Abstract
MicroRNA profiling in isogenic models of cellular transformation involving either breast epithelial cells or fibroblasts reveals that expression of miR-193a is lower in transformed cells than in non-tranformed cells. The transcription factors RXRα and Max bind directly to the miR-193a promoter and inhibit miR-193a expression during transformation. MiR-193a inhibits cellular transformation by directly targeting the 3′UTRs of PLAU and K-Ras. Interestingly, miR-193a controls anchorage-independent growth in soft agar through K-Ras, while it affects invasive growth through PLAU. MiR-193a overexpression inhibits the tumorigenicity of developmentally diverse, but not all cancer cell types, and it inhibits tumor growth in colon- and breast-derived xenografts. Lastly, expression of miR-193a is inversely correlated with PLAU and K-Ras in human colon adenocarcinomas. Thus, a pathway in which Max and RXRα inhibit miR-193a expression, thereby activating the PLAU and K-Ras oncogenes is important for distinct aspects of cellular transformation, as well as tumor growth and colon (and perhaps other types of) cancer.
INTRODUCTION
Comparative transcriptional profiling is a common way to identify genes important for carcinogenesis, and a variety of approaches have been used. Transcriptional profiles are compared among 1) groups of patients with different types of cancer, 2) primary tumor samples and normal samples from the same patient, 3) transformed and non-transformed cell lines, 4) cell lines that do or do not overexpress an oncogene. Each of these approaches has advantages and disadvantages, but all of them have contributed to our understanding of cellular transformation and cancer.
In previous work, we identified a cancer gene signature based on the identification of genes that are differentially expressed in two isogenic models of cellular transformation (1). One model involves non-transformed mammary epithelial cells (MCF-10A)(2) containing ER-Src, a derivative of the Src kinase oncoprotein (v-Src) that is fused to the ligand-binding domain of the estrogen receptor (3). Treatment of such cells with tamoxifen rapidly induces Src, and morphological transformation is observed within 24–36 hours (4, 5), thereby making it possible to kinetically follow the transition between non-transformed and transformed cells. The other model consists of three isogenic cell lines derived from primary fibroblasts in a serial manner (6): EH is immortalized by overexpression of telomerase (hTERT) and exhibits normal morphology; EL also expresses SV40 T antigens and displays an altered morphology but is not transformed; ELR also expresses oncogenic Ras (H-RasV12), and it is transformed by numerous criteria including tumor formation in mouse xenografts.
The availability of transcriptional profiles for isogenic, but biologically unrelated, models of cellular transformation makes it possible to distinguish between genes that play a relatively general role in transformation as opposed to those affected only by the specific experimental model. Indeed, the 343-gene signature derived from the combined analysis of these isogenic models is highly correlated with a wide variety of human cancers, thereby validating the clinical relevance of our experimental models (1). Of particular interest, this gene signature links cancer with a variety of inflammatory and metabolic diseases, suggesting that a common transcriptional program is involved is phenotypically disparate diseases (1). In the breast epithelial model, transient activation of Src causes an epigenetic switch from non-transformed to transformed cells that involves a positive feedback loop that is associated with a chronic inflammatory state (5, 7).
MicroRNAs (miRNAs) play critical roles in cancer and other biological processes by directly interacting with specific mRNAs through base pairing and then inhibiting expression of the target genes through a variety of molecular mechanisms (8–10). In the breast epithelial (ER-Src) model, 29 miRNAs are differentially regulated during the process of transformation, and many of these are important for transformation (5, 7). In particular, down-regulation of the Let-7 family, and induction of miR-21 and miR-181b are part of the inflammatory feedback loop necessary for the establishment and maintenance of the transformed state. In addition, 22 miRNAs are differentially expressed in cancer stem cells and non-stem cancer cells that arise during the transformation process (11), with the miR-200 family being particularly important for the function of cancer stem cells via repression of polycomb complexes (12–14).
Here, we perform miRNA profiling in the fibroblast model of cellular transformation and identify 22 differentially regulated miRNAs, 7 of which are similarly regulated in the breast epithelial system. We focus here on miR-193a, which is highly down-regulated during transformation of both the ER-Src and fibroblast models. MiR-193a is down-regulated in oral cancer via DNA hypermethylation (15), and it is poorly expressed in melanomas containing a B-Raf mutation (16). The related miR-193b is 88% identical to miR-193a, but is encoded by a separate gene that is regulated differently. MiR-193b has been implicated as a tumor suppressor in breast (17), prostate (18), and hepatocellular cancers (19), but high levels of miR-193b are associated with poor prognosis of malignant melanomas (16).
We define a molecular pathway in which expression of miR-193a is directly inhibited by RXRα and Max, thereby leading to increased expression of PLAU and K-Ras, which are direct targets of miR-193a. Furthermore, we show that PLAU and K-Ras have distinct roles in transformation, with PLAU affecting invasiveness, while K-Ras affects tumorigenicity. MiR-193a acts as a tumor suppressor in developmental diverse cancer cell lines, and miR-193a expression is inversely correlated with both PLAU and K-Ras in human colon adenocarcinomas, suggesting that this pathway is important for some types of human cancer.
MATERIALS AND METHODS
Cell culture
MCF-10A cells containing the ER-Src fusion protein were grown in DMEM/F12 medium supplemented with 5% donor HS, 20 ng/ml epidermal growth factor (EGF), 10 mg/ml insulin, 100 mg/ml hydrocortisone, 1 ng/ml cholera toxin, and 50 units/ml pen/strep, with the addition of puromycin (4, 5). Cells were passaged fewer than three months from resuscitation from the originally described cells(4, 5). To induce transformation, the Src oncogene was activated by the addition of 1 mM tamoxifen (Sigma, St. Louis) to confluent cell cultures. BJ fibroblast cell lines were grown in KO-DMEM media containing 14% FBS, Medium 199 glutamine, and pen/strep (6). All other cancer cell lines were obtained from ATCC and grown in DMEM, 10% FBS, and pen/strep; cells were passaged fewer than three months from resuscitation.
MicroRNA expression analysis
Expression levels of 365 microRNAs at various times after TAM addition and from non-transformed and transformed fibroblasts were evaluated with microRNA profiling assays (TLDA human miRNA v1.0) in the Dana Farber Molecular Diagnostics Facility. Validation of these results was performed using the mirVana qRT-PCR miRNA Detection Kit and qRT-PCR Primer Sets, according to the manufacturer’s instructions (Ambion Inc.; Austin, TX). RNU48 expression was used as an internal control.
Soft agar colony and invasion assays for the effect of miRNAs on transformation
For the genetic screen to identify miRNAs that act as tumor suppressors, the transformed fibroblast line (ELR) was treated individually with 365 miRNAs (100 nM) for 24 hr. For other experiments, miR-193a (PM11123), miR-193b (PM12383) or a miR-negative control (AM17110) from Ambion, Inc, TX were introduced at 100 nM into a variety of breast and other types of cancer cell lines for 24 hr. In the case of ER-Src cells, TAM was added and cellular transformation was assessed 36 hr later (total time 60 hr). The soft agar colony and MATRIGEL invasion assays were performed as described previously (1, 5). In all cases, experiments were repeated thrice, and the statistical significance was calculated using Student’s t test.
RNA analysis
For samples generated in this work, RNA was purified by the Trizol method (Invitrogen, Carlsbad, CA), reverse transcribed to generate cDNA, and analyzed by SYBR Green-based real-time PCR, with the level of β-actin used as a loading control. RNAs from colon adenocarcinomas (Origene and Biochain Inc.; Rockville, MD) were analyzed for levels of PLAU, K-Ras and miR-193a. Each sample was run in triplicate, and the data represent the mean ± SD. Correlation coefficients between PLAU, K-Ras and miR-193a expression levels in these colon adenocarcinomas were determined.
3′UTR luciferase assay
Firefly luciferase reporter constructs containing the 3′ UTR of PLAU (s207136 from Switchgear Genomics) and K-Ras (HmiT010133 from Genecopoeia Inc) were transfected in MCF10A ER-Src cells along with 100 nM miR-193a or a miRNA control. Cell extracts were prepared 24h after transfection, and luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega, WI, USA).
siRNA experiments
Cells seeded in 6-well plates were transfected with siRNAs against PLAU (s10610), K-Ras (s15602), RXRα (s12386, s12384), Max (s224030. s8540), or a negative control (AM4611) together with antisense against miR-193a (AM11123), miR-193 (AM12383) or a control antisense (AM17010) using the siPORT NeoFX transfection agent. Oligonucleotides (Ambion Inc, TX, USA) were used at 50–100 nM per well. No cell toxicity was detected due to the transfection agent. The resulting cells were analyzed for transformation or RNA levels as described above. All oligonucleotide transfection experiments were performed in triplicate.
Chromatin immunoprecipitation
ChIP was carried out as described previously (Yang et al., 2006). Briefly, the chromatin fragments, derived from untreated and TAM-treated (36 hours) MCF10A ER-Src cells, were immunoprecipitated with 6 mg of antibody against Max (ab53570, Abcam Inc.), and RXRα (sc553, Santa Cruz Biotechnology Inc.; Santa Cruz, CA). DNA extraction was performed using QIAGEN Purification Kit. The samples were analyzed by quantitative PCR in real time, and the results are presented as the mean ± SD of three independent experiments.
Xenograft experiments
MDA-MB-231, HCT-116 and HT-29 cancer cells were injected subcutaneously in the right flank of athymic nude mice (Charles River Laboratories; Wilmington, MA), and tumors were allowed to develop for 10 days (tumor volume ~100 mm3). Mice bearing tumors were randomly distributed in six groups (four mice/group) and treated i.p. with miR-193a (120 nM), or a negative control miR (mixed with liposomes; Altogen Biosystems Inc. 5010) for 3 cycles (days 10, 15, 20). Tumor growth was monitored every 5 days. Nude mice were maintained in accordance with the Animal Care and Use Committee procedures and guidelines of Tufts University.
RESULTS
Differentially regulated miRNAs during transformation in the isogenic fibroblast model
In previous work, we performed microRNA profiling in an inducible model cellular transformation in breast epithelial cells and identified 29 differentially expressed miRNAs during the transformation process (7). Here, we performed a similar miRNA profiling analysis in 3 cell isogenic cell lines derived from primary fibroblasts that are immortalized (h-TERT), predisposed (h-TERT + T antigen), and transformed (h-TERT + T antigen + v-Ras). A comparison between immortalized (EH) vs. transformed (ELR) cell lines identifies 7 miRNAs that are up-regulated and 15 miRNAs that are down-regulated in the transformed state (Fig. 1A). Most of these miRNAs are not differentially regulated in the predisposed (EL) cell line, although there are some exceptions.
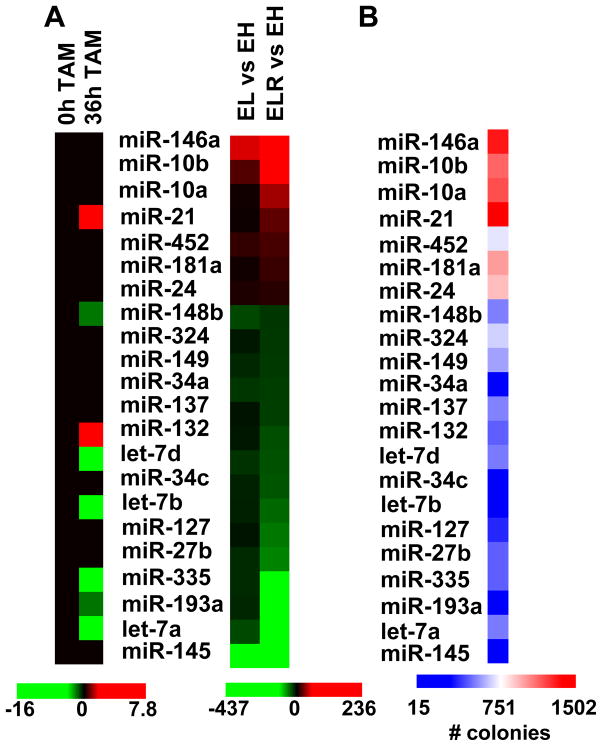
Figure 1.
Identification of miRNAs that are differentially expressed and show tumor suppressor activity in the isogenic fibroblast model. A, Heat map representation of differentially expressed miRNAs in EH (immortalized), EL (pre-disposed), and ELR (transformed) fibroblasts are shown on the right. Expression of these miRNAs in non-transformed or transformed (tamoxifen-treated for 36h) breast epithelial (MCF-10A-ER-Src) cells, as determined previously (7), are shown on the left. B, Heatmap representation of tumorigenicity (number of colonies in soft agar) of transformed (ELR) fibroblasts after transfection of miRNAs. All miRNAs that significantly increase (> 1100 colonies) or decrease (< 300 colonies) tumorigenicity with respect the ~800 colonies generated by cells transfected by control miRNAs) and/or are differentially expressed are included in the heat maps. The values represent the mean of three independent experiments.
Of the 22 miRNAs differentially regulated in the fibroblast model, 7 are regulated in the same manner during Src-induced transformation in the MCF-10A system (Fig. 1A). All but one of the remaining 15 miRNAs are not regulated in the breast epithelial system (the exceptional miR-132 is regulated in the opposite manner). MiR-21 is up-regulated in both isogenic models, whereas miR-148b, miR-335, miR-193a and several members of the Let-7 family are down-regulated in both models.
MiRNAs that inhibit tumorigenicity of transformed fibroblasts
As a genetic screen to identify miRNAs that function as tumor suppressors in the fibroblast model, we individually overexpressed 365 miRNAs in the transformed fibroblasts (ELR line) and examined the effect on tumorigenicity as assayed by colony formation in soft agar. This screen identified 18 miRNAs that inhibit tumorigenicity and 4 miRNAs that slightly enhance tumorigenicity of the transformed fibroblasts (Figs. 1B; S1). All miRNAs down-regulated in the transformed cells cause reduced tumorigenicity, and in most cases the effect is substantial, suggesting that these miRNAs function as tumor suppressors in the fibroblast model. Importantly, miR-148b, miR-335, miR-193a and the Let-7 family members also inhibit transformation in the breast epithelial system (7), suggesting that these miRNAs function as tumor suppressors in multiple cell types. In this paper, we focus on miR-193a, because molecular pathways involving this miRNA are essentially unknown.
MiR-193a down-regulation is important for tumorigenicity and invasive growth in genetically distinct breast cancer cell lines
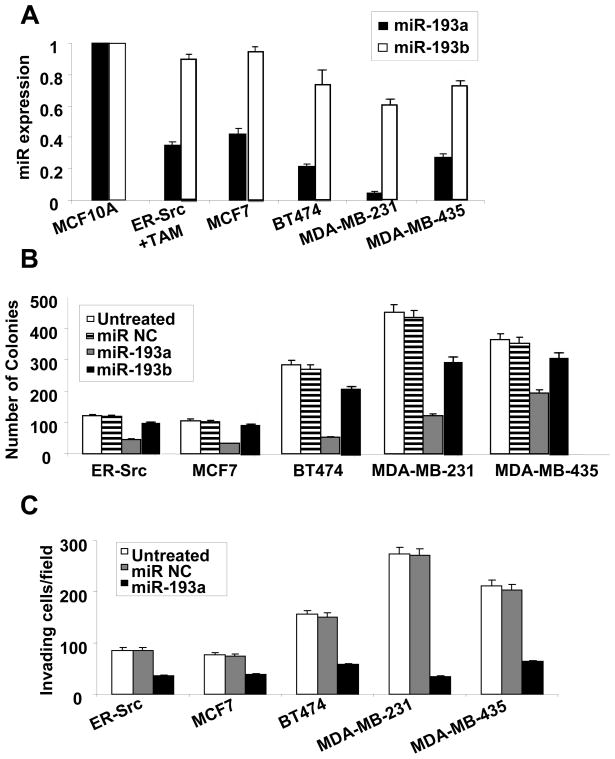
As shown here (Fig. 1) and elsewhere (7), miR-193a is down-regulated during transformation and important for tumorigenicity in both the fibroblast and breast epithelial models. To extend these findings, we analyzed miR-193a and miR-193b expression and function in genetically distinct breast cancer cell lines. In non-transformed MCF-10A cells, miR-193a and miR-193b levels are roughly comparable, with perhaps slightly higher levels for miR-193b (Fig. 2A). In all breast cancer cell lines tested, miR-193a is expressed in lower levels than in immortalized, non-transformed breast cells (Fig. 2A). Furthermore, in all breast cancer cell lines tested, miR-193a overexpression significantly reduces colony formation in soft agar (Fig. 2B). Lastly, over-expression miR-193a in these breast cancer cells reduces the number of invading cells in a MATRIGEL assay (Fig. 2C). In comparison, levels of miR-193b are more consistent (although not identical) among the cell lines, and the effects on colony formation are less pronounced (Figs. 2A, B). Thus, miR-193a inhibits cellular transformation by limiting anchorage-independent growth and reducing invasiveness.
Figure 2.
MiR-193a and miR-193b expression levels and effects on tumorigenicity and invasive growth in breast cancer cell lines. A, Relative MiR-193a and miR-193b RNA levels in the indicated cell lines. Values for miR-193a and miR-193b in the non-transformed MCF-10A cells are arbitrarily set to 1. Based on comparable PCR amplification efficiencies determined experimentally, miR-193b appears to be approximately 3-fold more abundant than miR-193a in MCF-10A cells. B, Colony formation in soft agar of the indicated breast cancer cell lines transfected with miR-193a, miR-193b, or control miRNAs. C, Invasive growth (invading cell/field after wounding) of the indicated breast cancer cell lines transfected with miR-193a control miRNAs.
MiR-193a regulates directly the expression of PLAU and K-RAS in breast cancer cells
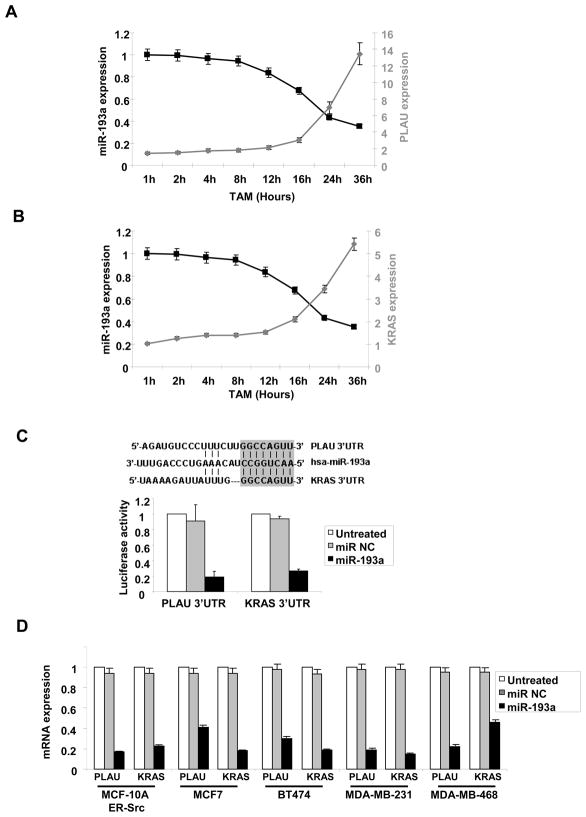
To investigate how miR-193a exerts its anti-cancer effects, we generated a list of candidate target genes using three prediction algorithms (PicTar, TargetScan, Sanger) based on sequence complementarity to 3′-untranslated regions (3′ UTR). Among these candidate target genes, PLAU and K-Ras expression is up-regulated, and hence inversely correlated, with miR-193a expression in both transformation models. Furthermore, during the process of cellular transformation in the inducible breast epithelial model, expression of both PLAU (Fig. 3A) and K-Ras (Fig. 3B) gradually increases in parallel to decreasing expression of miR-193a. In the transformed state (36 hr after tamoxifen induction), PLAU is expressed 7-fold higher and K-Ras is expressed 5-fold higher than the level in cells prior to transformation. The inverse expression of miR-193a with both PLAU and K-Ras is consistent with these genes being relevant targets during transformation.
Figure 3.
MiR-193a targets PLAU and K-Ras. A, Relative expression levels of PLAU and miR-193a during cellular transformation of ER-Src cells at the indicated times points after treatment with tamoxifen. B, Relative expression levels of K-Ras and miR-193a during cellular transformation of ER-Src cells at the indicated times points after treatment with tamoxifen. C, Sequence complementarity (vertical lines) between miR-193a and the 3′ UTRs of PLAU and K-Ras, with the 8 nt seed sequence shown as a gray box. Luciferase activity of reporters containing the 3′UTR of PLAU or K-Ras 24h after transfection with miR-193a or miR negative control. D, PLAU or K-Ras mRNA levels in the indicated breast cancer cell lines transfected with miR-193a or control miRNA.
The 3′ UTRs of both PLAU and K-Ras have an 8-bp sequence that is perfectly complementary with miR-193a (Fig. 3C), and the PLAU 3′ UTR is required for inhibition by the related miR-193b (17). Over expression of miR-193a in MCF-10A ER-Src cells down regulates the luciferase activity of reporter construct containing either the PLAU or K-Ras 3-UTR (Fig. 3C). This shows that miR-193a binds directly to these target RNAs and inhibits PLAU and K-Ras expression through 3′UTR base pairing. In addition, overexpression of miR-193a in all breast cancer cell lines tested (which all have low levels of miR-193a; Fig. 2A) significantly inhibits both PLAU and K-Ras expression (Fig. 3D). These results indicate that miR-193a directly inhibits PLAU and K-Ras expression.
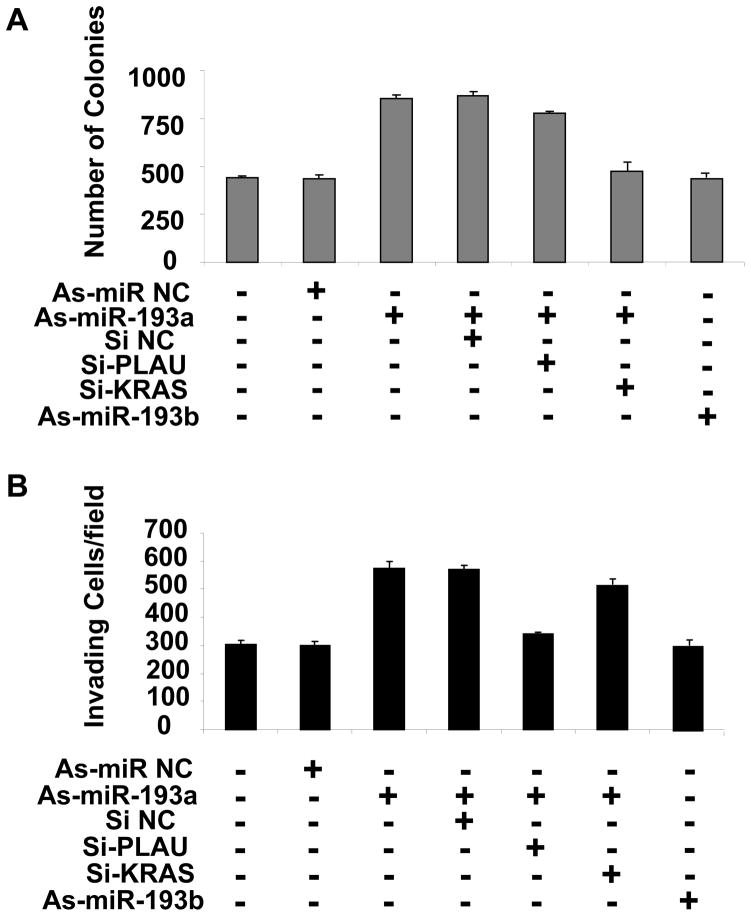
MiR-193a controls tumorigenicity through K-Ras and invasiveness through PLAU
PLAU is a metastasis-associated protein that supports cell migration and matrix-proteases activity (20), and K-Ras is involved in many oncogenic functions, such as anti-apoptotic activity, angiogenesis, motility, and cell growth (21). To address the roles PLAU and K-Ras play in breast cell transformation, we inhibited the expression of either one these genes (by siRNA; Fig. S2) and miR-193a or miR-193b (by antisense RNA) in MDA-MB-231 breast cancer cells and measured tumorigenicity and invasiveness. As expected, inhibition of miR-193a results in both increased tumorigenicity (Fig. 4A) and increased invasiveness, (Fig. 4B) whereas inhibition of miR-193b has little effect. In the context of reduced levels of miR-193a, inhibition of K-Ras reduced tumorigenicity (colony formation in soft agar), whereas inhibition of PLAU had no significant effect (Fig. 4A). In contrast, inhibition of PLAU blocked the increased invasiveness due to inhibition of miR-193a, while inhibition of K-Ras has no effect (Fig. 4B). These observations suggest that miR-193a controls the tumorigenicity of breast cancer cells through regulation of K-Ras while it affects their invasiveness through regulation of PLAU.
Figure 4.
Combined effects of miR-193a and PLAU or K-Ras tumorigenicity and invasive growth. Tamoxifen-treated ER-Src cells transfected with the indicated oligonucleotides - anti-sense (As) against miR-193a or miR-193b, siRNAs against PLAU and K-Ras, and negative controls – were assayed for A, number of colonies in soft agar and B, invasive growth.
Max and RXRα bind directly to the miR-193a promoter and inhibit miR-193a expression during transformation
The reduced expression of miR-193a expression during transformation is likely due to a transcription factor(s) that bind to specific DNA sequences in the miR-193a promoter and acts, formally, as a transcriptional repressor. We previously used the Lever algorithm (22) to predict putative transcription factor binding sites associated with promoters of differentially expressed miRNAs (7). Among the 466 human transcription factor binding site motifs from the TRANSFAC database and the 272 mouse transcription factor binding motifs derived by universal protein binding microarrays, the combination of RXRα and Max ranked among the top combinations for differentially regulated miRNAs including miR-193a.
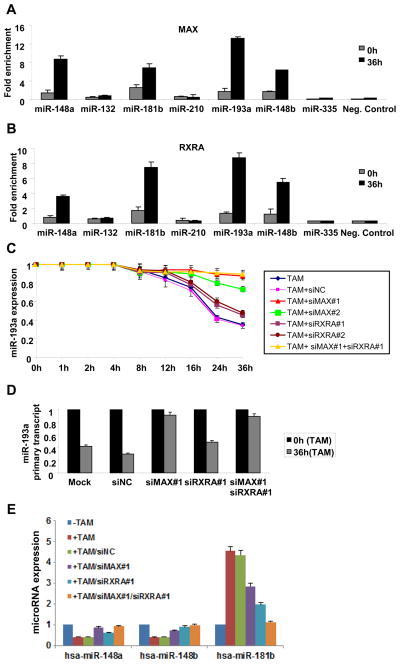
In accord with these predictions, chromatin immunoprecipitation experiments indicate that Max (Fig. 5A) and RXRα (Fig. 5B) bound to the miR-193a regulatory region in ER-Src transformed cells; minimal binding was observed in the non-transformed cells. Similarly, both Max and RXRα bind the miR-148a, miR-148b, and miR-181b promoters in transformed, but not non-transformed ER-Src cells. Thus, the combination of Max and RXRα bind to the miR-193a and other promoters that are regulated during the transformation process.
Figure 5.
Max and RXRα bind the miR-193a and other promoters and inhibit their expression. A, MAX and B, RXRα occupancy (fold enrichment) at the miR-148a, miR-132, miR-181b, miR-210, miR-193a, miR-148b and miR-335 loci as determined by chromatin immunoprecipitation of ER-Src cells that were or were not treated with TAM. C, miR-193a expression in ER-Src cells treated with tamoxifen for the indicated amount of time in the presence or absence of two siRNAs against MAX and two siRNAs against RXRA or their combination. D, Expression levels of miR-193a primary transcript in non-transformed and transformed ER-Src cells treated with siRNA control or siRNA against Max and/or RXRα. E, Expression of the indicated miRNAs in ER-Src cells treated with tamoxifen for 36h in the presence or absence of siRNAs against MAX, RXRα, or their combination.
Using siRNA-mediated inhibition, we examined the effect of Max and RXRα on transcription of miR-193a at various times during the transformation process (Fig. 5C). Importantly, the decreased level of miR-193a during the late stage of the transformation (24 and 36 hours after induction) is blocked by inhibition of Max (completely by one siRNA and strongly by another siRNA). In contrast, inhibition of RXRα has a modest, but significant (p < 0.001), effect on the down-regulation of miR-193a levels. Similar results for Max and RXRα are observed on the level of the miR-193a primary transcript (Fig. 5D), indicating that repression occurs via a transcriptional mechanism as opposed to a post-transcriptional mechanism involving processing of the miR-193a transcript. Thus, miR-193a transcription is directly repressed by Max and (to a much lesser extent) RXRα at later stages of transformation.
We also examined the effect of Max and RXRα on expression of the other miRNAs whose promoters are bound by these transcription factors (Fig. 5E). As observed for miR-193a, repression of miR-148a and miR-148b in transformed cells (36 hours after TAM treatment) is blocked by inhibition of Max and mildly alleviated by inhibition of RXRα. In contrast, miR-181b is induced in transformed cells, and this induction is blocked by the combined inhibition of Max and RXRα. Individually, both Max and RXRα are important for miR-181b induction, with RXRα having a stronger influence on expression levels (Fig. 5E). Thus, the combination of Max and RXRα directly regulate expression of target miRNA promoters, although regulation can be either positive or negative depending on the target.
MiR-193a inhibits tumorigenicity of multiple, but not all, cancer cell types
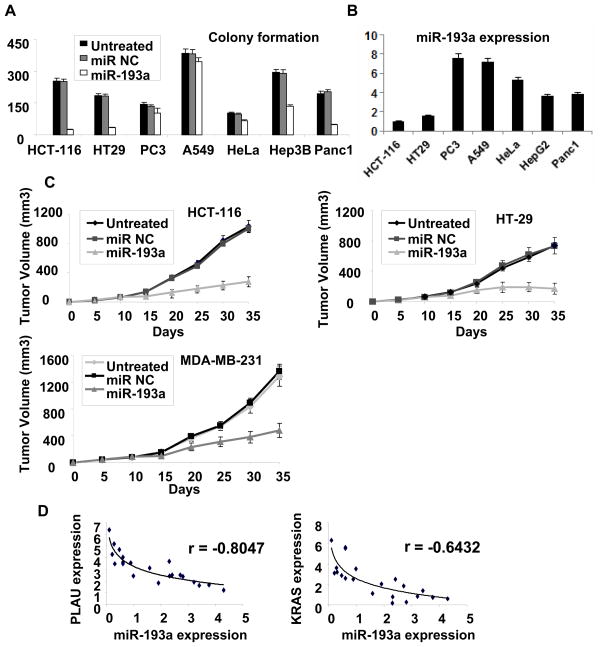
To examine whether miR-193a affects tumorigenicity of non-breast cancer cell types, we over-expressed miR-193a in a variety of cancer cells from diverse developmental lineages. In addition to its effects on all breast cancer cell lines tested (Fig. 2), miR-193a over-expression dramatically affects tumorigenicity of colorectal (HCT-116 and HT29) and pancreatic (Panc1) cells as assayed by colony formation in soft agar (Fig. 6A). MiR-193a has a modest effect on tumorigenicity of hepatcocellular cancer cells (Hep3B), but it has little if any effect on prostate (PC3), lung (A549), or cervical (HeLa) cancer cells. Thus, miR-193a appears to act as a tumor suppressor in breast, colorectal and pancreatic cancer cells, but it has limited effects in a variety of other cancer cell types. Interestingly, the cell lines showing a tumor suppressive effect of miR-193a typically have lower miR-193a expression levels than cell lines not showing this effect (Fig. 6B). This observation suggests that miR-193a is a tumor suppressor in most (and perhaps all) cancer cell types, but that detection of this function occurs only in cell types with reduced miR-193a levels that can be compensated by ectopic expression of miR-193a.
Figure 6.
Importance of MiR-193a in other cancer cell types, mouse xenografts, and human cancer patients. A, Soft agar colony formation in lung (A549), hepatocellular (Hep3B), pancreatic (Panc-1), prostate (PC3), cervical (HeLa) and two colon (HT29, HCT-116) cancer cell lines treated with miR-193a or control miRNA. B, Expression of miR-193a in the indicated cancer cell lines. C, Tumor growth (mean ± SD) of colon (HT29 and HCT-116) and breast (MDA-MB-231) cancer cells after intraperitoneal treatment with miR-193a or control miRNA (days 10, 15, 20 after the initial injection of cancer cells). D, miR-193a, PLAU and K-Ras expression in colon cancer tissues. Each data point represents an individual sample, and correlation coefficients (r) indicated.
MiR-193a acts as a tumor suppressor in mouse xenografts
To examine whether miR-193a acts as tumor suppressor in vivo, we injected nude mice subcutaneously with colorectal (HCT-116 and HT-29) and breast (MDA-MB-231) cancer cells and allowed tumors to develop for 10 days. We then performed i.p. injection of miR-193a or a negative control miRNA near these small tumors (3 cycles on days 10, 15, 20) and monitored tumor size every five days. Over the course of a month, mice treated with miR-193a show reduced tumor growth as compared to mice treated with the control (Fig. 6C). Thus, miR-193a suppresses tumor growth in mouse xenografts generated by breast and colorectal cancer cells, indicating that low levels of miR-193a are required to maintain transformation in these cell types.
MiR-193a levels are negatively correlated with PLAU and K-Ras in cancer tissues
In previous studies, we established the cancer relevance of miRNA-target interactions by determining the relationship between their RNA levels in human cancer patients (5, 7, 12). Specifically, an inverse correlation between miRNA and target mRNA levels in a set of cancer patients indicates that the miRNA-target interaction (as opposed to the miRNA or target individually) plays an important role in human cancer. In this regard, there are strong inverse relationships between miR-193a and PLAU RNA levels as well as miR-193a and K-Ras RNA levels in colon cancer patients (Fig. 6D). Thus, miR-193a regulation of PLAU and K-Ras is relevant for at least some types of cancer.
DISCUSSION
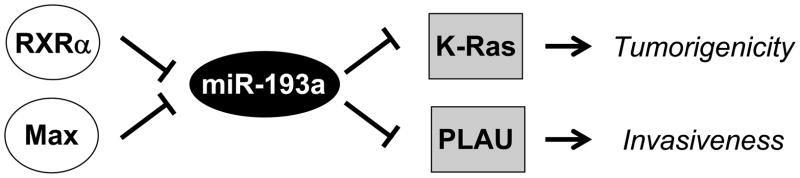
In this work, we describe a gene regulatory pathway involved in cellular transformation (Fig. 7). In this pathway, Max and (to a lesser extent) RXRα, directly binds the miR-193a promoter and inhibits transcription of the gene. The resulting reduction in miR-193a levels leads to increased expression of PLAU and K-Ras, which are direct targets of miR-193a. Increased levels of PLAU and K-Ras contribute to distinct aspects of cellular transformation, with PLAU being important for the invasive growth and K-Ras being important for tumorigenicity.
Figure 7.
Pathway involved in cellular transformation. Max and RXRα inhibit expression of miR-193a, which results in up-regulation of PLAU and K-Ras that respectively increase invasive growth and tumorigenicity.
The combination of Max and RXRα binding motifs is strongly enriched in miRNA promoters whose expression is differentially regulated during transformation (7). Indeed, both Max and RXRα bind to at least 4 miRNA promoters and affect transcription of the target genes. This observation strongly suggests that this combination of transcription factors contributes to the process of cellular transformation by regulating multiple miRNAs and presumably mRNAs. Interestingly, the combination of Max and RXRα can either repress or activate expression of miRNA genes, with Max appearing to be more important for repression, and RXRα appearing to be more important for activation. Max binds DNA in association with Myc and other Myc-family proteins, and the resulting heteromeric complexes can activate or repress transcription. In this regard, Myc represses transcription of a variety of miRNAs (23, 24). RXRα is a member of a family of nuclear receptors that mediate breast and prostate cancer cell growth (25–28). Our results do not address which Myc family members associate with Max at the miR-193a promoters, nor do they address the molecular mechanism of repression. In addition, the role of Max and RXRα in repressing miR-193a expression has been established only in the breast epithelial (ER-Src) model, and this mechanism differs from the down-regulation of miR-193a by DNA methylation in oral cancer (15). Nevertheless, we suspect that Max and RXRα will be important for regulating miR-193a in other breast cell lines and perhaps other cell types.
MiR-193a has been suggested to be a tumor suppressor because the gene is inactivated by DNA methylation in oral cancer (15) and poorly expressed in B-Raf melanomas (16). Our observations that miR-193a is down-regulated during the cellular transformation process and that miR-193a down-regulation is required for transformation in two different isogenic models provide direct evidence that miR-193a is a tumor suppressor. Furthermore, the tumor suppressive effects of miR-193a are observed in genetically distinct breast cancer cell lines, multiple colon cancer cell lines, a pancreatic cancer cell line, and in mouse xenografts generated by colon or breast cancer cells, indicating a widespread role of miR-193a in cellular transformation and tumor growth. However, consistent with the general idea that cancer types differ greatly in terms of the genetic and epigenetic events responsible for the diseased state, over-expression of miR-193a does not reduce tumorigenicity in all cancer cell types. In general, the effect of miR-193a is observed in cancer cell lines that have low levels of miR-193a, suggesting that miR-193a overexpression compensates for these low levels and reverses the transformed state. These observations suggest that miR-193a is a tumor suppressor in many, and perhaps all cell types, but that only some cancer cell types are associated with low miR-193a expression.
MiR-193a directly targets PLAU and K-Ras and inhibits expression of these genes in genetically distinct breast cancer cell lines. Furthermore, under conditions were miR-193a levels are reduced via an antisense RNA, PLAU and K-Ras are required, respectively, for invasive growth and tumorigenicity, indicating that these miRNA-target interacts are relevant for distinct aspects of cellular transformation. More importantly, expression of miR-193a is inversely correlated with both PLAU and K-Ras in samples from human colon cancer patients, indicating that the miR-193a interactions with PLAU and K-Ras are relevant for human colon cancer. We suspect that inhibition of PLAU and K-Ras by miR-193a will be relevant for other types of cancer, but this remains to be established directly by analysis of patient samples.
As indicated by their names, miR-193a and miR-193b are related miRNAs that possess the identical 8-nt seed sequence and share 18 out of 22 nucleotides (miR-193c is a 21-bp version of miR-193b). MiR-193a and miR-193b are encoded by separate genes located on different chromosomes, and their promoter regions are essentially unrelated. As a consequence, miR-193a and miR-193b are not coordinately regulated. In the two isogenic models employed here, miR-193a levels are significantly reduced during transformation, whereas miR-193b levels remain essentially unchanged.
The similarity between miR-193a and miR-193b suggests that these miR-193 paralogs have redundant functions, and indeed both paralogs inhibit expression of PLAU. However, in both isogenic models, reduced levels of miR-193a cause transformation, even though miR-193b appears to be expressed at comparable, and perhaps slightly higher, levels than miR-193a, suggesting that these miR-193 paralogs may have some non-redundant functions. Conversely, non-redundant functions are likely to explain why miR-193b can act as a tumor suppressor in cases where miR-193a is also expressed. For example, while miR-193a and miR-193b affect some common targets (e.g. PLAU), there may be other targets that are differentially affected by these miR-193 paralogs. Even in cases like PLAU (or other genes affected by both miR-193a and miR-193b), inhibition by one miR-193 paralog might be more robust than inhibition by the other paralog.
The idea that miR-193a and miR-193b have non-redundant functions may explain some previous observations. As shown here, miR-193a appears to play a role in distinguishing non-transformed cells from transformed cells, whereas previous studies suggest that miR-193b is important in distinguishing metastatic from hyper-metastatic cancer cells. In addition, while low levels of miR-193a are associated with melanoma, high levels of miR-193b are associated with poor prognosis of this disease (16). This situation in melanoma may mimic the situation in the isogenic models studied here where the transformed state is associated with low levels of miR-193a in the context of substantial levels of miR-193b. While details of functional distinctions between miR-193a and miR-193b remain to be resolved, our results identify a molecular pathway specifically involving miR-193a that is important for cellular transformation.
Supplementary Material
Acknowledgments
We thank Philip N. Tsichlis for very kindly providing facilities for performing the xenograft experiments. This work was supported by a grant to K.S. from the National Institutes of Health (CA 107496).
References
- 1.Hirsch HA, Iliopoulos D, Joshi A, Zhang Y, Jaeger SA, Bulyk M, et al. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17:348–61. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soule HD, Maloney TM, Wolman SR, Peterson WD, Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortallized human breast epithelial cell line, MCF10. Cancer Res. 1990;50:6075–86. [PubMed] [Google Scholar]
- 3.Aziz N, Cherwinski H, McMahon M. Complementation of defective colony-stimulating factor 1 receptor signaling and mitogenesis by Raf and v-Src. Mol Cell Biol. 1999;19:1101–15. doi: 10.1128/mcb.19.2.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells and acts together with chemotherapy to blocks tumor growth and prolong remission. Cancer Res. 2009;69:7507–11. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kB, lin 28, let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–8. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 7.Iliopoulous D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b, via PTEN and CYLD, are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–91. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nature reviews. 2009;10(10):704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci USA. 2011;108:1397–402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iliopoulos D, Lindahl-Allen M, Polytarchou CHAH, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–72. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 15.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 16.Carmuta S, Egyhazi S, Rodolfo M, Witten D, Hansson J, Larsson C, et al. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–70. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 17.Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28:3937–48. doi: 10.1038/onc.2009.245. [DOI] [PubMed] [Google Scholar]
- 18.Rauhala HE, Jalava SE, Isotalo J, Bracken H, Lehmusvaara S, Tammela TL, et al. miR-193b is an epigenetically regulated putative tumor suppressor in prostate cancer. Int J Cancer. 2010;127:1363–72. doi: 10.1002/ijc.25162. [DOI] [PubMed] [Google Scholar]
- 19.Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J, et al. MicroRNA-193b regulates proliferation, migration, and invasion in human hepatocellular carcinoma cells. Eur J Cancer. 2010;46:2828–36. doi: 10.1016/j.ejca.2010.06.127. [DOI] [PubMed] [Google Scholar]
- 20.Blasi F, Sidenius N. The urokinase receptor: focused cell surfact proteolysis, cell adhesion, and signaling. FEBS Lett. 2010;584:1923–30. doi: 10.1016/j.febslet.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 21.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 22.Warner JB, Philippakis AA, Jaeger SA, He FS, Lin J, Bulyk ML. Systematic identification of mammalian regulatory motifs’ target genes and functions. Nat Methods. 2008;5(4):347–53. doi: 10.1038/nmeth.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–12. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 25.Mu YM, Yanase T, Nishi Y, Hirase N, Goto K, Takayanagi R, et al. A nuclear receptor system constituted by RAR and RXR induces aromatase activity in MCF-7 human breast cancer cells. Mol Cell Endocrinol. 2000;166:137–45. doi: 10.1016/s0303-7207(00)00273-2. [DOI] [PubMed] [Google Scholar]
- 26.Pandey KK, Batra SK. RXRalpha: a novel target for prostate cancer. Cancer Biol Ther. 2003;2:185–6. [PubMed] [Google Scholar]
- 27.Peng X, Yun D, Christov K. Breast cancer progression in MCF10A series of cell lines is associated with alterations in retinoic acid and retinoid X receptors and with differential response to retinoids. Int J Oncol. 2004;25:961–71. [PubMed] [Google Scholar]
- 28.Zeisig BB, Kwok C, Zelent A, Shankaranarayanan P, Gronemeyer H, Dong S, et al. Recruitment of RXR by homotetrameric RARalpha fusion proteins is essential for transformation. Cancer Cell. 2007;12(1):36–51. doi: 10.1016/j.ccr.2007.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.