Abstract
Yersiniabactin (Ybt) is a siderophore-dependent iron uptake system encoded on a pathogenicity island that is widespread among pathogenic bacteria including the Yersiniae. While biosynthesis of the siderophore has been elucidated, the secretion mechanism and a few components of the uptake/utilization pathway are unidentified. ybt genes are transcriptionally repressed by Fur but activated by YbtA, likely in combination with the siderophore itself. The Ybt system is essential for the ability of Y. pestis to cause bubonic plague and important in pneumonic plague as well. However, the ability to cause fatal septicemic plague is independent of Ybt.
Keywords: Yersiniabactin, Yersinia pestis, iron transport, siderophore, plague
1. Yersinia pestis and plague
Within the Enterobacteriaceae family, the genus Yersinia is composed of environmental species, a fish pathogen, two enteropathogenic species (Yersinia enterocolitica and Yersinia pseudotuberculosis) and the plague bacillus Yersinia pestis which has caused three pandemics. However, plague is primarily a disease of rodents and their associated fleas. In nature, fleas feeding on a septicemic mammal become infected and the bacteria grow in the mid-gut of the flea. The infected flea transmits Y. pestis to mammals during subsequent blood meals via an early phase (blockage/biofilm-independent) mechanism and/or by a blockage/biofilm-dependent mechanism. From the bite wound, Y. pestis spreads via the lymphatics system to a regional lymph node and multiplies to high numbers causing necrosis and architecture destruction. This results in a swollen lymph node or bubo from which the name bubonic plague is derived. Bacteria spread through the bloodstream to the liver and spleen where they multiply and initiate a septicemia. High numbers of bacteria in the bloodstream are needed to ensure infection of a naïve flea feeding on the infected mammal. Untreated, fatality rates from bubonic plague can reach 50%. A small proportion of mammals develop septicemia with no bubo formation. This primary septicemic plague bypasses the lymphatic stage of the bubonic disease [1–4]. Although epidemic strains of Y. pestis (biovars Antiqua, Mediaevalis, and Orientalis) cause disease in animals and humans, enzootic strains cause disease in rodents but not humans [5–6].
During the course of bubonic plague, the lungs can become infected causing secondary pneumonic plague. Humans and non-human primates can then spread the disease via respiratory droplets causing primary pneumonic plague that is rapidly and nearly 100% fatal without treatment [2, 4].
2. Early studies leading to the discovery of the siderophore-dependent yersiniabactin (Ybt) iron transport system
A number of virulence determinants are important for the pathogenesis of bubonic, septicemic, and/or pneumonic plague. Some early observations noted as being important for the development of bubonic plague are now known to be due to the siderophore-dependent Ybt iron transport system. In 1956 Jackson and Burrows observed that spontaneous Y. pestis nonpigmented mutants (Pgm−; unable to bind hemin at room temperature) were avirulent in mice via subcutaneous injection unless supplemented with iron or hemin [7]. Although beyond the genetic analysis of that day, this mutant was likely a spontaneous deletion of the 102-kb pigmentation (pgm) locus that includes the High Pathogenicity Island (HPI) that encodes genes of the Ybt system (see section 5.1 below). Almost 20 years passed before Wake et al proposed that Y. pestis produced siderophores (which they called siderochromes). Pgm+ populations of Y. pestis had more siderophore producers than Pgm− populations and the secreted siderophore inhibited the activity of the bacteriocin pesticin [8]. Only Pgm+ cells of Y. pestis are sensitive to pesticin, a bacteriocin produced by Y. pestis whose activity is repressed by growth with iron or hemin [9–10]. It was subsequently shown that the outer membrane (OM) receptor for pesticin (designated Psn in Y. pestis or FyuA in Y. enterocolitica) mapped to the pgm locus in Y. pestis and serves as the receptor for the Ybt siderophore [11–13].
In 1987, Heesemann et al. identified a siderophore-like activity in Y. enterocolitica and Y. pseudotuberculosis which was later named yersiniabactin [14–15]. Haag et al. purified Ybt from Y. enterocolitica and showed that it was taken up through the pesticin receptor, FyuA [16]. Two different groups determined the structure of Ybt from Y. enterocolitica [17–18]. Perry et al later confirmed that Ybt from Y. pestis had the same structure [19] (Fig. 1). In 1987, Carniel et al. identified two iron-regulated proteins (HMWP1 and HMWP2 encoded by irp1 and irp2, respectively) that were expressed only in high virulence or pathogenic Yersiniae strains and determined that a portion of the irp2 gene was absent in low virulence or nonpathogenic strains [20–21]. HMWP1 and HMWP2 are required for Ybt biosynthesis [22]. Thus by the end of 1990, the Ybt structure and some components involved in its biosynthesis and uptake had been identified.
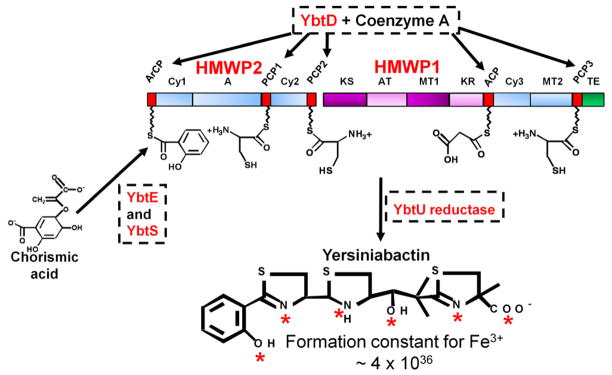
Fig. 1.
Biosynthesis of Yersiniabactin (Ybt). YbtD, a phosphopantetheinyl transferase, transfers the 4′-phosphopantetheine moiety of coenzyme A to carrier domains (red boxes) ArCP, PCP1, PCP2, ACP, and PCP3 which are attachment sites on HMWP1 and HMWP2 for the substrates adenylated salicylic acid, three cysteines, and malonate. Chorismic acid is converted to salicylic acid by YbtS. Salicylic acid is then adenylated by YbtE for attachment to the ArCP site of HMWP2. HMWP2 domains cyclize and condense two cysteines to form two thiazoline rings linked to the salicylate moiety while HMWP1 domains add the malonyl linker and convert a third cysteine to the final thiazoline ring. YbtU reduces the middle thiazolidine ring to a thiazoline ring to yield the final structure. Nonribosomal peptide synthetase (NRPS) domains are represented by blue boxes while polyketide synthase (PKS) domains are in purple. Aberrant or mischarged molecules on the enzyme complex are removed by YbtT, an editing thioesterase (not shown in diagram). The completed siderophore is released from the enzyme complex by the thioesterase (TE; green box) domain of HMWP1. The completed Ybt structure is shown with red asterisks identifying coordination sites for one Fe3+ atom. The disassociation constant (KD) for Fe3+ is shown. HMWP1 and HMWP2 enzymatic domains: A, adenylation; ACP, acyl carrier protein; ArCP, aryl carrier protein; AT, acyltransferase; Cy, condensation/cyclization; KR, β-ketoreductase; KS, ketoacyl synthase; PCP, peptidyl carrier protein; MT, methyltransferase; TE, thioesterase. The diagram is reproduced with modifications from Miller et al [31] with the permission of the Society for General Microbiology.
3. Biosynthesis of Ybt
Ybt is a member of a siderophore class whose Fe3+-binding groups include thiazoline, oxazoline, or methyloxazoline rings. Its synthesis proceeds by a mixed nonribosomal peptide synthetase (NRPS)/polyketide synthase (PKS) mechanism (Fig. 1). The siderophore is assembled from salicylate, three cysteines, a malonyl linker group and three methyl groups donated by S-adenosylmethionine to yield a four-ring structure composed of salicylate, one thiazolidine, and two thiazoline rings with a malonyl linker separating the final thiazoline from the thiazolidine ring (Fig. 1) [17–19, 23–24]. The requirement for seven gene products (HMWP1, HMWP2, YbtD, YbtE, YbtS, YbtT, and YbtU) for in vivo Ybt synthesis has been clearly demonstrated. Activated Ybt components are tethered to HMWP1 and HMWP2 by the 4′-phosphopantetheine moiety of coenzyme A which is added to sites in the carrier protein domains of these enzymes by the YbtD phosphopantetheinyl (P-pant) transferase. YbtS synthesizes salicylate from chorismate while YbtE adenylates salicylate and transfers this activated compound to HMWP2. HMWP1 and HMWP2 serve as a scaffold for the synthesis of the siderophore (Fig. 1). NRPS enzymatic domains in HMWP2 cyclize and condense two cysteines to form two thiazoline rings linked to the salicylate moiety. The first four domains of HMWP1 are a PKS module that bis-methylates and reduces a malonyl linker moiety. Two NRPS domains of HMWP1 cyclize and condense the third cysteine molecule to form the final thiazoline ring. YbtU reduces the middle thiazoline ring to thiazolidine and the terminal thioesterase (TE) domain of HMWP1 releases the completed siderophore (Fig. 1). The YbtT putative type II thioesterase likely removes aberrant molecules from the enzyme complex (not depicted in Fig. 1) [22–30].
In Y. pestis, a ybtT mutation reduces Ybt synthesis by ~75% compared to the Ybt+ parent strain. An alanine substitution for S or H in the putative thioesterase catalytic triad (S94-D172-H230) of YbtT yielded a phenotype similar to a ybtT deletion. In addition, reconstitution of the Y. pestis Ybt system in E. coli without YbtT resulted in 2.5-fold less Ybt than reconstitution with YbtT. However, in vitro, Ybt synthesis is efficient without YbtT and addition of the purified protein did not increase Ybt synthesis. This is likely because only correct precursor molecules were provided in the in vitro reaction mix, making mischarging of molecules onto HMWP1 and HMWP2 unlikely [27, 30–32]. Taken together these results support the contention that YbtT is an editing thioesterase that removes misprimed or aberrant intermediates from the Ybt synthetase complex. It also suggests that in vivo Ybt synthesis is relatively error prone.
The formation constant of Ybt with ferric iron is 4 × 1036 and the proposed iron binding sites are identified by asterisks in Fig. 1. The middle thiazolidine ring may provide needed flexibility in the molecule that allows binding of iron with high affinity in a 1:1 ratio. The crystal structure of the Fe-Ybt complex has been solved and confirms that iron is bound by the three nitrogen electron pairs and three negatively charged oxygen atoms depicted in Fig. 1 [19, 33].
4. Ybt transport
4.1 Export of Ybt
After synthesis of the siderophore is completed, Ybt is secreted from the bacterial cell by an undetermined mechanism. YbtX, an inner membrane (IM) protein with 12 predicted transmembrane domains, is tentatively assigned as part of the Ybt efflux system (Fig. 2) due to modest similarities to EntS and AlcS, exporters of enterobactin and alcaligin siderophores in E. coli and Bordetella, respectively [34–35]. However, YbtX also has similarities to RhtX and FptX which import the siderophores rhizobactin and pyochelin in Sinorhizobium meliloti and P. aeruginosa, respectively [36–37]. Experimentally, a Y. pestis ybtX mutant is not defective in Ybt secretion, uptake of iron from Ybt, or growth under iron-restrictive conditions [38]. Therefore the function of YbtX remains elusive. If it is involved in Ybt secretion, it is not essential to that process.
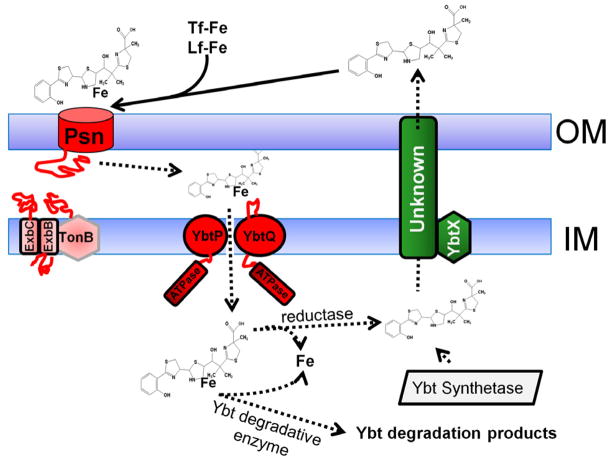
Fig. 2.
Ybt secretion and uptake. After release of the completed siderophore from the biosynthetic enzyme complex, Ybt is secreted from the cell by an unknown mechanism (green boxes). While there is evidence suggesting that YbtX is involved in secretion, it is not essential to this process. After release, Ybt can remove iron from transferrin (Tf-Fe) and lactoferrin (Lf-Fe). Although only uptake of radiolabeled iron has been experimentally demonstrated, our model favors uptake of the Fe-Ybt complex into the cell. The first step is binding to the TonB-dependent receptor Psn followed by translocation through the OM. Once in the periplasm Fe-Ybt is transported into the cytoplasm by YbtP/YbtQ. Whether additional components for this ABC transporter (e.g., a PBP or membrane spanning protein) are required for this step is undetermined. In the cytoplasm, Fe could be released from the siderophore by a reduction of Fe3+ to Fe2+ or by degradation of Ybt. Uptake components are in red with red lines denoting periplasmic and cytoplasmic domains. Dashed arrows indicate substrates or steps that are undetermined experimentally. The diagram is reproduced with modifications from Miller et al [31] with the permission of the Society for General Microbiology.
4.2 Uptake of iron from Ybt
Iron uptake from Ybt is better, but not completely, characterized (Fig. 2). The OM protein Psn (termed FyuA in Y. enterocolitica) is required for sensitivity to the bacteriocin pesticin and iron uptake via Ybt. Psn functions have been shown to be TonB-dependent. Although not proven in Y. pestis, in other Gram-negative bacteria TonB requires ExbB and ExbD for its function. Homologues of these genes appear to form an operon distal from tonB in Y. pestis. Y. pestis strains with mutations in either psn or tonB exhibit similar defects in iron-deficient growth and iron uptake from submicromolar levels. Although Y. pestis has a second TonB-like gene (hasB), HasB cannot substitute for TonB in iron uptake via the Ybt system [11, 13, 39–41].
In a deferrated, defined medium, a Y. pestis psn mutant shows greater defects in growth (Fig. 3) and uptake of iron when compared to a Y. pestis irp2 mutant. The psn mutant makes and secretes Ybt siderophore but it is unable to use it while the irp2 mutant cannot synthesize the siderophore [19, 42]. Secreted Ybt likely binds residual iron in the medium (~0.3 μM) making it unavailable for uptake by other functional Y. pestis iron uptake systems causing transport mutants to be more defective in iron acquisition than Ybt biosynthetic mutants. This is supported by the observation that a psn irp2 double mutant has a growth phenotype similar to the irp2 mutant (Fig. 3). Growth of this double mutant is inhibited by the addition of exogenous Ybt [42].
Fig. 3.
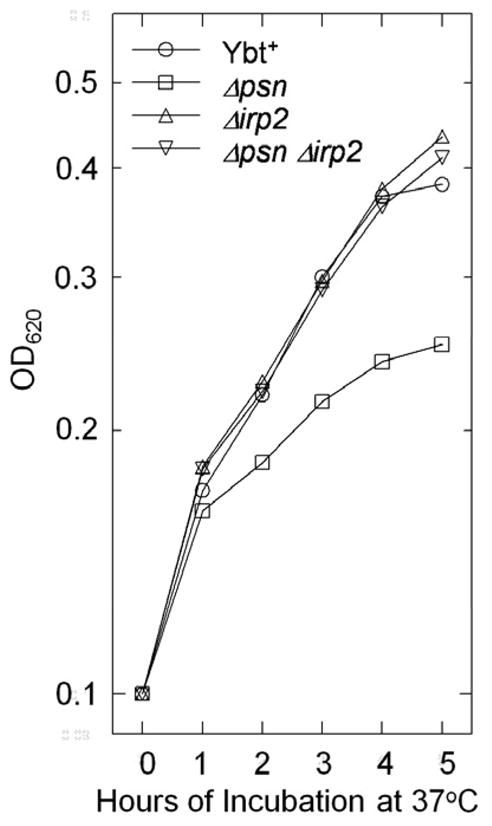
Growth of Y. pestis KIM strains in deferrated PMH2 at 37°C. A Δpsn mutant produces and secretes Ybt but cannot use the siderophore to obtain iron. A Δirp2 mutant cannot make the Ybt siderophore. The Δpsn Δirp2 mutant is defective in both biosynthesis and uptake. Y. pestis cells were acclimated to growth under iron-deficient conditions as previously described by Fetherston et al [42] which shows a similar experiment.
After passage through the OM into the periplasm is accomplished, the YbtP-YbtQ ABC transporter is required for uptake of Ybt-bound iron into the bacterial cell. YbtP and YbtQ are similar IM proteins with both permease and ATPase domains [38]. These fused-function types of ABC transporters are usually components of Type I secretion systems [43]. Nevertheless, strains with mutations in ybtP or ybtQ have an in vitro phenotype similar to a psn mutant – reduced growth and iron uptake without a significant reduction in Ybt siderophore secretion [38]. It is unclear whether a periplasmic binding protein (PBP) is required for this uptake system. No identifiable candidate lies within the HPI (see section 5.1) that encodes nearly all other ybt genes [23, 44]. However, studies with the Y. enterocolitica Ybt system suggest that additional component(s) are required. Brem et al. demonstrated that a Y. enterocolitica serotype O:5, biogroup IA strain (which does not encode the Ybt system) is unable to use Fe-Ybt even when expressing Psn, YbtP and YbtQ (FyuA, Irp6, and Irp7, respectively, in Y. enterocolitica) while a similarly transformed E. coli strain lacking the Ybt pathogenicity island was able to use Fe-Ybt. They speculated that O:5, biogroup IA strain either lacks the PBP essential for uptake or was unable to remove iron from the siderophore [45].
Since transport studies have used radiolabeled iron [19, 38], uptake of a Fe-Ybt complex into the bacterial cell has not been demonstrated. It remains a possibility that iron could be removed at the cell surface, in the periplasm, or after transport into the cell cytoplasm. While Fig. 2 shows transport of the Fe-Ybt complex into the cytoplasm and release of iron there, the actual mechanism and site of iron release from Ybt remains to be characterized.
For other siderophores, iron release is achieved either by reduction to ferrous iron and release due to the low affinity of most siderophores for ferrous iron or by degradation of the siderophore to release the bound ferric iron. Genes encoding either type of mechanism do not lie within the HPI that encodes most other ybt genes.
5. Genetics and transcriptional regulation of the Ybt system
5.1 The ybt locus and high pathogenicity island (HPI)
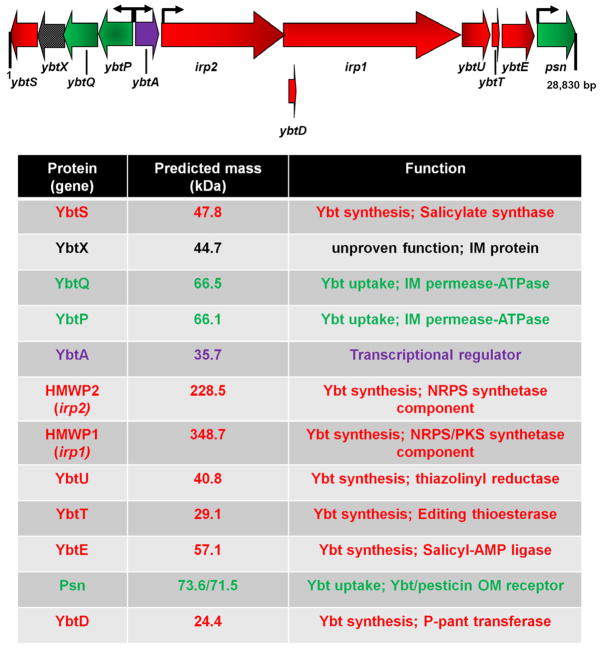
The ybt loci in Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis have been sequenced and found to be 97–100% identical. The locus consists of 4 operons; two are monocystronic, one contains 5 genes that encode biosynthetic enzymes, while the fourth operon has genes encoding proteins involved in Ybt uptake, biosynthesis, and possibly export (Fig. 4). Except for ybtD, fur, and additional transport components (i.e., tonB, exbB, and exbC) all other genes shown to be involved in the function or regulation of the Ybt system are encoded within this locus [23, 25, 46].
Fig. 4.
Genetic organization of the ybt locus and ybtD in Y. pestis. The ~29 kb ybt locus encoding 11 genes in four operons (promoters are designated by small arrows) lies within the 102-kb pgm locus. The monocystronic ybtD gene is located outside of the pgm locus. Arrows designating genes indicate the direction of transcription/translation. Red arrows indicate a biosynthetic function, green arrows are for uptake components, and the black checkered arrow denotes the potential secretion component YbtX. The transcriptional regulator YbtA is represented by a purple arrow. Protein masses and functions are indicated in the chart below the genetic maps. The processed and unprocessed masses are given for Psn which has a signal sequence.
The ybt locus lies within the HPI which was first identified in the three mammalian-pathogenic species of Yersinia. In turn, the HPI of Y. pestis lies within and comprises approximately one third of the pgm locus. The HPIs in Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica vary in size, associated insertion sequences, genomic insertion sites, and nucleotide sequences outside of the ybt locus in the “left-hand” portion of the HPI [46]. Excision and horizontal transfer of the HPI has been demonstrated in Y. pseudotuberculosis [47–48].
The HPI is widely distributed among the members of the Enterobacteriaceae family being found in species of Citrobacter, Enterobacter, Klebsiella, Photorhabdus, Salmonella, Serratia, and all of the E. coli pathotypes [46]. The IS100 associated with the Y. pestis HPI is absent in the other Enterobacteriaceae. Recently, the ybt locus has been detected in Pseudomonas syringae and found to be functional in 10 of 15 pathovars of this organism. However, this ybt locus may have been acquired prior to insertion into the HPI [49]. Most of the organisms that possess the HPI and have been tested produce the HMWP1 and HMWP2 proteins in an iron-repressible manner and synthesize the Ybt siderophore [46].
5.2 Fur regulation
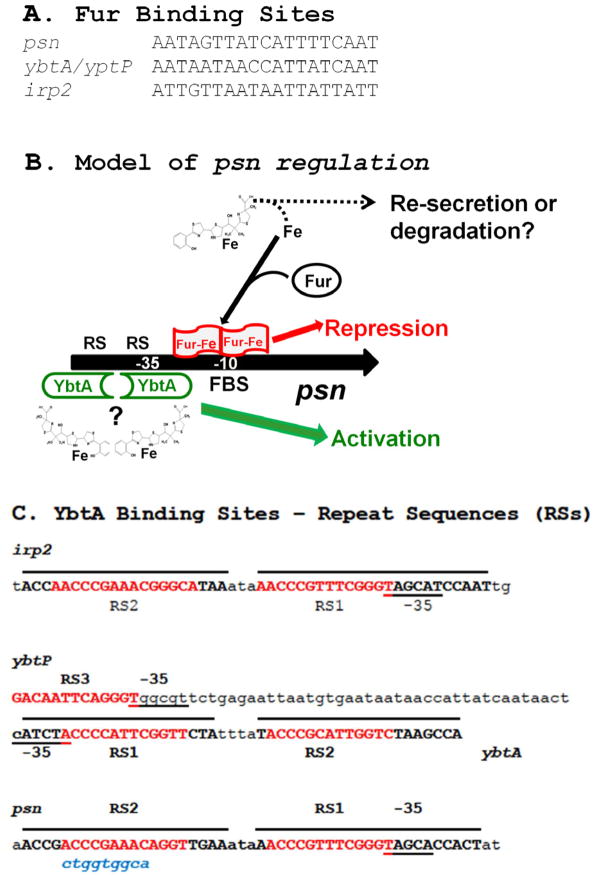
Y. pestis produces a typical Gram-negative Fur [Iron (Fe) Uptake Regulation] protein that represses the transcription of promoters with a Fur binding sequence (FBS or Fur box) when cells are grown with excess iron [50–52]. Each of the four ybt operons (Fig. 4) within the HPI have promoters with FBSs (Fig. 5A and 5B) and are repressed through Fur approximately 12-fold (psn), 8-fold (irp2-irp1-ybtUTE), 11-fold (ybtA) and 55-fold (ybtPQXS) during growth with 10 μM iron compared to growth in deferrated, defined media (PMH/PMH2) without additional iron [38, 53–54]. In contrast, ybtD, which is encoded outside of the HPI and pgm locus, is not regulated by Fur, iron, YbtA or the Ybt siderophore. The lack of regulation by Fur or YbtA suggests that YbtD may have been co-opted for use as the P-pant transferase that allows attachment of activated substrates to HMWP1 and HMWP2 [25]. Using the enzootic 201 strain, Gao et al demonstrated high-affinity binding of purified Y. pestis recombinant Fur-Mn (routinely used for in vitro analyses) to the four ybt locus promoter regions. Fig. 5A shows the FBSs derived from these studies [50].
Fig. 5.
Transcriptional regulation by Fur and YbtA. (A) Fur binding sites (FBSs) shown were mapped by Gao et al [50] and the sequences shown are from Y. pestis strain 201. The ybtP FBS is part of the ybtP/ybtA intergenic region and identical to the ybtA FBS. (B) Model of psn regulation. During iron-sufficient conditions, the psn promoter is repressed through Fur-Fe binding to an FBS that overlaps the −10 region. Under iron-deficient conditions this repression is relieved due to a shift to primarily iron-free Fur. If Ybt siderophore is present, YbtA activates transcription of this promoter by binding to repeat sequences (RSs), one of which overlaps the −35 region. The irp2 and ybtP promoters are regulated in the same manner. In contrast, YbtA represses transcription of its own promoter (not shown). Question marks indicate unresolved questions in our model. Our regulatory model favors a mechanism in which Fe-Ybt binds to YbtA for regulatory activity. However, binding of Fe-Ybt or iron-free Ybt to YbtA has not been demonstrated. The model is reproduced with modifications from Miller et al [31] with the permission of the Society for General Microbiology. (C) YbtA binding sites in the irp2, ybtP, ybtA, and psn promoter regions. Repeat sequences (RSs) are involved in YbtA binding. RS1, RS2, and RS3 as defined by Animisov et al are denoted in red, uppercase text [57–58]. Extended RS (overlined uppercase text) are proposed based on comparison of RSs among the ybt promoters. Underlined text indicates −35 regions of the promoter regions. The lowercase blue text below the psn sequence shows the mutated sequence that lowered expression of a psn::lacZ reporter [53].
5.3 Transcriptional regulation by YbtA and the Ybt siderophore
The earliest indication that the Ybt siderophore served as a signal to activate transcription of ybt genes was the observation that an irp2 mutant had a lower level of Psn protein. Normal Psn levels were restored by adding supernatant containing Ybt but not supernatant lacking the siderophore to an irp2 mutant culture [11]. Subsequent studies used transcriptional reporters to demonstrate a role for the siderophore in regulation. Compared to a Ybt-producing strain, an irp2::kan mutant had 17-, 2-, and 22-fold losses in transcription from the irp2, psn, and ybtP promoters respectively [38, 53–54]. Time course studies revealed that transcription from a ybtP::lacZ reporter was increased 4-fold 10 min after Ybt addition. Furthermore, activation was caused by adding Ybt at a concentration 500-fold lower than that required to stimulate growth of a biosynthetic mutant. Thus Ybt is a potent signaling molecule [54]
Ybt likely acts in concert with YbtA, a transcriptional regulator in the AraC family. YbtA is a negative regulator of its own expression and a transcriptional activator of the irp2, psn, and ybtP promoters (Fig. 4 and 5B). Under iron-deficient conditions, a ybtA::kan mutation caused a 50-fold drop in transcription from the psn promoter compared to the parent strain. Similar studies showed 109- and 16-fold losses in activation of the ybtP and irp2 promoters, respectively for the ybtA::kan mutant compared to its parent strain. In contrast, this same mutation caused a 2.8-fold increase in transcription from the ybtA promoter [38, 53–54].
Comprehensive analysis of the effect of mutations in 11 of the 12 known ybt genes (Fig. 4) and tonB on transcription from the ybtP promoter revealed a more complex story. The ybtA mutant had the lowest level of ybtP expression while mutations in genes required for uptake (tonB, psn, and ybtP) had modestly increased transcription (1.7-fold) compared to the Ybt+ strain. As described earlier (Section 4.2), these mutants are likely more iron starved due to siderophore secretion without subsequent iron acquisition. Mutations in ybtD, ybtE, ybtU and irp1 caused a loss of expression similar to that of the irp2 mutation (~18-fold) while a mutation in ybtX had no effect on transcription. In contrast, mutations in the thioesterase domain of HMWP1 (irp1-2086), ybtS, or YbtT caused a slight increase in ybtP expression (1.4-fold). These mutants, like the other Ybt biosynthetic mutants, failed to produce Ybt by a growth-stimulation bioassay yet they have a very different regulatory phenotype. Subsequent HPLC analysis demonstrated that the ybtT and irp1-2086 mutants produced 23% and 3%, respectively, of authentic Ybt compared to their Ybt+ parent. These lower levels of Ybt are apparently sufficient for regulatory signaling but insufficient for providing iron for growth. The ybtS mutant produced no detectable authentic Ybt. However it is possible that this mutant makes a “Ybt-like” molecule with another phenolic group substituted for salicylic acid. While this, and a possible second “Ybt-like” molecule, are biologically irrelevant for normal regulation, their identification and structural comparison with Ybt will help identify key chemical structures necessary for transcriptional activation. [31, 54].
It is puzzling that mutations in genes involved in Ybt uptake (ybtP, psn, tonB) have no apparent defect in transcriptional activation. We hypothesized that the Fe-Ybt complex enters the cytoplasm and interacts with YbtA to activate transcription from the irp2, psn, and ybtP promoters (Fig. 5B). In some systems, binding of the external siderophore to the siderophore receptor transmits a signal through the membranes. However, in that case, mutations in the OM receptor or TonB prevent signal transduction [55–56]. In Y. pestis, mutations in tonB or psn slightly increase transcription rather than causing a loss of transcriptional activation [54]. In these mutants, sufficient Fe-Ybt might enter through alternate, non-specific transport components. Alternatively, increased Ybt production by uptake mutants could saturate the secretion system artificially allowing unsecreted, cytoplasmic Ybt to activate transcription.
The promoter regions of psn (fyuA in Y. enterocolitica), irp2, and the intergenic region of the divergent promoters for ybtA and ybtP (irp6 in Y. enterocolitica) have similar inverted repeat sequences (RSs) with the promoter-proximal RS partially overlapping the −35 region for ybtP, psn, and irp2 (Fig. 5C) [13, 28, 53]. Mutation of the first 9 bp to disrupt the promoter-distal RS of psn (Fig. 5C) caused a 4-fold loss of transcription compared to the parent promoter. This suggested that the RSs are required for full expression possibly through binding of YbtA [53]. Anisimov et al demonstrated that purified YbtA binds to the promoter regions of Y. enterocolitica irp2 and fyuA and protects an ~50 bp region that includes both RSs (RS1 and RS2). Addition of either deferrated Ybt or Fe-Ybt did not affect mobility shifts or the protected promoter regions. Binding of YbtA to the Y. pestis and Y. enterocolitica ybtA-irp6/ybtP intergenic region differs from binding to the irp2 and fyuA/psn promoters in that there were two shifted bands and the protected region was twice as large, including both transcription starts, −10, −35, and FBSs. This extended protection lead Anisimov et al to suggest a third RS (RS3; Fig. 5C) although the sequence similarity to other RSs is modest. Deletion of RS2 or RS3 eliminated the upper shifted band and an ~2-fold increase in transcription from irp6/ybtP. Deletion of RS1 alone resulted in almost no binding of YbtA and loss of YbtA inhibition of ybtA transcription [57–58]. Overall, these results clearly show that YbtA binds to the ybt promoter regions and that this interaction involves the RS sequences.
The Y. enterocolitica ybtA/irp6 intergenic promoter region contains an ERIC sequence not found in Y. pestis or Y. pseudotuberculosis. This sequence does not affect the RS consensus sequence or the overall regulation of the Y. enterocolitica ybt genes. However a Y. enterocolitica ybtA promoter fusion had significantly higher transcriptional activity than the analogous Y. pestis ybtA reporter [11, 22, 38, 59].
6. Iron use and plague pathogenesis
6.1 Iron sources used by Ybt
Similar to other siderophores, Ybt has no affinity for ferrous iron. Its affinity for ferric iron (~4 × 1036) is higher than a number of other siderophores and indicates that Ybt should be able to remove iron from a variety of host iron-binding proteins [19]. A Ybt+ strain but not a Ybt− mutant will grow in the presence of partially iron-saturated transferrin or lactoferrin even when the cells are separated from these iron sources by a dialysis membrane which prevents contact between cell surfaces and transferrin or lactoferrin. Under these conditions, Ybt is necessary for the use of iron from transferrin and lactoferrin. The direct removal of iron from transferrin by Ybt was demonstrated using urea-polyacrylamide gels. Transferrin mobility in these gels is altered by the removal of iron from transferrin. Thus, Ybt removes iron from transferrin as well as other mammalian iron-binding proteins for nutritional use by the bacterium [42].
6.2 Bubonic plague and Ybt
Jackson and Burrows observation that spontaneous Pgm− Y. pestis mutants (unable to bind hemin at room temperature) were avirulent in mice via subcutaneous injection unless supplemented with iron or hemin [7] implicated iron acquisition as important for the progression of plague. Later studies using defined ybt mutations demonstrated that the Ybt system is essential for bubonic plague. Y. pestis strains with mutations in biosynthetic or uptake genes in a mildly attenuated background had virulence losses of >660-fold by the subcutaneous route of infection. This degree of virulence loss is an underestimate since the highest bacterial doses tested were less than 105 cells [22, 38]. More recently, we have found that mutations in psn or irp2 (in anotherwise wild-type background) result in subcutaneous LD50s of >107 – a virulence loss of >4 × 106 [42]. Thus loss of either Ybt uptake or Ybt synthesis results in a nearly complete inability to cause fatal bubonic disease [42].
6.3 Septicemic plague and Ybt
Despite being expressed in vitro in serum [60], studies suggest that the Ybt system is not required for septicemic plague. Une and Brubaker demonstrated that a Δpgm mutant is fully virulent in mice via an intravenous route of infection [7, 61]. Since the pgm locus includes the ybt locus [23, 40, 62–63], this clearly eliminates a role for the Ybt system in septicemic plague. This conclusion was recently confirmed using a defined irp2 mutation. In addition, using a flea-to-mouse infection model, Sebbane et al showed that the irp2 mutant caused fatal plague in 20% of the mice compared to 90% for the Ybt+ parent. Of the fatal irp2 infections, histological examination of proximal lymph nodes indicated that one mouse had primary septicemic plague while the second had septicemic plague with a mild lympyhadenitis [64].
These results indicate that the Ybt system is critical only during the early lymphatics stage of bubonic plague. In a rat model of bubonic plague, ybt and other genes encoding iron transport systems were highly expressed in bubos [65]. In Y. enterocolitica infections, fyuA (the gene encoding the Ybt receptor) and hemR (encoding an OM hemin receptor) were more highly expressed in the spleens of mice compared to the livers [66]. While this result indicates that the liver is less iron-restrictive than the spleen, overall these studies demonstrate that the Ybt system is expressed in liver and spleen. Consequently, other systems are likely more effective in acquiring iron once bacteria have disseminated via the bloodstream.
6.4 Pneumonic plague and Ybt
In contrast to bubonic plague where Ybt is essential and septicemic plague where Ybt is dispensable, in pneumonic plague, mutations in various components of the Ybt system have different effects on virulence. Two different mutations in psn had an LD50 of ~104 compared to the parent strain’s LD50 of ~300 – an ~33-fold loss of virulence. In this infection model two different irp2 mutations, causing loss of Ybt biosynthesis, had an LD50 of ~3 × 105 – approximately 790- and 24-fold virulence losses compared to the parent and psn mutants, respectively. Time-to-death analyses with infectious doses similar to the calculated LD50s showed a two-day delay in the 50% endpoint for the psn mutant compared to the parent strain (4 vs 6 days) and a 4-day delay for the irp2 mutant (4 vs 8 days) [42]. This suggests either that pneumonic disease progresses more slowly in ybt mutants or that mouse fatalities are due to systemic spread and septicemic disease. The latter possibility correlates with the findings of Lee-Lewis et al [67].
In contrast to infection with a Pgm+ strain, Lee-Lewis et al found that mice infected intranasally with a Δpgm strain failed to show histological signs of pneumonia. In addition, iron supplementation of mice decreased the time-to-death from a high dose of Δpgm cells compared to Pgm+ cells while the lungs still showed no histological signs of pneumonic disease. Instead mice appeared to die from septicemic plague [67]. Even with intraperitoneal injection of iron, BABL/c and C57BL/6 mice were more susceptible to the Δpgm strain than C3H mice. Both of the more susceptible mouse strains lack the natural resistance-associated macrophage protein 1 (NRAMP1) while the less susceptible C3H strain has this gene. NRAMP1 is involved in activating innate immune responses and killing intracellular bacteria, likely through the transport of iron, manganese, and zinc [67–68]
Our results and those of Lee-Lewis et al suggest not only that the pgm locus encodes additional virulence factor(s) important for pneumonic plague, but also that the Ybt siderophore may serve a virulence role in addition to iron acquisition [42, 67]. In vitro, growth defects under iron-deficient conditions are worse for mutants producing but unable to use Ybt compared to a mutant unable to produce the siderophore (Fig. 3). Secretion of the siderophore by the uptake mutant allows binding of residual iron (~0.3 μM) likely making it unavailable to the other Y. pestis iron transporters. Our in vivo results are the opposite of this in vitro phenotype. Thus we proposed that the Ybt siderophore also serves a non-Fe-acquisition role during lung infections [42]. We have suggested three possibilities: 1) Ybt may act as a signal molecule to transcriptionally activate unidentified, virulence factor genes in addition to ybt genes. 2) The Ybt molecule (possibly in conjunction with chelated iron) may have a direct toxic effect in the lung environment. Pyochelin is structurally similar to Ybt and has been shown to damage endothelial and epithelial cells by generating hydroxyl radicals. The siderophores desferrioxamine and enterobactin are cytotoxic for T cells in vitro. 3) Ybt affects immune cell function(s). Addition of purified Ybt has been shown to reduce the level of reactive oxygen species in J774A.1 cells, PMNs, and human monocytes in vitro. In vitro T-cell proliferation is inhibited by desferrithiocin, which has a structure similar to pyochelin and Ybt. Finally, a number of siderophores affect cytokine production in a variety of host cells in vitro [42]. These possibilities are not mutually exclusive. Thus, the non-Fe-acquisition role(s) of Ybt remain(s) to be elucidated.
7. Conclusions and future directions
Since the identification of the first Ybt protein over twenty years ago, great progress has been made identifying components of the biosynthetic and transport systems and in understanding how the system is regulated. Intriguing differences in the requirement for Ybt in bubonic, septicemic, and pneumonic plague have been uncovered. However a number of questions remain.
While the mechanisms involved in synthesis of Ybt have been largely elucidated, some aspects of Ybt transport, utilization and regulation remain obscure. Although YbtX appears to be involved in siderophore export, no essential component of the secretion apparatus has been identified. YbtP and YbtQ are essential for uptake of iron from Ybt, but it is unclear whether these fused-function permease-ATPases also require a PBP, a membrane spanning protein, or no other components. Whether the Fe-Ybt complex enters the periplasm and/or cytoplasm has not been experimentally determined. Indeed the mechanism used to release iron from the siderophore is unknown.
While the overall regulatory mechanisms have been identified, it remains to be determined whether or not YbtA binds the siderophore to affect transcription of ybt genes. Anisimov et al demonstrated binding of purified YbtA protein to promoter regions in the absence of siderophore suggesting that Ybt is not required for binding of the regulator to DNA [58]; however, the protein was purified from inclusion bodies and solubility issues may have affected its conformation. In P. aeruginosa, pyochelin (a siderophore structurally similar to Ybt) is required for binding of the related PchR transcriptional regulator to regulated promoters [69]. If YbtA does bind the siderophore, does it bind deferrated or Fe-Ybt? If the siderophore transmits a regulatory signal without binding YbtA, what are the intermediary components of this signaling pathway? In the absence of components required for uptake, how does regulatory signaling occur? Finally, in the case of a ybtS mutant, what is the nature of the molecule involved in activating transcription?
Acknowledgments
RDP and JDF are supported by Public Health Service grant AI-33481.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eisen RJ, Gage KL. Adaptive strategies of Yersinia pestis to persist during inter-epizootic and epizootic periods. Vet Res. 2009;40:01. doi: 10.1051/vetres:2008039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perry RD, Fetherston JD. Yersinia pestis - etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinnebusch BJ, Erickson DL. Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr Top Microbiol Immunol. 2008;322:229–248. doi: 10.1007/978-3-540-75418-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inglesby TV, Dennis DT, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Koerner JF, Layton M, McDade J, Osterholm MT, O’Toole T, Parker G, Perl TM, Russell PK, Schoch-Spana M, Tonat K. Plague as a biological weapon: medical and public health management. J Amer Med Assoc. 2000;283:2281–2290. doi: 10.1001/jama.283.17.2281. [DOI] [PubMed] [Google Scholar]
- 5.Anisimov AP, Lindler LE, Pier GB. Intraspecific diversity of Yersinia pestis. Clin Microbiol Rev. 2004;17:434–464. doi: 10.1128/CMR.17.2.434-464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou D, Tong Z, Song Y, Han Y, Pei D, Pang X, Zhai J, Li M, Cui B, Qi Z, Jin L, Dai R, Du Z, Wang J, Guo Z, Wang J, Huang P, Yang R. Genetics of metabolic variations between Yersinia pestis biovars and the proposal of a new biovar, microtus. J Bacteriol. 2004;186:5147–5152. doi: 10.1128/JB.186.15.5147-5152.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson S, Burrows TW. The virulence-enhancing effect of iron on non-pigmented mutants of virulent strains of Pasteurella pestis. Br J Exp Pathol. 1956;37:577–583. [PMC free article] [PubMed] [Google Scholar]
- 8.Wake A, Misawa M, Matsui A. Siderochrome production by Yersinia pestis and its relation to virulence. Infect Immun. 1975;12:1211–1213. doi: 10.1128/iai.12.5.1211-1213.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brubaker RR, Surgalla MJ. Pesticins. I. Pesticin-bacterium interrelationships and environmental factors influencing activity. J Bacteriol. 1961;82:940–949. doi: 10.1128/jb.82.6.940-949.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brubaker RR. Mutation rate to nonpigmentation in Pasteurella pestis. J Bacteriol. 1969;98:1404–1406. doi: 10.1128/jb.98.3.1404-1406.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fetherston JD, Lillard JW, Jr, Perry RD. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177:1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kutyrev VV, Filippov AA, Oparina OS, Protsenko OA. Analysis of Yersinia pestis chromosomal determinants Pgm+ and Psts associated with virulence. Microb Pathog. 1992;12:177–186. doi: 10.1016/0882-4010(92)90051-o. [DOI] [PubMed] [Google Scholar]
- 13.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65 000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 15.Heesemann J. Chromosomal-encoded siderophores are required for mouse virulence of enteropathogenic Yersinia species. FEMS Microbiol Lett. 1987;48:229–233. [Google Scholar]
- 16.Haag H, Hantke K, Drechsel H, Stojiljkovic I, Jung G, Zähner H. Purification of yersiniabactin: a siderophore and possible virulence factor of Yersinia enterocolitica. J Gen Microbiol. 1993;139:2159–2165. doi: 10.1099/00221287-139-9-2159. [DOI] [PubMed] [Google Scholar]
- 17.Chambers CE, McIntyre DD, Mouck M, Sokol PA. Physical and structural characterization of yersiniophore, a siderophore produced by clinical isolates of Yersinia enterocolitica. Biometals. 1996;9:157–167. doi: 10.1007/BF00144621. [DOI] [PubMed] [Google Scholar]
- 18.Drechsel H, Stephan H, Lotz R, Haag H, Zähner H, Hantke K, Jung G. Structure elucidation of yersiniabactin, a siderophore from highly virulent Yersinia strains. Liebigs Ann. 1995;1995:1727–1733. [Google Scholar]
- 19.Perry RD, Balbo PB, Jones HA, Fetherston JD, DeMoll E. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology. 1999;145:1181–1190. doi: 10.1099/13500872-145-5-1181. [DOI] [PubMed] [Google Scholar]
- 20.Carniel E, Mazigh DD, Mollaret HHHH. Expression of iron-regulated proteins in Yersinia species and their relation to virulence. Infect Immun. 1987;55:277–280. doi: 10.1128/iai.55.1.277-280.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carniel E, Mercereau-Puijalon O, Bonnefoy S. The gene coding for the 190,000-dalton iron-regulated protein of Yersinia species is present only in the highly pathogenic strains. Infect Immun. 1989;57:1211–1217. doi: 10.1128/iai.57.4.1211-1217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bearden SW, Fetherston JD, Perry RD. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehring AM, DeMoll E, Fetherston JD, Mori I, Mayhew GF, Blattner FR, Walsh CT, Perry RD. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol. 1998;5:573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 24.Walsh CT, Marshall CG. In: Iron Transport in Bacteria. Crosa JH, Mey AR, Payne SM, editors. ASM Press; Washington, D.C: 2004. pp. 18–37. [Google Scholar]
- 25.Bobrov AG, Geoffroy VA, Perry RD. Yersiniabactin production requires the thioesterase domain of HMWP2 and YbtD, a putative phosphopantetheinylate transferase. Infect Immun. 2002;70:4204–4214. doi: 10.1128/IAI.70.8.4204-4214.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gehring AM, Mori I, Perry RD, Walsh CT. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry. 1998;37:11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- 27.Geoffroy VA, Fetherston JD, Perry RD. Yersinia pestis YbtU and YbtT are involved in synthesis of the siderophore yersiniabactin but have different effects on regulation. Infect Immun. 2000;68:4452–4461. doi: 10.1128/iai.68.8.4452-4461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guilvout I, Mercereau-Puijalon O, Bonnefoy S, Pugsley AP, Carniel E. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLoughlin SM, Kelleher NL. Monitoring multiple active sites on thiotemplate enzymes in parallel: a molecular movie of yersiniabactin bioassembly. J Am Chem Soc. 2005;127:14984–14985. doi: 10.1021/ja0555264. [DOI] [PubMed] [Google Scholar]
- 30.Miller DA, Luo L, Hillson N, Keating TA, Walsh CT. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem Biol. 2002;9:333–449. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 31.Miller MC, Fetherston JD, Pickett CL, Bobrov AG, Weaver RH, DeMoll E, Perry RD. Reduced synthesis of the Ybt siderophore or production of aberrant Ybt-like molecules activates transcription of yersiniabactin genes in Yersinia pestis. Microbiology. 2010;156:2226–2238. doi: 10.1099/mic.0.037945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeifer BA, Wang CCC, Walsh CT, Khosla C. Biosynthesis of yersiniabactin, a complex polyketide-nonribosomal peptide, using Escherichia coli as a heterologous host. Appl Environ Microbiol. 2003;69:6698–6702. doi: 10.1128/AEM.69.11.6698-6702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller MC, Parkin S, Fetherston JD, Perry RD, DeMoll E. Crystal structure of ferric-yersiniabactin, a virulence factor of Yersinia pestis. J Inorg Biochem. 2006;100:1495–1500. doi: 10.1016/j.jinorgbio.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Furrer JL, Sanders DN, Hook-Barnard IG, McIntosh MA. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol Microbiol. 2002;44:1225–1234. doi: 10.1046/j.1365-2958.2002.02885.x. [DOI] [PubMed] [Google Scholar]
- 35.Brickman TJ, Armstrong SK. Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J Bacteriol. 2005;187:3650–3661. doi: 10.1128/JB.187.11.3650-3661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuív PÓ, Clarke P, Lynch D, O’Connell M. Identification of rhtX and fptX, novel genes encoding proteins that show homology and function in the utilization of the siderophores rhizobactin 1021 by Sinorhizobium meliloti and pyochelin by Pseudomonas aeruginosa, respectively. J Bacteriol. 2004;186:2996–3005. doi: 10.1128/JB.186.10.2996-3005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel L, Bachelard A, Reimmann C. Ferripyochelin uptake genes are involved in pyochelin-mediated signalling in Pseudomonas aeruginosa. Microbiology. 2007;153:1508–1518. doi: 10.1099/mic.0.2006/002915-0. [DOI] [PubMed] [Google Scholar]
- 38.Fetherston JD, Bertolino VJ, Perry RD. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol Microbiol. 1999;32:289–299. doi: 10.1046/j.1365-2958.1999.01348.x. [DOI] [PubMed] [Google Scholar]
- 39.Ferber DM, Fowler JM, Brubaker RR. Mutations to tolerance and resistance to pesticin and colicins in Escherichia coli. J Bacteriol. 1981;146:506–511. doi: 10.1128/jb.146.2.506-511.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fetherston JD, Perry RD. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 41.Perry RD, Shah J, Bearden SW, Thompson JM, Fetherston JD. Yersinia pestis TonB: role in iron, heme and hemoprotein utilization. Infect Immun. 2003;71:4159–4162. doi: 10.1128/IAI.71.7.4159-4162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fetherston JD, Kirillina O, Bobrov AG, Paulley JT, Perry RD. The yersiniabactin transport system is critical for the pathogenesis of bubonic and pneumonic plague. Infect Immun. 2010;70:2045–2052. doi: 10.1128/IAI.01236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchanan SK. Type I secretion and multidrug efflux: transport through the TolC channel-tunnel. Trends Biochem Sci. 2001;28:3–6. doi: 10.1016/s0968-0004(00)01733-3. [DOI] [PubMed] [Google Scholar]
- 44.Deng W, Burland V, Plunkett G, III, Boutin A, Mayhew GF, Liss P, Perna NT, Rose DJ, Mau B, Zhou S, Schwartz DC, Fetherston JD, Lindler LE, Brubaker RR, Plano GV, Straley SC, McDonough KA, Nilles ML, Matson JS, Blattner FR, Perry RD. Genome sequence of Yersinia pestis KIM. J Bacteriol. 2002;184:4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brem D, Pelludat C, Rakin A, Jacobi CA, Heesemann J. Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology. 2001;147:1115–1127. doi: 10.1099/00221287-147-5-1115. [DOI] [PubMed] [Google Scholar]
- 46.Lesic B, Carniel E. In: Yersinia Molecular and Cellular Biology. Carniel E, Hinnebusch BJ, editors. Horizon Bioscience; Norfolk, UK: 2004. pp. 285–306. [Google Scholar]
- 47.Lesic B, Bach S, Ghigo J-M, Dobrindt U, Hacker J, Carniel E. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol Microbiol. 2004;52:1337–1348. doi: 10.1111/j.1365-2958.2004.04073.x. [DOI] [PubMed] [Google Scholar]
- 48.Lesic B, Carniel E. Horizontal Transfer of the High-Pathogenicity Island of Yersinia pseudotuberculosis. J Bacteriol. 2005;187:3352–3358. doi: 10.1128/JB.187.10.3352-3358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bultreys A, Gheysen I, de Hoffmann E. Yersiniabactin production by Pseudomonas syringae and Escherichia coliand description of a second yersiniabactin locus evolutionary group. Appl Environ Microbiol. 2006;72:3814–3825. doi: 10.1128/AEM.00119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao H, Zhou D, Li Y, Guo Z, Han Y, Song Y, Zhai J, Du Z, Wang X, Lu J, Yang R. The iron-responsive Fur regulon in Yersinia pestis. J Bacteriol. 2008;190:3063–3075. doi: 10.1128/JB.01910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perry RD, Fetherston JD. In: Yersinia Molecular and Cellular Biology. Carniel E, Hinnebusch BJ, editors. Horizon Bioscience; Norfolk, U.K: 2004. pp. 257–283. [Google Scholar]
- 52.Staggs TM, Perry RD. Fur regulation in Yersinia species. Mol Microbiol. 1992;6:2507–2515. doi: 10.1111/j.1365-2958.1992.tb01427.x. [DOI] [PubMed] [Google Scholar]
- 53.Fetherston JD, Bearden SW, Perry RD. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 54.Perry RD, Abney J, Mier I, Jr, Lee Y, Bearden SW, Fetherston JD. Regulation of the Yersinia pestis Yfe and Ybt iron transport systems. Adv Exp Med Biol. 2003;529:275–283. doi: 10.1007/0-306-48416-1_53. [DOI] [PubMed] [Google Scholar]
- 55.Braun V, Braun M, Killmann H. In: Iron Transport in Bacteria. Crosa JH, Mey AR, Payne SM, editors. ASM Press; Washington D.C: 2004. pp. 158–177. [Google Scholar]
- 56.Poole K. In: Iron Transport in Bacteria. Crosa JH, Mey AR, Payne SM, editors. ASM Press; Washington, D.C: 2004. pp. 293–310. [Google Scholar]
- 57.Anisimov R, Brem D, Heesemann J, Rakin A. Molecular mechanism of YbtA-mediated transcriptional regulation of divergent overlapping promoters ybtA and irp6 of Yersinia enterocolitica. FEMS Microbiol Lett. 2005;250:27–32. doi: 10.1016/j.femsle.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 58.Anisimov R, Brem D, Heesemann J, Rakin A. Transcriptional regulation of high pathogenicity island iron uptake genes by YbtA. Int J Med Microbiol. 2005;295:19–28. doi: 10.1016/j.ijmm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 59.Pelludat C, Rakin A, Jacobi CA, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chauvaux S, Rosso M-L, Frangeul L, Lacroix C, Labarre L, Schiavo A, Marceau M, Dillies M-A, Foulon J, Coppée Y, Médigue C, Simonet M, Carniel E. Transcriptome analysis of Yersinia pestis in human plasma: an approach for discovering bacterial genes involved in septicaemic plague. Microbiology. 2007;153:3112–3124. doi: 10.1099/mic.0.2007/006213-0. [DOI] [PubMed] [Google Scholar]
- 61.Une T, Brubaker RR. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect Immun. 1984;43:895–900. doi: 10.1128/iai.43.3.895-900.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buchrieser C, Prentice M, Carniel E. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol. 1998;180:2321–2329. doi: 10.1128/jb.180.9.2321-2329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fetherston JD, Schuetze P, Perry RD. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 64.Sebbane F, Jarrett C, Gardner D, Long D, Hinnebusch BJ. Role of the Yersinia pestis yersiniabactin iron acquisition system in the incidence of flea-borne plague. PLoS ONE. 2010;5:e14379. doi: 10.1371/journal.pone.0014379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sebbane F, Lemaître N, Sturdevant DE, Rebeil R, Virtaneva K, Porcella SF, Hinnebusch BJ. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc Natl Acad Sci USA. 2006;103:11766–11771. doi: 10.1073/pnas.0601182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jacobi CA, Gregor S, Rakin A, Heesemann J. Expression analysis of the yersiniabactin receptor gene fyuA and the heme receptor hemR of Yersinia enterocolitica in vitro and in vivo using the reporter genes for green fluorescent protein and luciferase. Infect Immun. 2001;69:7772–7782. doi: 10.1128/IAI.69.12.7772-7782.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee-Lewis H, Anderson DM. Absence of inflammation and pneumonia during infection with nonpigmented Yersinia pestis reveals a new role for the pgm locus in pathogenesis. Infect Immun. 2009;78:220–230. doi: 10.1128/IAI.00559-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cellier MF, Courville P, Campion C. Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect. 2007;9:1662–1670. doi: 10.1016/j.micinf.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 69.Michel L, González N, Jagdeep S, Nguyen-Ngoc T, Reimmann C. PchR-box recognition by the AraC-type regulator PchR of Pseudomonas aeruginosa requires the siderophore pyochelin as an effector. Mol Microbiol. 2005;58:495–509. doi: 10.1111/j.1365-2958.2005.04837.x. [DOI] [PubMed] [Google Scholar]