Abstract
It has been proposed that microbial translocation might play a role in chronic immune activation during HIV/SIV infection. Key roles in fighting bacterial and fungal infections have been attributed to Th17 and Tc17 cells. Th17 cells can be infected with HIV/SIV, however whether effective vaccination leads to their maintenance following viral challenge has not been addressed. Here we retrospectively investigated if a vaccine regimen that potently reduced viremia post-challenge preserved Th17 and Tc17 cells, thus adding benefit in the absence of sterilizing protection. Rhesus macaques were previously vaccinated with replication-competent Adenovirus recombinants expressing HIVtat and HIVenv followed by Tat and gp140 protein boosting. Upon SHIV89.6P challenge, the vaccinees exhibited a significant 4 log reduction in chronic viremia compared to sham vaccinated controls which rapidly progressed to AIDS (Demberg et al. J. Virol. 81:3414, 2007). Plasma and cryopreserved PBMC samples were examined pre-challenge and during acute and chronic infection. Control macaques exhibited a rapid loss of CD4+ cells, including Th17 cells. Tc17 cells tended to decline over the course of infection although significance was not reached. Immune activation, assessed by Ki-67 expression, was associated with elevated chronic viremia of the controls. Significantly increased plasma IFN-γ levels were also observed. No increase in plasma LPS levels were observed suggesting a lack of microbial translocation. In contrast, vaccinated macaques had no evidence of immune activation within the chronic phase and preserved both CD4+ T-cells and Tc17 cells in PBMC. Nevertheless, they exhibited a gradual, significant loss of Th17 cells which concomitantly displayed significantly higher CCR6 expression over time. The gradual Th17 cell decline may reflect mucosal homing to inflammatory sites and/or slow depletion due to ongoing low levels of SHIV replication. Our results suggest that potent viremia reduction during chronic SHIV infection will delay but not prevent the loss of Th17 cells.
Keywords: Th17 cells, Tc17 cells, simian/human immunodeficiency virus, chronic immune activation, microbial translocation, vaccine
Introduction
The main route of HIV transmission worldwide is via heterosexual contact across mucosal epithelia. The virus first establishes a small localized infection in the submucosa before rapidly spreading systemically (Haase, 2005; Miller et al., 2005). Despite the rapid systemic dissemination, the majority of infected cells reside in the gut (Ling et al., 2010; Mattapallil et al., 2005). One hallmark of HIV infection of people and SIV infection of macaques is early rapid depletion of CD4+ T-cells in the gut mucosa, which also occurs in natural non-human primate hosts of SIV, although the natural hosts generally do not progress to simian AIDS (Brenchley and Douek, 2008a; Brenchley et al., 2004; Gordon et al., 2007; Mattapallil et al., 2005). In addition to CD4+ T cell loss, chronic immune activation of CD8+ T-cells is associated with disease progression, and in fact can be used to distinguish elite controllers from HIV progressors (Bello et al., 2009; Loke et al., 2010; Vollbrecht et al., 2010). HIV infection also leads to mucosal epithelial barrier damage, in part driven by the viral envelope protein, gp120 (Nazli et al., 2010). This damage to the epithelium can lead to enhanced microbial translocation, which has been associated with disease progression and may contribute to chronic immune activation (Brenchley et al., 2006; Klatt et al., 2010; Marchetti et al., 2008; Troseid et al., 2010).
IL-17 producing cells are of current interest due to their association with control of fungal and bacterial infections. These cells can be divided into three major phenotypes: Th17 cells (CD4+), Tc17 cells (CD8+) and IL-17+ γδ T-cells. The most commonly studied and best characterized are Th17 cells. They play a distinct role in the clearance of bacterial and fungal infections and have been shown to be involved in autoimmune disease in several animal models and in humans (Crome, Wang, and Levings, 2010; Damsker, Hansen, and Caspi, 2010; Hashimoto et al., 2010; Kamada et al., 2010; Wu et al., 2010; Zhao et al., 2009). Tc17 cells on the other hand seem to play a role similar to Th1 cells in anti-tumor immunity (Kuang et al., 2010) and also function in the clearance and control of viral infection (Hamada et al., 2009). IL-17+ γδ T-cells (Vδ1, Vδ2 cells) have been associated with responses to Candida albicans (Vδ1 cells) and to mycobacterium (Vδ2 cells) in HIV infected patients (Fenoglio et al., 2009) and with clearance of Listeria monocytogenes infection in mice (Xu et al., 2010). The role of γδ T-cells in protective immunity is further reviewed by Cua and Tato (Cua and Tato, 2010).
The origin of Th17 cells has not been definitively established; however the current consensus is that murine Th17 cells are related to Tregs, both originating from the same precursors, whereas human Th17 cells are more closely related to Th1 cells (Annunziato et al., 2008; de Jong, Suddason, and Lord, 2010; Romagnani et al., 2009; Torchinsky and Blander, 2010). Presumably the origin of non human primate Th17 cells will resemble that of humans, but to date this is not known. In addition to IL-17, Th17 and Tc17 cells produce a vast array of other cytokines including TNF-α, IL-1, IL-2, IL-10, IL-21, IL-22 and IFN-γ (Klatt and Brenchley, 2010; Kuang et al., 2010; Ndhlovu et al., 2008; Torchinsky and Blander, 2010). A few reports have shown that retroviral infection, including HIV (Fenoglio et al., 2009; Maek et al., 2007) and HTLV-1 (Dodon et al., 2004), leads to IL-17+ cell expansion. However, most studies have found Th17 cells to be lost or declining during SIV and HIV infections, either by homing to the mucosa or due to their susceptibility to infection (Brenchley et al., 2008; Hunt, 2010; Ndhlovu et al., 2008; Prendergast et al., 2010). The majority of these studies have compared IL-17+ cells in SIV-infected natural hosts and susceptible macaque species, or in healthy volunteers, HIV-infected non-progressors (elite controllers) and disease progressors. A recently published paper by Nigam et al. (2011) shows the distribution of Th17 and Tc17 populations in different tissues in healthy and SIV infected unvaccinated macaques. However, the question of whether vaccines that succeed in significantly reducing viral loads following challenge can prevent the loss of Th17 and Tc17 cells has not been addressed.
Our vaccine approach is based on priming with replication-competent Adenovirus vectors and boosting with envelope protein. This strategy has elicited strong protection in both SIV and SHIV challenge models in rhesus macaques (Demberg et al., 2007; Malkevitch et al., 2006; Patterson et al., 2008; Patterson et al., 2011; Patterson et al., 2004) and in the HIV challenge model in chimpanzees (Lubeck et al., 1997). Recently, priming of rhesus macaques with adenovirus 5 host range mutant (Ad5hr) recombinants encoding HIVenv and HIVtat followed by boosting with Tat and Env protein led to strong protection against a SHIV89.6P challenge (Demberg et al., 2007). While peak viremia in the vaccine and control groups was similar, the vaccinated animals controlled infection rapidly thereafter, leading to a significant 4-log decrease in chronic phase viremia compared to sham-vaccinated controls. CD4+ cells were preserved in contrast to control animals that lost CD4+ T-cells almost completely within two weeks. Here, using cryopreserved PBMC and plasma samples from this study and from six additional control animals similarly sham-vaccinated and challenged (Patterson et al., 2008), we asked whether the vaccinated macaques preserved their IL-17-secreting cells, and avoided chronic immune activation in contrast to the controls. Such an outcome would add benefit to partially protective vaccines by potentially reducing viral-induced mucosal damage, preventing the occurrence of opportunistic infections, slowing disease progression, and extending the time period before HAART therapy has to be initiated.
Material and methods
Peripheral blood and plasma specimens
Cryopreserved ficoll-paque plus (GE Healthcare, Piscataway, NJ, USA) gradient derived PBMC from rhesus macaques of Indian origin were used in this study. The PBMC were from two groups of animals from a previously published vaccine study (Demberg et al., 2007). One group of 8 macaques was vaccinated with replication competent Ad5hr-HIVIIIBtat and Ad5hr-HIV89.6pgp140ΔCFI followed by boosting with gp140ΔCFI and Tat proteins adjuvanted with MPL-SE and alum, respectively. The other group of 3 controls was supplemented with 6 similarly treated control animals from another study (Patterson et al., 2008). All controls received empty Ad5hr vector and were boosted with adjuvant only or with PBS (3 with MPL-SE plus alum, 3 with MPL-SE only, and 3 with PBS). Vaccinated and 3 control animals were challenged intravenously with SHIV89.6P at week 50 (12 weeks following the last boost; 30 MID50). The supplemental controls were challenged with the same SHIV89.6P stock at week 44 (8 weeks following the last adjuvant boost; 90 MID50). Cells were evaluated by flow cytometry and real time PCR at three different time points: pre-challenge/time of challenge (wk48 or wk44), 4 weeks post challenge (wk54/wk48) and during the chronic phase (24 weeks post challenge). Matched plasma samples, derived from EDTA-treated blood, were obtained at similar time points.
PBMC stimulation
PBMC from all animals were thawed, washed with R10 (RPMI1640 supplemented with penicillin/streptomycin, L-glutamine and 10% FBS, all Life Technologies, Carlsbad, CA, USA) counted and plated into two wells of a 24-well plate (Nunc, Thermo-Scientific, Waltham, MA, USA). One well was stimulated for 4 hours in 1 ml of R10 containing 50 ng/ml of PMA (Sigma-Aldrich, St Louis, MO, USA) and 250 ng/ml of Ionomycin (Sigma-Aldrich) to evaluate IL-17-producing cells. One hour into the stimulation, 1 µl of Golgi Stop (BD Bioscience, San Jose, CA, USA) was added. The unstimulated well was used to evaluate CD8+ and CD4+ T cell activation measured by Ki-67 and background cytokine secretion by flow cytometry.
Limulus Amoebocyte Lysate (LAL) assay
Plasma LPS levels were evaluated using the Hycult chromogenic endpoint LPS detection assay (Limulus amoebocyte lysate assay, Hycult Biotech, Uden, The Netherlands) according to the manufacturer’s instructions. Plasma samples were diluted 1:30 in DPBS (Lonza, Basel, Switzerland) and heat inactivated for 5 min at 75°C prior to assay. EU levels obtained were converted to pg/ml with 1 EU = 100 pg/ml.
16s rDNA quantitative real time PCR
Bacterial DNA was purified from 500 µl of non-inactivated plasma from macaques in the original study (Demberg et al., 2007) by a modified alkaline lysis protocol. Reagents from a GeneJet plasmid purification kit (Fermentas, Glen Burnie, MD, USA) were used under aseptic conditions to avoid bacterial contamination. Plasma samples were spun at 4000×g for 10 min to pellet any free or complexed (opsonized by complement and/or antibody) bacteria. The supernatant was aspirated, the pellet was resuspended in kit buffer using pulse vortexing, and the suspension was lysed using the SDS containing kit lysis buffer. The mixture was vigorously pulse vortexed for 10s to free genomic DNA. Neutralizing buffer was added and the lysate was again pulse vortexed. The mixture was applied onto Nucleospin Tissue XS DNA spin columns (Macherey-Nagel, Bethlehem, PA, USA) and centrifuged according to the manufacturer’s instructions. Pellets were washed with kit buffer, and samples were dissolved in 30 µl TrisHCL-buffer (pH=8) without EDTA, brought up to 70 µl with PCR grade water (Fermentas), and used directly in triplicate in the PCR assay. In parallel, free bacterial DNA was isolated from 200 µl of plasma using the Qiagen Blood Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. No carrier DNA was added.
For PCR detection of bacteria in plasma samples, we used universal primers for 16s RNA: p201 and p1370 (Table 1) obtained following a two-step HPLC purification from Eurofins MWG Operon (Huntsville, AL, USA). We used Invitrogen SYBR greenER premix reagents (Life Technologies) and pre-treated the mastermix minus primer with PCR grade DNAse I (Life Technologies) as described (Tseng et al., 2003). All samples were run in triplicate. E. coli DNA standards (0.25, 0.5, 1 and 2 pg) were included on each plate. DNAse I pre-treatment enhanced the sensitivity of the assay but did not eliminate all background. A consistent lower detection limit of 0.125 pg per reaction, or approximately 160 copies of E.coli 16s rDNA was achieved based on calculations reported elsewhere (Farrelly et al., 1995).
Table 1.
Real-time PCR primers.
| Target | Forward primer 5’ to 3’ | Reverse Primer 5’ to 3’ | Amplicon size (bp) |
Accession number | Reference |
|---|---|---|---|---|---|
| IFN-γ | GCAACAAAAAGAAACGGGATGAC | CTGACTCCTTTTTCGCTTCC | 148 | NM_001032905 | In house |
| MxA | AGGAGTTGCCCTTCCCAGA | TCGTTCACAAGTTTCTTCAGTTTCA | 78 | AF135187.1 | Dr. Fenizia |
| TNF-α | AGCCCATGTTGTAGCAAACC | GCTGGTTATCTGTCAGCTCCA | 104 | DQ902483 | In house |
| CCL-3 (MIP-1α) | CCTCCTGCTGCTTCAGCTAC | CTCCTTACTGGGGTCAGCAC | 146 | AF457195 | In house |
| CCL-4 (MIP-1β) | CTTCCTCGCAACTTTGTGGT | GCTTGCTTCTTTTGGTTTGG | 88 | NM_002984 | In house |
| CCL-5 (RANTES) | AGTGGCAAGTGCTCCAACC | CGAACCCATTTCTTCTCTGG | 86 | DQ913730 | (Hofmann-Lehmann et al., 2002) |
| CXCL8 (IL-8) | GAGTGGACCACACTGTGCCA | AAACTTCTCCACAACCCTCTGC | 108 | NM_001032965 | (Hardstedt et al., 2005) |
| IL-10 | AGAACCACGACCCAGACATC | GGCCTTGCTCTTGTTTTCAC | 119 | DQ890063 | In house |
| 18s | GCCCGAAGCGTTTACTTTGA | TCCATTATTCCTAGCTGCGGTATC | 81 | NR_003286 | (Medeiros et al., 2003) |
| p201 | GAGGAAGGIGIGGAIGACGT | Tseng et al., 2003 | |||
| p1370 | AGICCCGIGAACGTATTCAC | Tseng et al., 2003 | |||
| CD14 | GGTTCCTGCTCAGCTACTGG | TGGTGCCGGTTATCTCTAGG | 95 | XP_001087125.1 | In house |
Accession numbers were obtained from http://www.ncbi.nlm.nih.gov/nuccore.
Cytokine/chemokine and CD14 real time PCR
RNA was isolated from week 48 (pre), 56 and 70 cryopreserved PBMC using the Qiagen RNeasy Plus mini kit according to the manufacturer’s directions, followed by the immediate generation of cDNA using the Qiagen QuantiTect kit with an extension time of 1 hour at 42°C. Primers (Table 1) except for CD14 were designed to be exon spanning using the UCSC Genome Browser (http://genome.ucsc.edu/). Gene expression levels were normalized against 18s RNA as reference gene. For the calculation of expression levels we used the efficiency corrected (Pfaffl) equation (Pfaffl, 2001) except for CD14 and the interferon-α/β induced gene MxA (myxovirus resistance A gene). Samples were run in triplicate in a 25 µl reaction using Invitrogen SYBR greenER premix with ROX (Life Technologies) on an Applied Biosytems ABI7000 cycler (Life Technologies) under the following conditions: 2 min 50°C, 10 min 95°C and 40 cycles of 30s 95°C, 15s 59°C and 30s 72.5°C followed by standard melting curve analysis.
Multiplex bead assays (Luminex xMAP)
Previously frozen undiluted plasma samples from the original study (Demberg et al., 2007) were evaluated for the expression of cytokines and chemokines at four different time points: before challenge (wk48), at peak viremia (wk52), close to viral set point (wk61) and during the chronic phase (wk70). The non-human primate specific Invitrogen 5-plex chemokine kit (IL-8, MCP-1, MIP-1α, MIP-1β and RANTES) and the 5-plex cytokine kit (IFN-γ, IL-2, IL-4, IL-10 and TNF-α) (both Life Technologies) were used according to the manufacturer’s instructions. Duplicate samples were read on a Luminex 100 machine (Luminex Corporation, Austin, TX, USA).
Flow Cytometry
PBMC were cultured as stated above and transferred into 5 ml flow tubes, washed once with DPBS and incubated with the viability dye Aqua (Life Technologies) to discriminate between live and dead cells (done on 6 animals at all time points). After washing, cells were further stained for 25 min at RT while protected from light with a cocktail of the following surface antibodies: CD3, CD4, CD8, CD20, CD27, CD28, CD95 and CD196 (CCR6) (Table 2). Cells were washed in DPBS and fixed with 150 µl of BD Cytofix/Cytoperm solution (BD Bioscience) for 15 min at RT in the dark. After fixation, cells were washed in BD Perm/Wash solution and stained intracellularly for 25 min at RT in the dark with a cocktail of the following antibodies: IFN-γ, IL-17, Ki-67 or Isotype control (Table 2). Cells were washed again with BD Perm/Wash solution and resuspended in 150 µl of DPBS with 2% Formaldehyde (Tousimis, Rockville, MD, USA). A minimum of 250,000 events in the lymphocytic gate were acquired on the same day on a custom 4 laser LSR II cytometer (BD Biosciences) using DIVA 6.2 software. Only samples with at least 100 events in the central and effector memory compartments were included in the analyses. Results were analyzed in FlowJo version 9.0.2 for Macintosh (Tree Star, Inc., Ashland, OR, USA) and graphs were generated in GraphPad Prism 5.0.3 for PC (GraphPad Software, Inc., La Jolla, CA, USA).
Table 2.
Antibodies used in Flow Cytometry
| Antigen and color | Clone | Supplier |
|---|---|---|
| KI-67 FITC | B56 | BD Biosciences |
| IL-17 PE | eBio64CAP17 | eBioscience |
| CD28 Pe-Cy5 | CD28.2 | BD Biosciences |
| CD196 (CCR6) PerCP-Cy5.5 | 11A9 | BD Biosciences |
| CD95 Pe-Cy7 | DX2 | eBioscience |
| CD3 Pacific Blue | SP34-2 | BD Biosciences |
| CD4 Qdot605 | L200 | Invitrogen (Custom conjugate) |
| CD8 eFlour650 | RPA-T8 | eBioscience |
| IFN-γ APC | B27 | BD Biosciences |
| CD27 Alexa700 | 0323 | eBioscience |
| CD20 APC-Cy7 | 2H7 | Biolegend |
Statistical analysis
Differences in viral loads over time between control and vaccinated macaques were tested by the Wilcoxon rank sum test. Changes in IL-17+ and CCR6+ expressing cells over time and differences between the groups in the expression of Ki-67 were analyzed on square root or cube root-transformed values respectively, using repeated measures ANOVA corrected for multiple comparisons. Differences in viral loads at single time points and in LPS levels were evaluated by the exact Wilcoxon rank sum test. PCR data were assessed by ANOVA corrected for multiple comparisons on log-transformed data.
Results
Strong control of viremia in vaccinees does not prevent gradual loss of Th17 cells
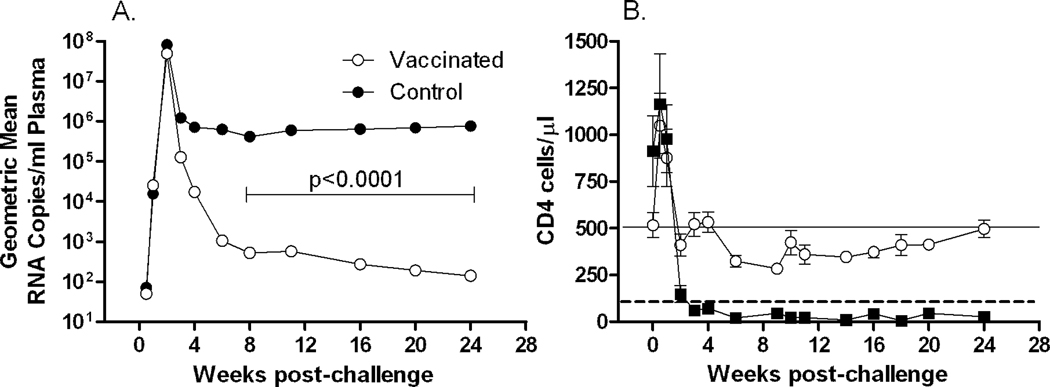
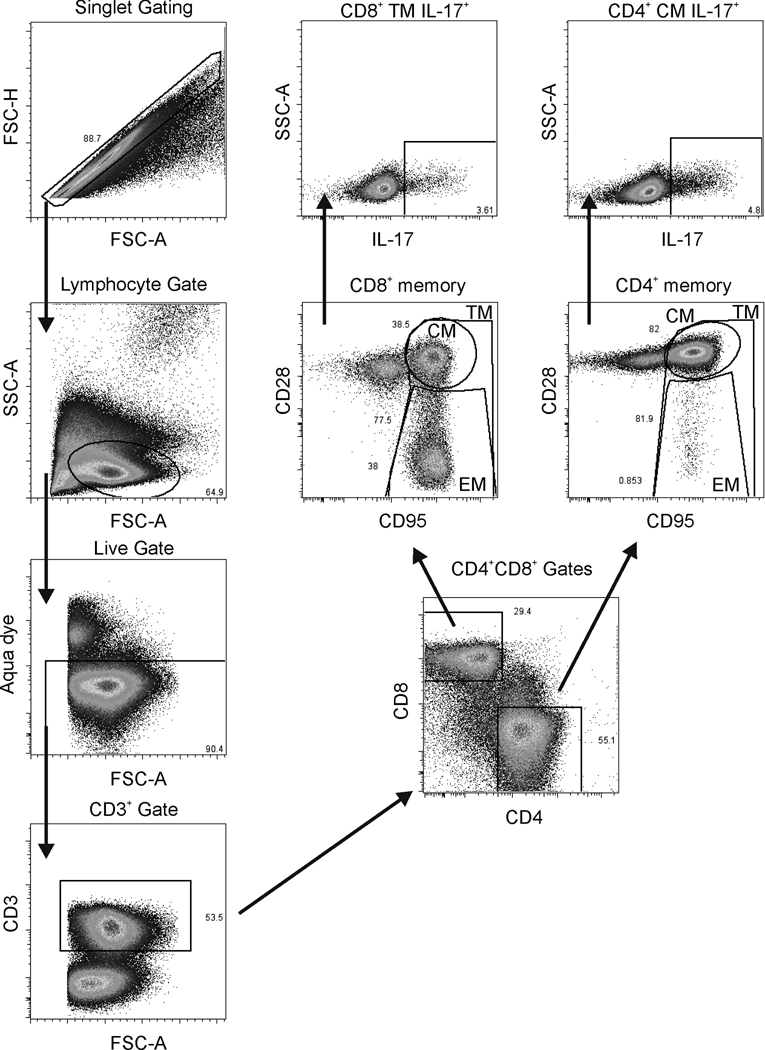
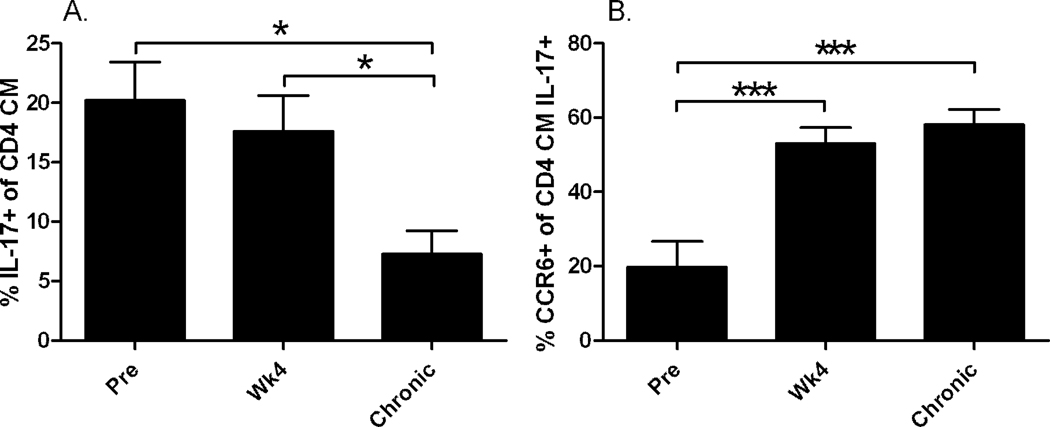
We investigated the preservation of IL-17-producing cells among CD4+ and CD8+ memory T cells in peripheral blood in vaccinated and control animals. As previously published, following intravenous SHIV89.6P challenge the vaccinated macaques exhibited a significant 4-log decrease in chronic viremia compared to controls and maintained peripheral blood CD4+ T-cells (Demberg et al., 2007). The addition of supplemental controls (Patterson et al., 2008) did not change this relationship appreciably. Macaques in the combined control group were unable to suppress viral replication and showed a significantly higher viral load of about 3.5 logs compared to the vaccinated animals (p<0.0001, Fig. 1A). In addition, CD4+ T-cells were rapidly depleted (Fig. 1B) after a short initial proliferative burst during the early phase of infection as described elsewhere (Kaur et al., 2000). For analysis of IL-17-producing cells, Th17 and Tc17 cells were gated as shown in Fig. 2. Th17 cells have a memory phenotype (Brenchley et al., 2008; Kader et al., 2009b). In our hands, at the pre-challenge time point the majority of Th17 cells in all the macaques displayed a significantly higher central memory (CM) phenotype (CD28+CD95+) than effector memory (EM) phenotype (CD28-CD95+) (data not shown). This observation is in accord with the distribution of memory populations among CD4 positive cells in healthy animals, where CM cells account for approximately 60% and EM cells only 9% of the CD4 population. We therefore define Th17 cells as CD3+CD4+ cells with a CM phenotype that are IL-17 positive. Pre-challenge, the total population of Th17 cells in all macaques ranged from 6% to 39%, but by 4 weeks post-challenge the cells were present at very low frequency in the control animals due to the rapid overall loss of CD4+ T-cells (Fig. 1B). Further, a loss of CD4 CM cells in the control macaques post-challenge, with a concomitant rise in CD4 EM cells, is illustrated in Table 3. In contrast, the CD4 CM population remained stable in the vaccinated macaques. By 24 weeks post-challenge, only 3 of 9 control macaques had sufficient cells in the CD4 CM population to allow analysis of Th17 cells. Therefore, post-challenge dynamics of Th17 cells were investigated in the vaccinated group only. Here despite control of viremia to very low levels in the chronic phase of infection (Fig. 1A) and maintenance of close to pre-challenge CD4 counts (after initial rapid expansion and contraction of CD4 cells, Fig. 1B), we observed a significant decline in the population of Th17 cells in comparison to pre-infection levels (pre vs chronic and wk 4 vs chronic: p= 0.027 for both; Fig 3A). In addition we monitored the expression of CCR6 (a homing marker for inflamed tissue) over time on these cells. At the pre-challenge time point, approximately 33% of IL-17+ cells in the vaccinated macaques expressed the CCR6 receptor on the cell surface. Over the course of infection, the remaining IL-17+ cells significantly upregulated the expression of the CCR6 receptor. This was seen by wk4 post challenge and was maintained in the chronic infection phase (pre vs wk4 and vs chronic: p = 0.0003 for both; Fig. 3B).
Fig. 1. Viral load and CD4 count after SHIV89.6P challenge.
(A) Geometric mean viral loads post challenge. A 4-log significant difference in median chronic viremia between control and vaccinated macaques over weeks 6 to 24 was previously reported [37]. With 6 additional controls, the 3.5 log difference remained significant (p<0.0001) (B) Mean CD4+ T-cell counts in blood over the course of the study. The dashed line indicates 100 cells/µl. The solid line marks the restoration of CD4 T cells in vaccinated macaques to pre-challenge levels. Post challenge, CD4+ T-cell counts declined to <10 cells/µl in controls. Error bar = standard error of the mean (sem).
Fig. 2. Flow Cytometry gating strategy.
On the left hand side, singlets were further gated on the lymphocytic population followed by a live/dead gate and selection of CD3+ cells. CD3+ cells were gated on CD4 or CD8 with further gating for central memory (CM), effector memory (EM), and total memory (TM) using CD28 and CD95. As an example, CD4 CM cells and CD8 TM cells were gated as shown for IL-17 secretion.
Table 3.
CD4 central and effector memory cells in vaccinated and control macaques over time post-challenge.
| % Positive Cells ± sem | |||
|---|---|---|---|
| Vaccinated Macaques | Pre-challenge | Wk 4 post-challenge | Wk 24 post-challenge |
| CD4CM | 53.8 ± 3.5 | 50.2 ± 5.9 | 56.8 ± 3.0 |
| CD4EM | 9.2 ± 3.9 | 20.8 ± 8.5 | 12.7 ± 3.4 |
| Control Macaques | |||
| CD4CM | 65.2 ± 4.1 | 37.3 ± 6.9 | 25.7 ± 5.7 |
| CD4EM | 8.5 ± 3.4 | 55.5 ± 8.7 | 69.0 ± 7.9 |
Fig. 3. Loss of Th17 cells and up-regulation of CCR6 on Th17 cells during the course of infection in the vaccinated macaques.
(A) Successive decline in the total population of Th17 cells (pre vs wk 4 and pre vs chronic, p = 0.027 for both) in vaccinated, protected macaques. (B) Up-regulation of CCR6 during the course of SHIV89.6P infection. Significantly higher expression was seen in the total Th17 population (pre vs wk4 and pre vs chronic, p = 0.0003 for both). Plotted are mean values with sem.
Tc17 cells showed a trend towards depletion from PBMC over time in control animals, but were maintained in vaccinees controlling viremia
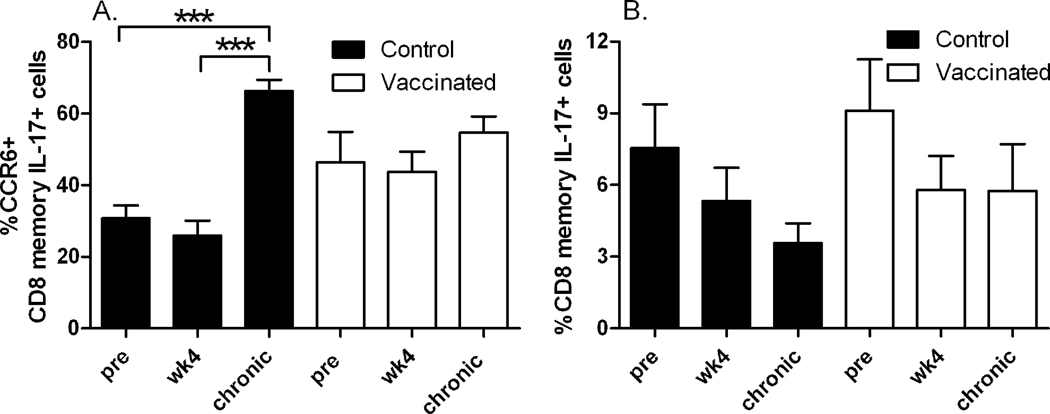
During SIV infection the balance between Th17 and Tc17 is altered in peripheral blood (Kader et al., 2009a; Nigam et al., 2011). Here we investigated the dynamics of the Tc17 cell population longitudinally in the vaccinated and control macaques. In contrast to Th17 cells which are mainly CM T-cells, Tc17 cells were more evenly divided between CM and EM phenotypes (data not shown). Therefore we report here data on total CD8 memory cells. Percentages of CCR6 expressing Tc17 cells were similar within control and vaccinated groups pre-challenge and at 4 weeks post-challenge. However, with disease progression Tc17 cells expressing CCR6 increased significantly compared to pre and wk4 levels (p = 0.0001 for both) but only among control animals (Fig. 4A). In comparison to the control animals, vaccinated macaques expressed slightly but not significantly higher levels of CCR6+ Tc17 cells at the pre and wk4 post-challenge time points. CCR6+ Tc17 cells in vaccinated animals were lower at the chronic phase time point when compared to controls, and overall did not increase over the course of infection (Fig. 4A). In comparison to the CCR6+ Th17 cells in the vaccinated group which increased over the course of infection (Fig. 3B), the CCR6+ Tc17 cells appeared to be more stable.
Fig. 4. CCR6 expression on Tc17 cells and population dynamics over the course of infection.
(A) Mean levels of CCR6 expression on Tc17 cells from the control animals (black bars) and vaccinated animals (open bars). Significantly increased expression on total memory cells (pre vs chronic and wk4 vs chronic, p = 0.0001 for both) was observed for control animals. (B) Tc17 memory population dynamics over the course of infection in control animals (black bars) and vaccinated animals (open bars). Error bar = sem.
The Tc17 population (total memory) varied from roughly 1% to 17% among all animals at the pre-challenge time point. Interestingly, the total Tc17 population tended to decline over time post-challenge in the control animals (Fig. 4B), but this effect did not reach significance. A similar decline in the Tc17 population was recently reported in unvaccinated, SIV infected macaques and was also observed in rectal tissue, supporting the trend observed here (Nigam et al., 2011). A similar trend was not seen in the vaccinated animals (Fig. 4B).
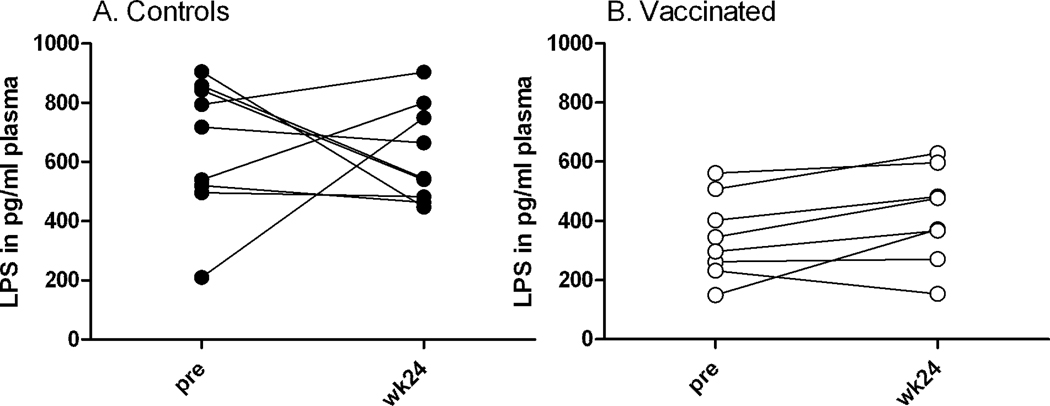
Despite rapid depletion of CD4+ cells in control macaques, enhanced LPS plasma levels in the chronic phase were not detected
As CD4 cells and thus also Th17 cells were depleted in control macaques following viral challenge, we assumed this loss might be associated with enhanced levels of lipopolysaccharides in plasma due to enhanced microbial translocation. This assumption was supported by the even more profound depletion of Th17 and Tc17 cells known to occur from the gut mucosa following pathogenic SIV infection (Nigam et al., 2011). We detected LPS in all plasma samples prior to challenge (Fig. 5). By chance, the control animals had significantly higher mean levels of LPS in plasma both prior to challenge (654 pg/ml) and at the chronic phase time point (622 pg/ml) compared to the vaccinees (345 pg/ml and 419 pg/ml, respectively; p = 0.015 and 0.036, respectively) (Fig. 5). However, post-challenge an expected increase in plasma LPS levels for the control animals was not seen, despite the rapid decline in total CD4 (and therefore Th17) cells and uncontrolled viremia (Fig. 1A, B). LPS levels remained similar for both groups in the chronic phase (wk24 post-challenge) compared to pre-challenge values in spite of the significant differences in challenge outcome (Fig. 5).
Fig. 5. Lipopolysaccharide (LPS) levels in plasma, measured by LAL assay.
Shown are LPS levels for paired samples before and after challenge (wk24, chronic phase).
To validate the lack of elevated LPS levels, we further tested the original study animals (Demberg et al., 2007) for free and pelleted bacterial DNA in plasma at the same time points. The PCR data confirmed our previous observations using the LAL assay; all samples evaluated for 16s rDNA by real time PCR were negative (data not shown).
To further validate the findings from the LAL and 16s rDNA PCR assays, we tested the same animals for CD14 mRNA by real-time PCR using total mRNA from PBMCs at wk48, wk56 and wk70. Increased levels of CD14 mRNA might suggest enhanced activation of monocytes and macrophages due to increased levels of bacteria or bacterial endotoxins (LPS) in the blood resulting from microbial translocation and/or mucosal barrier damage. The results were in concert with the LAL and 16s PCR data. CD14 mRNA levels were not different at wk56 post-challenge compared to pre-challenge values, either in the control group (0.8 fold change) or in the vaccinated group (non-significant 1.4 fold change). At wk70 CD14 mRNA levels further increased in the vaccinated group, however, the moderate 2.2 fold change was still not significantly different compared to pre-challenge (wk48) levels (data not shown). Similarly, the control group showed no elevation in CD14 mRNA levels at wk70 compared to pre-challenge values (1.2 fold change).
Chronic immune activation was associated with increased viremia in the absence of microbial translocation
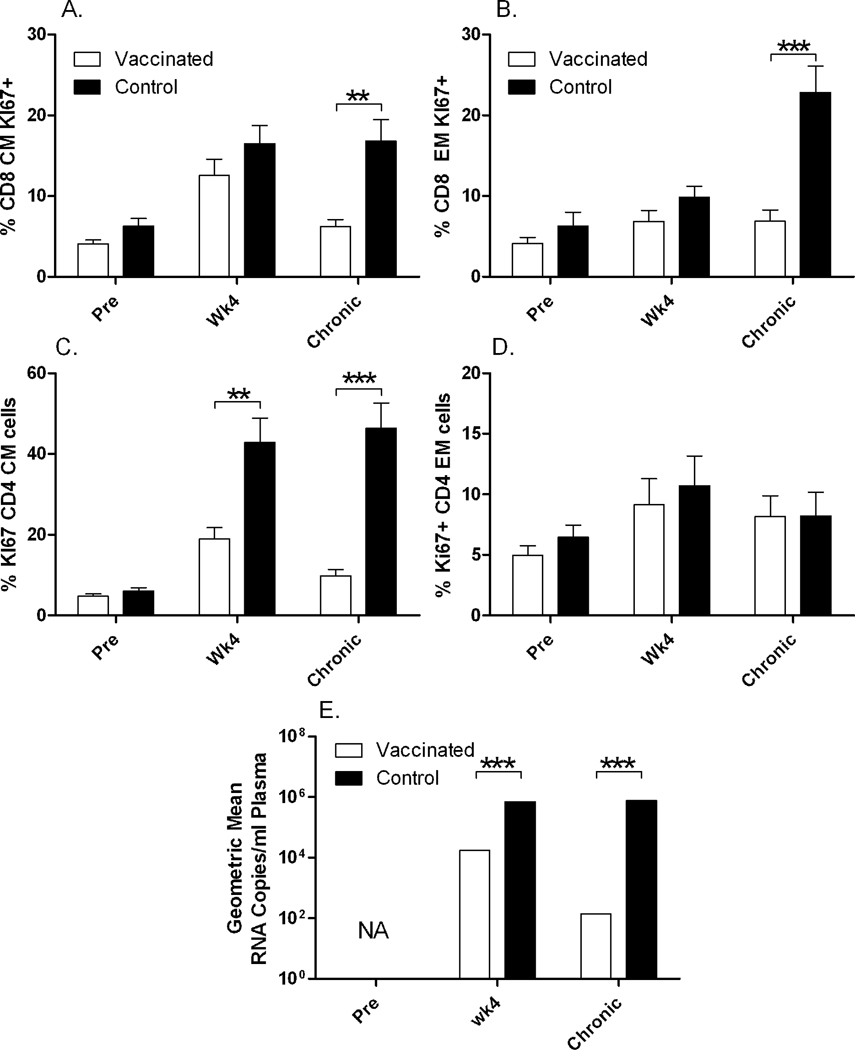
Chronic immune activation in HIV infected individuals has been associated with bacterial DNA plasma levels (Jiang et al., 2009). In spite of the lack of increase in LPS levels with time of infection, we nevertheless looked at chronic immune activation by Ki-67 and IFN-γ staining in all the control and vaccinated macaques pre-challenge, 4 weeks post-challenge and at wk24 post-challenge during the chronic phase. Overall IFN-γ levels were low (data not shown). We detected significantly elevated immune activation in the chronic phase of infection in control macaques, as shown by increased percentages of Ki-67+ CD8+ CM (p = 0.0043) and EM cells (p= 0.0006) at wk24 post challenge and CD4+ CM cells at wk4 and wk24 post-challenge (p = 0.014 and p<0.0001), compared to values in vaccinated animals in which activation increased by wk4 post challenge and dropped during the chronic phase back to almost pre challenge levels (Fig. 6A–C). In contrast, CD4+ EM cells were not elevated in control macaques at any time point (Fig. 6D). The increase in Ki-67+ activated cells was in concert with the increased viremia both at wk 4 and wk 24 post-challenge in the control macaques compared to the vaccinated macaques (p <0.0001 for both; Fig. 6E). Overall, the control animals exhibited a sustained high level of viremia throughout the chronic phase of infection (Fig. 1A).
Fig. 6. Chronic immune activation among CD8 and CD4 memory populations in control and vaccinated macaques.
(A) Mean Ki-67 expression among CD8+ CM and (B) EM cells pre- and post-challenge (wk4 and wk24). Ki-67+ cells were elevated in controls compared to vaccinated macaques during chronic infection (wk24) for both CD8+ CM (p = 0.0043) and EM (p = 0.0006) cells. (C) Mean Ki-67 expression among CD4+ CM and (D) EM cells pre- and post-challenge (wk4 and wk24). Ki-67+ cells were elevated in controls compared to vaccinated macaques at wk4 and wk24 for CD4+ CM cells (p = 0.014 and p<0.0001, respectively) (C), whereas no differences were observed for EM cells (D). (E) Geometric mean viral loads illustrating increased viral burdens in control compared to vaccinated macaques post-challenge at wk4 and wk24 (p<0.0001 for both).
Elevated plasma IFN-γ and elevated MxA mRNA are indicative of chronic inflammation in control macaques
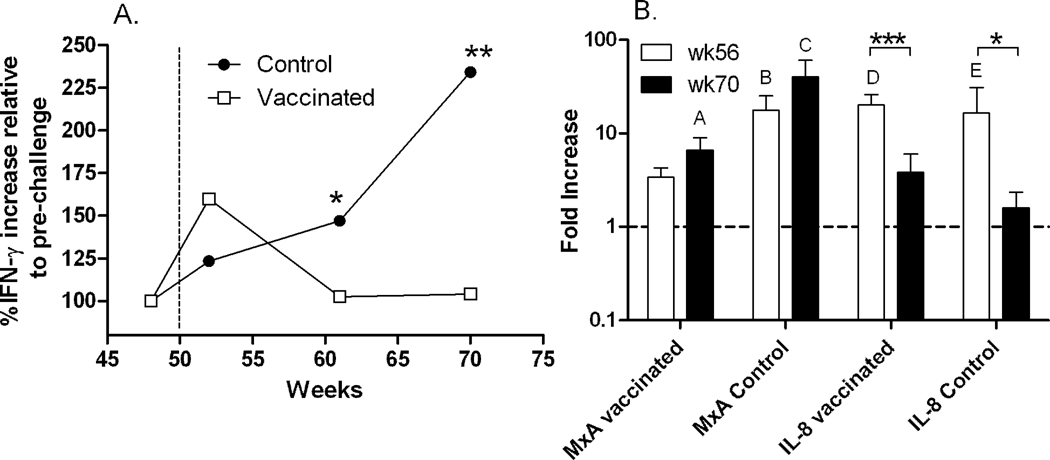
In addition to cellular markers of activation, we tried to identify a cytokine/chemokine activation signature in plasma at the protein level in the animals from the original vaccine study (3 controls, 8 vaccinated) (Demberg et al., 2007), examining plasma from both groups at pre- and post-challenge time points (pre, wk2, wk11, wk20) using the Luminex bead array. Surprisingly, the cytokines IL-2, IL-4, IL-10 and TNF-α were below the lower limit of detection in both groups. However, we observed a transient non-significant increase in the levels of five additional cytokines/chemokines (Rantes, MIP-1α, MIP-1β, MCP-1, and IL-8) during acute infection (wk2). Only IFN-γ was significantly elevated in control macaques at wk11 and wk20 compared to the vaccinated animals (p = 0.036 and 0.0036, respectively; Fig. 7A). This increase in plasma IFN-γ was marginally but not significantly correlated with the corresponding viral loads (p = 0.052; data not shown).
Fig. 7. Plasma IFN-γ levels post-challenge normalized to pre-challenge values and real time PCR analysis of MxA and IL-8 mRNA.
(A) Plasma levels of IFN-γ as indicated over the course of infection. The mean of duplicate assays are plotted. The dotted line indicates the time of challenge. IFN-γ levels in control macaques compared to the vaccinated group were elevated at wk11 and wk20 post-challenge (p = 0.036 and 0.0036, respectively). (B) Mean fold increases of MxA and IL-8 mRNA at wk6 and wk20 post-challenge compared to pre values for the vaccinated and control groups are shown. Significant elevations in MxA mRNA are indicated as follows: A: p = 0.047; B: p = 0.0026; C: p = 0.0021. No significant differences between levels in control and vaccinated macaques were observed. Significant elevations in IL-8 mRNA are indicated as follows: D: p<0.0001; E: p = 0.047. IL-8 mRNA dropped significantly in the vaccinated and control groups between wk6 and wk20 (p = 0.0005 and p = 0.048, respectively). Error bar = sem.
To further explore cytokine/chemokine expression, we conducted real-time PCR experiments using cDNA derived from total mRNA isolated from PBMC at pre, wk6 and wk20 post-challenge timepoints, assaying MxA, TNF-α, IFN-γ, MIP-1α, MIP1β, Rantes, IL-8 and IL-10. The PCR results differed somewhat from the protein expression data. Compared to pre-challenge levels, only mRNA of MxA, one of several IFN-α/β induced genes, was significantly upregulated post-challenge in controls at wk6 and wk20 (p = 0.0026, p = 0.0021, respectively) and in the vaccinated group at wk20 only (p = 0.047) (Fig. 7B). In addition to MxA, IL-8 expression post-challenge was transiently elevated in both control and vaccinated groups compared to pre-challenge values at wk6 (p<0.0001 and p = 0.047 for vaccinated and controls, respectively), but declined by wk20 (Fig. 7B). The decreases in IL-8 expression between wk6 and wk20 in the vaccinated and control groups were statistically significant (p = 0.0005 and p = 0.048, respectively; Fig. 7B).
Discussion
In this study we addressed the hypothesis that vaccinated macaques, partially protected against a SHIV89.6P challenge and showing strong control of chronic viremia, although not exhibiting sterilizing immunity, would exhibit enhanced preservation of Th17 and Tc17 cells and decreased microbial translocation compared to unprotected control animals. Such an outcome would suggest that partially protective vaccines might have a potential benefit by reducing immune activation, potentially preventing opportunistic bacterial infections and slowing disease progression. In addition to monocytes/macrophages, Th17 cells play a role in control of bacterial infections (Crome, Wang, and Levings, 2010; Cua and Tato, 2010; Damsker, Hansen, and Caspi, 2010). Here, for the first time to our knowledge, they were investigated longitudinally in a preclinical HIV vaccine study in non-human primates that achieved a highly protective outcome. We observed a very rapid decline in peripheral blood CD4 cells in unvaccinated control animals by 4 weeks post-challenge (Fig. 1B). In vaccinated macaques which exhibited strong control of viremia and maintained CD4 counts over time (Fig. 1A,B) we nevertheless observed a significant loss of Th17 cells by 24 weeks post-challenge (Fig. 3A). Th17 cells are targets for HIV/SIV replication (El Hed et al., 2010; Kader et al., 2009b; Klatt and Brenchley, 2010) and can be depleted during infection. Th17 cells express CXCR4 under inflammatory conditions and express CCR5 constitutively (Lim et al., 2008; Miller Sanders, Cruse, and Lewis, 2010). Both co-receptors are used by the dual-tropic SHIV89.6P challenge virus, perhaps contributing to the loss of the Th17 population even in conjunction with low levels of viremia.
In contrast to the loss of Th17 cells seen here in SHIV infected macaques; Maek et al. (2007) reported that HIV positive patients show an increase in both Th17 and CD3+CD4−IL-17+ populations compared to healthy volunteers. Further, Ndhlovu et al. (2008) reported that suppression or control of viral replication below 50 copies/ml preserves IL-17 producing cells in HIV infected children, whereas children with higher viral loads showed a significant decline in these cells. By 24 weeks post-challenge, in 4 animals with viral loads of <50 copies/ml and a fifth with <100 copies/ml, we observed a decline in Th17 cells compared to pre-challenge levels (data not shown), contradicting these findings. Further evidence contradicting the preservation of Th17 cells during infection was reported by Kader et al. (2009a) and Nigam et al. (2011) who showed that HAART treatment of SIV infected rhesus monkeys cannot restore the Th17 population to pre-infection levels.
Loss of Th17 cells during HAART might be due either to ongoing low level infection despite control of plasma viremia, or to homing of the cells to inflamed tissues. Together with the decline of Th17 cells over time in the protected macaques we observed an increase in CCR6 receptor expression (Fig. 3B), indicating potential homing to inflammatory sites (Potzl et al., 2008; Yamazaki et al., 2008). CCR6, a hallmark of human Th17 cells, may serve as an activation marker on rhesus macaque Th17 cells, and expression levels may change with homing of the cells to target tissues. Unfortunately due to limitations of PBMC samples and the flow cytometry staining panel, we were unable to investigate expression of the gut mucosal homing marker α4β7 which is expressed on a large proportion of Th17 cells (Kader et al., 2009b; Nigam et al., 2011). In fact cells that express CCR6 also express the gut homing markers α4β7 or αEβ7 to some extent (Nigam et al., 2011). It would be of interest to investigate whether these gut homing markers also increase in parallel with the higher expression of CCR6. Homing in the presence of low-level, persistent infection could explain in part the loss of these cells from the circulation over time as previously proposed (Klatt and Brenchley, 2010). In fact, Ciccone et al. (2011) recently reported higher levels of Th17 cells in the colon compared to the peripheral blood of HIV-infected long-term non-progressors and those on long-term ART therapy.
Here we also report for the first time investigation of Tc17 cells overtime in a highly effective preclinical vaccine study. Only a few reports have appeared on Tc17 cells with regard to SIV/HIV infection (Kader et al., 2009a; Maek et al., 2007; Ndhlovu et al., 2008; Nigam et al., 2011). Interestingly, the upregulation of CCR6 expression on Tc17 cells seems to be similar to that of Th17 cells. The downward trend of these cells over time in the unprotected control group but not in the vaccinated group might be due to enhanced migration of this population to inflamed tissue sites, potentially following the “footsteps” of Th17 cells (Fig. 4B). The significantly higher expression in control macaques of CCR6 at wk74 compared to the pre and wk4 time points (Fig. 4A) supports this assumption. Currently the role of Tc17 cells in vivo is unclear. In cancer patients Tc17 cells cluster around the edges of tumors, secrete high amounts of inflammatory cytokines, and express CCR6 and an effector phenotype, but exhibit limited cytotoxic capacity (Kuang et al., 2010; Nigam et al., 2011). These findings are in accord with our results showing that the cells have in part an effector phenotype and express CCR6, indicative of greater activation. Overall the function and contribution of Tc17 cells in SIV and HIV infection remain elusive and warrant further investigation.
Plasma LPS levels are routinely used as an indication of microbial translocation. Here, no increase in LPS levels was observed post-challenge during the chronic phase in either control or vaccinated macaques using a sensitive LAL assay. Further, bacterial DNA was not detected, and CD14 mRNA levels were not increased post-challenge. Overall microbial translocation might be slow in onset and require a sustained period of chronic infection rather than the rapid CD4 depletion and disease course seen in the SHIV model. This suggestion is supported by the recent report of Redd et al. (2011) which proposed that increased microbial translocation and elevated LPS levels are a consequence of advanced HIV disease and AIDS rather than a cause. Numerous discrepancies in the literature regarding microbial translocation and HIV-1 disease progression have been reported (Redd et al., 2011). Similar discrepancies have occurred in the SIV literature as well. LPS levels were not predictive of SIV disease progression in rhesus macaques (Leinert et al., 2010) whereas high levels of LPS prior to SIV infection were associated with faster disease progression in pigtailed macaques (Klatt et al., 2010). In some aspects, the course of SHIV89.6P infection seen here in control animals resembles SIV infection of natural hosts, where high viremia occurs in the absence of microbial translocation (Brenchley and Douek, 2008b; Pandrea et al., 2007). However, in contrast to our results, rhesus monkeys challenged with non-pathogenic SIVagm display a transient drop in CD4 counts but no preferential drop in mucosal Th17 cells occurs (Paiardini, 2010). Further, although increased apoptosis and immune activation are seen at mucosal sites in the rhesus/SIVagm model, only a transient, non-significant increase in LPS is seen before levels drop below pre-challenge values with control of viremia (Pandrea et al., 2007).
Several mechanisms might contribute to control of LPS during SHIV infection. During early stages of HIV infection, endotoxin core antibodies (EndoCab) can help control LPS blood levels (Brenchley et al., 2006) but may also interfere with the detection of LPS by the LAL assay (Bennett-Guerrero et al., 2001). The expansion of EndoCab antibody (IgG) responses has been shown in live attenuated Shigella vaccine trials (Simon et al., 2009) and in cystic fibrosis patients after infection with Pseudomonas cepacia (Nelson et al., 1993). Enhanced EndoCab antibodies have been reported in plasma of an HIV-infected African cohort and might have contributed to “protection” from increased LPS levels (Redd et al., 2009). Moreover, it was recently shown that plasma LPS levels correlated inversely with serum anti-LPS antibodies in untreated HIV patients, suggesting a possible role of the antibody in decreasing LPS levels (Lim et al., 2011). We did not measure EndoCab antibodies due to limited cross-reactivity of available reagents (only IgM reactive) with rhesus macaque proteins (Leinert et al., 2010). The role and influence of anti-LPS antibodies during HIV/SIV infection should be investigated in future studies.
While elevated LPS plasma levels were not observed in the SHIV89.6P model, we did observe chronic immune activation in CD8+ CM and EM, and CD4+ CM cells, evidenced by Ki-67+ cells, in association with high viremia in the control macaques, confirming previous reports that activation is correlated with viral load and disease progression (Leligdowicz et al., 2010; Redd et al., 2010; Tuaillon et al., 2009). In contrast to our findings (Fig. 6D), others have reported strong Ki-67 expression in the CD4+ EM compartment following CD8+ T cell depletion in the SHIV89.6P model (Mueller et al., 2009), however, this might be attributed to proliferation of CD4+ T-cells following the depletion. In the SIV model, strong Ki-67 expression has been seen on CD4+ CM cells (Okoye et al., 2007), similar to our findings, but also on CD4+ EM cells. The latter observation follows distinction of “transitional” EM and EM cells as measured by CCR7 and CCR5. Without this further division of memory subpopulations, almost all the reported Ki-67+ cells would have been phenotyped as CM cells, in agreement with our findings (Fig. 6C). Ki-67 has been correlated with expression of HLA-DR and CD38 in HIV infected individuals (Chattopadhyay and Roederer, 2010). Moreover, as markers of chronic immune activation, CD38 and HLA-DR have been recently reported to correlate well in monkeys (Manrique et al., 2011). The inclusion of these markers in future studies might provide a more detailed picture regarding chronic immune activation.
To examine immune activation in greater depth, we sought to define an activation profile based on cytokines/chemokines in plasma. In general strong IFN-γ responses are associated with control of viral infection. A recent report showed that higher IFN-γ levels during acute HIV infection were associated with lower viral loads 12 months later at set point (Roberts et al., 2010). Here, plasma IFN-γ was slightly (but not significantly) elevated in the vaccinated group of macaques compared to controls during acute infection (Fig. 7A), most likely reflecting recall of adaptive immunity, whereas IFN-γ levels rose rapidly thereafter in controls in response to continuing high viremia.
Our investigations of gene transcription by PCR revealed significant drops in IL-8 mRNA in the vaccinated and control groups at wk20 post-challenge (Fig. 7B). Overall, however, total IL-8 mRNA remained slightly higher in the vaccinated group (Fig. 7B). This might indicate that Th17 or Tc17 cells migrated to mucosal sites. IL-8 is an IL-17-regulated gene (Raffatellu et al., 2008), therefore higher IL-8 mRNA levels in vaccinated animals during the chronic phase might be explicable by the slower decline in Th17 cells and stable Tc17 population. In the control macaques the lower IL-8 mRNA levels in PBMC might be due to diminished gene activation attributed to loss or migration of Th17 cells and the potentially limited compensatory capacity of Tc17 cells which tended to decline or migrate into tissues (strong CCR6 upregulation) (Fig. 4B).
The type I interferons, IFN-α/β, are mainly produced by plasmacytoid dendritic cells (pDC) through activation by TLR9 and TLR7 (Fitzgerald-Bocarsly and Jacobs, 2010; Swiecki and Colonna, 2010). During in vitro HIV/SIV infection, pDCs secrete high amounts of IFN-α (Harris et al., 2010; Swiecki and Colonna, 2010) which activates the MxA gene (Haller, Staeheli, and Kochs, 2009). MxA is a well defined surrogate marker for the IFN-α family which exhibits high diversity in humans (Nyman et al., 1998) and rhesus macaques (Easlick et al., 2010). In early phase of chronic SIV infection the down regulation of an initial robust IFN-α response distinguishes nonpathogenic infection of the natural host from pathogenic infection of rhesus macaques (Harris et al., 2010). Here, using activation of the MxA gene as a surrogate marker IFN-α, we asked whether high levels of IFN-α are similarly associated with disease progression and low levels with control of viremia. MxA levels were elevated rapidly in control macaques with significantly enhanced expression levels seen at wk6 and wk20 post-challenge (Fig. 7B), whereas MxA expression in the vaccinated group was not significantly elevated until wk20. Stronger induction was seen in the control group, as expected for a pathogenic infection (Harris et al., 2010). However, the increased MxA expression in the vaccinated group by week 20 suggests eventual disease progression.
The lack of correlation observed between plasma cytokine protein levels and mRNA levels can be attributed to the different samples used for analysis. The cytokine/chemokine levels in plasma represent a pool to which many cell types from multiple tissues contributed. In contrast mRNA levels measured by real-time PCR were obtained on purified PBMC, thus representing only a fraction of potential cell contributors.
The dual-tropic SHIV89.6P is able to infect and deplete not only memory CD4 cells but also naïve cells in the rhesus macaque model (Zhang et al., 2000), leading to more rapid depletion of CD4+ T-cells compared to the SIV model. Thus the SHIV infected animals in general progress faster to AIDS. In this challenge model we show that a partially protective vaccine was unable to prevent the loss of Th17 over time, despite preservation of CD4 cells potent control of viremia. However no loss of Tc17 cells over the course of infection was seen in vaccinated animals, as has been reported for unvaccinated, SIV-infected animals (Nigam et al., 2011), similar to the trend observed here for the control group. Higher levels of immune activation as measured by Ki-67 on CD4 and CD8 cells, plasma IFN-γ and MxA mRNA levels were observed in the control group in accordance with uncontrolled viremia in these animals. Interestingly elevated plasma LPS levels were not observed in either group during SHIV89.6P infection calling for further investigation of microbial translocation in the mucosa and the role of Th17 and Tc17 cells. Overall, despite the control of viremia to the threshold of detection in the strongly protected vaccinated macaques, we detected a loss of Th17 cells from the PBMC. In control animals, in addition to the severe depletion of CD4 cells from the blood, a slow loss of Tc17 cells was seen over time potentially indicating that Tc17 and Th17 cells home to the gut mucosal tissue, where virus persists and where damage is most profound. Due to the classic role of Th17 cells in bacterial infection and the emerging role of Tc17 cells, it is likely that both cell types home to the inflamed tissue sites and engage to control bacterial, fungal and viral infections. Unfortunately no fresh tissue samples were available from these animals for cellular analyses nor were frozen tissues for immunohistochemical staining for Th17 and Tc17 cells and bacteria in the GI tract. Questions to address in the future include design of strategies to better preserve these cells, characterization of their role in the gut mucosa, and methods to harness their functions in vaccine approaches to prevent HIV induced epithelial damage and microbial translocation and to fight disease progression.
Acknowledgment
We thank Dr. Claudio Fenizia for providing the MxA primer sequences. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions.
Conceived and designed the experiments: TD, KM, LJP, TMP, MRG. Performed the experiments: TD, AAE, SA, KM, TK. Analyzed the data: TD, KM, DV, TMP, MRG. Wrote the paper: TD, MRG.
References
- Annunziato F, Cosmi L, Liotta F, Maggi E, Romagnani S. The phenotype of human Th17 cells and their precursors, the cytokines that mediate their differentiation and the role of Th17 cells in inflammation. Int Immunol. 2008;20(11):1361–1368. doi: 10.1093/intimm/dxn106. [DOI] [PubMed] [Google Scholar]
- Bello G, Velasco-de-Castro CA, Bongertz V, Rodrigues CA, Giacoia-Gripp CB, Pilotto JH, Grinsztejn B, Veloso VG, Morgado MG. Immune activation and antibody responses in non-progressing elite controller individuals infected with HIV-1. J Med Virol. 2009;81(10):1681–1690. doi: 10.1002/jmv.21565. [DOI] [PubMed] [Google Scholar]
- Bennett-Guerrero E, Barclay GR, Weng PL, Bodian CA, Feierman DE, Vela-Cantos F, Mythen MG. Endotoxin-neutralizing capacity of serum from cardiac surgical patients. J Cardiothorac Vasc Anesth. 2001;15(4):451–454. doi: 10.1053/jcan.2001.24980. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Douek DC. HIV infection and the gastrointestinal immune system. Mucosal Immunol. 2008a;1(1):23–30. doi: 10.1038/mi.2007.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Douek DC. The mucosal barrier and immune activation in HIV pathogenesis. Curr Opin HIV AIDS. 2008b;3(3):356–361. doi: 10.1097/COH.0b013e3282f9ae9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, Khoruts A, Frank I, Else J, Schacker T, Silvestri G, Douek DC. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112(7):2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12(12):1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200(6):749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay PK, Roederer M. Good cell, bad cell: flow cytometry reveals T-cell subsets important in HIV disease. Cytometry A. 2010;77(7):614–622. doi: 10.1002/cyto.a.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone EJ, Greenwald JH, Lee PI, Biancotto A, Read SW, Yao MA, Hodge JN, Thompson WL, Kovacs SB, Chairez CL, Migueles SA, Kovacs JA, Margolis LB, Sereti I. CD4+ T Cells, Including Th17 and Cycling Subsets, Are Intact in the Gut Mucosa of HIV-1-Infected Long-Term Nonprogressors. J Virol. 2011;85(12):5880–5888. doi: 10.1128/JVI.02643-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crome SQ, Wang AY, Levings MK. Translational mini-review series on Th17 cells: function and regulation of human T helper 17 cells in health and disease. Clin Exp Immunol. 2010;159(2):109–119. doi: 10.1111/j.1365-2249.2009.04037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol. 2010;10(7):479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- Damsker JM, Hansen AM, Caspi RR. Th1 and Th17 cells: adversaries and collaborators. Ann N Y Acad Sci. 2010;1183:211–221. doi: 10.1111/j.1749-6632.2009.05133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong E, Suddason T, Lord GM. Translational mini-review series on Th17 cells: development of mouse and human T helper 17 cells. Clin Exp Immunol. 2010;159(2):148–158. doi: 10.1111/j.1365-2249.2009.04041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demberg T, Florese RH, Heath MJ, Larsen K, Kalisz I, Kalyanaraman VS, Lee EM, Pal R, Venzon D, Grant R, Patterson LJ, Korioth-Schmitz B, Buzby A, Dombagoda D, Montefiori DC, Letvin NL, Cafaro A, Ensoli B, Robert-Guroff M. A replication-competent adenovirus-human immunodeficiency virus (Ad-HIV) tat and Ad-HIV env priming/Tat and envelope protein boosting regimen elicits enhanced protective efficacy against simian/human immunodeficiency virus SHIV89.6P challenge in rhesus macaques. J Virol. 2007;81(7):3414–3427. doi: 10.1128/JVI.02453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodon MD, Li Z, Hamaia S, Gazzolo L. Tax protein of human T-cell leukaemia virus type 1 induces interleukin 17 gene expression in T cells. J Gen Virol. 2004;85(Pt 7):1921–1932. doi: 10.1099/vir.0.79921-0. [DOI] [PubMed] [Google Scholar]
- Easlick J, Szubin R, Lantz S, Baumgarth N, Abel K. The early interferon alpha subtype response in infant macaques infected orally with SIV. J Acquir Immune Defic Syndr. 2010;55(1):14–28. doi: 10.1097/QAI.0b013e3181e696ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hed A, Khaitan A, Kozhaya L, Manel N, Daskalakis D, Borkowsky W, Valentine F, Littman DR, Unutmaz D. Susceptibility of human Th17 cells to human immunodeficiency virus and their perturbation during infection. J Infect Dis. 2010;201(6):843–854. doi: 10.1086/651021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrelly V, Rainey FA, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplivication of 16S rRNA genes from a mixture of bacterial species. Appl environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio D, Poggi A, Catellani S, Battaglia F, Ferrera A, Setti M, Murdaca G, Zocchi MR. Vdelta1 T lymphocytes producing IFN-gamma and IL-17 are expanded in HIV-1-infected patients and respond to Candida albicans. Blood. 2009;113(26):6611–6618. doi: 10.1182/blood-2009-01-198028. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Bocarsly P, Jacobs ES. Plasmacytoid dendritic cells in HIV infection: striking a delicate balance. J Leukoc Biol. 2010;87(4):609–620. doi: 10.1189/jlb.0909635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SN, Klatt NR, Bosinger SE, Brenchley JM, Milush JM, Engram JC, Dunham RM, Paiardini M, Klucking S, Danesh A, Strobert EA, Apetrei C, Pandrea IV, Kelvin D, Douek DC, Staprans SI, Sodora DL, Silvestri G. Severe depletion of mucosal CD4+ T cells in AIDS-free simian immunodeficiency virus-infected sooty mangabeys. J Immunol. 2007;179(5):3026–3034. doi: 10.4049/jimmunol.179.5.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AT. Perils at mucosal front lines for HIV and SIV and their hosts. Nat Rev Immunol. 2005;5(10):783–792. doi: 10.1038/nri1706. [DOI] [PubMed] [Google Scholar]
- Haller O, Staeheli P, Kochs G. Protective role of interferon-induced Mx GTPases against influenza viruses. Rev Sci Tech. 2009;28(1):219–231. doi: 10.20506/rst.28.1.1867. [DOI] [PubMed] [Google Scholar]
- Hamada H, Garcia-Hernandez Mde L, Reome JB, Misra SK, Strutt TM, McKinstry KK, Cooper AM, Swain SL, Dutton RW. Tc17, a unique subset of CD8 T cells that can protect against lethal influenza challenge. J Immunol. 2009;182(6):3469–3481. doi: 10.4049/jimmunol.0801814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardstedt M, Finnegan CP, Kirchhof N, Hyland KA, Wijkstrom M, Murtaugh MP, Hering BJ. Post-transplant upregulation of chemokine messenger RNA in non-human primate recipients of intraportal pig islet xenografts. Xenotransplantation. 2005;12(4):293–302. doi: 10.1111/j.1399-3089.2005.00228.x. [DOI] [PubMed] [Google Scholar]
- Harris LD, Tabb B, Sodora DL, Paiardini M, Klatt NR, Douek DC, Silvestri G, Muller-Trutwin M, Vasile-Pandrea I, Apetrei C, Hirsch V, Lifson J, Brenchley JM, Estes JD. Downregulation of robust acute type I interferon responses distinguishes nonpathogenic simian immunodeficiency virus (SIV) infection of natural hosts from pathogenic SIV infection of rhesus macaques. J Virol. 2010;84(15):7886–7891. doi: 10.1128/JVI.02612-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Hirota K, Yoshitomi H, Maeda S, Teradaira S, Akizuki S, Prieto-Martin P, Nomura T, Sakaguchi N, Kohl J, Heyman B, Takahashi M, Fujita T, Mimori T, Sakaguchi S. Complement drives Th17 cell differentiation and triggers autoimmune arthritis. J Exp Med. 2010;207(6):1135–1143. doi: 10.1084/jem.20092301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann-Lehmann R, Williams AL, Swenerton RK, Li PL, Rasmussen RA, Chenine AL, McClure HM, Ruprecht RM. Quantitation of simian cytokine and beta-chemokine mRNAs, using real-time reverse transcriptase-polymerase chain reaction: variations in expression during chronic primate lentivirus infection. AIDS Res Hum Retroviruses. 2002;18(9):627–639. doi: 10.1089/088922202760019329. [DOI] [PubMed] [Google Scholar]
- Hunt PW. Th17, gut, and HIV: therapeutic implications. Curr Opin HIV AIDS. 2010;5(2):189–193. doi: 10.1097/COH.0b013e32833647d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Lederman MM, Hunt P, Sieg SF, Haley K, Rodriguez B, Landay A, Martin J, Sinclair E, Asher AI, Deeks SG, Douek DC, Brenchley JM. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009;199(8):1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader M, Bixler S, Piatak M, Lifson J, Mattapallil JJ. Anti-retroviral therapy fails to restore the severe Th-17: Tc-17 imbalance observed in peripheral blood during simian immunodeficiency virus infection. J Med Primatol. 2009a;38 Suppl 1:32–38. doi: 10.1111/j.1600-0684.2009.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader M, Wang X, Piatak M, Lifson J, Roederer M, Veazey R, Mattapallil JJ. Alpha4(+)beta7(hi)CD4(+) memory T cells harbor most Th-17 cells and are preferentially infected during acute SIV infection. Mucosal Immunol. 2009b;2(5):439–449. doi: 10.1038/mi.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Hisamatsu T, Honda H, Kobayashi T, Chinen H, Takayama T, Kitazume MT, Okamoto S, Koganei K, Sugita A, Kanai T, Hibi T. TL1A produced by lamina propria macrophages induces Th1 and Th17 immune responses in cooperation with IL-23 in patients with Crohn's disease. Inflamm Bowel Dis. 2010;16(4):568–575. doi: 10.1002/ibd.21124. [DOI] [PubMed] [Google Scholar]
- Kaur A, Hale CL, Ramanujan S, Jain RK, Johnson RP. Differential dynamics of CD4(+) and CD8(+) T-lymphocyte proliferation and activation in acute simian immunodeficiency virus infection. J Virol. 2000;74(18):8413–8424. doi: 10.1128/jvi.74.18.8413-8424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt NR, Brenchley JM. Th17 cell dynamics in HIV infection. Curr Opin HIV AIDS. 2010;5(2):135-–140. doi: 10.1097/COH.0b013e3283364846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt NR, Harris LD, Vinton CL, Sung H, Briant JA, Tabb B, Morcock D, McGinty JW, Lifson JD, Lafont BA, Martin MA, Levine AD, Estes JD, Brenchley JM. Compromised gastrointestinal integrity in pigtail macaques is associated with increased microbial translocation, immune activation, and IL-17 production in the absence of SIV infection. Mucosal Immunol. 2010;3(4):387–398. doi: 10.1038/mi.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang DM, Peng C, Zhao Q, Wu Y, Zhu LY, Wang J, Yin XY, Li L, Zheng L. Tumor-activated monocytes promote expansion of IL-17-producing CD8+ T cells in hepatocellular carcinoma patients. J Immunol. 2010;185(3):1544–1549. doi: 10.4049/jimmunol.0904094. [DOI] [PubMed] [Google Scholar]
- Leinert C, Stahl-Hennig C, Ecker A, Schneider T, Fuchs D, Sauermann U, Sopper S. Microbial translocation in simian immunodeficiency virus (SIV)-infected rhesus monkeys (Macaca mulatta) J Med Primatol. 2010;39(4):243–251. doi: 10.1111/j.1600-0684.2010.00429.x. [DOI] [PubMed] [Google Scholar]
- Leligdowicz A, Feldmann J, Jaye A, Cotten M, Dong T, McMichael A, Whittle H, Rowland-Jones S. Direct relationship between virus load and systemic immune activation in HIV-2 infection. J Infect Dis. 2010;201(1):114–122. doi: 10.1086/648733. [DOI] [PubMed] [Google Scholar]
- Lim A, Amini A, D’Orsogna L, Rajasuriar R, Kramski M, Lewin SR, Purcell D, Price P, French MA. Antibody and B cell responses may control circulating lipopolysaccharide in patients with HIV infection. AIDS. 2011 doi: 10.1097/QAD.0b013e328348a789. in press. [DOI] [PubMed] [Google Scholar]
- Lim HW, Lee J, Hillsamer P, Kim CH. Human Th17 cells share major trafficking receptors with both polarized effector T cells and FOXP3+ regulatory T cells. J Immunol. 2008;180(1):122–129. doi: 10.4049/jimmunol.180.1.122. [DOI] [PubMed] [Google Scholar]
- Ling B, Mohan M, Lackner AA, Green LC, Marx PA, Doyle LA, Veazey RS. The large intestine as a major reservoir for simian immunodeficiency virus in macaques with long-term, nonprogressing infection. J Infect Dis. 2010;202(12):1846–1854. doi: 10.1086/657413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke P, Favre D, Hunt PW, Leung JM, Kanwar B, Martin JN, Deeks SG, McCune JM. Correlating cellular and molecular signatures of mucosal immunity that distinguish HIV controllers from noncontrollers. Blood. 2010;115(15):e20–e32. doi: 10.1182/blood-2009-12-257451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck MD, Natuk R, Myagkikh M, Kalyan N, Aldrich K, Sinangil F, Alipanah S, Murthy SC, Chanda PK, Nigida SM, Jr., Markham PD, Zolla-Pazner S, Steimer K, Wade M, Reitz MS, Jr., Arthur LO, Mizutani S, Davis A, Hung PP, Gallo RC, Eichberg J, Robert-Guroff M. Long-term protection of chimpanzees against high-dose HIV-1 challenge induced by immunization. Nat Med. 1997;3(6):651–658. doi: 10.1038/nm0697-651. [DOI] [PubMed] [Google Scholar]
- Maek ANW, Buranapraditkun S, Klaewsongkram J, Ruxrungtham K. Increased interleukin-17 production both in helper T cell subset Th17 and CD4-negative T cells in human immunodeficiency virus infection. Viral Immunol. 2007;20(1):66–75. doi: 10.1089/vim.2006.0063. [DOI] [PubMed] [Google Scholar]
- Malkevitch NV, Patterson LJ, Aldrich MK, Wu Y, Venzon D, Florese RH, Kalyanaraman VS, Pal R, Lee EM, Zhao J, Cristillo A, Robert-Guroff M. Durable protection of rhesus macaques immunized with a replicating adenovirus-SIV multigene prime/protein boost vaccine regimen against a second SIVmac251 rectal challenge: role of SIV-specific CD8+ T cell responses. Virology. 2006;353(1):83–98. doi: 10.1016/j.virol.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Manrique M, Kozlowski PA, Cobo-Molinos A, Wang SW, Wilson RL, Montefiori DC, Mansfield KG, Carville A, Aldovini A. Long-term control of simian immunodeficiency Virus mac251 viremia to undetectable levels in half of infected female rhesus macaques nasally vaccinated with simian immunodeficiency virus DNA/recombinant modified vaccinia virus Ankara. J Immunol. 2011;186(6):3581–3593. doi: 10.4049/jimmunol.1002594. [DOI] [PubMed] [Google Scholar]
- Marchetti G, Bellistri GM, Borghi E, Tincati C, Ferramosca S, La Francesca M, Morace G, Gori A, Monforte AD. Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS. 2008;22(15):2035–2038. doi: 10.1097/QAD.0b013e3283112d29. [DOI] [PubMed] [Google Scholar]
- Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434(7037):1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- Medeiros M, Sharma VK, Ding R, Yamaji K, Li B, Muthukumar T, Valderde-Rosas S, Hernandez AM, Munoz R, Suthanthiran M. Optimization of RNA yield, purity and mRNA copy number by treatment of urine cell pellets with RNAlater. J Immunol Methods. 2003;279(1–2):135–142. doi: 10.1016/s0022-1759(03)00237-0. [DOI] [PubMed] [Google Scholar]
- Miller CJ, Li Q, Abel K, Kim EY, Ma ZM, Wietgrefe S, La Franco-Scheuch L, Compton L, Duan L, Shore MD, Zupancic M, Busch M, Carlis J, Wolinsky S, Haase AT. Propagation and dissemination of infection after vaginal transmission of simian immunodeficiency virus. J Virol. 2005;79(14):9217–9227. doi: 10.1128/JVI.79.14.9217-9227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller Sanders C, Cruse JM, Lewis RE. Toll-like receptor and chemokine receptor expression in HIV-infected T lymphocyte subsets. Exp Mol Pathol. 2010;88(1):26–31. doi: 10.1016/j.yexmp.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Mueller YM, Do DH, Boyer JD, Kader M, Mattapallil JJ, Lewis MG, Weiner DB, Katsikis PD. CD8+ cell depletion of SHIV89.6P-infected macaques induces CD4+ T cell proliferation that contributes to increased viral loads. J Immunol. 2009;183(8):5006–5012. doi: 10.4049/jimmunol.0900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazli A, Chan O, Dobson-Belaire WN, Ouellet M, Tremblay MJ, Gray-Owen SD, Arsenault AL, Kaushic C. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PLoS Pathog. 2010;6(4) doi: 10.1371/journal.ppat.1000852. e1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndhlovu LC, Chapman JM, Jha AR, Snyder-Cappione JE, Pagan M, Leal FE, Boland BS, Norris PJ, Rosenberg MG, Nixon DF. Suppression of HIV-1 plasma viral load below detection preserves IL-17 producing T cells in HIV-1 infection. Aids. 2008;22(8):990–992. doi: 10.1097/QAD.0b013e3282ff884e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Butler SL, Brown PH, Greening AP, Govan JR. Serum IgG and sputum IgA antibody to core lipopolysaccharide antigen from Pseudomonas cepacia in patients with cystic fibrosis. J Med Microbiol. 1993;39(1):39–47. doi: 10.1099/00222615-39-1-39. [DOI] [PubMed] [Google Scholar]
- Nigam P, Kwa S, Velu V, Amara RR. Loss of IL-17-producing CD8 T cells during late chronic stage of pathogenic simian immunodeficiency virus infection. J Immunol. 2011;186(2):745–753. doi: 10.4049/jimmunol.1002807. [DOI] [PubMed] [Google Scholar]
- Nyman TA, Tolo H, Parkkinen J, Kalkkinen N. Identification of nine interferon-alpha subtypes produced by Sendai virus-induced human peripheral blood leucocytes. Biochem J. 1998;329(Pt 2):295–302. doi: 10.1042/bj3290295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye A, Meier-Schellersheim M, Brenchley JM, Hagen SI, Walker JM, Rohankhedkar M, Lum R, Edgar JB, Planer SL, Legasse A, Sylwester AW, Piatak M, Jr., Lifson JD, Maino VC, Sodora DL, Douek DC, Axthelm MK, Grossman Z, Picker LJ. Progressive CD4+ central memory T cell decline results in CD4+ effector memory insufficiency and overt disease in chronic SIV infection. J Exp Med. 2007;204(9):2171–2185. doi: 10.1084/jem.20070567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiardini M. Th17 cells in natural SIV hosts. Curr Opin HIV AIDS. 2010;5(2):166–172. doi: 10.1097/COH.0b013e328335c161. [DOI] [PubMed] [Google Scholar]
- Pandrea IV, Gautam R, Ribeiro RM, Brenchley JM, Butler IF, Pattison M, Rasmussen T, Marx PA, Silvestri G, Lackner AA, Perelson AS, Douek DC, Veazey RS, Apetrei C. Acute loss of intestinal CD4+ T cells is not predictive of simian immunodeficiency virus virulence. J Immunol. 2007;179(5):3035–3046. doi: 10.4049/jimmunol.179.5.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson LJ, Beal J, Demberg T, Florese RH, Malkevich N, Venzon D, Aldrich K, Richardson E, Kalyanaraman VS, Kalisz I, Lee EM, Montefiori DC, Robey FA, Robert-Guroff M. Replicating adenovirus HIV/SIV recombinant priming alone or in combination with a gp140 protein boost results in significant control of viremia following a SHIV89.6P challenge in Mamu-A*01 negative rhesus macaques. Virology. 2008;374(2):322–337. doi: 10.1016/j.virol.2007.12.037. [DOI] [PubMed] [Google Scholar]
- Patterson LJ, Daltabuit-Test M, Xiao P, Zhao J, Hu W, Wille-Reece U, Brocca-Cofano E, Kalyanaraman VS, Kalisz I, Whitney S, Lee EM, Pal R, Montefiori DC, Dandekar S, Seder R, Roederer M, Wiseman RW, Hirsch V, Robert-Guroff M. Rapid SIV Env-specific mucosal and serum antibody induction augments cellular immunity in protecting immunized, elite-controller macaques against high dose heterologous SIV challenge. Virology. 2011;411(1):87–102. doi: 10.1016/j.virol.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson LJ, Malkevitch N, Venzon D, Pinczewski J, Gomez-Roman VR, Wang L, Kalyanaraman VS, Markham PD, Robey FA, Robert-Guroff M. Protection against mucosal simian immunodeficiency virus SIV(mac251) challenge by using replicating adenovirus-SIV multigene vaccine priming and subunit boosting. J Virol. 2004;78(5):2212–2221. doi: 10.1128/JVI.78.5.2212-2221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RTPCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potzl J, Botteron C, Tausch E, Pedre X, Mueller AM, Mannel DN, Lechner A. Tracing functional antigen-specific CCR6 Th17 cells after vaccination. PLoS One. 2008;3(8):e2951. doi: 10.1371/journal.pone.0002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast A, Prado JG, Kang YH, Chen F, Riddell LA, Luzzi G, Goulder P, Klenerman P. HIV-1 infection is characterized by profound depletion of CD161+ Th17 cells and gradual decline in regulatory T cells. AIDS. 2010;24(4):491–502. doi: 10.1097/QAD.0b013e3283344895. [DOI] [PubMed] [Google Scholar]
- Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, Godinez I, Sankaran S, Paixao TA, Gordon MA, Kolls JK, Dandekar S, Baumler AJ. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14(4):421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd AD, Dabitao D, Bream JH, Charvat B, Laeyendecker O, Kiwanuka N, Lutalo T, Kigozi G, Tobian AA, Gamiel J, Neal JD, Oliver AE, Margolick JB, Sewankambo N, Reynolds SJ, Wawer MJ, Serwadda D, Gray RH, Quinn TC. Microbial translocation, the innate cytokine response, and HIV-1 disease progression in Africa. Proc Natl Acad Sci U S A. 2009;106(16):6718–6723. doi: 10.1073/pnas.0901983106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd AD, Eaton KP, Kong X, Laeyendecker O, Lutalo T, Wawer MJ, Gray RH, Serwadda D, Quinn TC. C-reactive protein levels increase during HIV-1 disease progression in Rakai, Uganda, despite the absence of microbial translocation. J Acquir Immune Defic Syndr. 2010;54(5):556–559. doi: 10.1097/QAI.0b013e3181e0cdea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd AD, Gray RH, Quinn TC. Is microbial translocation a cause or consequence of HIV disease progression? J Infect Dis. 2011;203:744–745. doi: 10.1093/infdis/jiq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L, Passmore JA, Williamson C, Little F, Bebell LM, Mlisana K, Burgers WA, van Loggerenberg F, Walzl G, Siawaya JF, Karim QA, Karim SS. Plasma cytokine levels during acute HIV-1 infection predict HIV disease progression. AIDS. 2010;24(6):819–831. doi: 10.1097/QAD.0b013e3283367836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnani S, Maggi E, Liotta F, Cosmi L, Annunziato F. Properties and origin of human Th17 cells. Mol Immunol. 2009;47(1):3–7. doi: 10.1016/j.molimm.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Simon JK, Wahid R, Maciel M, Jr., Picking WL, Kotloff KL, Levine MM, Sztein MB. Antigen-specific B memory cell responses to lipopolysaccharide (LPS) and invasion plasmid antigen (Ipa) B elicited in volunteers vaccinated with live attenuated Shigella flexneri 2a vaccine candidates. Vaccine. 2009;27(4):565–572. doi: 10.1016/j.vaccine.2008.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M, Colonna M. Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol Rev. 2010;234(1):142–162. doi: 10.1111/j.0105-2896.2009.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchinsky MB, Blander JM. T helper 17 cells: discovery, function, and physiological trigger. Cell Mol Life Sci. 2010;67(9):1407–1421. doi: 10.1007/s00018-009-0248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troseid M, Nowak P, Nystrom J, Lindkvist A, Abdurahman S, Sonnerborg A. Elevated plasma levels of lipopolysaccharide and high mobility group box-1 protein are associated with high viral load in HIV-1 infection: reduction by 2-year antiretroviral therapy. AIDS. 2010;24(11):1733–1737. doi: 10.1097/QAD.0b013e32833b254d. [DOI] [PubMed] [Google Scholar]
- Tseng C-P, Cheng J-C, Tseng C-C, Wang C, Chen Y-L, Chiu D, T-Y, Liao H-C, Chang S-S. Broad-range ribosomal RNA real-time PCR after removal of DNA from reagents: melting profiles for clinically important bacteria. Clin Chem. 2003;49:306–309. doi: 10.1373/49.2.306. [DOI] [PubMed] [Google Scholar]
- Tuaillon E, Al Tabaa Y, Baillat V, Segondy M, Picot MC, Reynes J, Vendrell JP. Close association of CD8+/CD38 bright with HIV-1 replication and complex relationship with CD4+ T-cell count. Cytometry B Clin Cytom. 2009;76B(4):249–260. doi: 10.1002/cyto.b.20467. [DOI] [PubMed] [Google Scholar]
- Vollbrecht T, Brackmann H, Henrich N, Roeling J, Seybold U, Bogner JR, Goebel FD, Draenert R. Impact of changes in antigen level on CD38/PD-1 co-expression on HIV-specific CD8 T cells in chronic, untreated HIV-1 infection. J Med Virol. 2010;82(3):358–370. doi: 10.1002/jmv.21723. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Han Y, Xu X, Bao Y, Zhang M, Cao X. IL-17A-producing gammadeltaT cells promote CTL responses against Listeria monocytogenes infection by enhancing dendritic cell cross-presentation. J Immunol. 2010;185(10):5879–5887. doi: 10.4049/jimmunol.1001763. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Yang XO, Chung Y, Fukunaga A, Nurieva R, Pappu B, Martin-Orozco N, Kang HS, Ma L, Panopoulos AD, Craig S, Watowich SS, Jetten AM, Tian Q, Dong C. CCR6 regulates the migration of inflammatory and regulatory T cells. J Immunol. 2008;181(12):8391–8401. doi: 10.4049/jimmunol.181.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lou B, Lal RB, Gettie A, Marx PA, Moore JP. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J Virol. 2000;74(15):6893–6910. doi: 10.1128/jvi.74.15.6893-6910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Balato A, Fishelevich R, Chapoval A, Mann DL, Gaspari AA. Th17/Tc17 infiltration and associated cytokine gene expression in elicitation phase of allergic contact dermatitis. Br J Dermatol. 2009;161(6):1301–1306. doi: 10.1111/j.1365-2133.2009.09400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]