Abstract
Chalcone synthase (CHS, EC 2.3.1.74) is a key enzyme of the flavonoid/isoflavonoid biosynthesis pathway. Besides being part of the plant developmental program the CHS gene expression is induced in plants under stress conditions such as UV light, bacterial or fungal infection. CHS expression causes accumulation of flavonoid and isoflavonoid phytoalexins and is involved in the salicylic acid defense pathway. This review will discuss CHS and its function in plant resistance.
Keywords: Chalcone synthase, Flavonoids, Plant resistance
Introduction
During their life cycle, plants respond actively to stress by producing phytoalexins and other stress metabolites. Such stress can result from injuries caused by the attack of insects and microbes or by mechanical wounding, and can induce many distinctive biochemical changes. These include the production of protective compounds either at the site of injury, or systemically in distant unwounded tissues (Kuhn 1988; Bowles 1990; Ryan 1990). In plants, phenylalanine is derived from the precursor chorismate and leads to the flavonoid, phenylpropanoid and stilbenoid biosynthetic pathways. All are interesting in connection with plant defense but in this review we will focus on the flavonoid biosynthesis pathway and its key enzyme chalcone synthase (CHS).
CHS is a member of the plant polyketide synthase superfamily, which also includes stilbene synthase (STS), acridone synthase, pyrone synthase, bibenzyl synthase, and p-coumaroyltriacetic acid synthase (Sanchez 2008). Chalcone synthases, the most well known representatives of this family, provide the starting materials for a diverse set of metabolites (flavonoids) which have different and important roles in flowering plants, such as providing floral pigments, antibiotics, UV protectants and insect repellents (Hahlbrock and Scheel 1989). Flavonoids also have benefits for human health, as they exhibit amongst others cancer chemopreventive (Jang et al. 1997), antimitotic (Edwards et al. 1990), estrogenic (Gehm et al. 1997) antimalarial (Li et al. 1995) antioxidant (Jang et al. 1997) and antiasthmatic (Zwaagstra et al. 1997) activities.
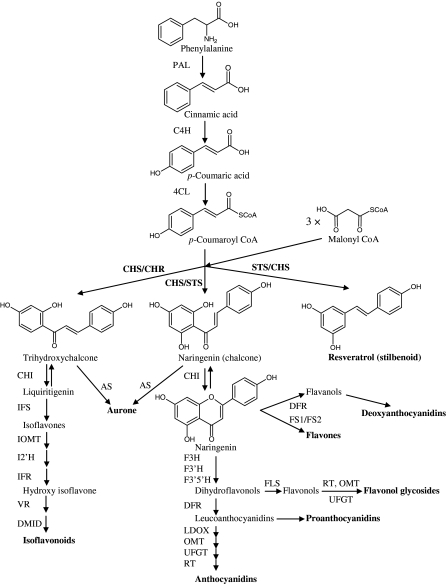
Flavonoids are synthesized via the phenylpropanoid and polyketide pathway, which starts with the condensation of one molecule of CoA-ester of cinnamic acid or derivatives such as coumaric or ferulic acid, and three molecules of malonyl-CoA, yielding a naringenin chalcone as major product. This reaction is carried out by the enzyme chalcone synthase (CHS). The chalcone is isomerised to a flavanone by the enzyme chalcone flavanone isomerase (CHI). From these central intermediates, the pathway diverges into several branches, each resulting in a different class of flavonoids. Flavanone 3-hydroxylase (F3H) catalyzes the stereospecific 3ß-hydroxylation of (2S)-flavanones to dihydroflavonols. For the biosynthesis of anthocyanins, dihydroflavonol reductase (DFR) catalyzes the reduction of dihydroflavonols to flavan-3,4-diols (leucoanthocyanins), which are converted to anthocyanidins by anthocyanidin synthase (ANS). The formation of glucosides is catalyzed by UDP glucose-flavonoid 3-O-glucosyl transferase (UFGT), which stabilizes the anthocyanidins by 3-O-glucosylation (Harborne and Grayer 1994; Bohm 1998). An overview of the flavonoid pathway is presented in Fig. 1. Flavonoids play an important role in plant defense, and CHS as the gatekeeper of flavonoid biosynthesis plays an important role in regulating the pathway. In fact CHS gene expression is influenced by many stress and environmental factors such as UV, wounding or pathogen attack (Dixon and Paiva 1995; Gläßgen et al. 1998; Ito et al. 1997).
Fig. 1.
Flavonoid biosynthetic pathway. ANS anthocyanidin synthase; AS aureusidin synthase; C4H cinnamate-4-hydroxylase; CHR chalcone reductase; DFR dihydroflavonol 4-reductase; DMID 7,2′-dihydroxy, 4′-methoxyisoflavanol dehydratase; F3H flavanone 3-hydroxylase; F3′H flavonoid 3′ hydroxylase; F3′5′H flavonoid 3′5′ hydroxylase; FS1/FS2 flavone synthase; I2′H isoflavone 2′-hydroxylase; IFR isoflavone reductase; IFS isoflavone synthase; IOMT isoflavone O-methyltransferase; LCR leucoanthocyanidin reductase; LDOX leucoanthocyanidin dioxygenase; OMT O-methyltransferase; PAL phenylalanine ammonia-lyase; RT rhamnosyl transferase; UFGT UDP flavonoid glucosyl transferase; VR vestitone reductase; STS stilbene synthase; FLS flavanol synthase. (Winkel 1999; Yamaguchi et al. 1999; KEGG pathways)
In this review we will evaluate the present understanding about CHS and its regulation in plant resistance.
Structure and mechanism of chalcone synthase
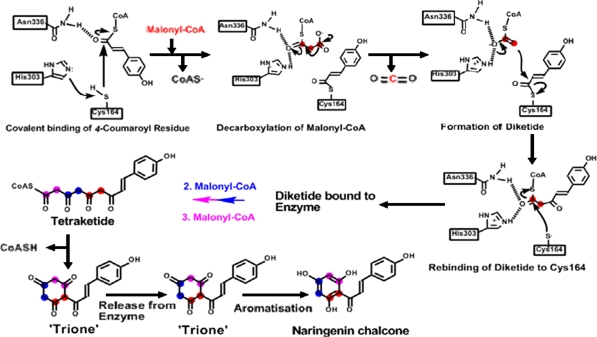
The chalcone synthase (CHS) enzyme—known as a type III polyketide synthase enzyme (PKS) is structurally and mechanistically the simplest PKS (Schröder 1997; Sanchez 2008). These enzymes function as homodimeric iterative PKS (monomer size of 42–45 kDa) with two independent active sites that catalyze a series of decarboxylation, condensation, and cyclization reactions (Tropf et al. 1995). The three dimensional structure of alfalfa CHS2 was studied intensively by Ferrer et al. (1999). The study revealed that each alfalfa CHS2 monomer consists of two structural domains. In the upper domain, there are four amino acids (Cys164, Phe215, His303, and Asn336) are present at the active site were defined as the catalytic machinery of CHS. The lower domain of CHS has a large active site providing space for the tetraketide required for chalcone formation (i.e., naringenin and resveratrol) from one p-coumaroyl-CoA and three malonyl-CoA (Fig. 2) (Jez et al. 2001a, b). Production of chalcone requires the condensation of one molecule of p-coumaroyl-CoA and three malonyl-CoA molecules which is catalyzed by CHS. It starts with the transfer of a coumaroyl moiety from a p-coumaroyl-CoA starter molecule to an active site cysteine (Cys164) (Lanz et al. 1991). Next, a series of condensation reactions of three acetate units derived from three malonyl-CoA molecules, each proceeding through an acetyl-CoA carbanion derived from malonyl-CoA decarboxylation, extends the polyketide intermediate. Following generation of the thioester-linked tetraketide, a regiospecific intramolecular Claisen condensation forms a new ring system to yield chalcone. In plants, chalcone isomerase (CHI) will convert the chalcone to (2S)-5,7,4′-trihydroxyflavanone (naringenin); however, spontaneous ring closure in vitro results in mixed enantiomers of naringenin (Hahlbrock et al. 1970 ; Jez et al. 2000). In vivo chalcone can convert to narigenin without need of CHI. Four amino acids (Cys164, Phe215, His303, and Asn336) situated at the intersection of the CoA-binding tunnel and the active site cavity play an essential and distinct role during malonyl-CoA decarboxylation and chalcone formation. Cys164 plays role as the active-site nucleophile in polyketide formation and elucidate the importance of His303 and Asn336 in the malonyl-CoA decarboxylation reaction. Phe215 may help orient substrates at the active site during elongation of the polyketide intermediate. (Jez et al. 2000). The general reaction mechanism of CHS is presented in Fig. 2.
Fig. 2.
Reaction catalyzed by chalcone synthase (CHS). In CHS, three amino acids play key roles in the catalytic functions of type III PKS: Cys164: active site, covalent binding site of starter residues and intermediates, His303 and Asn336: stabilization/activation of both starter (e.g. 4-coumarate) and extender units (malonyl-/acetyl-residues) (Ferrer et al. 1999; Bomati et al. 2005; modified by Schröder 2008)
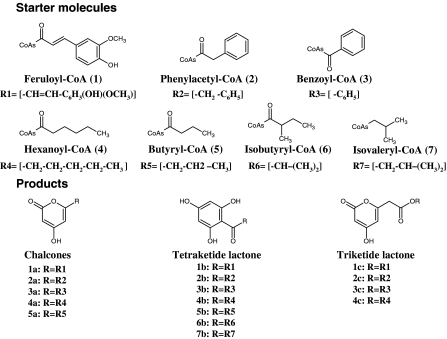
Several other cyclization reactions are possible besides the one yielding a chalcone. In addition to the starter molecule p-coumaroyl-CoA, in vitro alfalfa CHS accepts other CoA-linked thioesters as alternate starter molecules to generate corresponding chalcones, tetraketide lactone, and triketide lactone products (Fig. 3). The substrates can be feruloyl-CoA, hexanoyl-CoA, phenylacetyl-CoA, benzoyl-CoA, butyryl-CoA, isobutyryl-CoA and isovaleryl-CoA. With the starter substrates p-coumaroyl-CoA and malonyl-CoA, CHS catalyzes an intramolecular Claisen condensation yielding the chalcone naringenin. Alfalfa CHS2 and parsley CHS (Hrazdina et al. 1976), accept feruloyl-CoA as a starter molecule and produce the tetraketide lactone (1b) and methylpyrone as the major products with the triketide lactone (1c) generated as a minor product. With hexanoyl-CoA, alfalfa CHS2 yields the tetraketide lactone (4b) as the major product, triketide lactone (4c) and methylpyrone are minor products (Jez et al. 2001a). Parsley CHS accepts butyryl-CoA and hexanoyl-CoA as substrates in vitro, which yield, respectively, the chalcone analogues, phlorobutyrophenone (5b) and phlorocaprophenone (4b) at pH 6.5 (Schuez et al. 1983). Medicago sativa CHS2 accepts phenylacetyl-CoA as a starter molecule yielding a phlorobenzyl ketone (2a), the chalcone-like product, accounts for less than 10% and others like tetraketide lactone (2b), triketide lactone (2c), and methylpyrone comprise the other products. The overall product distribution with phenylacetyl-CoA is similar to Scutellaria baicalensis CHS (Morita et al. 2000). With benzoyl-CoA as the starter molecule, alfalfa CHS2 generates phlorobenzophenone (3a) and methylpyrone as the major product, and tetraketide lactone (3b) and triketide lactone (3c) as minor products (Jez et al. 2001a). The recombinant hop CHS1 expressed in E. coli showed activity with isobutyryl-CoA and isovaleryl-CoA substrates, which produced as main products phloroisobutyrophenone (6b) and phloroisovalerophenone (7b) (Zuurbier et al. 1998; Novák et al. 2006).
Fig. 3.
Alternate starter molecules and their in vitro reaction products catalyzed by CHS
The steady-state kinetic parameters of Medicago sativa CHS2 for p-coumaroyl-CoA, malonyl-CoA, feruloyl-CoA, hexanoyl-CoA, phenylacetyl-CoA and benzoyl-CoA have been determined, these are presented in Table 1 (Jez et al. 2001a; Novák et al. 2006).
Table 1.
Steady-state kinetic constants of Medicago sativa CHS2 with different starter substrates (Jez et al. 2001a; Novák et al. 2006)
| kcat (min−1) | Km (μM) | |
|---|---|---|
| p-Coumaroyl-CoA | 5.14 ± 0.30 | 6.1 ± 1.3 |
| Malonyl-CoA | 4.58 ± 0.24 | 4.7 ± 1.1 |
| Feruloyl-CoA | 1.04 ± 0.17 | 5.2 ± 0.9 |
| Hexanoyl-CoA | 2.52 ± 0.22 | 4.1 ± 1.2 |
| Phenylacetyl-CoA | 2.17 ± 0.35 | 5.1 ± 0.7 |
| Benzoyl-CoA | 1.73 ± 0.21 | 2.2 ± 0.2 |
| Isobutyryl-CoA | – | 14.9 ± 0.2 |
| Isovaleryl-CoA | – | 8.0 ± 0.2 |
Control of CHS activity
In plants, CHS is activated by a wide range of environmental and developmental stimuli. Theoretically, there are many ways to regulate CHS activity in vivo, from metabolic control to the control of initiation of transcription of the CHS gene (Martin 1993).
Metabolic control
There are many studies showing that CHS is inhibited noncompetitively by flavonoid pathway products like naringenin, chalcone naringenin and the other end products of CoA esters. For example, the parsley CHS is 50% inhibited by 100 μM naringenin and 10 μM CoA esters (Hinderer and Seitz 1985; Kreuzaler and Hahlbrock 1975), the flavonoids luteolin and apigenin are inhibitory to rye CHS in vitro (Peters et al. 1988), whereas in carrot, among the range of flavonoids tested, only naringenin and chalcone narigenin can inhibit CHS at 100 μM (Hinderer and Seitz 1985). It seems that flavonoids accumulate in the cytosol to a level that blocks CHS activity to avoid toxic levels for the plant (Whitehead and Dixon 1983), though there is no direct evidence that this inhibition happens in vivo.
Control of CHS turnover
In plants, CHS may always be present in the cells but is only activated under certain specific conditions. The statement “CHS may always be present in the cells but is only activated under certain specific conditions” means that CHS is activated at the protein level. However, it has been shown that UV light and biotic elicitors induce the flavonoid biosynthetic pathway at the transcriptional level and that CHS is not detectable before the onset of the various stress situations. Studies on parsley cell cultures showed that the induction of CHS activity by UV light was the result of de novo synthesis and active enzyme subsequently decayed with a half-life of 6 h, whereas inactive enzyme decayed more slowly with a half-life of 18 h (Schröder and Schäafer 1980). Inactive CHS could be detected by CHS antibodies and the size of the protein was not changed. In another study about accumulation of CHS during UV induction, Chappell and Hahlbrock (1984) concluded that the accumulation of flavonoid end products is presumably determined by activity of the rate-limiting step(s) in flavonoid biosynthesis and may not precisely reflect the dynamics of CHS activity in vivo.
Control of CHS through trans-genes
The activity of CHS can be controlled by antisense or sense genes. The studies on expression of antisense genes in Petunia (e.g. Van der Krol et al. 1988; Van der Meer et al. 1993), tobacco (Wang et al. 2006), Gerbera hybrida (Elomaa et al. 1996) and Arabidopsis (Le Gall et al. 2005) have shown that the presence of antisense CHS could inhibit the expression of the endogenous CHS in plants. In flowers of antisense CHS transgenic Petunia, the antisense construct was able to inhibit expression of the endogenous CHS genes to varying degrees, which is observed phenotypically as an inhibition of anthocyanin production to give completely acyanic or patterned flowers. In the cyanic sectors and flowers, transcripts of the endogenous CHS genes were under the detection limit, but the antisense transcripts were also barely detectable (Van der Krol et al. 1990b). The antisense effect most likely involves homologous pairing between the transcripts of endogenous CHS genes and transcripts of the introduced antisense CHS gene to form double stranded RNA that is very rapidly degraded, thus inhibiting CHS transcript accumulation and hence CHS activity.
Introducing a heterologous CHS gene in sense orientation can inhibit CHS activity in transgenic plants. This phenomenon is called co-suppression since it involves the reduction of transcriptional level of both endogenous and introduced genes in tissues where the endogenous gene is normally expressed (Napoli et al. 1990; Jorgensen 1995). This is known as gene silencing in which the transgene triggered not only its own silencing but also the endogenous chalcone synthase gene (Hammond et al. 2001). But on the other hand the introduced CHS gene may be expressed to high levels in tissue where the endogenous CHS genes are not expressed, such as in leaves of Petunia (Van der Krol et al. 1990a). Some studies have shown that co-suppression correlates with DNA methylation of the silenced sequences, presumably leading to a blockade at the transcriptional level or/and failure of transcript to accumulate in the cytoplasm resulting in a lack of enzyme activity (Ingelbrecht et al. 1994; Furner et al. 1998; Amedeo et al. 2000). Nowadays, the molecular mechanism of co-suppression of gene expression is thought to be related to the RNAi mechanism (Hannon 2002).
CHS localization and dynamics
The CHS protein in buckwheat (Fagopyrum esculentum) hypocotyls is located in the cytosol and associates with the cytoplasmic face of the rough endoplasmic reticulum (rER), but not with nuclei, plastids, mitochondria, Golgi, or tonoplasts (Hrazdina and Jensen 1992). Saslowsky and Winkel (2001) examined the subcellular location of CHS and CHI in Arabidopsis roots. High levels of both enzymes were found in the epidermal and cortex cells of the elongation zone and the root tip, consistent with the accumulation of flavonoid endproducts at these sites. Co-localization of CHS and CHI was observed at the endoplasmic reticulum and tonoplast in these cells.
However, there is evidence that flavonoids located in the nucleus may be synthesized in situ (Saslowsky and Winkel 2001). Several recent reports describe the accumulation of flavonoids in the nucleus in such diverse species as Arabidopsis thaliana, Brassica napus, Flaveria chloraefolia, Picea abies, Tsuga canadensis, and Taxus baccata (Buer and Muday 2004; Feucht et al. 2004; Grandmaison and Ibrahim 1996; Hutzler et al. 1998; Kuras et al. 1999; Peer et al. 2001). For the enzymes of the flavonoid pathway, several mechanisms may be involved. In the cytoplasm, flavonoid enzyme complexes are believed to assemble at the ER and in electron dense particles through the association of operationally-soluble enzymes such as CHS and CHI with the membrane-bound P450 hydroxylase, flavonoid 3′-hydroxylase (Saslowsky and Winkel 2001; Hrazdina and Wagner 1985). CHS possesses sequences resembling a classic nuclear localization signal (NLS). This signal is located on the surface, on the opposite side of the protein from the dimerization interface and could function to direct CHS, and perhaps associated enzymes into the nucleus. The localization of end products such as flavonol sulfate esters and flavan-3-ols to the nucleus suggests that additional flavonoid enzymes are also present in the nucleus (Grandmaison and Ibrahim 1996; Feucht et al. 2004).
There is an immuno gold-labeling study in grape berry showing that CHS was localized in rough endoplasmic reticulum (ER) and cytoplasm of the skin cells, while few gold particles were found on the cell wall. Besides, two novel sites of CHS were observed within cells of developing grape berry, one is in the plastids which remain unchanged throughout all stages of berry development. At the ripening stage of grape berry, CHS is present in the vacuole and in the vacuole membrane (tonoplast) (Tian et al. 2008). It is suggested that in grape berries, the synthesis of flavonoids in the ripening stage may occur in the vacuole.
Control of CHS gene expression
In Arabidopsis, parsley, and snapdragon only a single copy of the CHS gene has been found. In most angiosperms CHS has been shown to be encoded by a multigene family, such as in petunia (violet 30) (Koes et al. 1987), morning glories (Ipomoea) (Durbin et al. 2000), Gerbera (Helariutta et al. 1996), leguminous plants (Ryder et al. 1987; Wingender et al. 1989; Ito et al. 1997), and Cannabis sativa (Sanchez 2008).
Regulation of CHS gene expression
Many studies have shown that the CHS gene is constitutively expressed in flowers, but also its expression can be induced by light/UV light and in response to phytopathogens, elicitors or wounding in different parts of the plant, resulting in enhanced production of flavonoids (Koes et al. 1987; Ryder et al. 1984, 1987; Bell et al. 1986; Burbulis et al. 1996). CHS expression is also regulated by the circadian clock (Thain et al. 2002).
The level of CHS gene expression is reflected by the level of the CHS transcripts in plant cells. In order for transcription to take place, the RNA polymerase II must attach to specific DNA sequences in the CHS promoter in the vicinity of the TATA box and must be activated by specific DNA-binding proteins (transcription factors) binding to response elements further upstream in the promoter. The CHS promoter was studied extensively in Phaseolus vulgaris, Antirrhinum, Arabidopsis, and parsley (Dixon et al. 1994; Faktor 1997; Feinbaum et al. 1991; Lipphardt et al. 1988).
The CHS promoter contains the nucleotide sequence CACGTG regulatory motif known as G-box, which has been found to be important in the response to light/UV light (Kaulen et al. 1986; Staiger et al. 1989; Dixon et al. 1994; Schulze et al. 1989). Besides the G-box there are other domains in the CHS promoter involved in the light activation of CHS transcription. Those domains have been identified in the parsley CHS promoter as Box I, Box II, Box III, Box IV or three copies of H-box (CCTACC) in the Phaseolus vulgaris CHS15 promoter. These boxes play a role as core promoter together with the G-box and all are required for light inducibility (Block et al. 1990; Lawton et al. 1990; Weisshaar et al. 1991).
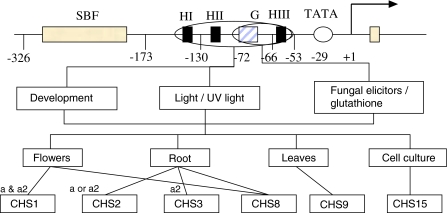
The environmental and developmental control of CHS transcription has been investigated for the CHS15 bean gene (Fig. 4) (Dixon et al. 1994; Harrison et al. 1991). The sequence elements required for transcriptional activation of the CHS15 gene in response to fungal elicitors and glutathione are contained in a 130 bp region of the promoter (Choudhary et al. 1990; Dron et al. 1988; Harrison et al. 1991]). This region contains a G-box and H-box III. There is a silencer element located between positions −326 and −173 of the CHS15 promoter (Dron et al. 1988). No trans-acting factors were found that could bind to cis elements in this region but the region reduced expression of CHS (Harrison et al. 1991). An enhancer element was found in the Antirrhinum CHS promoter. It is located in the region between −564 and −647 and increased CHS gene expression in roots, stems, leaves, and seeds but not in petal tissue (Fritze et al. 1991).
Fig. 4.
Bean CHS15 promoter and regulators. SBF silencer binding factor, H H-Box (CCTACC), G G-Box (CACGTG), a/a2 regulation loci
The Petunia CHSA promoter was studied by van der Meer et al. (1990, 1993) to understand the role of the promoter in tissue-specific CHS expression. The studies showed that the promoter sequence between +1 and −67 confers flower specific CHS gene expression. Another study on the Antirrhinum CHS promoter has shown that the sequences between +1 and −39 allow CHS expression in root and stems, whereas sequences between −39 and −197 are required for expression in petals and seeds (Fritze et al. 1991).
The regulators of CHS in plants are controlled by some specific loci. In maize, there are four loci, cl, r, vp, and clf, involved in the regulation of CHS expression (Dooner 1983). Multiple regulatory loci for CHS expression have also been described for the petunia regulatory mutant Red Star. The phenotype of this mutant of red and white sectors in the flower petals is thought to depend on at least four regulatory genes, all of which regulate CHS expression in trans (Mol et al. 1983). In the CHS gene family of Phaseolus vulgaris, the regulation is via the a and a2 loci though they regulate different CHS members in different ways. The CHS genes might have different combinations of cis elements that determine their response to the products of these regulatory loci. The expression of CHSl in flower tissue has an absolute requirement for the products of both the a and a2 loci, whereas, in root tissue, the products of these loci are not required. It is possible that the CHSl gene interacts with one or more factors present in roots, which are absent in flowers, that can substitute for the products of the a and a2 loci. CHS3 expression in flower tissue is more complicated: it requires the product of the a2 locus, but has a lower level of expression in a mutants compared with wild type. This suggests that CHS3 interacts with both the a2 and a locus products, but, unlike the CHSl gene, it may also interact with other products, allowing transcription at a low level in a mutants. CHS2 is expressed in roots but not in petal tissue, suggesting that it may not be able to interact with the products of a and a2 loci in petal tissue (Harker et al. 1990).
Transcription factors involved in of CHS gene expression
Trans-acting factors of bean CHS15 that bind to two short sequences centered on the G-box and H-box also make major contributions to the in vivo transcription of the promoter (Arias et al. 1993; Yu et al. 1993). Trans activation required both a MYB-binding site and a G-box like element (Sablowski et al. 1994). MYB305, one of the MYB-like proteins that have been implicated in the transcriptional control of tissue-specific CHS gene expression, is also recognized by a cis element of the light-regulatory unit 1 (LRUI) of CHS in parsley (Feldbrügge et al. 1997). G-box/H-box binding factor 1(G/HBF-1), a basic leucine zipper (bZIP) protein, that binds to both the G-box and the adjacent H-box in the proximal region of the CHS15 bean promoter, is rapidly phosphorylated in elicited soybean cells, this happen also to the CHS15, CHS7, and CHS1 promoter (Dröge et al. 1997; Yoshida et al. 2008). Protein and mRNA levels of G/HBF-l do not change during the induction of CHS genes following pathogen attack (Yoshida et al. 2008) but CHS gene expression is strongly stimulated following phosphorylation responding to fungal elicitor treatment in vitro (Dröge et al. 1997).
CHS activity in plant resistance
In nature plants are exposed to a variety of biotic and abiotic stresses. Viruses, bacteria, fungi, nematodes and other pests attacking plants are biotic stresses, while light, temperature, wounding, drought, etc. are abiotic stresses. During stress conditions a plant is expressing a number of genes as part of its defense. Among these genes, CHS is quite commonly induced in different plant species under different forms of stress like UV, wounding, herbivory and microbial pathogens resulting in the production of compounds that have e.g. antimicrobial activity (phytoalexins), insecticidal activity, and antioxidant activity or quench UV light directly or indirectly. The current knowledge about regulation of CHS in plant pathogen resistance is presented in Table 2.
Table 2.
Chalcone synthase expression in plant under stress conditions
| No. | Host | Pathogen/stresses | Metabolites | References | |
|---|---|---|---|---|---|
| 1 | Petroselinum crispum | Parsley | UV | Flavonoids | Schmelzer et al. (1988), Schulze et al. (1989) |
| 2 | Phaseolus vulgaris cells | French bean | Colletotrichum lindemuthianu | Ryder et al. (1984) | |
| 3 | Arabidopsis cells | UV-B and UV-A/blue light | Christie and Jenkins (1996) | ||
| 4 | Arabidopsis thaliana | Low temperature | Anthocyanins | Leyva et al. (1995) | |
| UV-B, UV-A, and blue Light | Fuglevand et al. (1996), Hartmann et al. (1998), Wade et al. (2001) | ||||
| High-intensity lights | Anthocyanins | Feinbaum and Ausubel (1988) | |||
| SA, ethylene, methyl jasmonate Alternaria brassicicola | Schenk et al. (2000) | ||||
| Pseudomonas syringae | Phenolic compounds | Soylu (2006) | |||
| 5 | Petunia hybrida | UV | Koes et al. (1989) | ||
| Low temperature | Anthocyanin | Shvarts et al. (1997) | |||
| 6 | Petroselinum hortense cells | UV | Kreuzaler et al. (1983) | ||
| 7 | Pinus sylvestris | Scots pine | UV-B | Phenolic compounds, flavonoids, catechin | Schnitzler et al. (1996) |
| 8 | Picea abies | Norway spruce | Ceratocystis polonica | Nagy et al. (2004) | |
| Ophiostoma polonicum and wounding | Catechin | Brignolas et al. (1995) | |||
| 9 | Secale cereale | UV | Haussuehl et al. (1996) | ||
| 10 | Hordeum vulgare | Barley | Blumeria graminis | Christensen et al. (1998) | |
| Erysiphe graminis | |||||
| UV | |||||
| 11 | Medicago truncatula | Alfalfa | Glomus versiforme | Isoflavonoid | Harrison and Dixon (1993) |
| Medicago sativa | |||||
| 12 | Antirrhinum majus | Snapdragon | Erwinia chrysanthemi | Junghans et al. (1993) | |
| Rhizobium meliloti | |||||
| CuCl2 | |||||
| Wounding | |||||
| Phoma medicaginis | |||||
| Colletotrichum lindemuthianum | Dalkin et al. (1990) | ||||
| UV | Lipphardt et al. (1988) | ||||
| Staiger et al. (1989) | |||||
| 13 | Lycopersicon esculentum | Tomato | |||
| 14 | Glycine max | Soybean | Pseudomonas syringae pv glycinea | Dhawale et al. (1989) | |
| Phytophthora megasperma f. sp. Glycinea | |||||
| 15 | Picea glauca | White Spruce | Wounding, JA, MeJ | Richard et al. (2000) | |
| 16 | Daucus carota | Carrot cell | UV, Pythium aphanidermatum | Anthocyanin | Gläßgen et al. (1998) |
| 17 | Brassica rapa | Turnip | UV | Anthocyanin | Zhou et al. (2007) |
| 18 | Sorghum bicolor | Sorghum mesocotyl, juvenile sorghum tissues | Colletotrichum graminicola | 3-Deoxyanthocyanidins, apigeninidin luteolinidin | Lue et al. (1989), Nicholson et al. (1987) |
| Helminthosporium maydis |
Phytoalexins
Phytoalexins are antimicrobial metabolites produced by plants in response to microbial attack (or biotic and abiotic elicitors) (Dixon 1986). Phytoalexins come from many different metabolite classes such as flavonoids, stilbenoids, sesquiterpenoids, steroids and alkaloids. CHS can help the plant to produce more flavonoids, isoflavonoid-type phytoalexins and other related metabolites to protect it against stress. Accumulation of flavonoids and isoflavonoids in response to pathogen attack is seen in many plant species, and their importance as antimicrobial phytoalexins is well established (Matthews and Matthews 1989; Van Etten and Pueppke 1976). Flavonoid phytoalexins have been described in legumes, cereals, sorghum, rice, Cephalocereus senilis, Beta vulgaris (Hipskind et al. 1990; Johnson et al. 1976; Kodama et al. 1992; Pare et al. 1992). Some isoflavonoids were increased in Lupin luteus after infection with Fusarium oxysporum such as genistein, wighteone and luteon (Morkunas et al. 2005). The isoflavones, daidzein, genistein and glycitein, in soybean were strongly increased after infection by Sclerotinia sclerotiorum (Wegulo et al. 2005). Stilbenes are known as the phytoalexins in peanut (Ingham 1976) and grapes (Langcake and Pryce 1977a, b). There is also evidence that stilbene synthase (STS) has developed from CHS several times in the evolution (Tropf et al. 1994).
Phytoanticipins
Van Etten et al. (1995) defined phytoanticipins as low molecular weight, antimicrobial compounds that are constitutively expressed in plants without the need for infection with fungal pathogens or are produced after infection solely from preexisting constituents. The distinction between phytoalexins and phytoanticipins is not always clear as some compounds may be phytoalexins in one species and phytoanticipins in another species. Phytoanticipins also are classed into several chemical groups such as flavonoids, terpenoids, steroids, glucosinolates, and alkaloids.
The flavonoid epicatechin plays an important role as phytoanticipin in avocado fruits (Guetsky et al. 2005) and antimicrobial isoflavones desmodianones A, B and C have been isolated from Desmodium canum (Monache et al. 1996). Anthocyanins as products of the flavonoid metabolism are, for example responsible for the red to purple and blue colors of many fruits, vegetables, flowers, and cereal grains. In plants they serve as attractants for pollination and seed dispersal, give constitutive protection against the harmful effects of UV irradiation, and as phytoanticipins provide antiviral and antimicrobial activities in plants (Wrolstad 2000). Genotypes of Ipomoea purpurea with nonfunctional copies of chalcone synthase (CHS) received greater herbivore damage and twice the intensity of infection by the fungal pathogen Rhizoctonia solani than the wild type (Zufall and Rausher 2001).
Light protection
Phenolic compounds like flavonoids strongly absorb UV light and thus are able to protect plants from DNA damage caused by UV. Anthocyanins belong to a class of flavonoids that accumulate in leaves and stems as plant sunscreen in response to light intensity (Leyva et al. 1995). Expression of CHS genes is known to be regulated by light through a photoreceptor-mediated mechanism (Koes et al. 1989). In several cases, it was found that the photoregulated production of flavonoids is at least in part due to the transcriptional induction of CHS (Chappell and Hahlbrock 1984; Feinbaum and Ausubel 1988; van Tunen et al. 1988; Taylor and Briggs 1990). Examination of CHS expression in parsley cell culture suggested that a UV-B light receptor, a blue light receptor and phytochrome may all play a role in light-induced CHS expression (Bruns et al. 1986; Ohl et al. 1989).
High intensity light and UV-A were found to regulate expression of chimeric chalcone synthase genes in transgenic Arabidopsis thaliana plants (Feinbaum et al. 1991). High-intensity light treatment of A. thaliana plants for 24 h caused a 50-fold increase in CHS enzyme activity and an accumulation of visibly detectable levels of anthocyanin pigments in the vegetative structures of these plants (Feinbaum and Ausubel 1988). The expression of CHS genes was increased with time during a 24 h exposure to UV-A on swollen hypocotyls of the red turnip ‘Tsuda’ and induced anthocyanin accumulation (Zhou et al. 2007). The flavonoids accumulate in epidermal cells of the leaves and it is specifically in these cells that CHS gene expression is induced by light stimuli (Schmelzer et al. 1988). However, in mustard the expression of two CHS genes is induced coordinately in seedlings grown in a dark environment for 36–42 h, though this induction is enhanced by supplying red or far red light (Ehmann et al. 1991).
Auxin and jasmonic acid signaling
In plant increase of CHS activity causes a high accumulation flavonoid level that inhibit polar auxin transport (Brown et al. 2001; Faulkner and Rubery 1992; Jacobs and Rubery 1988). Inhibitors of auxin transport could increase the resistance of tomato plants to Fusarium oxysporum (Davis 1954). Also other research showed that CHS is expressed in the nodule primordium and later primarily in uninfected cells of the nodule apex in Rhizobium infected legumes. This may explain the induction of nodule on infected legume roots, higher accumulation of flavonoids blocks auxin transport, causing a local accumulation of auxin, a growth hormone, which caused the induction of nodule growth and development (Estabrook and Sengupta 1991; Yang et al. 1992).
Jasmonic acid and its esters, such as methyl jasmonate (MeJA) are a group of plant hormones having a signaling role in insect and disease resistance (Xu et al. 1994). They could activate CHS in soybean and parsley cell cultures (Creelman et al. 1992) and Picea glauca (Richard et al. 2000). It is thought that volatile jasmonates are released from wounded tissue; thus elicitating plants to activate CHS which cause a production of phytoalexins in advance to resist an infection.
Conclusion
CHS is known as the key entry enzyme commited to the production of the polyketide phenylpropanoids in plants. It seems that all plants contain at least one CHS gene and often CHS gene families in plant with different expression patterns. In certain cases evolution into genes that encode enzymes with different substrate specificity, particularly for the starter molecule (e.g. aliphatic CoA ester instead of cinamic acid derivative) give different ring closure such as in stilbenes. The flavonoid pathway genes are highly diverted and have been found to be present from the earliest plants on land (the bryophytes, liverworts and hornworts) to the highly evolved flowering plants. Chalcones, flavonols and flavones were found in the earliest plants. Those flavonoids function as sunscreen protecting against UV radiation as plants began colonizing land and also play a regulation auxin transport (Markham, 1988; Shirley, 1996; Li et al. 1993; Brown et al. 2001). Later stage of plants such as the ferns and allies are known as oldest group of plants producing proanthocyanidins, procyanidin, prodelphinidin and flavanols. Anthocyanidin, a flavonoid, plays an important role in plant pigment action and is found in gymnosperms and angiosperms. These flavonoids serve diverse functions in different plant species, e.g. as pigments, phytoalexins, UV protectants, signal molecules in plant-microbe interactions, antioxidants, and pollinator attractants or feeding deterrents. In other words these unique plant compounds play a major role in the interaction of plants with their environment (De Bruyne et al. 1999; Kong et al. 2003: Marles et al. 2003; Yilmaz and Toledo 2004).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Amedeo P, Habu Y, Afsar K, Scheid OM, Paszkowski J. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature. 2000;405:203–206. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- Arias JA, Dixon RA, Lamb CJ. Dissection of the functional architecture of a plant defense gene promoter using a homologous in vitro transcription initiation system. Plant Cell. 1993;5:485–496. doi: 10.1105/tpc.5.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JN, Ryder TB, Wingate VPM, Bailey JA, Lamb CJ. Differential accumulation of plant defense gene transcripts in a compatible and an incompatible plant: pathogen interaction. Mol Cell Biol. 1986;6:1615–1623. doi: 10.1128/mcb.6.5.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A, Dangl JL, Hahlbrock K, Schulze LP. Functional borders, genetic fine structure, and distance requirements of cis elements mediating light responsiveness of the parsley chalcone synthase promoter. Proc Natl Acad Sci USA. 1990;87(14):5387–5391. doi: 10.1073/pnas.87.14.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm BA. Introduction to flavonoids. Amsterdam: Hardwood Academic Publishers; 1998. [Google Scholar]
- Bomati EK, Austin MB, Bowmann ME, Dixon RA, Noel JP. Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis. J Biol Chem. 2005;280:30496–30503. doi: 10.1074/jbc.M502239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D. Defense-related proteins in higher plants. Annu Rev Biochem. 1990;59:873–907. doi: 10.1146/annurev.bi.59.070190.004301. [DOI] [PubMed] [Google Scholar]
- Brignolas F, Lacroix B, Lieutier F, Sauvard D, Drouet A, Claudot AC, Yart A, Berryman AA, Christiansen E. Induced responses in phenolic metabolism in two Norway spruce clones after wounding and inoculations with Ophiostoma polonicum, a bark beetle-associated fungus. Plant Physiol. 1995;109:821–827. doi: 10.1104/pp.109.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Tague BW, Peer WA, Taiz L, Muday GK. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis thaliana. Plant Physiol. 2001;126:524–535. doi: 10.1104/pp.126.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns B, Hahlbrock K, Schafer E. Fluence dependente of the ultraviolet-light-induced accumulation of chalcone synthase mRNA and effects of blue and far-red light in cultured parsley cells. Planta. 1986;169:393–398. doi: 10.1007/BF00392136. [DOI] [PubMed] [Google Scholar]
- Buer CS, Muday GK. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell. 2004;16:1191–1205. doi: 10.1105/tpc.020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis IE, Iacobucci M, Shirley BW. A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell. 1996;8:1013–1025. doi: 10.1105/tpc.8.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, Hahlbrock K. Transcription of plant defense genes in response to UV light or funga1 elicitor. Nature. 1984;311:76–78. [Google Scholar]
- Choudhary AD, Lamb CJ, Dixon RA (1990) Stress responses in alfalfa (Medicago sativa L.). VI. Differential responsiveness of chalcone synthase induction to fungal elicitor or glutathione in electroporated protoplasts. Plant Physiol 94:1802–1807 [DOI] [PMC free article] [PubMed]
- Christensen AB, Gregersen PL, Schroder J, Collinge DB. A chalcone synthase with an unusual substrate preference is expressed in barley leaves in response to UV light and pathogen attack. Plant Mol Biol. 1998;37:849–857. doi: 10.1023/a:1006031822141. [DOI] [PubMed] [Google Scholar]
- Christie JM, Jenkins GI. Distinct UVB and UV-A/blue light signal transduction pathways induce chalcone synthase gene expression in Arabidopsis cells. Plant Cell. 1996;8:1555–1567. doi: 10.1105/tpc.8.9.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Tierney ML, Mullet JE. Jasmonic acid/methyl jasmonate accumulated in wounded soybean hypocotyls and modulate wound gene expression. Proc Natl Acad Sci USA. 1992;89:4938–4941. doi: 10.1073/pnas.89.11.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkin K, Edwards R, Edington B, Dixon RA. Stress responses in alfalfa (Medicago sativa L.). I. Induction of phenylpropanoid biosynthesis and hydrolytic enzymes in elicitor-treated cell suspension culture. Plant Physiol. 1990;92:440–446. doi: 10.1104/pp.92.2.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D. The use of intergeneric grafts to demonstrate toxins in the Fusarium wilt disease of tomato. Am J Bot. 1954;41:395–398. [Google Scholar]
- De Bruyne T, Pieters L, Deelstra H, Vlietinck A. Condensed vegetable tannins: biodiversity in structure and biological activities. Biochem Syst Ecol. 1999;27:445–459. [Google Scholar]
- Dhawale S, Souciet G, Kuhn DN. Increase of chalcone synthase mRNA in pathogen-induced soybeans with race-specific resistance is different in leaves and roots. Plant Physiol. 1989;91:911–916. doi: 10.1104/pp.91.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Harrison MJ, Lamb-C J. Early events in the activation of plant defense responses. Annu Rev Phytopath. 1994;32:479–501. [Google Scholar]
- Dixon RA. The phytoalexin response: elicitation, signaling, and control of host gene expression. Biol Rev Camb Philos Soc. 1986;61:192–239. [Google Scholar]
- Dixon RA, Paiva N. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK. Co-ordinate genetic regulation of flavonoid biosynthetic enzymes in maize. Genetics. 1983;91:309–315. [Google Scholar]
- Dröge LW, Kaiser A, Lindsay WP, Halkier BA, Loake GJ, Doerner P, Dixon RA, Lamb C. Rapid stimulation of a soybean protein-serine kinase that phosphorylates a novel bZIP DNA-binding protein, G/HBF-1, during the induction of early transcription-dependent defenses. EMBO J. 1997;16:726–738. doi: 10.1093/emboj/16.4.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron M, Clouse SD, Dixon RA, Lawton MA, Lamb CJ. Glutathione and fungal elicitor regulation of a plant defense gene promoter in electroporated protoplasts. Proc Natl Acad Sci USA. 1988;85(18):6738–6742. doi: 10.1073/pnas.85.18.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin ML, McCaig B, Clegg MT. Molecular evolution of the chalcone synthase multigene family in the morning glory genome. Plant Mol Biol. 2000;42:79–92. [PubMed] [Google Scholar]
- Edwards ML, Stemerick DM, Sunkara PS. Chalcones: a new class of antimitotic agents. J Med Chem. 1990;33:1948–1954. doi: 10.1021/jm00169a021. [DOI] [PubMed] [Google Scholar]
- Ehmann B, Ocker B, Schafer E. Development and light dependent regulation of the expression of two different chalcone synthase transcripts in mustard cotyledons. Planta. 1991;183:416–422. doi: 10.1007/BF00197741. [DOI] [PubMed] [Google Scholar]
- Elomaa P, Helariutta Y, Kotilainen M, Teeri TH. Transformation of antisense constructs of the chalcone synthase gene superfamily into Gerbera hybrida: differential effect on the expression of family members. Mol Breed. 1996;2:41–50. [Google Scholar]
- Estabrook EM, Sengupta GC. Differential expression of phenylalanine ammonia-lyase and chalcone synthase during soybean nodule development. Plant Cell. 1991;3:299–308. doi: 10.1105/tpc.3.3.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faktor O, Loake G, Dixon RA, Lamb CJ. The G-box and H-box in a 39 bp region of a French bean chalcone synthase promoter constitute a tissue-specific regulatory element. Plant J. 1997;11:1105–1113. [Google Scholar]
- Faulkner IJ, Rubery PH. Flavonoids and flavonoid sulphates as probes of auxin-transport regulation in Cucurbita pepo hypocotyl segments and vesicles. Planta. 1992;186:618–625. doi: 10.1007/BF00198044. [DOI] [PubMed] [Google Scholar]
- Feinbaum RL, Ausubel FM. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol. 1988;8:1985–1992. doi: 10.1128/mcb.8.5.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum RL, Storz G, Ausubel FM. High intensity and blue light regulated expression of chimeric chalcone synthase genes in transgenic Arabidopsis thaliana plants. Mol Gen Genet. 1991;226:449–456. doi: 10.1007/BF00260658. [DOI] [PubMed] [Google Scholar]
- Feldbrügge M, Sprenger M, Hahlbrock K, Weisshaar B. PcMYB1, a novel plant protein containing a DNA-binding domain with one MYB repeat, interacts in vivo with a light-regulatory promoter unit. Plant J. 1997;11:1079–1093. doi: 10.1046/j.1365-313x.1997.11051079.x. [DOI] [PubMed] [Google Scholar]
- Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- Feucht W, Treutter D, Polster J. Flavanol binding of nuclei from tree species. Plant Cell Rep. 2004;22:430–436. doi: 10.1007/s00299-003-0705-7. [DOI] [PubMed] [Google Scholar]
- Fritze K, Staiger D, Czaja I, Walden R, Schell J, Wing D. Developmental and UV light regulation of the snapdragon chalcone synthase promoter. Plant Cell. 1991;3:893–905. doi: 10.1105/tpc.3.9.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglevand G, Jackson JA, Jenkins GI. UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell. 1996;8:2347–2357. doi: 10.1105/tpc.8.12.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furner IJ, Sheikh MA, Collett CE. Gene silencing and homology-dependent gene silencing in Arabidopsis: genetic modifiers and DNA methylation. Genetics. 1998;149:651–662. doi: 10.1093/genetics/149.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehm BD, McAndrews JM, Chien PY, Jameson JL. Resveratrol, a polyphenolic compound found in grapes and wine, is an agonist for the estrogen receptor. Proc Natl Acad Sci USA. 1997;94:14138–14143. doi: 10.1073/pnas.94.25.14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gläßgen WE, Rose A, Madlung J, Koch W, Gleitz J, Seitz HU. Regulation of enzymes involved in anthocyanin biosynthesis in carrot cell cultures in response to treatment with ultraviolet light and fungal elicitors. Planta. 1998;204:490–498. doi: 10.1007/s004250050283. [DOI] [PubMed] [Google Scholar]
- Grandmaison J, Ibrahim RK. Evidence for nuclear protein binding of flavonol sulfate esters in Flaveria chloraefolia. J Plant Physiol. 1996;147:653–660. [Google Scholar]
- Guetsky R, Kobiler I, Wang X, Perlman N, Gollop N, Avila-Quezada G, Hadar I, Prusky D. Metabolism of the flavonoid epicatechin by laccase of Colletotrichum gloeosporioides and its effect on pathogenicity on avocado fruits. Phytopathology. 2005;95:1341–1348. doi: 10.1094/PHYTO-95-1341. [DOI] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:347–369. [Google Scholar]
- Hahlbrock K, Zilg H, Grisebach H. Stereochemistry of the enzymatic cyclisation of 4,2′, 4′-trihydroxychalcone to 7,4′-dihydroxyflavanone by isomerases from Mung Bean seedlings. Eur J Biochem. 1970;15:13–18. doi: 10.1111/j.1432-1033.1970.tb00969.x. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat Rev Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Harborne JB, Grayer RJ (1994) Flavonoids and insects. In: Harborne JB (ed) The flavonoids, advances in research since 1986. Chapman & Hall, London, pp 589–618
- Harker CL, Ellis THN, Coen ES. Identification and genetic regulation of the chalcone synthase multigene family in pea. Plant Cell. 1990;2:185–194. doi: 10.1105/tpc.2.3.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ, Dixon RA. Isoflavonoid accumulation and expression of defense gene transcripts during the establishment of vesicular-arbuscular mycorrhizal associations in roots of Medicago truncatula. Mol Plant Microb Interact. 1993;5:643–654. [Google Scholar]
- Harrison MJ, Lawton MA, Lamb CJ, Dixon RA. Characterization of a nuclear protein that binds to three elements within the silencer region of a bean chalcone synthase gene promoter. Proc Natl Acad Sci USA. 1991;88:2515–2519. doi: 10.1073/pnas.88.6.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Valentine WJ, Christie JM, Hays J, Jenkins GI, Weisshaar B. Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol Biol. 1998;36:741–754. doi: 10.1023/a:1005921914384. [DOI] [PubMed] [Google Scholar]
- Haussuehl KK, Rohde W, Weissenboeck G. Expression of chalcone synthase genes in coleoptiles and primary leaves of Secalecereale L. after induction by UV radiation: evidence for a UV protective role of the coleoptiles. Bot Acta. 1996;109:229–238. [Google Scholar]
- Helariutta Y, Kotilainen M, Elomaa P, Kalkkinen N, Bremer K, Teeri TH, Albert VA. Duplication and functional divergence in the chalcone synthase gene family of Asteraceae: evolution with substrate change and catalytic simplification. Proc Natl Acad Sci USA. 1996;93:9033–9038. doi: 10.1073/pnas.93.17.9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderer W, Seitz HU. Chalcone synthase from cell suspension cultures of Daucus carota L. Arch Biochem Biophys. 1985;240:265–272. doi: 10.1016/0003-9861(85)90032-3. [DOI] [PubMed] [Google Scholar]
- Hipskind J, Hanau R, Leite B, Nicholson RL. Phytoalexin synthesis in sorghum: identification of an apigeninidin acyl ester. Physiol Mol Plant Pathol. 1990;36:381–396. [Google Scholar]
- Hrazdina G, Jensen RA. Spatial organization of enzymes in plant metabolic pathways. Ann Rev Plant Physiol Plant Mol Biol. 1992;43:241–267. [Google Scholar]
- Hrazdina G, Kreuzaler F, Hahlbrock K, Grisebach H. Substrate specificity of flavanone synthase from cell suspension cultures of parsley and structure of release products in vitro. Arch Biochem Biophys. 1976;175:392–399. doi: 10.1016/0003-9861(76)90526-9. [DOI] [PubMed] [Google Scholar]
- Hrazdina G, Wagner GJ. Metabolic pathways as enzyme complexes: evidence for the synthesis of phenylpropanoids and flavonoids on membrane associated enzyme complexes. Annu Proc Phytochem Soc Eur. 1985;25:120–133. doi: 10.1016/0003-9861(85)90257-7. [DOI] [PubMed] [Google Scholar]
- Hutzler P, Rischbach R, Heller W, Jungblut TP, Reuber S, Schmitz R, Veit M, Weissenbck G, Schmitzler JP. Tissue localization of phenolic compounds in plants by confocal laser scanning microscopy. J Exp Bot. 1998;49:953–965. [Google Scholar]
- Ingelbrecht I, Van Houdt H, Van Montagu M, Depicker A. Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc Natl Acad Sci USA. 1994;91:10502–10506. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham JL. Induced and constitutive isoflavonoids from stems of chickpeas (Cicer arietinum L.) inoculated with spores of Helminthosporiumcarbonum Ullstrup. J Phytopathol. 1976;87:353–367. [Google Scholar]
- Ito M, Ichinose Y, Kato H, Shiraishi T, Yamada T. Molecular evolution and functional relevance of the chalcone synthase genes of pea. Mol Gen Genet. 1997;255:28–37. doi: 10.1007/s004380050471. [DOI] [PubMed] [Google Scholar]
- Jacobs M, Rubery PH. Naturally occurring auxin transport regulators. Science. 1988;241:346–349. doi: 10.1126/science.241.4863.346. [DOI] [PubMed] [Google Scholar]
- Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HHS, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, Pezzuto JM. Cancer chemo preventive activity of resveratrol, a natural product derived from Grape. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- Jez JM, Austin MB, Ferrer JL, Bowman ME, Schröder J, Noel JP. Structural control of polyketide formation in plant-specific polyketide synthases. Chem Biol. 2000;40:1–12. doi: 10.1016/s1074-5521(00)00041-7. [DOI] [PubMed] [Google Scholar]
- Jez JM, Bowman ME, Noel JP. Structure-guided programming of polyketide chain-length determination in chalcone synthase. Biochemistry. 2001;40:14829–14838. doi: 10.1021/bi015621z. [DOI] [PubMed] [Google Scholar]
- Jez JM, Ferrer JL, Bowman ME, Austin MB, Schröder J, Dixon RA, Noel JP. Structure and mechanism of chalcone synthase-like polyketide synthases. J Ind Microbiol Biotechnol. 2001;27:393–398. doi: 10.1038/sj.jim.7000188. [DOI] [PubMed] [Google Scholar]
- Johnson G, Maag DD, Johnson DK, Thomas RD. The possible role of phytoalexins in the resistance of sugarbeet (Beta vulgaris) to Cercospora beticola [Fungal pathogens] Physiol Plant Path. 1976;8:225–230. [Google Scholar]
- Jorgensen RA. Cosuppression, flower color patterns, and metastable gene expression states. Science. 1995;268:686–691. doi: 10.1126/science.268.5211.686. [DOI] [PubMed] [Google Scholar]
- Junghans H, Dalkin K, Dixon RA. Stress responses in alfalfa (Medicago sativa L.). 15. Characterization and expression patterns of members of a subset of the chalcone synthase multigene family. Plant Mol Biol. 1993;22:239–253. doi: 10.1007/BF00014932. [DOI] [PubMed] [Google Scholar]
- Kaulen H, Schell J, Kreuzaler F. Light-induced expression of the chimeric chalcone synthase-NPTII gene in tobacco cells. EMBO J. 1986;5:1–8. doi: 10.1002/j.1460-2075.1986.tb04169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama O, Miyakawa J, Akatsuka T, Kiyosawa S. Sakuranetin, a flavanone phytoalexin from ultraviolet-irradiated rice leaves. Phytochemistry. 1992;31:3807–3809. [Google Scholar]
- Koes RE, Spelt CE, Mol JNM. The chalcone synthase multigene family of Petunia hybrida (V30): differential, light-regulated expression during flower development and UV light induction. Plant Mol Biol. 1989;12:213–225. doi: 10.1007/BF00020506. [DOI] [PubMed] [Google Scholar]
- Koes RE, Spelt CE, Mol JNM, Gerats AGM. The chalcone synthase multigene family of Petunia hybrida: sequence homology, chromosomal localization and evolutionary aspects. Plant Mol Biol. 1987;10:159–169. doi: 10.1007/BF00016153. [DOI] [PubMed] [Google Scholar]
- Kong JM, Chia LS, Goh NK, Chia TF, Brouillard R. Analysis and biological activities of anthocyanins. Phytochemistry. 2003;64:923–933. doi: 10.1016/s0031-9422(03)00438-2. [DOI] [PubMed] [Google Scholar]
- Kreuzaler F, Hahlbrock K. Enzymatic synthesis of an aromatic ring from acetate units. Partia1 purification and some properties of flavanone synthase from cell-suspension cultures of Petroselinum hortense. Eur J Biochem. 1975;56:205–213. doi: 10.1111/j.1432-1033.1975.tb02223.x. [DOI] [PubMed] [Google Scholar]
- Kreuzaler F, Ragg H, Fautz E, Kuhn DN, Hahlbrock K. UV-induction of chalcone synthase mRNA in cell suspension cultures of Petroselinum hortense. Proc Natl Acad Sci USA. 1983;80:2591–2593. doi: 10.1073/pnas.80.9.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn DN. Plant stress responses: discussion of models for race-specific resistance. Recent Adv Phytochem. 1988;22:127–161. [Google Scholar]
- Kuras M, Stefanowska-Wronka M, Lynch JM, Zobel AM. Cytochemical localization of phenolic compounds in columella cells of the root cap in seeds of Brassica napus changes in the localization of phenolic compounds during germination. Ann Bot. 1999;84:135–143. [Google Scholar]
- Langcake P, Pryce RJ. A new class of phytoalexins from grapevines. Experientia. 1977;33:151–152. doi: 10.1007/BF02124034. [DOI] [PubMed] [Google Scholar]
- Langcake P, Pryce RJ. The production of resveratrol and the viniferins by grapevines in response to ultraviolet irradiation. Phytochemistry. 1977;16:1193–1196. [Google Scholar]
- Lanz T, Tropf S, Marner FJ, Schröder J, Schröder G. The role of cysteines in polyketide synthases: site-directed mutagenesis of resveratrol and chalcone synthases, two key enzymes in different plant-specific pathways. J Biol Chem. 1991;266:9971–9976. [PubMed] [Google Scholar]
- Lawton MA, Clouse SD, Lamb CJ. Glutathione-elicited changes in chromatin structure within the promoter of the defense gene chalcone synthase. Plant Cell Rep. 1990;8:561–564. doi: 10.1007/BF00820210. [DOI] [PubMed] [Google Scholar]
- Le Gall G, Metzdorff SB, Pedersen J, Bennett RN, Colquhoun IJ. Metabolite profiling of Arabidopsis thaliana (L.) plants transformed with an antisense chalcone synthase gene. Metabolomics. 2005;1:181–198. [Google Scholar]
- Leyva A, Jarillo TA, Salinas J, Martinez-Zapater JM. Low temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiol. 1995;108:39–46. doi: 10.1104/pp.108.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RS, Kenyon GL, Cohen FE, Chen XW, Gong BQ, Dominquez JN, Davidson E, Kurzban G, Miller RE, Nuzum EO, Rosenthal PJ, McKerrow JH. In vitro antimalarial activity of chalcones and their derivatives. J Med Chem. 1995;38:5031–5037. doi: 10.1021/jm00026a010. [DOI] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UV-B radiation. Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipphardt S, Brettschneider R, Kreuzaler F, Schell J, Dangl JL. UV-inducible transient expression in parsley protoplasts identifies regulatory cis-elements of a chimeric Antirrhinum majus chalcone synthase gene. EMBO J. 1988;7:4027–4033. doi: 10.1002/j.1460-2075.1988.tb03296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue WL, Kuhn D, Nicholson RL. Chalcone synthase activity in sorghum mesocotyls inoculated with Colletotrichumgraminicola. Physiol Mol Plant Pathol. 1989;35:413–422. [Google Scholar]
- Markham KR (1988) Distribution of flavonoids in the lower plants and its evolutionary significance. In: Harborne JB (ed) The flavonoids advance in research since 1980. Chapman and Hall, London, pp 427–468
- Marles MA, Ray H, Gruber MY. New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry. 2003;64:367–383. doi: 10.1016/s0031-9422(03)00377-7. [DOI] [PubMed] [Google Scholar]
- Martin CR. Structure, function and regulation of the chalcone synthase. Int Rev Cytol. 1993;147:233–284. doi: 10.1016/s0074-7696(08)60770-6. [DOI] [PubMed] [Google Scholar]
- Matthews DE, Matthews PS. Phytoalexin detoxification: Importance for pathogen city and practical implications. Annu Rev Phytopath. 1989;27:143–164. doi: 10.1146/annurev.py.27.090189.001043. [DOI] [PubMed] [Google Scholar]
- Mol NJJ, Schram AW, de Vlaming P, Gerats AGM, Kreuzaler F, Hahlbrock K, Reif HJ, Veltkamp E. Regulation of flavonoid gene expression in Petunia hybrida: description and partial characterization of a conditional mutant in chalcone synthase gene expression. Mol Gen Genet. 1983;192:424–429. [Google Scholar]
- Monache GD, Botta B, Vinciguerra V, de Mello JF, Andrade Chiapetta de A. Antimicrobial isoflavanones from Desmodium canum. Phytochemistry. 1996;41:537–544. doi: 10.1016/0031-9422(95)00653-2. [DOI] [PubMed] [Google Scholar]
- Morita H, Takahashi Y, Noguchi H, Abe I. Enzymatic formation of unnnatural aromatic polyketides by chalcone synthase. Biochem Biophys Res Commun. 2000;279:190–195. doi: 10.1006/bbrc.2000.3920. [DOI] [PubMed] [Google Scholar]
- Morkunas I, Marczak L, Stachowiak J, Stobiecki M. Sucrose-induced lupine defense against Fusarium oxysporum: Sucrose-stimulated accumulation of isoflavonoids as a defense response of lupine to Fusarium oxysporum. Plant Physiol Biochem. 2005;43:363–373. doi: 10.1016/j.plaphy.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Nagy NE, Fossdal CG, Krokene P, Krekling T, Lonneborg A, Solheim H. Induced responses to pathogen infection in Norway spruce phloem: changes in polyphenolic parenchyma cells, chalcone synthase transcript levels and peroxidase activity. Tree Physiol. 2004;24:505–515. doi: 10.1093/treephys/24.5.505. [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen RA. Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson RL, Kollipara SS, Vincent JR, Lyons PC, Cadena-Gomez G. Phytoalexin synthesis by the sorghum mesocotyl in response to infection by pathogenic and nonpathogenic fungi. Proc Natl Acad Sci USA. 1987;84:5520–5524. doi: 10.1073/pnas.84.16.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Krofta K, Matoušek J. Chalcone synthase homologues from Humulus lupulus: some enzymatic properties and expression. Biol Plant. 2006;50:48–54. [Google Scholar]
- Ohl S, Hahlbrock K, Schafer E. A stable blue-lightderived signal modulates ultraviolet-light-induced activation of the chalcone-synthase gene in cultured parsley cells. Planta. 1989;177:228–236. doi: 10.1007/BF00392811. [DOI] [PubMed] [Google Scholar]
- Pare PW, Mischek CF, Edwards R, Dixon RA, Norman HA, Mabry TJ. Induction of phenylpropanoid pathway enzymes in elicitro-treated cultures of Cephalocereus senilis. Phytochemistry. 1992;31:149–153. [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS. Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol. 2001;126:536–548. doi: 10.1104/pp.126.2.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Schneider-Poetsch HJ, Schwarz H, Weissenböck G. Biochemical and immunological characterization of chalcone synthase from rye leaves. J Plant Physiol. 1988;133:178–182. [Google Scholar]
- Richard S, Lapointe G, Rutledge RG, Seguin A. Induction of chalcone synthase expression in white spruce by wounding and jasmonate. Plant Cell Physiol. 2000;41:982–987. doi: 10.1093/pcp/pcd017. [DOI] [PubMed] [Google Scholar]
- Ryan C. Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu Rev Phytopathol. 1990;28:425–449. [Google Scholar]
- Ryder TB, Cramer CL, Bell JN, Robbins MP, Dixon RA, Lamb CJ. Elicitor rapidly induces chalcone synthase mRNA in Phaseolus vulgaris cells at the onset of the phytoalexin defense response. Proc Natl Acad Sci USA. 1984;81:5724–5728. doi: 10.1073/pnas.81.18.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder TB, Hedrick SA, Bell JN, Liang X, Clouse SD, Lamb CJ. Organization and differential activation of a gene family encoding the plant defense enzyme chalcone synthase in Phaseolus vulgaris. Mol Gen Genet. 1987;210:219–233. doi: 10.1007/BF00325687. [DOI] [PubMed] [Google Scholar]
- Sablowski R, Moyano E, Culianez-Macia F, Schuch W, Martin C, Bevan M. A flower-specific Myb protein activates transcription of phenylpropanoid synthetic genes. EMBO J. 1994;13:128–137. doi: 10.1002/j.1460-2075.1994.tb06242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez IJF (2008) Polyketide synthase in Cannabis sativa L. PhD thesis, Leiden University, Leiden, The Netherlands
- Saslowsky D, Winkel SB. Localization of flavonoid enzymes in Arabidopsis roots. Plant J. 2001;27:37–48. doi: 10.1046/j.1365-313x.2001.01073.x. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer E, Jahnen W, Hahlbrock K. In situ localization of light-induced chalcone synthase mRNA, chalcone synthase, and flavonoid end products in epidermal cells of parsley leaves. Proc Natl Acad Sci USA. 1988;85:2989–2993. doi: 10.1073/pnas.85.9.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzler JP, Jungblut TP, Heller W, Hutzler P, Heinzmann U, Schmelzer E, Ernst D, Langebartels C, Sandermann H. Tissue localisation of UV-B screening pigments and chalcone synthase mRNA in Scots pine (Pinus sylvestris L.) needles. New Phythol. 1996;132:247–258. [Google Scholar]
- Schröder J. A family of plant—specific polyketide synthases: facts and predictions. Trends Plant Sci. 1997;2:373–378. [Google Scholar]
- Schröder J (2008) http://www.biologie.unifreiburg.de/data/bio2/schroeder/CHS_Reaction_Mechanism.html
- Schröder J, Schäafer E. Radioiodinated antibodies, a tool in studies on the presence and role of inactive enzyme forms: regulation of chalcone synthase in parsley cell suspension cultures. Arch Biochem Biophys. 1980;203:800–808. doi: 10.1016/0003-9861(80)90241-6. [DOI] [PubMed] [Google Scholar]
- Schuez R, Heller W, Hahlbrock K. Substrate specificity of chalcone synthase from Petroselinum hortense. J Biol Chem. 1983;258:6730–6734. [PubMed] [Google Scholar]
- Schulze LP, Becker AM, Schulr W, Hahlbrock K, Dangl JL. Functional architecture of the light-responsive chalcone synthase promoter from parsley. Plant Cell. 1989;1:707–714. doi: 10.1105/tpc.1.7.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvarts M, Borochov A, Weiss D. Low temperature enhances petunia flower pigmentation and induces chalcone synthase gene expression. Physiol Plant. 1997;99:67–72. [Google Scholar]
- Soylu S. Accumulation of cell-wall bound phenolic compounds and phytoalexin in Arabidopsis thaliana leaves following inoculation with pathovars of Pseudomonas syringae. Plant Sci. 2006;170:942–952. [Google Scholar]
- Shirley BW. Flavonoid biosynthesis: “new” functions for an “old” pathway. Trends Plant Sci. 1996;1:377–382. [Google Scholar]
- Staiger D, Kaulen H, Schell J (1989) A CACGTG motif of the Antirrhinum majus chalcone synthase promoter is recognised by an evolutionary conserved nuclear protein. Proc Natl Acad Sci USA 86:6930–6934 [DOI] [PMC free article] [PubMed]
- Taylor LP, Briggs WR. Genetic regulation and photocontrol of anthocyanin accumulation in maize seedlings. Plant Cell. 1990;2:115–127. doi: 10.1105/tpc.2.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ. The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol. 2002;130:102–110. doi: 10.1104/pp.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Wan SB, Pan QH, Zheng YJ, Huang WD. A novel plastid localization of chalcone synthase in developing grape berry. Plant Sci. 2008;175:431–436. [Google Scholar]
- Tropf S, Kärcher B, Schröder G, Schröder J. Reaction mechanisms of homodimeric plant polyketide synthase (stilbenes and chalcone synthase). A single active site for the condensing reaction is sufficient for synthesis of stilbenes, chalcones, and 6′-deoxychalcones. J Biol Chem. 1995;270(14):7922–7928. doi: 10.1074/jbc.270.14.7922. [DOI] [PubMed] [Google Scholar]
- Tropf S, Lanz T, Rensing SA, Schröder J, Schröder G. Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution. J Mol Evol. 1994;38:610–618. doi: 10.1007/BF00175881. [DOI] [PubMed] [Google Scholar]
- Van der Krol AR, Mur LA, Beld M, MoI JNM, Stuitje AR. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Krol AR, Mur LA, de Lange P, Gerats AGM, MOI JNM, Stuitje AR. Antisense chalcone synthase genes in petunia: visualization of variable of transgene expression. Mol Gen Genet. 1990;220:204–212. [Google Scholar]
- Van der Krol AR, Lenting PE, Veenstra J, van der Meer IM, Koes RE, Gerats AGM, MOI JNM, Stuitje AR. An anti-sense chalcone synthase gene in transgenic plants inhibits flower pigmentation. Nature. 1988;333:866–869. [Google Scholar]
- Van der Meer IM, Spelt CE, Mol JNM, Stuitje AR. Promoter analysis of the chalcone synthase (chsA) gene of Petunia hybrida: a 67 bp promoter region directs flower-specific expression. Plant Mol Biol. 1990;15:95–109. doi: 10.1007/BF00017727. [DOI] [PubMed] [Google Scholar]
- Van der Meer IM, Stuitje AR, Mol JNM. Regulation of general phenylpropanoid and flavonoid gene expression. In: Verma DPS, editor. Control of plant gene expression. Boca Raton: CRC Press; 1993. pp. 125–155. [Google Scholar]
- Van Etten HD, Pueppke SG. Isoflavonoid phytoalexins. In: Friend J, Threlfall DR, editors. Biochemical aspects of plant–parasite relationships. New York: Academic Press; 1976. pp. 239–289. [Google Scholar]
- Van Etten HD, Sandrock RW, Wasmann CC, Soby SD, McCluskey K, Wang P (1995) Detoxification of phytoanticipins and phytoalexins by phytopathogenic fungi. Can J Bot 73:518–525
- Van Tunen AJ, Koes RE, Spelt CE, van der Krol AR, Stuitje AR, Mol JNM. Cloning of the two chalcone flavanone isomerase genes from Petunia hybfida: coordinate, light-regulated, and differential expression of flavonoid genes. EMBO J. 1988;7:1257–1263. doi: 10.1002/j.1460-2075.1988.tb02939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade HK, Bibikova TN, Valentine WJ, Jenkins GI. Interactions within a network of phytochrome, cryptochrome and UV-B phototransduction pathways regulate chalcone synthase gene expression in Arabidopsis leaf tissue. Plant J. 2001;25:675–685. doi: 10.1046/j.1365-313x.2001.01001.x. [DOI] [PubMed] [Google Scholar]
- Wang CK, Chen PY, Wang HM, To KY. Cosuppression of tobacco chalcone synthase using Petunia chalcone synthase construct results in white flowers. Bot Stud. 2006;47:71–82. [Google Scholar]
- Wegulo SN, Yang XB, Martinson CA, Murphy PA. Effects of wounding and inoculation with Sclerotinia sclerotiorum on isoflavones concentrations in soybean. Can J Plant Sci. 2005;85:749–760. [Google Scholar]
- Weisshaar B, Armstrong GA, Block A, e Silva O, Hahlbrock K. Light inducible and constitutively expressed DNAbinding proteins recognising a plant promoter element with functional relevance in light responsiveness. EMBO J. 1991;10:1777–1786. doi: 10.1002/j.1460-2075.1991.tb07702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead JM, Dixon RA (1983) Chalcone synthase from cell suspension cultures of Phaseolus vulgaris. Biochem Biophys Acta 747:298–303
- Wingender R, Röhrig H, Höricke C, Wing D, Schell J. Differential regulation of soybean chalcone synthase genes in plant defence, symbiosis and upon environmental stimuli. Mol Gen Genet. 1989;218:315–322. doi: 10.1007/BF00331284. [DOI] [PubMed] [Google Scholar]
- Winkel SB (1999) Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol Plant 107:142–149
- Wrolstad RE. Anthocyanins. In: Francis FJ, Lauro GJ, editors. Natural food colorants. New York: Marcel Dekker; 2000. pp. 237–252. [Google Scholar]
- Xu Y, Chang PFL, Liu D, Narasimhan ML, Raghothama KG, Hasegawa PM, Bressan RA. Plant defence genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell. 1994;6:1077–1085. doi: 10.1105/tpc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Kurosaki F, Suh DY, Sankawa U, Nishioka M, Akiyama T, Shibuya M, Ebizuka Y. Cross-reaction of chalcone synthase and stilbene synthase overexpressed in Escherichia coli. FEBS Lett. 1999;460:457–461. doi: 10.1016/s0014-5793(99)01403-9. [DOI] [PubMed] [Google Scholar]
- Yang WC, Canter CHCJ, Hogendijk P, Katinakis P, Wijffelman CA, Franssen H, van Kammen A, Bisseling T. In situ localization of chalcone synthase mRNA in pea root nodule development. Plant J. 1992;2:143–152. [Google Scholar]
- Yilmaz Y, Toledo RT. Major flavonoids in grape seeds and skins: antioxidant capacity of catechin, epicatechin, and gallic acid. J Agric Food Chem. 2004;52:255–260. doi: 10.1021/jf030117h. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Wakamatsu S, Sakuta M. Characterization of SBZ1, a soybean bZIP protein that binds to the chalcone synthase gene promoter. Plant Biotechnol. 2008;25:131–140. [Google Scholar]
- Yu LM, Lamb CJ, Dixon RA. Purification and biochemical characterization of proteins which bind to the H-box cis-element implicated in transcriptional activation of plant defense genes. Plant J. 1993;3:805–816. doi: 10.1111/j.1365-313x.1993.00805.x. [DOI] [PubMed] [Google Scholar]
- Zhou B, Li Y, Xu Z, Yan H, Homma S, Kawabata S. Ultraviolet A-specific induction of anthocyanin biosynthesis in the swollen hypocotyls of turnip (Brassica rapa) J Exp Bot. 2007;58:1771–1781. doi: 10.1093/jxb/erm036. [DOI] [PubMed] [Google Scholar]
- Zufall R, Rausher MD (2001) Diffuse coevolution and anthocyanin production. Botany 2001: “plants and people”, Albuquerque, New Mexico
- Zuurbier KWM, Lesser J, Berger T, Hofte AJP, Schröder G, Verpoorte R, Schröder J (1998) 4-hydroxy-2- pyrone formation by chalcone synthase and stylbene synthase with nonphysiological substrates. Phytochemistry 49:1945–1951 [DOI] [PubMed]
- Zwaagstra ME, Timmerman H, Tamura M, Tohma T, Wada Y, Onogi K, Zhang MQ. Synthesis and structure-activity relationships of carboxylated chalcones: a novel series of CysLT1 (LTD4) receptor antagonists. J Med Chem. 1997;40:1075–1089. doi: 10.1021/jm960628d. [DOI] [PubMed] [Google Scholar]