Abstract
Mutations in FBN1 cause Marfan syndrome, a heritable disorder of connective tissue. FBN1 encodes the extracellular matrix protein, fibrillin. Our objective was to elucidate the extent that variation in RNA splicing contributes to FBN1 isoforms. To identify FBN1 splice variants, we scanned each of its 64 internal exons in a set of pooled human brain cDNA samples. FBN1 splicing is generally efficient as we identified only two variants. Neither variant has previously been reported in the literature and include (i) an isoform which contains a cryptic 105 basepair exon between exons 54–55 (54A-FBN1) and (ii) an isoform which contains a cryptic 62 basepair exon between exons 57 and 58 (57A-FBN1). We compared 57A-FBN1 and FBN1 expression in multiple human tissues, including adult skeletal muscle and brain, as well as fetal skeletal muscle, brain, liver, aorta, lung, skin, and heart. 57A-FBN1 represents 8–44% of FBN1 mRNA and varies in a tissue- and development-specific fashion. In adult brain, 57A-FBN1 represented 39 ± 3 (%, mean ± SD) of total FBN1 expression. In contrast, 57A-FBN1 represented 19 ± 2 (%, mean ± SD) of FBN1 expression in skeletal muscle. In fetal tissue, the 57A-FBN1 proportion was highest in brain (27%) and low elsewhere, e.g., skin, aorta and lung (9–13%). In summary, a significant proportion of FBN1 is expressed as 57A-FBN1 and this proportion varies in a tissue- and development-specific fashion. Since the 57A insertion creates a premature stop codon that mimics Marfan-associated mutations, the protein encoded by 57A-FBN1 is likely to not be functional. These results suggest that altered splicing may modulate disease severity, regulate FBN1 expression, and potentially represent a therapeutic target.
Keywords: (6) Marfan syndrome, fibrillin, alternative splicing, isoforms
1. Introduction
Mutations in FBN1 cause Marfan syndrome, a heritable disorder of connective tissue. FBN1 encodes the extracellular matrix protein, fibrillin [1]. In the extracellular matrix, fibrillin acts as a reservoir for transforming growth factor-β (TGF-β). Decreased deposition of fibrillin in the extracellular matrix leads to enhanced TGF- β signaling and the resultant Marfan phenotype (reviewed in [2]). FBN1 expression is widespread based upon in situ hybridization and immunocytochemistry [3, 4].
Variation in RNA splicing is emerging as a mechanism that modulates disease susceptibility and severity (reviewed in [5, 6]). This emergence has lead to a nascent pharmacological approach to ameliorate disease phenotypes by using small molecules to alter splicing (reviewed in [7]). Here, we test the hypothesis that atypical FBN1 splicing represents a significant proportion of FBN1 expression. We identify two previously unreported FBN1 splice variants that incorporate cryptic exons from introns 54 and 57 (designated 54A-FBN1 and 57A-FBN1, respectively). Moreover, we show that 57A-FBN1 expression varies among tissues and between the fetal and adult stages. Since 57A-FBN1 expression ranges from 10–40% of total FBN1 expression and the protein encoded by 57A-FBN1 is similar to that associated with a Marfan-associated mutation, decreasing 57A-FBN1 as a proportion of FBN1 expression may offer a future therapeutic approach.
2. Materials and Methods
2A. Human tissue
The human brain and liver samples used for this study have been described in detail elsewhere [8, 9, 10, 11]. Briefly, the brain samples were obtained from the University of Kentucky Alzheimers Center while the liver samples were obtained from the Brain and Tissue Bank for Developmental Disorders in Baltimore, MD. The brain and liver samples were from deceased individuals with an average age at death of 83 years (range of 68–105) and 27 years (range of 13–46), respectively. Skeletal muscle samples were also obtained from the Brain and Tissue Bank for Developmental Disorders and were from deceased male individuals with an average age at death of 38 years (range of 20–61). Total RNA from human fetal skeletal muscle, brain, liver, aorta, lung, skin, and heart was obtained from Stratagene. Gestational ages ranged from 16–22 weeks.
2B. Cell Culture
HepG2 (human hepatocellular carcinoma), SH-SY5Y (human neuroblastoma), HEK293 (human embryonic kidney), H4 (human neuroglioma), and MCF-7 (human breast adenocarcinoma) cell lines were maintained in Dulbecoo’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 50 U/ml penicillin and 50 μg/ml streptomycin at 37°C in a humidified 5% CO2-95% air atmosphere.
2C. PCR Amplification
RNA was extracted and converted to cDNA in one microgram aliquots using SuperScriptIII reverse transcriptase (Invitrogen) and random hexamers (Invitrogen) as described previously [8, 10, 12]. Primer pairs were designed to produce overlapping products; this enabled evaluation of splicing efficiency of each internal exon (Table 1). These primer pairs were then used to screen for splice variants in cDNA pooled from 20 brain samples. The pooled cDNA was amplified with each primer pair by using Platinum Taq (Invitrogen) and cycling conditions of a pre-incubation for 3 minutes at 95° C, followed by 34 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 60 seconds (Perkin Elmer 9600). PCR products were separated by electrophoresis in an 8% polyacrylamide gel, detected by SYBR-gold staining, and visualized by using a fluorescence imager (Fuji FLA-2000). PCR products that were different than the expected size were identified by direct sequencing (Davis Sequencing).
Table 1.
PCR primers.
| Exon Amplified | Primer | Primer Sequence |
|---|---|---|
| 2–5 | 2F | CCGTGCTTTTAGCGTCCTAC |
| 5R | ACTGCAGCTACCTCCATTCA | |
| 4–9 | 4F | CTGTGGGGATGGATTTTGTT |
| 9R | TTCACCCCCTTCACAGATTC | |
| 8–13 | 8F | GGCTCTGTCAGGGAGGAAAT |
| 13R | GGCACTGACAGGTGTACGAA | |
| 12–16 | 12F | AGTGCAACAAAGGGTTCCAG |
| 16R | GTCCAGGGAAGCATTCACAT | |
| 15–19 | 15F | TGGATGAATGCAGCATAAGG |
| 19R | CCATTTGGGCAAATATCAGG | |
| 18–24 | 18F | CGGAATATCAGGCACTCTGC |
| 24R | TCATTCCACTGGGACACTGA | |
| 23–26 | 23F | TCCCATATGTGGTAAAGGGTACT |
| 26R | TGCACTTAAAGCTGCCAATG | |
| 25–29 | 25F | CTGGAAACCTGCTTCCTGAG |
| 29R | TCCTCGCATTCCTCAGTACC | |
| 28–32 | 28F | ATCCTCTCCTATGCCGAGGT |
| 32R | GCCTTTCGTGTTTTCACAGG | |
| 31–35 | 31F | TGGATTCATGGCATCTGAAG |
| 35R | GCATTCACAGCGGTATCCTC | |
| 34–38 | 34F | CATGGGATCTTACCGCTGTC |
| 38R | AGCAGGAAGCTTTGGAAACA | |
| 37–42 | 37F | GTGACTGCCCACCTGATTTT |
| 42R | GCAGCACATCTTCTTGGTCA | |
| 41–45 | 41F | TGTGGTCCAGGGACATGTTA |
| 45R | GTAGCTGCCTGCAGTGTTGA | |
| 44–48 | 44F | AGCTTCCGATGTGAATGTCC |
| 48R | CACTGGAAAGACCCCACTGT | |
| 47–51 | 47F | AAGAGATGCCTGTGGGAATG |
| 51R | GCATTCCTGCTTGGAGTGAT | |
| 50–55 | 50F | CTGGGCACATGCAGTAACAC |
| 55R | GAAGGCACAGAGCAGAGGAT | |
| 54–58 | 54F | CCTGCAAGAATGTGATTGGA |
| 58R | AAGGGCAGATCTCACAGTGG | |
| 57–61 | 57F | AAGCCAGGGATCTGTGAGAA |
| 61R | GGTAAATCCGGGAGGACATT | |
| 60–64 | 60F | TCCCAAACCCTGCAATTTTA |
| 64R | CGGGACACATGCACTTGTAG | |
| 63–66 | 63F | ATGGCTGCCAGAACATCATT |
| 66R | CCCAACTTGCAAGACTCACA |
2D. Real-time PCR (RT-PCR)
The expression level of the 57A-FBN1 isoform, which contains a cryptic exon within intron 57, as well as the FBN1 isoform without this cryptic exon (FBN1) were quantified by using RT-PCR. Each assay used a common antisense primer corresponding to sequence within exon 59, i.e., 5′-TTCGGCAAACATCGTGAATA-3′. The sense primer for FBN1 detection corresponded to sequence within exon 57, i.e., 5′-CTTGACAATCGGGAAGGGTA-3′ while the sense primer for 57A-FBN1 quantification corresponded to sequence within exon 57A, i.e., 5′-ACCCAGGACAATCGGGAAG-3′. The 20 μl reaction mixture, which contained 20 ng of cDNA, 1 μM of each primer and 1× PerfecTaqPCR Supermix (Quanta Biosciences, Gaithersburg, MD) was subjected to RT-PCR (MJ Research PTC-200 with Chromo4 detector) with conditions of a preincubation at 95°C for 120 seconds, followed by 40 cycles of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 20 seconds. Reaction specificity was evaluated by subjecting the PCR products to gel electrophoresis and by performing a melting curve analysis after PCR amplification. The copy numbers of PCR product in each sample were determined relative to standard curves that were amplified in parallel and were based upon purified and quantified PCR products. For the brain and fetal tissue RNAs, copy numbers were then normalized to the geometric mean of the copy numbers of hypoxanthine-guanine phosphoribosyltransferase 1 (HPRT) and ribosomal protein L32 (RPL32) as previously described [13]. Since the hypoxanthine-guanine phosphoribosyltransferase 1 and ribosomal protein L32 geometric mean showed unacceptable variation among the adult skeletal muscle samples, the expression of FBN1 in these samples was normalized to the 18S rRNA copy number. All real-time PCR assays were repeated twice.
3. Results
3A. Identification of FBN1 Splice Variants
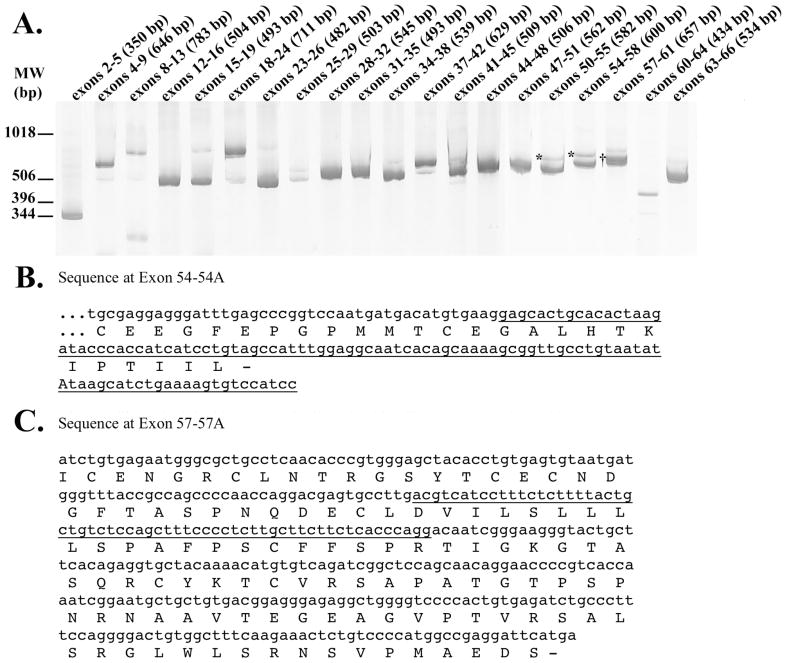
To detect alternatively spliced FBN1 exons, we scanned the 64 internal exons of FBN1 in human brain cDNA for inefficient splicing or the presence of cryptic exons. FBN1 exons were generally spliced efficiently because a single PCR product of the expected size was produced in most reactions (Figure 1A). However, several PCR reactions amplified two products that sequencing established as representing FBN1 splice variants. In particular, sequencing of the PCR products generated from exons 50–55 and from 54–58 revealed the expected isoform as well as a novel isoform that includes a cryptic 105 bp exon from within intron 54. This 105 bp exon is predicted to introduce a premature stop codon, resulting in a truncated fibrillin protein of 2,217 amino acids as compared to the standard 2,871 amino acid fibrillin (Figure 1B). Sequencing of the PCR products amplified between exons 57–61 revealed the expected isoform, which contained exons 57–61, as well as an isoform which contained a 62 bp cryptic exon from intron 57; this isoform has not been reported in the literature but is in the Ensembl database (http://uswest.ensembl.org/Homo_sapiens/Transcript/Summary?db=core;g=ENSG00000166147;r=15:48700505-48937918;t=ENST00000389087). The insertion of exon 57A is predicted to cause a frameshift and subsequent premature termination codon, producing a truncated 2,417 amino acid fibrillin (Figure 1C). Since the standard 2,871 amino acid fibrillin polymerizes in a head-to-tail fashion in the extracellular matrix [14], the truncated proteins encoded by 54A-FBN1 and 57A-FBN1 may disrupt the structure of fibrillin in the extracellular matrix.
Figure 1. FBN1 Splicing Patterns in Human Brain Tissue.
The products of PCR amplification of human brain cDNA across the indicated exons were separated by polyacrylamide gel electorphoresis and visualized by SYBR gold fluorescence; the sizes of the expected products are shown in parentheses (A). Since this is a composite image of multiple gels, the molecular weight markers are approximate. The nucleotide and protein sequence corresponding to the products marked with an * show that retention of a cryptic exon within intron 54 (underlined) introduces a premature translation termination codon (B). Similarly, the nucleotide and predicted protein sequence of the PCR product marked with a † shows retention of a cryptic exon within intron 57 (underlined) that causes a frameshift and premature termination codon (C).
3B. Comparison of 57A-FBN1 between individuals and tissues
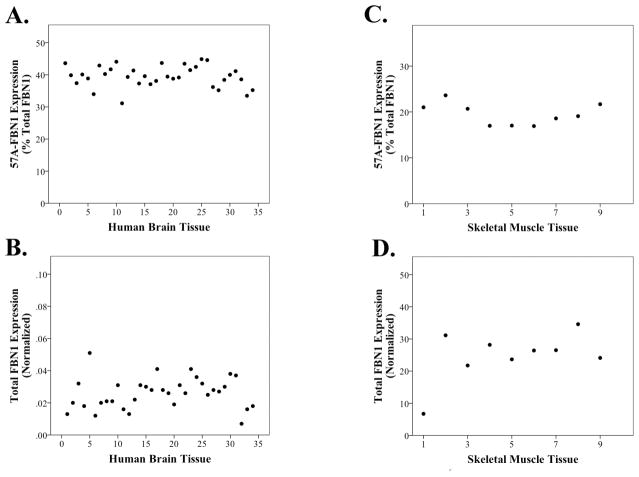
To investigate expression of these isoforms, we focused upon the 57A-FBN1 prototypic isoform because of its prior listing in Ensembl. When we quantified the expression of 57A-FBN1 and FBN1 in a series of 34 brain samples by quantitative RT-PCR, we found that 57A-FBN1 was expressed consistently among individual samples and accounted for 39 ± 3 (%, mean ±SD, n=34, Figure 2A–B) of total FBN1 expression. Hence, 57A-FBN1 represents a major FBN1 isoform in the brain. We next compared the expression of FBN1 and 57A-FBN1 in a series of adult skeletal muscle samples, a tissue in which Marfan pathology is observed. When we compared across the cohort, we found that 57A-FBN1 represented a consistent 19 ± 2 (%, mean ±SD, n=9, Figure 2C–D) of total FBN1 mRNA. Hence, the proportion of 57A-FBN1 in adult tissues varies strikingly between tissues but varies little between individuals.
Figure 2. Quantitation of 57A-FBN1 and Total FBN1 in Human Adult Tissue.
Real time PCR quantification of 57A-FBN1 and FBN1 shows that 57A-FBN1 represents a consistent proportion of total FBN1 expression in brain (A–B) and skeletal muscle (C–D). A greater proportion of FBN1 is expressed as 57A-FBN1 in the brain. Each data point represents a single human sample analyzed. Since HPRT and RPL32 were not consistent in the muscle samples, FBN1 expression was normalized to 18S rRNA in the muscle samples.
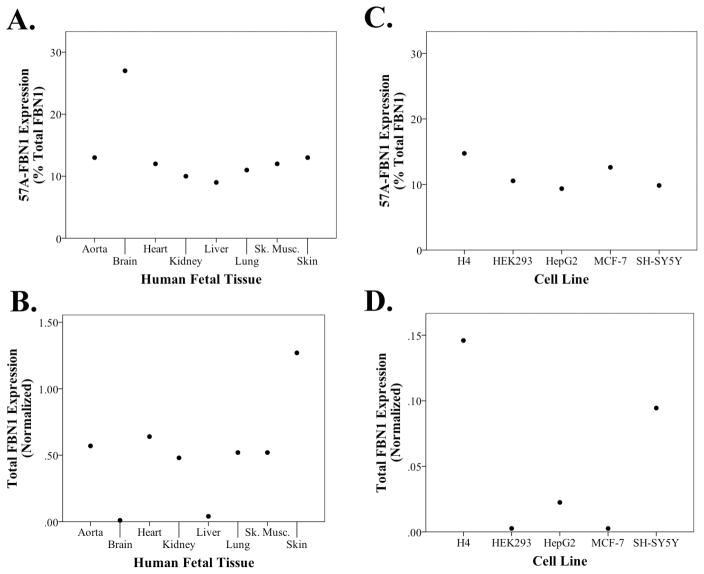
Since FBN1 mutations alter tissue formation during development, especially in the fetal aorta, skeletal muscle, heart, lung and skin (reviewed in [15, 16]), we proceeded to compare FBN1 and 57A-FBN1 in these fetal tissues as well as fetal brain, liver and kidney (Figure 3A–B). We found that the proportion of 57A-FBN1 was highest in the fetal brain and relatively uniform in aorta, heart, lung, liver, kidney, skeletal muscle and skin (Figure 3A). Furthermore, total FBN1 expression was highest in skin, moderate in aorta, heart, lung, kidney, and skeletal muscle, and low in brain and liver (Figure 3B). Interestingly, this pattern of FBN1 expression recapitulates the phenotype of Marfan syndrome in that tissues with higher total FBN1 expression, i.e., skin, aorta, heart, lung and skeletal muscle, generally have significant pathology in Marfan syndrome relative to two tissues with low expression, brain and liver [15, 16]. Since FBN1 function is often studied in cell lines [17], we also investigated the proportion of 57A-FBN1 in several cell lines. The proportion of 57A-FBN1 ranged from 9–15% of total FBN1 expression (Figure 3C), which was similar to the proportion of 57A-FBN1 in the majority of fetal tissues examined. FBN1 expression was low in these cell lines, relative to the fetal tissues, with H4 neuroglioma cells being highest and human embryonic kidney (HEK) 293 and MCF-7 breast cancer cells being lowest (Figure 3D).
Figure 3. Quantitation of 57A-FBN1 and Total FBN1 in Embryonic Human Tissues and Commonly Used Human Cell Lines.
Real time PCR quantification shows that 57A-FBN1 represents a consistent ~12% (A, C) of total FBN1 expression (B, D) across a series of human embryonic tissues and cell lines, with the exception of brain tissue, which has a higher proportion of 57A-FBN1.
4. Discussion
Our primary findings are that (i) the exons of FBN1 are generally spliced with high efficiency, (ii) two FBN1 isoforms not previously described in the literature are generated by the inclusion of cryptic exons from introns 54 and 57 and (iii) the quantities of one isoform, 57A-FBN1, are sufficiently large to account for a significant proportion of FBN1 expression. Moreover, the proportion of FBN1 expression as 57A-FBN1 varies in a tissue- and development-specific fashion. If the fibrillin variant protein encoded by 57A-FBN1 is not functional, optimizing FBN1 splicing to minimize 57A-FBN1 may represent a therapeutic approach to ameliorate the Marfan syndrome phenotype.
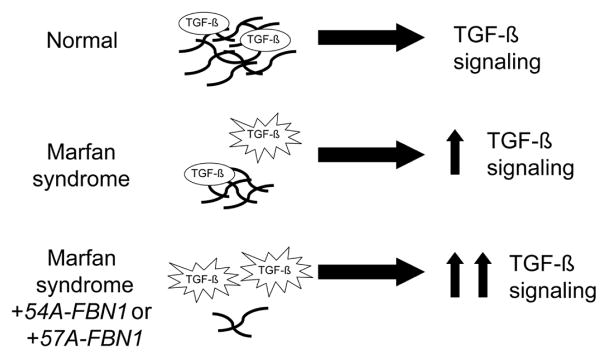
Although the functional impact of FBN1 truncation after exons 54 or 57 is unclear, in vivo and in vitro evidence suggests that these truncated fibrillin proteins will not be functional. First, at the human level, many mutations that cause Marfan syndrome are the result of similarly truncated fibrillin proteins (FBN1 mutation list can be found at http://www.umd.be:2030/W_FBN1/indexR_FBN1.shtml). For example, a single basepair deletion within exon 57 (at amino acid 2347) and the resulting frameshift and stop codon eighteen amino acids later, causes Marfan syndrome [18]. Second, in vitro studies have found that the portion of fibrillin encoded after exon 62 is necessary for fibrillin multimerization [14]. Hence, the truncated proteins encoded by 54A-FBN1 and 57A-FBN1 are highly likely to be nonfunctional. Individuals with a greater proportion of 57A-FBN1 due to variation in splicing would be predicted to have a more severe Marfan syndrome phenotype (Figure 4) because lower fibrillin protein results in greater TGF-β signaling. Hence, variations in FBN1 splicing may offer one possible mechanism for the intra-familial variability seen in people with Marfan syndrome.
Figure 4. Model for FBN1 Isoforms in Marfan Syndrome.
Fibrillin polymers (black lines) in the extracellular matrix serve to maintain a TGF-β reservoir. In Marfan syndrome, a decrease in fibrillin polymer results in increased TGF-β signaling and the Marfan phenotype. The two isoforms identified here are predicted to produce truncated fibrillin proteins that mimic the effects of Marfan-associated mutations. Hence, these isoforms are likely to exacerbate Marfan syndrome.
Nonsense mediated decay (NMD) has been reported to degrade some abnormally spliced FBN1 variants, including a variant wherein exon 51 is skipped [19]. Whether NMD regulates the levels of 54A-FBN1 or 57A-FBN1 is unclear. We interpret the finding that 57A-FBN1 represents 10–40% of FBN1 mRNA as suggesting that at least 57A-FBN1 is likely not subjected to NMD; the alternative interpretation would require that 57A-FBN1 represents the major FBN1 isoform prior to NMD for 10–40% of FBN1 to be present as 57A-FBN1 after NMD. Alternatively, if 57A-FBN1 does undergo NMD, expression of the 57A-FBN1 variant and subsequent NMD may represent a mechanism regulating total FBN1 expression. In this event, identifying splicing regulatory proteins that minimize exon 57A inclusion may be an especially robust means to enhance FBN1 expression.
In summation, FBN1 exons are generally included with high efficiency in mature FBN1. However, we have identified two previously unreported FBN1 isoforms, and note that one isoform, 57A-FBN1, represents a significant portion of total FBN1 expression. Since the protein encoded by 57A-FBN1 is likely to be non-functional, efforts to decrease the proportion of FBN1 expressed as 57A-FBN1 represents a potential therapeutic modality.
Highlights.
We evaluate splicing in FBN1, mutations in which cause Marfan syndrome.
We report two FBN1 splice variants that are both predicted to produce non-functional protein.
The expression of one isoform, 57A-FBN1, varies between tissues and with development.
57A-FBN1 represents a significant proportion of FBN1 expression.
57A-FBN1 may regulate fibrillin expression and disease severity.
Acknowledgments
The authors gratefully acknowledge tissue supplied by the University of Kentucky Alzheimers Disease Center, which is supported by P30AG028383, NIH for grant support (R01AG026147 and P01AG030128), and Dr. Hal Dietz (Johns Hopkins University) for helpful discussion.
Abbreviations
- RT-PCR
Real-time PCR
- HEK
human embryonic kidney
- NMD
Nonsense mediated decay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Mary E. Burchett, Email: mary.burchett@uky.edu.
I-Fang Ling, Email: ifang.ling@gmail.com.
Steven Estus, Email: steve.estus@uky.edu.
References
- 1.Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, Puffenberger EG, Hamosh A, Nanthakumar EJ, Curristin SM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–9. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 2.Dietz HC. TGF-beta in the pathogenesis and prevention of disease: a matter of aneurysmic proportions. J Clin Invest. 2010;120:403–7. doi: 10.1172/JCI42014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, Hu W, Ramirez F. Developmental expression of fibrillin genes suggests heterogeneity of extracellular microfibrils. J Cell Biol. 1995;129:1165–76. doi: 10.1083/jcb.129.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quondamatteo F, Reinhardt DP, Charbonneau NL, Pophal G, Sakai LY, Herken R. Fibrillin-1 and fibrillin-2 in human embryonic and early fetal development. Matrix Biol. 2002;21:637–46. doi: 10.1016/s0945-053x(02)00100-2. [DOI] [PubMed] [Google Scholar]
- 5.Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol. 2010;220:152–63. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poulos MG, Batra R, Charizanis K, Swanson MS. Developments in RNA Splicing and Disease. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietz HC. New therapeutic approaches to mendelian disorders. N Engl J Med. 2010;363:852–63. doi: 10.1056/NEJMra0907180. [DOI] [PubMed] [Google Scholar]
- 8.Zhu H, Tucker HM, Grear KE, Simpson JF, Manning AK, Cupples LA, Estus S. A common polymorphism decreases low-density lipoprotein receptor exon 12 splicing efficiency and associates with increased cholesterol. Hum Mol Genet. 2007;16:1765–72. doi: 10.1093/hmg/ddm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou F, Gopalraj RK, Lok J, Zhu H, Ling IF, Simpson JF, Tucker HM, Kelly JF, Younkin SG, Dickson DW, Petersen RC, Graff-Radford NR, Bennett DA, Crook JE, Estus S. Sex-dependent association of a common low-density lipoprotein receptor polymorphism with RNA splicing efficiency in the brain and Alzheimer’s disease. Hum Mol Genet. 2008;17:929–35. doi: 10.1093/hmg/ddm365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grear KE, Ling IF, Simpson JF, Furman JL, Simmons CR, Peterson SL, Schmitt FA, Markesbery WR, Liu Q, Crook JE, Younkin SG, Bu G, Estus S. Expression of SORL1 and a novel SORL1 splice variant in normal and Alzheimers disease brain. Mol Neurodegener. 2009;4:46. doi: 10.1186/1750-1326-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling IF, Estus S. Role of SFRS13A in low-density lipoprotein receptor splicing. Hum Mutat. 2010;31:702–9. doi: 10.1002/humu.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocynate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 13.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubmacher D, El-Hallous EI, Nelea V, Kaartinen MT, Lee ER, Reinhardt DP. Biogenesis of extracellular microfibrils: Multimerization of the fibrillin-1 C terminus into bead-like structures enables self-assembly. Proc Natl Acad Sci U S A. 2008;105:6548–53. doi: 10.1073/pnas.0706335105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramirez F, Dietz HC. Marfan syndrome: from molecular pathogenesis to clinical treatment. Curr Opin Genet Dev. 2007;17:252–8. doi: 10.1016/j.gde.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Judge DP, Dietz HC. Marfan’s syndrome. Lancet. 2005;366:1965–76. doi: 10.1016/S0140-6736(05)67789-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Summers KM, Bokil NJ, Baisden JM, West MJ, Sweet MJ, Raggatt LJ, Hume DA. Experimental and bioinformatic characterisation of the promoter region of the Marfan syndrome gene, FBN1. Genomics. 2009;94:233–40. doi: 10.1016/j.ygeno.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Korkko J, Kaitila I, Lonnqvist L, Peltonen L, Ala-Kokko L. Sensitivity of conformation sensitive gel electrophoresis in detecting mutations in Marfan syndrome and related conditions. J Med Genet. 2002;39:34–41. doi: 10.1136/jmg.39.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietz HC, Kendzior RJ., Jr Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat Genet. 1994;8:183–8. doi: 10.1038/ng1094-183. [DOI] [PubMed] [Google Scholar]