 5′ UTR usage is indicative of alternative promoter usage. Hence to determine small intestinal region-specific alternative Bcrp1 promoter usage, Bcrp1 mRNA isoform expression in the mouse duodenum, jejunum and ileum was characterized using real-time qRT-PCR. The Bcrp1 E1b mRNA isoform accounted for more than 50% of the total Bcrp1 mRNA expression in each intestinal section studied (Figure 1) except for the high expression of E1c observed from the duodenum of one mouse. Since the sum of the individual isoforms failed to add up to the total Bcrp1 expressed in this particular mouse, it was classified as an outlier. Bcrp1 E1c and E1a mRNA isoforms were also detected in the mouse small intestine, although the median expression levels were much lower than that of the E1b isoform (Figure 1). The E1u mRNA isoform was not detected in any part of the mouse small gut (Figure 1). Taken together, these data suggest that the alternative promoter upstream of the Bcrp1 E1b 5′ UTR predominantly regulates Bcrp1 expression in the mouse small intestine.
Figure 1. Real-time quantification of Bcrp1 5′ UTR expression in different regions of the mouse intestine.
E1b was the major Bcrp1 5′ UTR expressed in the mouse duodenum, jejunum and ileum accounting for at least 50% of the total Bcrp1 mRNA expression. Each data point in the figure is the average of duplicate PCR reactions from individual mice. Median values are represented by horizontal bar. E1u, E1a, E1b and E1c represent Bcrp1 alternative first exons 72kb, 58kb, 10kb and 5kb respectively upstream from the translation start site of Bcrp1 in Exon 2.
3.2. Utilization of variable transcription start sites by Bcrp1 E1b promoter
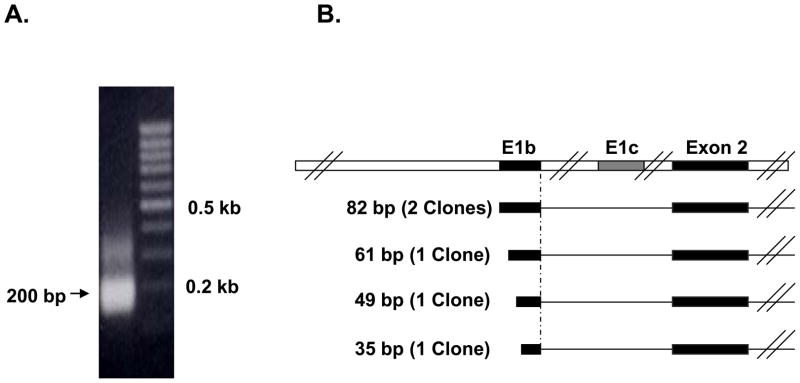
In order to characterize the transcription start site of the mouse intestinal Bcrp1 E1b mRNA isoform, 5′ RACE PCR was performed on a representative mouse intestinal tissue, the jejunum epithelium. Gel electrophoresis of the mouse intestinal 5′ RACE products yielded an intense 200 bp band (Figure 2A). This band was cloned and five clones were randomly selected and sequenced. These sequences have been deposited in GenBank (supplemental Table S1). On subsequent sequence comparison with the genomic region upstream of Bcrp1 Exon 2, the five clones aligned exclusively with the Bcrp1 E1b 5′ UTR supporting the real-time qRT-PCR data for predominance of the Bcrp1 E1b mRNA isoform expression in the mouse intestine. While all E1b clones shared a common 3′ end, the intestinal Bcrp1 E1b 5′ UTRs from five clones were mostly of variable lengths (82 bp or 61 bp or 45 bp or 35 bp), demonstrating variable transcription start site utilization in the mouse intestinal E1b promoter (Figure 2B).
Figure 2. RACE-PCR analysis of mouse jejunum RNA.
A) Agarose gel electrophoresis of the final RACE product yielded a 200 bp band. B) Schematic representation of the mouse jejunum 5′ RACE products in relation to Exon 2 of Bcrp1 mRNA. E1b 5′ UTR with multiple transcription start sites was identified as the major 5′ UTR in the mouse jejunum
3.3. Bcrp1 mRNA and protein expression along the mouse intestine
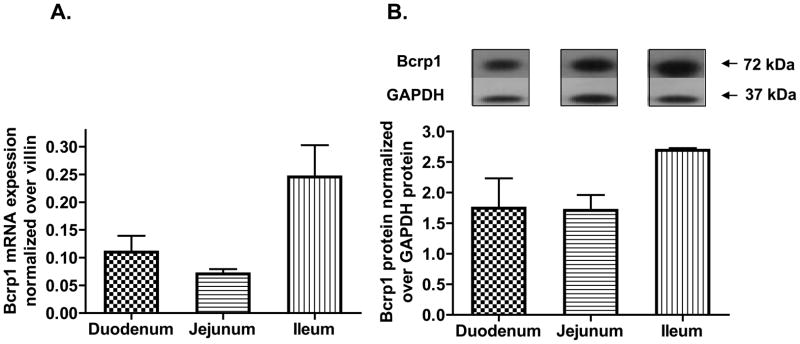
Real-time PCR quantification identified high expression for Bcrp1 mRNA in the mouse duodenum and jejunum and higher Bcrp1 mRNA expression in the mouse ileum (Figure 3A). Immunoblotting followed by densitometric quantification established maximum Bcrp1 protein expression in the mouse ileum as well (Figure 3B).
Figure 3. Mouse intestine region-specific Bcrp1 mRNA and protein expression.
A) Real-time RT-PCR quantification of total Bcrp1 mRNA expression in the different regions of the mouse intestine. Highest Bcrp1 mRNA expression was observed in the ileum. B) A representative immunoblot of Bcrp1 and GAPDH (housekeeping gene) from the various intestinal regions and the densitometric analysis of the relative Bcrp1 expression (n=3) in the different regions of the intestine using GAPDH as control is shown below the immunoblot. Highest Bcrp1 expression was observed in the ileum
3.4. Reporter activity for alternative Bcrp1 promoters in the MSIE cell line
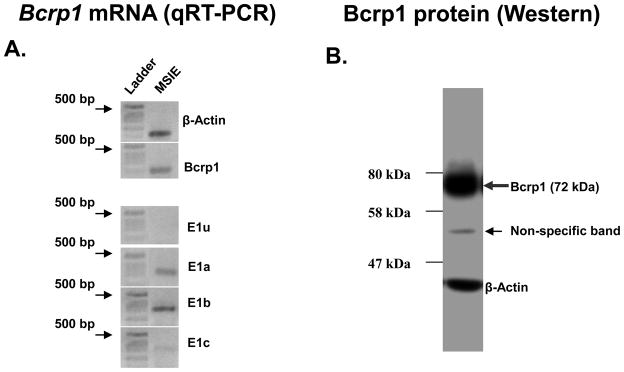
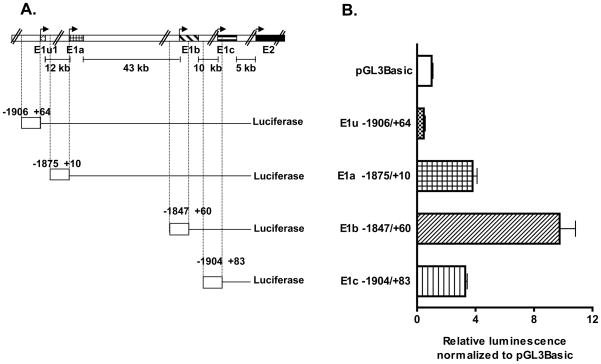
To test the ability of the 5′ region upstream of the altenative Bcrp1 5′ UTRs to promote gene transcription in mouse intestine using reporter assays we chose the conditionally immortalized MSIE cells as a model of mouse intestinal epithelium. First, the MSIE cell line was characterized for Bcrp1 alternative mRNA isoform expression using qRT-PCR. The MSIE cell line, like the small intestine, predominantly expressed the Bcrp1 E1b mRNA isoform (Figure 4A). Comparatively lower levels of both Bcrp1 E1a and E1c isoform were also expressed in the MSIE cell line, but not the E1u isoform (Figure 4A). Bcrp1 protein expression in the MSIE cell line was confirmed by the appearance of a 72 kDa immunoreactive band on Western blotting (Figure 4B). To prepare luciferase reporter constructs, the 5′ regions upstream of each Bcrp1 alternative first exon was cloned into pGL3-Basic vector. A schematic showing the location of E1u −1906/+64, E1a −1875/+10, E1b −1847/+60 and E1c −1904/+83 promoter constructs in relation to the TSS of their respective longest 5′ UTRs identified, as well as in relation to Bcrp1 Exon 2, is given in Figure 5A. As expected from the expression of Bcrp1 E1a, E1b and E1c mRNA isoforms in the MSIE cell line (Figure 4A), significant (Kruskal-Wallis non-parametric test, p<0.005) promoter activity over the empty vector (pGL3-Basic) was observed only for deletion constructs upstream of the E1a, E1b and E1c Bcrp1 5′ UTRs (Figure 5B) but not for E1u, the 5′ UTR not expressed in the MSIE cell line. The highest promoter activity was observed for the promoter upstream of E1b 5′ UTR (Fig 5B) which was the major Bcrp1 5′ UTR expressed in the MSIE cell line (Figure 4A).
Figure 4. Bcrp1 5′ UTR and protein expression in MSIE cell line.
A) Bcrp1 mRNA was expressed in the MSIE cell line and E1b 5′ UTR expression was higher than either E1a or E1c 5′ UTR expression. B) Immunoblotting identified the 72 kDa Bcrp1 protein monomer to be expressed in the MSIE cell line. The faint band of ~52 kDa is non-specific.
Figure 5. Promoter activity for the alternative Bcrp1 5′ UTRs in the MSIE cell line.
A) Schematic representation of the four Bcrp1 5′ UTRs in relation to Exon 2 and the promoter deletion constructs for each 5′ UTR in relation to the TSS (indicated by arrows) of the respective 5′ UTRs. The distance between each 5′ UTR is also shown. B) Promoter activity was higher than the empty vector control (pGL3-Basic) for E1a, E1b and E1c promoter constructs while E1u promoter activity was lower than pGL3-Basic promoter activity in the MSIE cell line. The experiment was repeated at least three times with triplicate samples per experiment.
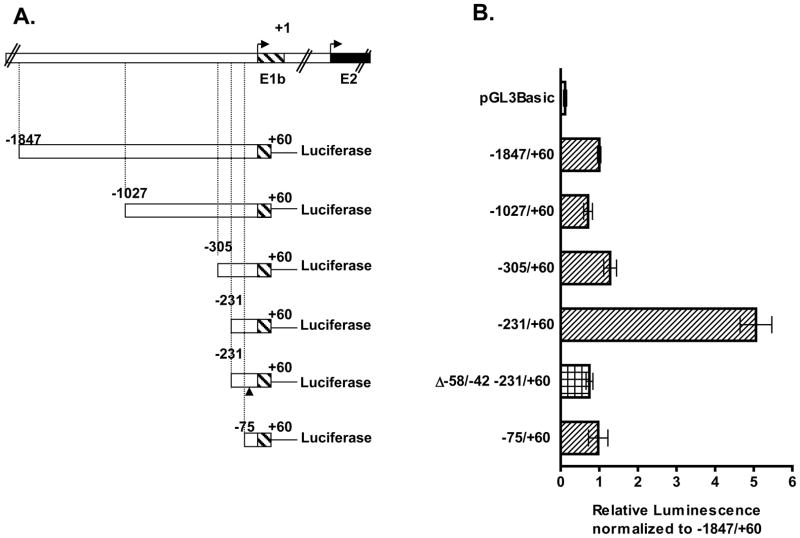
3.5. Identification of the core promoter region upstream of Bcrp1 E1b 5′ UTR
The core promoter region upstream of the major Bcrp1 5′ UTR expressed in the MSIE cell line (E1b) was then characterized using deletion analysis (Figure 6). We made five progressively 5′ end shortened Bcrp1 E1b deletion constructs, −1847/+60, −1027/+60, −305/+60, −231/+60, and −75/+60 (Figure 6A) which all exhibited significant promoter activity (p<0.001) compared to the empty vector control, pGL3-Basic (Figure 6B). The promoter activity remained comparable for the −1847/+60, −1027/+60, −305/+60 deletion constructs but increased significantly on additional 5′ end shortening to the −231/+60 deletion construct (Figure 6B). The comparatively high reporter activity observed for the −231/+60 construct was lost on the successive 5′ end shortened −75/+60 construct. Of note, the reporter activity observed for the −231/+60 construct was also lost when ~16 bps −58/−42 were deleted in the −231/+60 construct (−231/+60 Δ −58/−42 construct) (Figure 6B). Hence we conclude the regions between −231 and −75 bp and between −58 and −42 bp contain areas for binding of positive trans acting regulatory elements. We deduce the core promoter region occupies the region between −231 and −42 bps.
Figure 6. Promoter activity of Bcrp1 E1b deletion constructs.
A) Schematic representation of E1b deletion constructs in relation to the transcription start site of the longest E1b 5′ UTR sequenced. The position of the deleted 17 bp in the −231/+60 bp Δ −58/−42 is indicated by (▲). B) Reporter activity of the mouse Bcrp1 E1b deletion constructs in the MSIE cells. Cells were grown in the absence of interferon-γ for at least a week before the experiments. The mean ± S.E.M. is shown for each deletion constructs. The experiments were repeated at least three times with duplicate samples per experiment.
3.6. In silico characterization of E1b promoter region
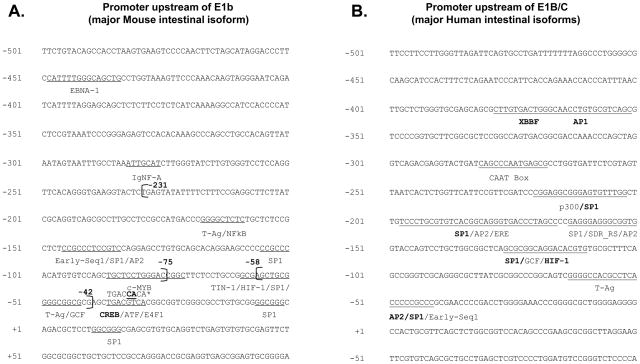
The cis/trans factor prediction programs identified several transcription factor binding sites within the E1b core promoter region spanning −231 to +60 bps (Figure 7A). Most of the transcription factor binding sites predicted in silico lie within the −58 to −42 bps and −231 to −75 bps, regions identified by reporter assays to contain important cis-regulatory elements (Figure 7A). Several Sp1 sites with varying weights (increasing weight indicates preferential binding of the transcription factor to promoter regions compared to non-promoter regions) were predicted in the core promoter region by the PROSCAN promoter prediction program (Table 2). Interestingly, the Sp1 binding site predicted with weight = 34.423 (Table 2), occupied the region between −58 and −42 bp, the area deleted in the Δ −58/−42 variant of the −231/+60 construct. T-Ag, hypoxia-inducible factor (HIF) and GC-binding factor (GCF) binding sites were also identified in this region. The region between −231 and −75 bp contained two additional Sp1 binding sites predicted by PROSCAN (weight = 3 and 8) and also a T-Ag, CREB, NF-kB and AP2 binding sites. IgNF-A, ATF and E4F1 were additional sites predicted in the −231/+60 E1b promoter region. The mouse E1b promoter region and the human E1B/C promoter share only the Sp1, HIF, AP2, GCF and T-Ag transcription factor binding sites (Figure 7A, B). The CCAAT box and the estrogen response element [29] are unique to the human E1B/C promoter while NF-kB, CREB, IGNF-A, c-MYB, ATF and TIN-1 binding sites are unique to the mouse E1b promoter. The position of the cis elements, software program utilized and their weights are given in Table 2.
Figure 7. Comparison of predicted and identified cis-elements on Bcrp1 E1b and BCRP E1B/C promoter regions.
Several SP1, Hif-1, T-Ag, AP2 sites were common to both the mouse and human promoter regions. A) The start site of the −231/+60 and −75/+60 deletion constructs as well as the −58/−42 spontaneous deletion region are shown in bold in the figure. The CREB binding site, selected for mutational analysis, is indicated by *. The position of the point mutations and the mutant nucleotides are given in bold face above the original nucleotide sequence. B) The previously predicted transcription factor binding sites on the human promoter region are in BOLD type face while sites predicted using PROSCAN, PAGEN and TRRD are in normal font color.
Table 2. Predicted cis/trans element location on E1b Bcrp1 promoter.
All PROSCAN identified signals on the positive strand within 500 bps upstream of the TSS of E1b 5′ UTR is shown below.
| Cis binding factor | Trans regulatory element | Position | Promoter Prediction Program | Weight | |
|---|---|---|---|---|---|
| Start | End | ||||
| EBNA-1 | CATTTTGGGCAGCTG | −449 | -435 | TRRDsite | 3.8* |
| IgNF-A | ATTGCAT | −282 | −276 | PROSCAN | 1.434 |
| NF-kB | GGGRNTYYC | −167 | −159 | PROSCAN | 1.08 |
| T-Ag | GGGGC | −167 | −164 | PROSCAN | 1.086 |
| EARLY-SEQ1 | YYCCGCCC | −147 | −140 | PROSCAN | 6.322 |
| AP-2 | YCSCCMNSSS | −145 | −136 | PROSCAN | 1.355 |
| Sp1 | CCGCCC | −145 | −140 | PROSCAN | 3.292 |
| AP-2 | CCCMNSSS | −109 | −102 | PROSCAN | 1.108 |
| (Sp1) | CCCCGCCC | −107 | −100 | PROSCAN | 8.117 |
| Sp1 | CCCGCC | −106 | −101 | PROSCAN | 2.755 |
| TIN-1 | TGCTCCTGGGACCGGC | −87 | −72 | TRRDsite | 3.8* |
| c-MYB | TGCTCCTGGGACCGGC | −87 | −72 | TRRDsite | 3.8* |
| HIF-1 | GCGAGCTGCGGGGCGG | −60 | −45 | TRRDsite | 3.8* |
| GCF | SCGSSSC | −52 | −46 | PROSCAN | 2.361 |
| Sp1 | CGGGGCGGCG | −51 | −42 | PROSCAN | 34.423 |
| T-Ag | GGGGC | −50 | −46 | PROSCAN | 1.086 |
| Sp1 | GGGGCGGCGC | −50 | −41 | PROSCAN | 3.442 |
| Sp1 | GGGCGG | −49 | −44 | PROSCAN | 3.013 |
| beta-pol_CS | NTGACGTCAN | −37 | −28 | PROSCAN | 8.603 |
| CREB | TGACGTC | −36 | −31 | PROSCAN | 3.442 |
| CREB | TGACGTCA | −36 | −29 | PROSCAN | 8.603 |
| CREB | TGACGTYW | −36 | −29 | PROSCAN | 3.886 |
| ATF | TGACGYMR | −36 | −29 | PROSCAN | 3.721 |
| ATF | TGACGT | −36 | −31 | PROSCAN | 1.157 |
| CREB | KWCGTCA | −35 | −29 | PROSCAN | 1.912 |
| E4F1 | ACGTCAC | −34 | −28 | PROSCAN | 3.824 |
| E4F1 | ACGTMAC | −34 | −28 | PROSCAN | 3.764 |
| ATF/CREB | ACGTCA | −34 | −29 | PROSCAN | 1.564 |
| CREB | CTGACGTCACGG | −38 | −27 | PAGEN | 94%** |
Transcription factor binding sites with an E value less than 10 for TRRDsites indicated by* and a score higher than 90% for PAGEN indicated by ** are given below.
3.7 Characterization of the cAMP response element (CRE) on the E1b −1847/60 promoter in NIH3T3 cells
We used NIH3T3 murine fibroblasts for these studies because they obviated the potential interference of T-Ag/interferon-γ observed in the MSIE cell line. Also, NIH3T3 cells express significant amount of Bcrp1 E1b mRNA isoform and exhibit E1b promoter reporter activity (Supplemental figure S1).
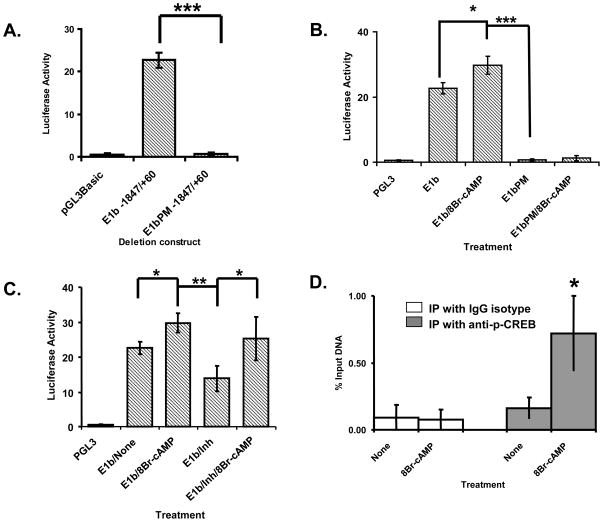
To understand intestine-specific regulation of mouse Bcrp1 expression further, we introduced two point mutations in the CRE (indicated by * in Figure 7A), a CIS site predicted exclusively on the mouse E1b promoter, the major intestinal promoter. We then compared the reporter activity of the mutated E1bPM −1847/+60 construct with its non-mutated parent construct. The reporter activity of the −1847/+60 E1b construct was completely lost in the mutated E1bPM −1847/+60 construct and was comparable with the reporter activity of the negative control pGL3-Basic (Figure 8A). This confirmed the presence of a critical, positive cis regulatory element in the mutated region. Next we treated the mutated and non-mutated constructs with 8Br-cAMP, an analog of cAMP and compared their reporter activities with their respective non-treated controls. We observed that the reporter activity of the non-mutated construct but not the point mutant increased on treatment with 8Br-cAMP suggesting the cis regulatory element is responsive to cAMP (Figure 8B). We then tested the response of the non-mutated −1847/+60 deletion construct to inhibitors of cAMP (Figure 8C). The reporter activity of the −1847/+60 E1b deletion construct was significantly inhibited when treated with the cAMP inhibitor mix. The cAMP inhibitor mix contained a combination of inhibitors of the PI3K, PKA as well as the ERK pathways, the signal transduction pathways downstream of cAMP. This inhibition of reporter activity observed was reversed on inhibitor washout and subsequent treatment with 8Br-cAMP, substantiating the presence of a cAMP response element in the E1b promoter region at the site of the two-point mutation. Finally we performed ChiP assays for p-CREB, the trans-regulatory element of CRE, in the presence and absence of 8Br-cAMP. Phospho-CREB interaction with the mouse Bcrp1 CRE was significantly increased on treatment with 8Br-cAMP when compared with untreated or with IgG isotype control immunoprecipitations (Figure 8D).
Figure 8. Analysis of interaction of p-CREB with the CRE on the mouse Bcrp1 E1b core promoter region in NIH3T3 cells.
p-CREB directly interacts with the CRE activating the E1b Bcrp1 core promoter. A) Two adjacent point mutations were introduced in the CRE site on the E1b Bcrp1 promoter. The promoter activity of the CRE site mutated (E1bPM −1847/+60) construct was then compared with the promoter activity of the non-mutated (E1b −1847/+60) construct. Shown is the representation of three independent experiments (n=3). **, p< 0.01; ***, p<0.001 compared to pGL3Basic control group. B) The promoter activity of the mutated (E1bPM −1847/+60) construct was compared with the non-mutated construct (E1b −1847/+60) in the presence and absence of treatment with 8Br-cAMP (2mM), an analog of cAMP. Shown is the representation of three independent experiments (n=3). *, p<0.05; **, p<0.01; ***, p< 0.001 compared to non-treated control group. C) The promoter activity of the non-mutated E1b −1847/+60 deletion construct was measured after treating NIH3T3 cells either individually with the cAMP analog, 8Br-cAMP or cAMP inhibitor mix or sequentially with cAMP inhibitor mix followed by inhibitor washout and subsequent treatment with 8Br-cAMP. Shown is the representation of three independent experiments (n=3). *, p< 0.05; **, p<0.01 compared to non-treated control group. D) ChiP analysis of p-CREB interaction with CRE was performed in the presence of 8Br-cAMP under serum staved conditions. ChiP assays with p-CREB (in the absence of 8Br-cAMP) and with IgG isotype control were performed to exclude non-specific interactions. The results are expressed as percentage of immunoprecipitated (IP) p-CREB-DNA to total DNA input (input). Shown is the representation of three independent experiments (n=3, *, p< 0.05) compared to non-treated control group.
3.8. Alternative BCRP 5′ UTR expression in human intestine
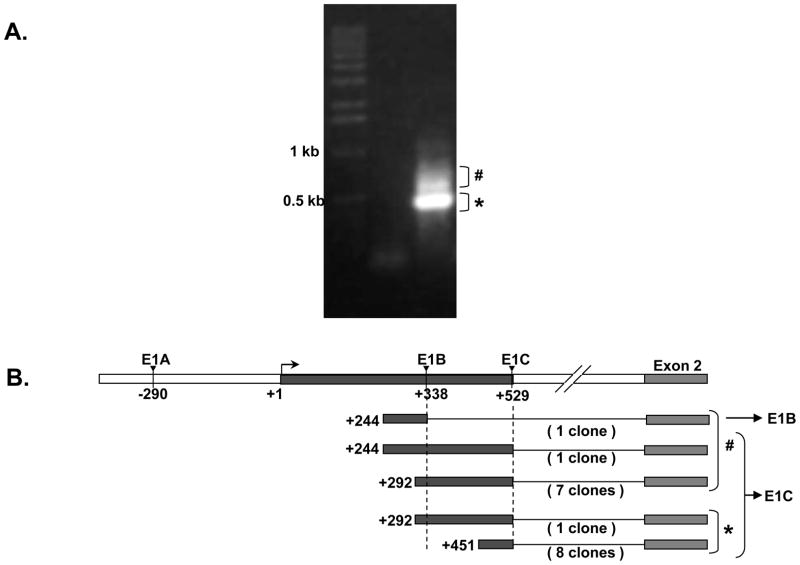
Using RT-PCR our lab at the University of Maryland has shown preliminarily that the promoter associated with the E1B and E1C alternative BCRP 5′ UTRs may control BCRP expression in the human small intestine [14]. To investigate this further, we performed 5′ RACE for BCRP mRNA using human intestinal RNA, and gel electrophoresed the final RACE products. A prominent 500 bp band (*) and disperse higher bands (#, ~550 to 850 bp) were observed (Figure 9A). Eighteen clones were selected from these areas, and sequenced (Figure 9B), and then the sequences were aligned with the genomic region upstream of BCRP Exon 2. Among the 18 clones sequenced, 17 clones aligned with E1C 5′ UTR while only one clone aligned with E1B 5′ UTR (Figure 9B) therefore clarifying that the E1C 5′ UTR is the major human intestinal BCRP 5′ UTR (Figure 9A and 9B). These sequences have been deposited in GenBank (supplemental Table S1).
Figure 9. Human intestinal 5′ UTR analysis by RACE-PCR.
A) * denotes the 500 bp band observed on agarose gel electrophoresis of the nested RACE PCR product from human small intestinal RNA while # indicates disperse higher bands observed B) Diagrammatic alignment of the human small intestinal BCRP 5′ RACE sequences with the human E1B and E1c 5′ UTRs and Exon 2 of Human BCRP gene. E1C 5′ UTR was the highest 5′ UTR identified by 5′ RACE in the human small intestine. # and * indicate the 5′ RACE clones obtained from the disperse higher bands and the prominent 500bp band respectively.
4.0 Discussion
In the current study, we propose that Bcrp1 mRNA expression in all regions of the mouse small intestine is regulated from a single functional alternative promoter (E1b). We further clarified transcription of BCRP mRNA isoforms in the human small intestine from a single alternative promoter (E1C). Based on the predominant expression of a single alternative promoter for Bcrp1 and BCRP in mouse and human small intestine respectively, we hypothesized that similar pre-transcriptional mechanisms regulate Bcrp1/BCRP expression in mouse/human small intestine. However, based on in silico analysis, we report the existence of significant differences between the mouse intestinal alternative promoter E1b and the human intestinal promoter E1C. The qRT-PCR and 5′ RACE data from the current study established a predominant expression pattern for the Bcrp1 E1b isoform in the mouse small intestine. The predominance of a single 5′ UTR in the intestine of both mice and humans suggests conservation of intestine-specific alternative promoter usage for BCRP across two species.
Alternative transcription initiation can occur from variable transcription start sites within a single promoter. Drug-resistant and drug-sensitive phenotypes were shown to initiate transcription from alternative transcription start sites from the same promoter for BCRP [14]. Tissue-specific variable transcription start site utilization has also been reported in the E1b promoter for Bcrp1 in mouse fetal liver and bone marrow cells [23]. Likewise, we found variation in transcriptional start sites for the E1b promoter in mouse intestine. From these findings it is reasonable to conclude that the 82 bp E1b 5′ UTR is the longest E1b 5′ UTR and both the 82 bp and 49 bp E1b 5′ UTRs have ubiquitous expression. BLAST analysis of dbEST identified a Bcrp1 E1b expressed sequence tag with 35 bp from the mouse kidney but with a splice donor site from an 82 bp E1c 5′ UTR (NCBI Accession: AW611052).
High Bcrp1 mRNA as well as protein expression have been reported in the mouse ileum [18, 19]. However, Bcrp1 mRNA and protein expression levels have not been studied simultaneously in different mouse intestinal regions. Hence a relationship between intestinal Bcrp1 mRNA expression and protein expression, measured simultaneously in the different intestinal regions has never been reported. We measured both Bcrp1 protein and Bcrp1 mRNA levels from the same intestinal homogenates. A similar pattern of expression along the mouse small intestine was observed for both total Bcrp1 mRNA and Bcrp1 protein (Figure 3A and 3B). This suggests that intestinal Bcrp1 mRNA expression for the most part might be regulated transcriptionally. Expression of a specific 5′ UTR is accompanied by functional activity of the promoter upstream of the same 5′ UTR. The reporter activity of the promoter constructs containing the 5′ upstream region of the alternative first exons generally correlated with the expression of the Bcrp1 mRNA isoforms in MSIE cells (Figure 4 and 5). We found a similar correlation between Bcrp1 promoter construct reporter activity and mRNA isoforms in NIH3T3 cells, where the E1a and E1b alternative first exons are most abundantly expressed (Supplementary data Figure S1). Reporter activity was not observed for the promoter upstream of E1u, the Bcrp1 mRNA isoform not expressed in either cell line.
In silico secondary structure analysis of the deleted −58/−42 bp region (Figure 7A) using DINAmelt server [30, 31] indicated a stem loop structure. DNA polymerase mediated slippage across this stem-loop structure might explain why the spontaneous deletion of −58 to −42 bp was observed [32]. The −58/−42 bp region encompasses several cis factor binding sites suggesting the secondary stem-loop structure formed by this region might facilitate binding of these factors. The fortuitously prepared Δ−58/−42 Bcrp1 E1b −231/+60 deletion construct therefore helped us identify this as a significant region within the core promoter spanning −243/−42bp.
Several of the in silico predicted sites on the E1b promoter were also predicted in the other alternative promoters of mouse Bcrp1. Hence to identify an intestine-specific regulator of mouse Bcrp1 expression, the CRE, one of the cis-elements unique to the mouse E1b promoter was selected for further analysis. Introduction of two point mutations on the predicted CRE site resulted in complete loss of reporter activity for the -1847/+60 E1b deletion construct suggesting the CRE site is essential for transcriptional regulation of the E1b promoter (Figure 8A). The CRE or the cAMP response element in Bcrp1 −1847/+60 E1b deletion construct was activated when treated with either 8Br-cAMP, an analog of cAMP or inhibited by cAMP inhibitor mix (Figure 8B,C). The downstream effect of cAMP treatment is phosphorylation of CREB, its subsequent binding with and activation from the CRE’s in the promoter region of its effector genes. Subsequent ChiP assays revealed enhanced binding of p-CREB with mouse Bcrp1 CRE in NIH3T3 cells treated with 8Br-cAMP but not in untreated cells (Figure 8D). Taken together these data indicate that Bcrp1 expression in the mouse intestine is at least in part regulated by cAMP associated pathways.
Identification of Sp1 binding sites in the −231 to −42 bp promoter region indicated that the alternative promoter regulating Bcrp1 expression in the mouse intestine is a TATA-less promoter similar to the human intestinal promoter [15]. Several identical and unique cis-regulatory elements were predicted in the mouse and human intestinal TATA-less promoters. Further, comparison of the human E1C 5′ UTR and promoter region with genomic sequences across-species using BLASTN revealed 90% homology with chimpanzee and monkey. The mouse Bcrp1 E1b 5′ UTR and its genomic upstream region was conserved only in rat [33]. This suggests that although a single alternative promoter regulates intestinal BCRP expression in mouse and humans, differences might exist in regulation of BCRP expression from these promoters. The identification of dissimilarities in BCRP regulation in both species might help us understand any difference that may arise when data from pre-clinical studies are translated to clinical studies in the future.
We have been able to establish for the first time that a distinct functional alternative promoter (E1b) regulates Bcrp1 expression in the mouse intestine. We have also been able to show regulation of Bcrp1 E1b mRNA expression from a CRE by its direct interaction and activation by p-CREB. We were also able to confirm the utilization of a distinct alternative promoter in the human intestine. To date, transcription factors shown to regulate human and mouse BCRP/Bcrp1 intestinal promoters remain divergent. However, human–mouse species cross-comparison studies of the transcription factors regulating BCRP/Bcrp1 intestinal promoter activity is currently underway in our laboratory and will provide the data necessary to develop the mouse as an in vivo model to study the effects of diet or xenobiotics on the transcriptional regulation of intestinal Bcrp1 expression.
Supplementary Material
A) E1a was the major isoform Bcrp1 mRNA isoform expressed in the NIH3T3 cell line as determined by qRT-PCR. B) Significant promoter activity over pGL3-Basic was observed only for Bcrp1 E1a (p <0.001) and E1b (P<0.01) deletion constructs but not for the E1c isoform which was barely detected in the NIH3T3 cell line or E1u Bcrp1 mRNA isoform not detected in the NIH3T3 cell line.
Research Highlights.
The goals were to identify and characterize the major alternative promoter(s) regulating mouse intestinal Bcrp1 expression and to compare it with the alternative promoter(s) controlling human intestinal BCRP expression;
A single alternative promoter (E1b) regulates mouse intestinal Bcrp1 expression; in humans, the E1C promoter controls intestinal BCRP expression;
The promoter elements predicted in silico to control mouse and human intestinal Bcrp1/BCRP expression appear to be somewhat different.
The murine intestinal specific promoter for Abcg2, E1b, contains a cAMP response element (CRE). Functional and ChiP assays reveal that this CRE binds phospho-CREB; hence cAMP plays a role in the transcriptional regulation of Bcrp1/Abcg2 in the mouse intestine.
Acknowledgments
This work was supported in part by a VA Merit Review grant awarded to Dr. Douglas D. Ross and by the National Cancer Institute grant CA40570 awarded to Dr. William T. Beck.
Footnotes
English lowercase letters are used throughout the manuscript to describe murine Bcrp1 mRNA isoforms. The human BCRP mRNA isoforms are indicated by capital letters.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
5.0 References
- 1.Doyle LA, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci U S A. 1998;95(26):15665–70. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyake K, et al. Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistant cells: demonstration of homology to ABC transport genes. Cancer Res. 1999;59(1):8–13. [PubMed] [Google Scholar]
- 3.Allikmets R, et al. A human placenta-specific ATP-binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res. 1998;58(23):5337–9. [PubMed] [Google Scholar]
- 4.Kruijtzer CM, et al. Increased oral bioavailability of topotecan in combination with the breast cancer resistance protein and P-glycoprotein inhibitor GF120918. J Clin Oncol. 2002;20(13):2943–50. doi: 10.1200/JCO.2002.12.116. [DOI] [PubMed] [Google Scholar]
- 5.Maliepaard M, et al. Subcellular localization and distribution of the breast cancer resistance protein transporter in normal human tissues. Cancer Res. 2001;61(8):3458–64. [PubMed] [Google Scholar]
- 6.van Loevezijn A, et al. Inhibition of BCRP-mediated drug efflux by fumitremorgin-type indolyl diketopiperazines. Bioorg Med Chem Lett. 2001;11(1):29–32. doi: 10.1016/s0960-894x(00)00588-6. [DOI] [PubMed] [Google Scholar]
- 7.Ee PL, et al. Modulation of breast cancer resistance protein (BCRP/ABCG2) gene expression using RNA interference. Mol Cancer Ther. 2004;3(12):1577–83. [PubMed] [Google Scholar]
- 8.Kowalski P, et al. Selection and characterization of a high-activity ribozyme directed against the antineoplastic drug resistance-associated ABC transporter BCRP/MXR/ABCG2. Cancer Gene Ther. 2001;8(3):185–92. doi: 10.1038/sj.cgt.7700294. [DOI] [PubMed] [Google Scholar]
- 9.Priebsch A, et al. Complete reversal of ABCG2-depending atypical multidrug resistance by RNA interference in human carcinoma cells. Oligonucleotides. 2006;16(3):263–74. doi: 10.1089/oli.2006.16.263. [DOI] [PubMed] [Google Scholar]
- 10.Allen JD, et al. Potent and specific inhibition of the breast cancer resistance protein multidrug transporter in vitro and in mouse intestine by a novel analogue of fumitremorgin C. Mol Cancer Ther. 2002;1(6):417–25. [PubMed] [Google Scholar]
- 11.Cooray HC, et al. Interaction of the breast cancer resistance protein with plant polyphenols. Biochem Biophys Res Commun. 2004;317(1):269–75. doi: 10.1016/j.bbrc.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 12.Zhou XF, et al. Effects of dihydropyridines and pyridines on multidrug resistance mediated by breast cancer resistance protein: in vitro and in vivo studies. Drug Metab Dispos. 2005;33(8):1220–8. doi: 10.1124/dmd.104.003558. [DOI] [PubMed] [Google Scholar]
- 13.Henrich CJ, et al. Botryllamides: natural product inhibitors of ABCG2. ACS Chem Biol. 2009;4(8):637–47. doi: 10.1021/cb900134c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakanishi T, et al. Novel 5′ untranslated region variants of BCRP mRNA are differentially expressed in drug-selected cancer cells and in normal human tissues: implications for drug resistance, tissue-specific expression, and alternative promoter usage. Cancer Res. 2006;66(10):5007–11. doi: 10.1158/0008-5472.CAN-05-4572. [DOI] [PubMed] [Google Scholar]
- 15.Bailey-Dell KJ, et al. Promoter characterization and genomic organization of the human breast cancer resistance protein (ATP-binding cassette transporter G2) gene. Biochim Biophys Acta. 2001;1520(3):234–41. doi: 10.1016/s0167-4781(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 16.Jonker JW, et al. The breast cancer resistance protein protects against a major chlorophyll-derived dietary phototoxin and protoporphyria. Proc Natl Acad Sci U S A. 2002;99(24):15649–54. doi: 10.1073/pnas.202607599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou S, et al. Bcrp1 gene expression is required for normal numbers of side population stem cells in mice, and confers relative protection to mitoxantrone in hematopoietic cells in vivo. Proc Natl Acad Sci U S A. 2002;99(19):12339–44. doi: 10.1073/pnas.192276999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Y, Sugiyama Y. Expression and regulation of breast cancer resistance protein and multidrug resistance associated protein 2 in BALB/c mice. Biol Pharm Bull. 2006;29(5):1032–5. doi: 10.1248/bpb.29.1032. [DOI] [PubMed] [Google Scholar]
- 19.Enokizono J, Kusuhara H, Sugiyama Y. Regional expression and activity of breast cancer resistance protein (Bcrp/Abcg2) in mouse intestine: overlapping distribution with sulfotransferases. Drug Metab Dispos. 2007;35(6):922–8. doi: 10.1124/dmd.106.011239. [DOI] [PubMed] [Google Scholar]
- 20.Gutmann H, et al. Distribution of breast cancer resistance protein (BCRP/ABCG2) mRNA expression along the human GI tract. Biochem Pharmacol. 2005;70(5):695–9. doi: 10.1016/j.bcp.2005.05.031. [DOI] [PubMed] [Google Scholar]
- 21.Englund G, et al. Regional levels of drug transporters along the human intestinal tract: co-expression of ABC and SLC transporters and comparison with Caco-2 cells. Eur J Pharm Sci. 2006;29(3-4):269–77. doi: 10.1016/j.ejps.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Urquhart B, et al. Breast cancer resistance protein (ABCG2) and drug disposition: intestinal expression, polymorphisms and sulfasalazine as an in vivo probe. Pharmacogenet Genomics. 2008;18(5):439–48. doi: 10.1097/FPC.0b013e3282f974dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zong Y, et al. Expression of mouse Abcg2 mRNA during hematopoiesis is regulated by alternative use of multiple leader exons and promoters. J Biol Chem. 2006;281(40):29625–32. doi: 10.1074/jbc.M606314200. [DOI] [PubMed] [Google Scholar]
- 24.Radanovic T, et al. Regulation of intestinal phosphate transport. I. Segmental expression and adaptation to low-P(i) diet of the type IIb Na(+)−P(i) cotransporter in mouse small intestine. Am J Physiol Gastrointest Liver Physiol. 2005;288(3):G496–500. doi: 10.1152/ajpgi.00167.2004. [DOI] [PubMed] [Google Scholar]
- 25.Whitehead RH, et al. Establishment of conditionally immortalized epithelial cell lines from both colon and small intestine of adult H-2Kb-tsA58 transgenic mice. Proc Natl Acad Sci U S A. 1993;90(2):587–91. doi: 10.1073/pnas.90.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prestridge DS. Predicting Pol II promoter sequences using transcription factor binding sites. J Mol Biol. 1995;249(5):923–32. doi: 10.1006/jmbi.1995.0349. [DOI] [PubMed] [Google Scholar]
- 27.Kamalakaran S, Radhakrishnan SK, Beck WT. Identification of estrogen-responsive genes using a genome-wide analysis of promoter elements for transcription factor binding sites. J Biol Chem. 2005;280(22):21491–7. doi: 10.1074/jbc.M409176200. [DOI] [PubMed] [Google Scholar]
- 28.Kolchanov NA, et al. Transcription Regulatory Regions Database (TRRD): its status in 2002. Nucleic Acids Res. 2002;30(1):312–7. doi: 10.1093/nar/30.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huls M, et al. The breast cancer resistance protein transporter ABCG2 is expressed in the human kidney proximal tubule apical membrane. Kidney Int. 2008;73(2):220–5. doi: 10.1038/sj.ki.5002645. [DOI] [PubMed] [Google Scholar]
- 30.Markham N, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33(Web Server issue):W577–81. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markham N, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 32.Trinh T, Sinden R. The influence of primary and secondary DNA structure in deletion and duplication between direct repeats in Escherichia coli. Genetics. 1993;134(2):409–22. doi: 10.1093/genetics/134.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) E1a was the major isoform Bcrp1 mRNA isoform expressed in the NIH3T3 cell line as determined by qRT-PCR. B) Significant promoter activity over pGL3-Basic was observed only for Bcrp1 E1a (p <0.001) and E1b (P<0.01) deletion constructs but not for the E1c isoform which was barely detected in the NIH3T3 cell line or E1u Bcrp1 mRNA isoform not detected in the NIH3T3 cell line.