SUMMARY
Epidemiologic and clinical evidence points to an increased risk of cancer when coupled with chronic inflammation. However, the molecular mechanisms that underpin this interrelationship remain largely unresolved. Herein we show that the inflammation-derived cholesterol 5,6-secosterol aldehydes, atheronal-A (KA) and –B (ALD), but not the PUFA-derived aldehydes 4-hydroxynonenal (HNE) and 4-hydroxyhexenal (HHE), induce misfolding of wild-type p53 into an amyloidogenic form that binds thioflavin T and Congo Red dyes but cannot bind to a consensus DNA sequence. Treatment of lung carcinoma cells with KA and ALD leads to a loss of function of extracted p53, as determined by analysis of extracted nuclear protein and in activation of p21. Our results uncover a plausible chemical link between inflammation and cancer and expands the already pivotal role of p53 dysfunction and cancer risk.
INTRODUCTION
Inflammation, which functions at all three stages of tumour development: initiation, progression and metastasis, is a host’s response to acute tissue damage (Balkwill and Mantovani, 2001, Coussens and Werb, 2002, Lu et al., 2006). It is generally considered to play a role in tumour suppression by triggering an anti-tumour immune response (Philip et al., 2004), but can also stimulate tumour development (Balkwill and Mantovani, 2001, Philip, et al., 2004). In this regard, many chronic inflammatory conditions have associated neoplastic risk, for example the inflammatory bowel diseases, ulcerative colitis and Crohn’s disease, are increased risk factors for colorectal carcinomas (Coussens and Werb, 2002). Similarly, chronic lung inflammation or infection has been associated with the development of lung cancer (Borm and Driscoll, 1996, Keeley and Rees, 1997). Basic research has shown that chronic inflammation appears to contribute to tumour progression by establishing a long-term milieu conducive to neoplasia (Coussens and Werb, 2002). However, the precise mechanisms by which inflammation achieves this remain to be determined.
The tumour suppressor protein p53 is a sequence-specific transcription factor that functions to maintain the integrity of the genome (Lane, 1992). On its induction/activation in response to DNA damage, p53 promotes cell cycle arrest in G1 phase (Kastan et al., 1991) and apoptosis if DNA repair is not possible. When p53 is inactivated, most commonly by a single missense mutation or by binding of viral oncoproteins to p53, cell growth can proceed without regulation, leading to tumour growth (Vogelstein and Kinzler, 1992). The critical nature of maintaining functional p53 within the cell is emphasized by the fact that mutations in the P53 gene are detected in ~50 % of human cancers (Hollstein et al., 1991) and that 90 % of these mutations are in the core domain that is responsible for DNA binding (Hernandez-Boussard et al., 1999). In addition to mutant p53 being inactive, several types of malignant and pre-malignant tissues harbour a genetically wild-type, but transcriptionally inactive form of p53, often localized in their cytoplasm, hinting at epigenetic environmental triggers for p53 inactivation (Bosari et al., 1995, Isaacs et al., 1998, Moll et al., 1995, Moll et al., 1992, Tominaga et al., 1993).
Recently we and others have discovered a process that is being studied in the context of a number of disease-related sporadic amyloidoses. We have shown that in vitro, certain inflammatory-derived lipidic aldehydes, when adducted to pro-amyloidogenic proteins in their native state, can induce misfolding and aggregation of native protein sequences (Wentworth et al., 2003, Zhang et al., 2004) (Fig. 1). Thus, we have shown that the cholesterol 5,6-seco-sterols atheronal-A and –B (KA and ALD respectively), which are derived from oxidation of cholesterol by activated leukocytes during chronic inflammation, accelerate the in vitro misfolding of apoB100, the protein component of low-density lipoprotein (LDL), under physiologically-relevant conditions (Fig. 1) (Wentworth, et al., 2003). We and others have also shown that 4-hydroxynonenal (HNE, a major lipid peroxidation product of membrane lipids), KA and ALD accelerate the in vitro amyloidogenesis of β-amyloid peptides (Aβ 1-40 and 1-42) leading to pre-fibrillar assemblies of Aβ(Zhang, et al., 2004) via a process that involves a reversible site-specific modification of Aβ (Scheinost et al., 2008) and that KA and ALD accelerate the aggregation of α-synuclein (Bosco et al., 2006) leading to fibrillar aggregates. In addition, we have shown that antibody light chains can be induced to aggregate by lipid aldehydes, both to form amyloid or amorphous aggregates, in a process dependant upon the specific aldehyde structure (Nieva et al., 2008). Most recently, we have shown that adduction of KA and ALD to a murine prion protein actually stabilizes the non-scrapie form of the prion and prevents/inhibits scrapie formation (Scheinost et al., 2009), showing that aldehyde adduction can be both pro- and anti-amyloidogenic dependant upon which protein and which aldehyde is being studied.
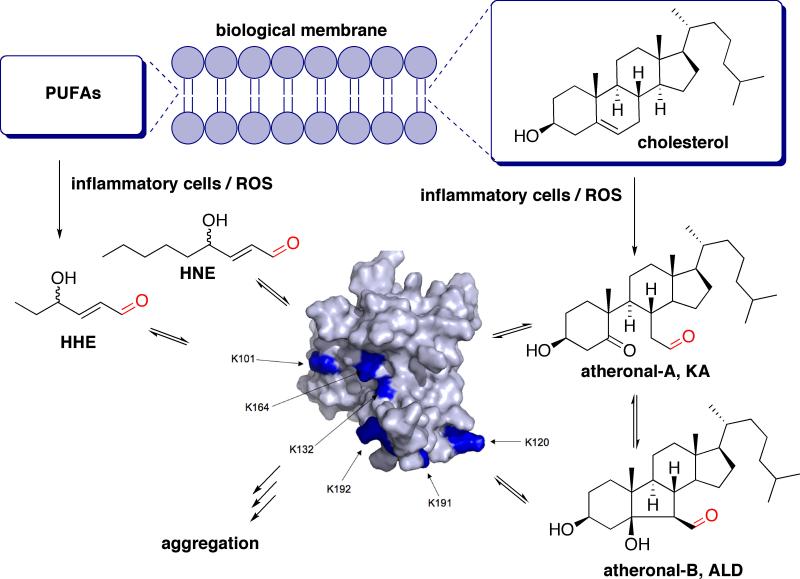
Figure 1. A Model for Lipid-Aldehyde Initiated p53 Inactivation.
Scheme showing the concept of how membrane lipids, upon oxidation by reactive oxygen species (ROS), generate lipid-derived aldehydes such as KA, ALD, HNE and HHE, which then adduct reversibly to lysine residues of cellular proteins, such as p53, which can then lead to misfolding/aggregation and dysfunction. The protein shown is a surface rendering of the p53 tetramerization domain (pdb reference 1tub), with surface lysines highlighted in blue. PUFAs (polyunsaturated fatty acids).
Herein we report that the cholesterol derived aldehydes KA and ALD, but not the PUFA-derived aldehydes HNE and HHE, can induce amyloidogenesis of wild type-p53 in a process that leads to a loss-of-function of p53. Given the known associations between inflammation and cancer, we see this as potential new chemical link between the two syndromes and further highlights the potential for p53 misfolding, dysfunction and cancer risk.
RESULTS AND DISCUSSION
Lipid Derived Aldehydes Adduct to p53 and Induce Misfolding
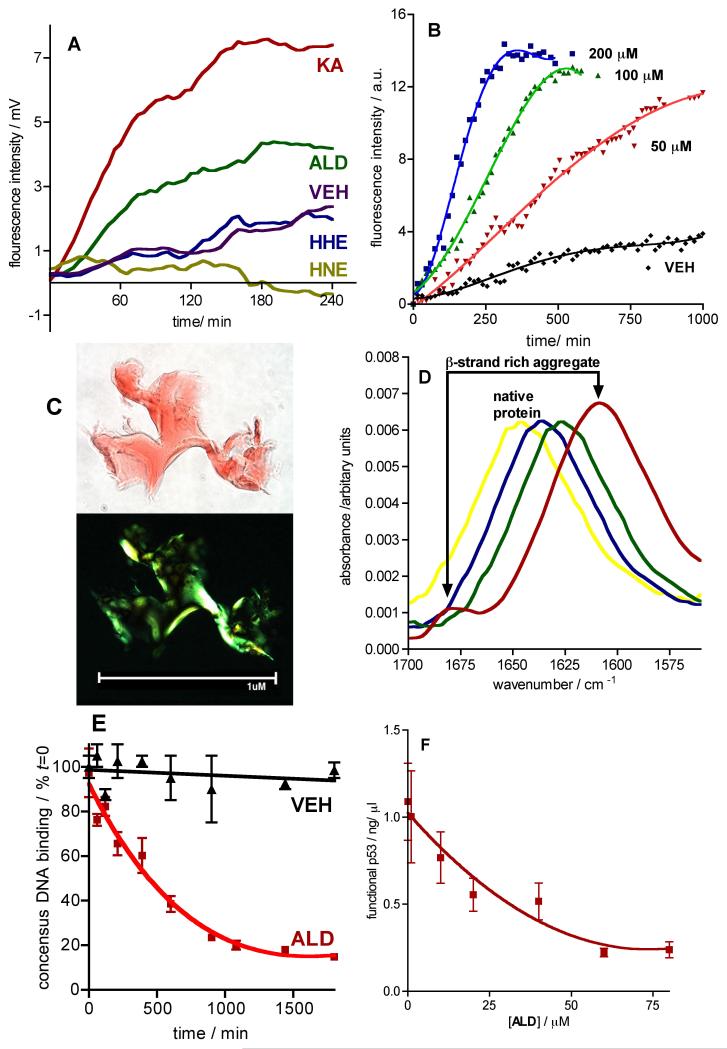
Quiescent incubation of full-length recombinant wild-type hexahistidine-tagged human p53 (His6-p53, 0.8 mg/mL) with the inflammatory aldehydes KA, ALD, HNE and HHE (each at 100 μM) in phosphate buffered saline (PBS, pH 7.4) with ethanol (0.1 % v/v) as a co-solvent, leads to the acceleration in formation of ThT-positive aggregates with the cholesterol seco-sterol aldehydes (time to half maximal fluorescence, KA t50 ~ 80 min; ALD, t50 ~ 100 min) but not with the α,β-unsaturated inflammatory aldehydes HHE and HNE, relative to vehicle (VEH, PBS pH 7.4, containing 0.1 % v/v ethanol) (Fig. 2A). The fluorescence emission of the dye ThT is increased upon binding to protein aggregates with a cross-β-sheet structure (Levine and Ronald, 1999), either fibrillar or non-fibrillar in morphology (Hurshman et al., 2004, Zhang, et al., 2004). The time-course of ThT fluorescence observed for the atheronal-A (KA) and atheronal-B (ALD)-initiated His6-p53 amyloid formation, a rapid rise in ThT fluorescence to a plateau phase with no lag-phase, is indicative of a ‘seeded’ or so-called down-hill polymerization that is thermodynamically favoured from the outset (Ferrone, 1999), and is in-line with what we have observed previously for the atheronal-initiated polymerization of amyloid-β and α-synuclein (Bosco, et al., 2006, Zhang, et al., 2004).
Figure 2. Lipid Aldehydes KA and ALD, but not HNE and HHE, Induce Amyloidogenesis of and Dysfunction of Recombinant His6-p53.
a Graph of ThT fluorescence vs time during quiescent incubation of His6-p53 (0.8 mg/ml) in PBS (pH 7.4) with ethanol (0.1 % v/v) co-solvent in the presence of either KA, ALD, HNE or HHE (each at 100 μM). Data is reported as the mean fluorescence emission (Ex: 440 nm; EM: 485 nm) of duplicate experiments. b Graph of KA concentration-dependent (0-200 μM) quiescent aggregation of His6-p53 (0.8 mg/ml) in PBS (pH 7.4) with ethanol (0.1 % v/v) co-solvent, as measured by ThT fluorescence. Data is reported as the mean fluorescence (Ex: 440 nm; EM: 485 nm) of duplicate experiments. Data is fitted to a third order polynomial using Graphpad Prism v4.0 for MAC. c Optical microscope images (100 x) obtained under normal (upper) and cross-polarized (lower) light of aggregates generated by incubation of His6-p53 (0.8 mg/mL) with KA (25 μM) for 5 h, followed by centrifugation and staining with Congo Red (100 μM). d FTIR spectra at times [0 min (yellow), 300 min (blue), 800 min (green) and 1200 min (red)] during the quiescent aggregation of His6-p53 (0.5 mg/ml) in D2O with KA (100 μM). e Graph of functional p53 (as a percent of t = 0) vs. time during the quiescent aggregation of His6-p53 (0.8 mg/ml) in the presence of ALD (50 μM) or VEH (ethanol 0.1 % v/v). Data are means ± SEM of at least duplicate determinations. Data for VEH is fitted to a linear regression analysis (r2 = 0.992); data for ALD is fitted to a single-phase exponential decay using Graphpad prism v 4.0. f Graph of functional p53 (in ng/ml) vs. [ALD] during a 2 h quiescent aggregation of His6-p53 (0.8 mg/ml) in the presence of ALD (0-80 μM). Data are reported as the mean ± SEM of at least duplicate determinations. Data is fitted to a single-phase exponential decay using Graphpad prism v 4.0. See also Figure S1.
Atheronal-Induced Misfolding of p53 Leads to Amyloid Aggregates
The atheronal-induced aggregation of His6-p53 was studied in some detail, and is concentration-dependent, exhibiting an increase in the initial rate of ThT fluorescence as the ratio of ALD to p53 increases (Fig. 2B). In addition, the plateau phase appears to be independent of ALD concentration, but is reached faster at higher concentrations of lipid aldehyde. Incubation of His6-p53 with KA generates aggregates that exhibit classic apple-green birefringence under polarizing light, when stained with Congo Red dye, further strengthening the notion that incubation with cholesterol seco-sterols generates amyloid fibrillar p53 aggregates (Fig. 2C). In addition TEM analysis of His6-p53 incubated with ALD reveals ribbon-like unbranched fibrillar assemblies (Fig. S1).
To gain insight into the structural transition from native wild-type His6-p53 to its atheronal-A-induced misfolded state we recorded FT-IR spectra during the incubation of His6-p53 with KA (Fig. 2D). In the absence of atheronal-A, the FT-IR of the native structure of His6-p53 is unchanged. However, the FT-IR spectrum of His6-p53 during incubation with atheronal-A (100 μM) at 37 °C in PBS pH 7.4 exhibits a time-dependant shift of the amide-I absorbance band (t = 0 min, λmax−1 = 1647 cm−1; t = 300 min, λmax−1 = 1635 cm−1; t = 800 min, λmax−1 = 1626 cm−1; t = 1200 min, λmax−1 = 1609 cm−1). Thus, the FT-IR spectrum of His6-p53 at t = 0 and 37 °C in PBS is dominated by a strong absorbance at 1647 cm−1, corresponding to α-helices. As quiescent incubation with KA progresses, this absorbance band shifts to lower wave-numbers, consistent with unfolding of α-helices and increased random peptide conformations. After incubation for 1200 min, the absorbance is increased and is centred at 1609 cm−1, characteristic of β-strands in amyloid fibrils. There is also an accompanying weaker absorbance band at 1683 cm−1, also indicative of β-strand containing structures with strong intermolecular hydrogen bonds, classic evidence of amyloid formation.
The process by which amyloid fibrillization occurs from natively folded proteins is constrained by aggregation from partially unfolded intermediates (Uversky and Fink, 2004). Therefore, proteins such as p53 require partial unfolding prior to the onset of fibril formation. To explain the molecular basis of amyloid fibril formation, it has been proposed that fibrillization can occur when the rigid native structure of a protein is destabilized, favoring partial unfolding and culminating in formation of a partially unfolded conformation or intermediate (Dobson, 1999, Kelly, 1998). In the context of p53 very few studies have been performed on full-length protein, however a number of biophysical studies of the p53 tetramerization domain have revealed that at acidic pH (4.0) and 37°C a mutant tetramerization domain p53tet-R337H forms fibrils much faster than p53-tetwt, as monitored by ThT fluorescence. Interestingly, the time-dependent profile of ThT fluorescence p53tet-R33H incubated at pH 4.0 and KA-treated His6-p53 are remarkably similar. Specifically there is no apparent lag phase in the aggregation, with peak ThT fluorescence occurring between 2 and 4 h. The rationale proposed for the atheronal-induced aggregation of p53 in our work therefore is inline with what has been proposed for p53-tet-R337H, which involves partial unfolding following by assembly into proamyloid oligomers and fibrilization.
Atheronal-Misfolded p53 Cannot Bind to DNA
To investigate whether atheronal treatment of His6-p53 leads to a loss of DNA binding, we utilized an ELISA assay that detects p53 when bound to a consensus duplex deoxyoligonucleotide sequence. Thus, the ELISA is a primary readout of DNA-bound p53, and does not detect p53 that is not bound to DNA. Quiescent incubation of His6-p53 (0.8 mg/ml) with ALD (50 μM) in PBS, pH 7.4 leads to a time-dependant loss of DNA binding (Fig. 2E). Thus, incubation of His6-p53 for 30 h with ALD (50 μM), leads to an ~ 85 % drop in DNA-binding ability of p53, as determined by ELISA. Whereas, incubation with VEH, under the same conditions, leads to a drop of < 10 % in DNA-binding. The impact of atheronal incubation on His6-p53 function is also concentration dependent. Incubation of His6-p53 (0.8 mg/ml) for 24 h with ALD (0-80 μM) leads to increased reduction in p53-DNA binding (Fig. 2F).
Interestingly, a comparison of the time-dependent changes induced by incubation of His6-p53 with ALD on ThT fluorescence and DNA binding, reveals that the ThT fluorescence plateau is reached at ~ 240 min (Fig. 2A), whereas the maximum impact on DNA binding is not reached until ~ 1300 min (Fig. 2E). While the reasons for this disparity are unresolved, what is clear is that ThT-fluorescence is a precise measure of amyloid structural changes, whereas the ELISA assay is a read-out for any structural change, amyloid and non-amyloid that impairs DNA binding. Therefore, an explanation of the two sets of data, ThT-fluorescence and DNA-binding, is that ALD induces amyloid changes in p53 structure first followed by, or in concert with, longer duration non-amyloid like changes in p53 and it is the non-amyloid changes that impact p53 DNA binding. This explanation of amyloid and non-amyloid changes in His6-p53 induced by ALD is supported by the fact that the solution turbidity of ALD-treated p53, under conditions in Fig. 2A, is visibly increased after 24 h, whereas at 4 h (where ThT fluorescence has reached a plateau) the solution is not visibly turbid.
Lipid-Derived Aldehydes Induce Misfolding of p53 in Cultured Cells
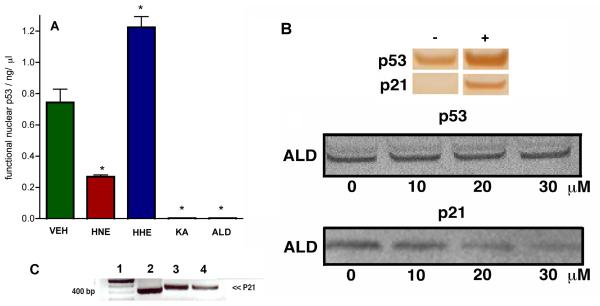
To investigate the effect of lipid aldehydes on p53 in a cell-based system, we treated the lung carcinoma cell line A-427, known to up-regulate wild-type p53 during u.v. or radiation-based stress. Thus, cultured A-427 cells were exposed to γ-irradiation (5000 rads) and then media was replaced with either KA, ALD, HNE, HHE (each at 20 μM) or VEH in media with 0.1 % ethanol (co-solvent). The cells were then incubated at 37 °C for a further 24 h. After incubation of the cells with aldehydes, nuclear protein was extracted and the DNA-binding of p53 was assessed by ELISA as described vide supra (Fig. 3A). The levels of p53 extracted from all treated cells was essentially unchanged, however the DNA-binding ability of p53 levels were significantly different in all aldehyde-treated cells (p<0.05) relative to VEH. HHE-treated cells had an elevated level of functional p53 relative to VEH (1.6 × VEH, p < 0.05) and while the mechanism by which this effect is under investigation, this may be due to a redistribution of functional p53 from the cytosol to the nucleus. HNE-treated cells have a significantly reduced level of DNA-binding nuclear p53 relative to VEH (33 % of VEH, p < 0.05) and, given that HNE does not induce p53 misfolding (Fig. 2A), this hints at an indirect off-target mechanism by which HNE is causing p53 dysfunction. Previous work by Moos and co-workers (Cassidy et al., 2006) has shown that certain endogenous αβ-unsaturated prostaglandin-derived aldehydes, such as PGA1 and PGA2, can cause p53 misfolding in cells, by a process that involves the chemical modification and inactivation of the selenoprotein, thioredoxin reductase (TrxR) (Moos et al., 2003, Moos et al., 2000). It seems plausible therefore that that HNE, which is an αβ-unsaturated aldehyde and is known to inactive TrxR (Cassidy, et al., 2006), is inducing p53 dysfunction indirectly via this mechanism.
Figure 3. Lipid Aldehydes Affect Wild-type p53 DNA Binding and p21 Activation in Cells.
A Bar chart representing level of DNA binding nuclear p53 extracted from A-427 cells incubated at 37 °C for 24 h with either VEH (0.1 % ethanol), HNE, HHE, KA or ALD (each at 40 μM) after receiving a dose of γ-radiation (5000 rads). After the incubation, nuclear extracts were taken, total protein (BCA), p53 (Western blot) and functional p53 (ELISA) were measured. Data are reported as the mean ± SEM of triplicate determinations. Data was analyzed using a student two-tail t test and was considered significantly different from VEH (*) if P<0.05. B Upper Western blot analysis of p53 and p21 in nuclear protein extracts from A-427 cells before (−) and after (+) receiving γ-radiation (5000 rads). Lower Western blot analysis of p53 and p21 in nuclear protein extracts from A-427 cells. Cells incubated at 37 °C for 24 h with ALD (0-30 μM) after receiving a dose of γ-radiation (5000 rads). After the incubation, nuclear extracts were taken, total protein (BCA), p53 and p21 (Western blot) were measured. C Agarose gel analysis of p21 PCR DNA derived from p53 immunoprecipitation (IP) of A-427 genomic DNA. Lane 1, 100 bp ladder; Lane 2, A-427 genomic DNA with p53 IP (after irradiation); Lane 3, A-427 genomic DNA with p53 IP (after irradiation and incubated with VEH for 20 h); Lane 4, A-427 genomic DNA with p53 IP (after irradiation, ALD, 30 μM for 20 h).
The most dramatic disruption of p53 DNA-binding in nuclear extracts of A-427 cells occurs after incubation with the cholesterol seco-sterols, KA and ALD (Fig. 3A). There is very little DNA binding of p53 in nuclear extracts from A-427 incubated with KA and ALD (3.4 %, p<0.05 and 4.2 %, p<0.05 of VEH respectively). Extrapolation of the misfolding studies in Fig. 2 to this p53 extraction data is complicated by potential off-target effects in cells, however it seems clear that the atheronals which induce direct misfolding of p53, are far more impactful, on a mole for mole equivalence, at disrupting functional p53 in cells than HNE and HHE, indirect, off-target, processes by which certain aldehydes, such as HNE, PGA1 and PGA2 influence p53 function, the direct induction of p53 misfolding is a potent mechanism by which inflammatory aldehydes can modulate p53 binding to DNA.
Atheronals Impair p53 Chromatin Binding and p53-Mediated Transcription
Genome damage usually increases the cellular levels of wild-type p53 tumour suppressor and p53-dependent transcription (Lane, 1992, Levine et al., 1991, Vogelstein and Kinzler, 1992, Zambetti and Levine, 1993). Consistent with this phenomenon, exposure of A-427 cells to γ-irradiation (5000 rads) leads to a proportional increase in p53 protein and its transcriptionally-regulated protein, p21, relative to untreated cells (Fig. 3B upper). Western blot analysis of p53 and p21 extracted from A-427 cells that had been exposed to γ-irradiation (5000 rads) and subsequently incubated with ALD (0-30 μM) reveals that nuclear p53 levels remain unchanged with increasing levels of atheronals (Fig. 3B, middle). However, ALD treatment of A-427 cells causes an atheronal concentration-dependent reduction in the expression of p21 (Fig. 3B, lower). This atheronal-induced reduction in p21 expression is supported by chromatin immunoprecipitation studies from atheronal-treated A-427 cells. Purification of sheared chromatin, immunoprecipitated by an anti-p53 antibody, followed by PCR of the purified chromatin fragments, with p21 primers (forward 5′-CCAGCCCTTGGATGGTTT-3′, reverse (5′-GCCTCCTTTCTGTGCCTGA-3′) reveals that atheronal treatment leads to a reduction in p21 expression (Fig. 3C). These combined data suggest that the atheronals do not directly impact the nuclear level of p53, but by inducing p53 misfolding, may cause a direct reduction in p53 transactivation within the cell.
For the aldehydes KA and ALD to impact p53 misfolding in the cells, they must come into direct contact with the protein and to do this, have to either enter the nucleus or cytoplasm, as p53 exists in equilibrium between the nucleus and cytoplasm. While we have yet to perform an in-depth study of the trafficking of the atheronals in cells, what we have shown previously is that a fluorescently-tagged atheronal-B analogue is rapidly taken up (within 5 min) into cultured macrophages (J774), by a process of diffusion, and trafficks to all compartments of the cell, excluding the nucleus (Takeuchi et al., 2006). Therefore, the cellular effects we observe on the A-427 cell line here are most likely initiated by interaction between ALD and p53 in the cytoplasm, which then in its misfolded form is either sequestered in the cytoplasm preventing its movement into the nucleus, or moves into the nucleus where it is unable to bind to DNA.
The concentration of ALD that we show achieves clear DNA-binding reduction in A-427 cells is ~ 20 μM (Figure. 2F). The concentration of secosterols in the systemic circulation is ~ 30 nM, while patients with inflammatory artery disease have almost a log higher level of circulating aldehyde (~ 200 nM) (Wentworth, et al., 2003). However, what is not yet known is the effective local concentrations of atheronals in tissues that are undergoing a chronic inflammatory response, as would be the case in tumorogenic systems, which may be much higher than in systemic circulation. An example of such a difference is observed with HNE, which has nanomolar circulating concentrations but cell membrane levels as high as millimolar (Esterbauer, 1982, Esterbauer et al., 1991, Esterbauer et al., 1990). Therefore, the concentrations of ALD required to reduce p53 binding in cells is not beyond the plausible levels achievable under the pathological syndrome being studied.
We propose that endogenous lipid-derived aldehydes are able to adduct to surface-exposed lysine residues on wild-type p53 protein, and in doing so specific aldehydes, such as the cholesterol seco-sterol aldehydes, (KA and ALD), but not the PUFA-derived α,β-unsaturated aldehydes (HNE and HHE), can induce a direct misfolding p53 event that is amyloidogenic. This KA and ALD-induced misfolding event renders the p53 unable to bind to DNA in vitro and exposure of cultured lung carcinoma cells to the atheronals leads to a dramatic (p<0.05) reduction in the amount of DNA-binding nuclear p53 protein (but not total nuclear p53) and a reduction in transactivation of p21.
Finally, the observation that only certain aldehydes induce the misfolding of p53 suggests there is a molecular recognition aspect to the aggregation process. This observation of molecular recognition between an aggregation-prone protein and the atheronal aldehydes, has also been observed with lipid aldehyde-initiated aggregation of amyloid-β peptides (Scheinost, et al., 2008). The importance of the recognition of the lipid’s structure to protein aggregation, is that this non-covalent recognition should be able to be mimicked, leading to the development of molecular chaperones that will protect p53 from the aldehyde-initiated protein misfolding event and lead to potential new anticancer drugs.
SIGNIFICANCE
What we show for the first time is that direct modification of p53 by certain endogenous aldehydes, is sufficient to cause structural changes in p53 that may contribute to epigenetic silencing. This direct inactivation of p53 by cholesterol-derived aldehydes maybe a hitherto unknown chemical link between inflammation and cancer and may contribute in part to the known cancers where wild-type inactivation of p53 has been reported such as breast cancer (Moll, et al., 1992), neuroblastoma (Isaacs, et al., 1998, Moll, et al., 1995), colon adenocarcinoma (Bosari, et al., 1995), and colon adenoma (Tominaga, et al., 1993).
EXPERIMENTAL PROCEDURES
Cells and proteins
The human A-427 lung carcinoma cell line carries wild-type p53. The hexa-histidine tagged recombinant p53 (His6-p53) was purified from E. coli BL21 (DE3) transformed with pET-15b plasmid containing the open reading frame for p53.
Recombinant His6-p53 Expression, purification and refolding
The open reading frame encoding wild type p53 was cloned by PCR into pET-15b (Novagen) between Nde I and BamH I sites. PCR was performed on pCMV-XL5-p53 from OriGene (Rockville, MD) using two primers: 5′-GAG AGA CAT ATG GAG GAG CCG CAG TCA GAT CC-3′, 5-GAG AGA GGA TCC TCA TCA GTC TGA GTC AGG CCC TTC TG-3′. The resulting plasmid, pET-15b-p53, was transformed into E. coli BL21 (DE3) (Invitrogen) for expression. The overnight culture from a single colony was diluted 1:100 into 1 L Luria Bertani media containing 100 mg/ml ampicillin, which then was incubated with shaking at 37 °C until an OD600 of 0.6 – 0.8. Recombinant protein expression was induced overnight at OD600nm of 0.4 by addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 25°C. After centrifugation at 5000 rpm for 15 min, the cell pellets (2 – 4g) were frozen overnight at −20 °C.
More than 70 % of the recombinant protein, expressed as detailed above, deposits into inclusion bodies and isolation was carried out after bacterial cell lysis using 5 ml of BugBuster (Novagen) per gram of wet pellet, benzonase nuclease and EDTA free protease inhibitors, followed by centrifugation at 5000 × g for 30 min. The inclusion bodies were washed again with 10 % BugBuster to further remove cell debris and other proteins. For solubilization, the inclusion bodies were resuspended in 1 ml of 100mM Tris-HCl, 6 M Guanidinium Chloride (GdmCl), 50 mM DTT at pH 8.0. Renaturation of the recombinant proteins isolated from the inclusion bodies were performed in refolding buffer consisting of 50 mM sodium phosphate, 1M L-arginine, 2mM DTT, 0.2 mM ZnCl2 at pH 8.0. The samples were incubated for 10 hours at 4°C. For large-scale production (200 ml refolding buffer), pulse renaturation of p53 was followed (solubilisate was added to the renaturation buffer six times with approximately 150 μg/ml per step and an interval of 90 min between each addition). After an overnight dialysis against 50 mM NaPPhos, 5% (v/v) glycerol, 4 mM DTT, pH 8.0 at 4°C, the renatured proteins were precipitated using 30 000 MWCO Vivaspin centricons (Sartorius).
Refolded protein samples were applied to the Qiagen Ni-NTA Fast Start 2 mL purification columns. All fractions were collected and were analyzed with 10% SDS-PAGE and Western blot using a murine monoclonal antibody for p53 (Abcam, Catalogue number ab1101)
Thioflavin-T binding and fluorescence of His6-p53 with lipid aldehydes
Recombinant, human wild type His6-tagged p53 protein (His6-p53, 0.8 mg/mL), was incubated in at least duplicate with lipid aldehydes KA, ALD, HNE, HHE (final concentration 100 μM) or vehicle (VEH) in PBS, pH 7.4 and ethanol (0.1 % v/v co-solvent) at 37 °C and in the presence of Th-T (10 μM) in a 384 well microtiter plate (final assay volume 50 ml). Time-dependent changes in the fluorescence (Ex: 440 nm; Em: 485 nm) of the assays was measured on a SpectraMax Gemini XS fluorescence detector (Molecular Devices, Union City, CA). Note 1. All the non-cell-based assays reported in this study were performed with the His6-p53 recombinant protein. However, we have performed ThT aggregation assays with human wild-type p53 (Calbiochem, San Diego CA) and KA and the time-course of fluorescence, both with VEH and KA (100 μM) were identical with the wild-type p53 to that we observed with our recombinant His6-p53 protein construct. Note 2. For ease of assay, the ThT was present during the aggregation assay. However, we have performed ThT aggregation assays where assay aliquots are removed and added to a ThT solution (10 μM) in PBS, pH 7.4, both with VEH and KA (100 μM) and the profiles were identical with to that we observed with ThT in the aggregation assay. Also note incubation of ThT with VEH, KA, ALD, HNE, or HHE had no measurable effect on ThT fluorescence excitation or emission.
Congo Red binding and birefringence
Wild-type native His6-p53 (0.7 mg/ml), prepared and purified as detailed above, was incubated with KA (25 μM) in phosphate buffered saline (pH 8.0) with ethanol (4.0 % v/v) for 5 h at room temperature (assay volume 100 μl) in a protein concentration tube (Millipore). Congo red (100 μM in 1 % ethanol in water) was added and the mixture was allowed to stand at room temperature for 30 min. The resultant sample was then centrifuged (Millipore) at 3600 rpm for 30 min. The concentrated supernatant and aggregates were then added to a glass slide and warmed at 37 °C for 10 min. Glycerol and a cover-slip were then applied. Aggregates were analyzed under normal and phase-contrast light microscopy conditions (100 × magnification, Olympus BX51 microscope). Images were recorded on an Olympus Microfire camera.
FTIR studies
In a typical experiment, His6-p53 (0.5 mg/ml) in refolding buffer, prepared as detailed above, was dialyzed overnight in deuterium oxide at 4°C. Prior to FTIR experiments, the binding activity of the His6-p53 was tested using the ELISA-based transbinding assay kit (Panomics) detailed below.
The misfolding process was started by addition of KA (100 μM final concentration) or VEH to the solution of His6-p53 (0.5 mg/ml) in D2O with incubation at room temperature. FTIR spectra were recorded throughout the assay at room temperature on a BioRad FTS 6000 FTIR machine, equipped with diamond attenuated total reflectance (ATR) with a He-Ne laser detector. Each spectra was the average of 100 spectra recorded over the course of 100 min. The raw absorbance data was exported into Microsoft Excel and Graphpad Prism 4.0 for Mac for analysis.
Effect of ALD on DNA binding of His6-p53
Human wild type His6-p53 protein (final concentration 0.8 mg/mL), prepared above, was incubated in at least duplicate with lipid aldehyde ALD, (final concentration 50 μM) or vehicle (VEH) in PBS, pH 7.4 and ethanol (0.1 % v/v co-solvent) at 37 °C. At times during the incubation, aliquots were removed and DNA binding measured by the PANOMICS ELISA transbinding assay. Data are reported as the mean ± SEM of the functional p53 as a % of t = 0. Data analysis was using Graphpad Prism v 4.0 for MAC.
Recombinant wild type His6-p53 protein (final concentration 0.8 mg/mL), prepared as described above, was incubated in at least duplicate with lipid aldehyde ALD at various concentrations (0-80 μM) in PBS, pH 7.4 and ethanol (0.1 % v/v co-solvent) at 37 °C. After 2 h of incubation, DNA binding of the p53 was measured by the Panomics ELISA assay described above. Data are reported as the mean ± SEM of the functional p53 as ng/ml. Data analysis was performed using Graphpad Prism v 4.0 for MAC.
Incubation of A-427 cells with lipid aldehdyes
Human lung carcinoma (ATCC# HTB-53, A-427) cells were routinely cultured in fetal calf serum (FCS, 10 %) with minimum essential medium and L-glutamine (2 mM), sodium bicarbonate (1.5 g/L), non-essential amino acids (0.1 mM), and sodium pyruvate (1 mM), to 80-90 % confluency in 75 cm2 flasks. A-427 cells were then exposed to γ-irradiation (5000 cGy) from a cesium source, media was replaced and the cells were incubated for 24 h growth medium. At this time, media was replaced with fresh media (15 mL) containing either KA, ALD, HNE, HHE (each at 40 μM), or VEH (ethanol, 0.1 % v/v) in at least duplicate and incubated for a further 24 h at 37 °C.. The cells were then pelleted by centrifugation and washed with PBS (pH 7.4). Nuclear extracts were obtained using a nuclear extraction kit (Panomics, Redwood City, CA). Protein concentration of the extracts was determined using a BCA protein assay kit (Pierce, Rockford, IL) and p53 concentration was measured using Western blot analysis. Quantification of functional p53 was assessed by DNA binding using the Panomics transbinding p53 assay kit as detailed above. 5 and 2.5 μg of nuclear extract was added per well of the ELISA in duplicate for each condition respectively. The data for functional p53 (reported as mean ± SEM) was determined by taking the raw absorbance data and interpolating from a standard curve of known concentrations of wild-type p53 using Graphpad Prism v 4.0 for MAC.
ALD treatment of A-427 cells
A-427 cells were exposed to γ-irradiation (5000 cGy) from a cesium source, media was replaced and the cells were incubated for 24 h growth medium. At this time, media was replaced with fresh media (15 mL) containing ALD (40 μM) and VEH (ethanol, 0.1 % v/v) in at least duplicate and incubated for varying time up to 30 h at 37 °C. At various times throughput the incubation, cells were pelleted by centrifugation and washed with PBS (pH 7.4). Nuclear extracts were obtained using a nuclear extraction kit (Panomics, Redwood City, CA). Protein concentration of the extracts was determined using a BCA protein assay kit (Pierce, Rockford, IL) and p53 concentration was measured using Western blot analysis. Quantification of functional p53 was assessed by DNA binding using the Panomics transbinding p53 assay kit as detailed above. Nuclear extract (5 and 2.5 μg) was added per well of the ELISA in duplicate for each condition respectively. The data for functional p53 (reported as mean ± SEM) was determined by taking the raw absorbance data and interpolating from a standard curve of known concentrations of wild-type p53 using Graphpad Prism v 4.0 for MAC.
Measurement of p21 WAF transcription
A-427 human lung carcinoma (ATCC# HTB-53) cells were grown in FCS (10 %) minimum essential medium (MEM) with L-glutamine (2mM), sodium bicarbonate (1.5 g/L), non-essential amino acids (0.1 mM), and sodium pyruvate (1 mM), to 80-90% confluency in 75 cm2 flasks. The cells were exposed to γ-irradiation (5000 cGy) from a cesium source, media was replaced and the cells were incubated for 24 h with VEH (0.1 % ethanol) or ALD at 10-30 μM in at least duplicate. The cells were then incubated for a further 16 h. Nuclear extracts were obtained from each condition using the Panomics nuclear extraction kit (Cat # AY2002). Protein concentration was determined for each extract using the Pierce BCA protein assay kit (Cat # 23225). The nuclear extracts were frozen at −80 C and then analyzed by western blots. he PVDF membranes were developed using the western Breeze chromogenic immunodetection kit (Invitrogen Cat # WB7103). Total protein was visualized with Ponceu S solution (Sigma Cat # P-7170). The primary antibodies, mouse anti-human p53 (BD Cat # 554167) and mouse anti-human p21 (BD Cat # 554262) were diluted 1:500 and incubated overnight.
P21 CHIP assay protocol
A-427 cells were cultured to 80 % confluence as detailed above. The cells were exposed to γ-irradiation (5000 cGy) from a cesium source, media was replaced and the cells were incubated for 20 h with VEH (0.1 % ethanol) or ALD at 30 μM in at least duplicate. Cells were harvested by trypsinization, counted and lysed using standard protocols.
Chromatin shearing
Cell lysates were sonicated with a Misonix 3000 ultrasonicating bath at 4 °C (270 watts, 20 cycles: 30 sec pulse with 2 min cooling). A 1 kb DNA smear confirmed successful shearing.
P53 immunoprecipitation
Sheared lysates were diluted 2:1 with antibody dilution buffer. 5 μg/sample of biotinylated goat anti human P53 ab was then added to (note: biotinylated goat anti-IgG added to as a negative control). Samples were then incubated for 17 h at 4 °C, with shaking at 250 rpm. Streptavidin agarose was then added to the immunoprecipitation mixture and the mixture incubated for 30 min at 4 °C with shaking, followed by addition of chelating resin and elution.
Purification and PCR of immunoprecipitated DNA
The IP DNA was purified using Qiagen quick PCR purification kit (# 28106) and PCR was performed using P21 primers.
Supplementary Material
Highlights.
Lipid-derived aldehydes cause misfolding and amyloidogenesis of p53
Aldehyde-misfolded p53 is transcriptionally inactive.
Lipid-aldehydes maybe a chemical link between cancer and inflammation
Acknowledgements
P.W. would like to thank all members of the Wentworth groups, in Oxford and La Jolla their support. This work was supported by the NIH (AG028300), the ALSAM foundation (J.N.) and the Skaggs Institute for Research (P.W.) and a grant from The Scripps Research Institute (P.W.).
Abbreviations
- PBS
phosphate-buffered saline
- ThT
thioflavin-T
- PUFA
polyunsaturated fatty acids
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- Borm PJ, Driscoll K. Particles, inflammation and respiratory tract carcinogenesis. Toxicol. Lett. 1996;88:109–113. doi: 10.1016/0378-4274(96)03725-3. [DOI] [PubMed] [Google Scholar]
- Bosari S, Viale G, Roncalli M, Graziani D, Borsani G, Lee AK, Coggi G. p53 gene mutations, p53 protein accumulation and compartmentalization in colorectal adenocarcinoma. Am. J. Pathol. 1995;147:790–798. [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Fowler DM, Zhang Q, Nieva J, Powers ET, Wentworth P, Jr., Lerner RA, Kelly JW. Elevated levels of oxidized cholesterol metabolites in Lewy body disease brains accelerate alpha-synuclein fibrilization. Nat. Chem. Biol. 2006;2:249–253. doi: 10.1038/nchembio782. [DOI] [PubMed] [Google Scholar]
- Cassidy PB, Edes K, Nelson CC, Parsawar K, Fitzpatrick FA, Moos PJ. Thioredoxin reductase is required for the inactivation of tumor suppressor p53 and for apoptosis induced by endogenous electrophiles. Carcinogenesis. 2006;27:2538–2549. doi: 10.1093/carcin/bgl111. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- Esterbauer H. Aldehydic products of lipid peroxidation. In: McBrien DCH, Slater TF, editors. Free Radicals, Lipid Peroxidation and Cancer. Academic Press; London: 1982. p. 101. [Google Scholar]
- Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Zollner H, Schaur RJ. Aldehydes formed by lipid peroxidation: mechanisms of formation, occurrence, and determination. In: Vigo-Pelfrey C, editor. Membrane Lipid Oxidation. Volume I. CRC Press; Boston: 1990. pp. 239–268. [Google Scholar]
- Ferrone F. Analysis of protein aggregation kinetics. Methods Enzymol. 1999;309:256–274. doi: 10.1016/s0076-6879(99)09019-9. [DOI] [PubMed] [Google Scholar]
- Hernandez-Boussard T, Rodriguez-Tome P, Montesano R, Hainaut P. IARC p53 mutation database; a relational database to compile and analyze p53 mutations in human tumors and cell lines. International Agency for Research on Cancer. Hum. Mutat. 1999;14:1–8. doi: 10.1002/(SICI)1098-1004(1999)14:1<1::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Hurshman AR, White JT, Powers ET, Kelly JW. Transthyretin aggregation under partially denaturing conditions is a downhill polymerization. Biochemistry. 2004;43:7365–7381. doi: 10.1021/bi049621l. [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Hardman R, Carman TA, Barrett JC, Weissman BE. Differential subcellular p53 localization and function in N- and S-type neuroblastoma cell lines. Cell Growth Differ. 1998;9:545–555. [PubMed] [Google Scholar]
- Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- Keeley D, Rees J. New guidelines in asmtha management. Br. Med. J. 1997;314:315–316. doi: 10.1136/bmj.314.7077.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- Lane DP. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- Levine IIIH, Ronald W. [18] Quantification of [beta]-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- Lu H, Ouyang W, Huang C. Inflammation, a key event in cancer development. Mol. Cancer Res. 2006;4:221–233. doi: 10.1158/1541-7786.MCR-05-0261. [DOI] [PubMed] [Google Scholar]
- Moll UM, LaQuaglia M, Benard J, Riou G. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not differentiated tumors. Proc. Natl. Acad. Sci. (USA) 1995;92:4407–4411. doi: 10.1073/pnas.92.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Riou G, Levine AJ. Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc. Natl. Acad. Sci. (U.S.A.) 1992;89:7262–7266. doi: 10.1073/pnas.89.15.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moos PJ, Edes K, Cassidy P, Massuda E, Fitzpatrick FA. Electrophilic prostaglandins and lipid aldehydes repress redox-sensitive transcription factors p53 and hypoxia-inducible factor by impairing the selenoprotein thioredoxin reductase. J. Biol. Chem. 2003;278:745–750. doi: 10.1074/jbc.M211134200. [DOI] [PubMed] [Google Scholar]
- Moos PJ, Edes K, Fitzpatrick FA. Inactivation of wild-type p53 tumor suppressor by electrophilic prostaglandins. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9215–9220. doi: 10.1073/pnas.160241897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J, Shafton A, Altobell LJ, Tripuraneni S, Rogel JK, Wentworth AD, Lerner RA, Wentworth P. Lipid-Derived Aldehydes Accelerate Light Chain Amyloid and Amorphous Aggregation. Biochemistry. 2008;47:7695–7705. doi: 10.1021/bi800333s. [DOI] [PubMed] [Google Scholar]
- Philip M, Rowley DA, Schreiber H. Inflammation as a tumor promoter in cancer induction. Semin. Cancer Biol. 2004;14:433–439. doi: 10.1016/j.semcancer.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Scheinost JC, Wang H, Boldt GE, Offer J, Wentworth PJ. Cholesterol seco-sterol-induced aggregation of methylated amyloid-β peptides – Insights into aldehyde-initiated fibrillization of amyloid-β. Angew. Chemie Int. Ed. 2008 doi: 10.1002/anie.200705922. in press. [DOI] [PubMed] [Google Scholar]
- Scheinost JC, Witter DP, Boldt GE, Offer J, Wentworth PJ. Cholesterol secosterol adduction inhibits the misfolding of a mutant prion protein fragment that induces neurodegeneration. Angew. Chemie Int. Ed. 2009;48:9469–9472. doi: 10.1002/anie.200904524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi C, Galve R, Nieva J, Witter DP, Wentworth AD, Troseth RP, Lerner RA, Wentworth P., Jr. Proatherogenic effects of the cholesterol ozonolysis products, atheronal-A and atheronal-B. Biochemistry. 2006;45:7162–7170. doi: 10.1021/bi0604330. [DOI] [PubMed] [Google Scholar]
- Tominaga O, Hamelin R, Trouvat V, Salmon RJ, Lesec G, Thomas G, Remvikos Y. Frequently elevated content of immunochemically defined wild-type p53 protein in colorectal adenomas. Oncogene. 1993;8:2653–2658. [PubMed] [Google Scholar]
- Uversky VN, Fink AL. Conformational constraints for amyloid fibrillization: the importance of being unfolded. Biochimica et Biophysica Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. p53 function and dysfunction. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- Wentworth P, Jr., Nieva J, Takeuchi C, Galve R, Wentworth AD, Dilley RB, DeLaria GA, Saven A, Babior BM, Janda KD, Eschenmoser A, Lerner RA. Evidence for ozone formation in human atherosclerotic arteries. Science. 2003;302:1053–1056. doi: 10.1126/science.1089525. [DOI] [PubMed] [Google Scholar]
- Zambetti GP, Levine AJ. A comparison of the biological activities of wild-type and mutant p53. FASEB J. 1993;7:855–865. doi: 10.1096/fasebj.7.10.8344485. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Powers ET, Nieva J, Huff ME, Dendle MA, Bieschke J, Glabe CG, Eschenmoser A, Wentworth P, Jr., Lerner RA, Kelly JW. Metabolite-initiated protein misfolding may trigger Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4752–4757. doi: 10.1073/pnas.0400924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.