SUMMARY
Macroautophagy is a lysosomal degradative pathway that maintains cellular homeostasis by turning over cellular components. Here, we demonstrate a role for autophagy in hypothalamic agouti-related peptide (AgRP) neurons in the regulation of food intake and energy balance. We show that starvation-induced hypothalamic autophagy mobilizes neuron-intrinsic lipids to generate endogenous free fatty acids, which in turn regulate AgRP levels. The functional consequences of inhibiting autophagy are the failure to upregulate AgRP in response to starvation, and constitutive increases in hypothalamic levels of pro-opiomelanocortin and its cleavage product α-melanocyte stimulating hormone that typically contribute to a lean phenotype. We propose a new conceptual framework for considering how autophagy-regulated lipid metabolism within hypothalamic neurons may modulate neuropeptide levels to have immediate effects on food intake, as well as long-term effects on energy homeostasis. Regulation of hypothalamic autophagy could become an effective intervention in conditions such as obesity and the metabolic syndrome.
INTRODUCTION
Chronic overnutrition contributes to metabolic disturbances that predispose to the development of obesity and insulin resistance (Lionetti et al., 2009), hallmark of the metabolic syndrome (de Luca and Olefsky, 2008). The molecular mechanisms and cell-intrinsic pathways that govern neuronal regulation of food intake are unclear. The hypothalamic arcuate nucleus consists of neurochemically discrete and functionally antagonistic neurons, including agouti-related peptide (AgRP), and pro-opiomelanocortin (POMC) neurons (Sainsbury and Zhang, 2010) that form a focal point for the integration of nutritional and metabolic cues, central and peripheral neural afferents (Morris and Rui, 2009), and action of adiposity hormones such as leptin and insulin (Belgardt and Bruning, 2010). Hypothalamic neurons release specific neuropeptides (Sainsbury and Zhang, 2010) that generate behavioral responses including food seeking and the initiation or cessation of feeding. AgRP increases food intake by acting as a natural antagonist for the melanocortin receptors (Garfield et al., 2009). POMC neurons express POMC that is processed further to secrete α-melanocyte stimulating hormone (α-MSH) for regulating food intake and energy expenditure (Mountjoy, 2010). In addition to extrinsic nutritional signals, levels of intracellular metabolites, particularly the neuronal availability of free fatty acids have been proposed to play a role in the hypothalamic regulation of food intake (Andrews et al., 2008). However, the intra-neuronal mechanisms that regulate endogenous levels of free fatty acids within the hypothalamus, as well as mechanisms that link fatty acid availability to AgRP secretion are not known.
Macroautophagy (hereafter autophagy) is a cellular process that recycles organelles and proteins to maintain cellular homeostasis (He and Klionsky, 2009). In addition, autophagy serves as an alternative energy source to sustain cellular function during starvation. Induction of autophagy requires the de novo formation of a double-walled limiting membrane that elongates and seals to form an autophagosome. This vesicle engulfs and targets cargo destined for degradation by fusing with the lysosome (He and Klionsky, 2009). An upstream negative regulator of autophagy is the nutrient-sensor mammalian target of rapamycin (mTOR) (Pattingre et al., 2008). Activation of hypothalamic mTOR has been shown to regulate food intake and energy homeostasis (Cota et al., 2006), but whether part of the mTOR effect is via its modulatory effect on autophagy remains unknown. We have recently reported a novel lipophagic function of autophagy, by which the activation of this pathway during starvation induces mobilization of lipid droplets to generate free fatty acids in liver (Singh et al., 2009a). Fasting increases circulating free fatty acids that are rapidly taken up by organs such as liver and esterified to triglycerides within lipid droplets. Lipolytic mechanisms, for instance lipophagy (Singh et al., 2009a), then break down these cellular lipid stores to provide endogenous free fatty acids for energy under nutrient-deficient conditions. In addition, lipophagy can be upregulated in response to increased fatty acids (Singh et al., 2009a). We hypothesized that a similar process of starvation-induced lipophagy may occur within the hypothalamus. In fact, a recent study has demonstrated lipid accumulation within striatal neurons defective for autophagy (Martinez-Vicente et al., 2010). In the present study, using AgRP-expressing hypothalamic GT1–7 cells and primary hypothalamic neurons we show that starvation-induced increase in circulating free fatty acids promotes hypothalamic fatty acid uptake and induction of hypothalamic autophagy. Hypothalamic autophagy helps mobilize neuronal lipid stores to increase the availability of endogenous free fatty acids that, in turn, regulate AgRP levels. We show that compromising autophagy in the cultured hypothalamic cells or in mice with AgRP neuron-specific deletion of the essential autophagy gene atg7 reduces AgRP levels, food intake and adiposity. Our work thus reveals a critical function for autophagy in regulation of food intake and energy balance through the modulation of hypothalamic AgRP expression.
RESULTS
Starvation induces autophagy in the hypothalamus
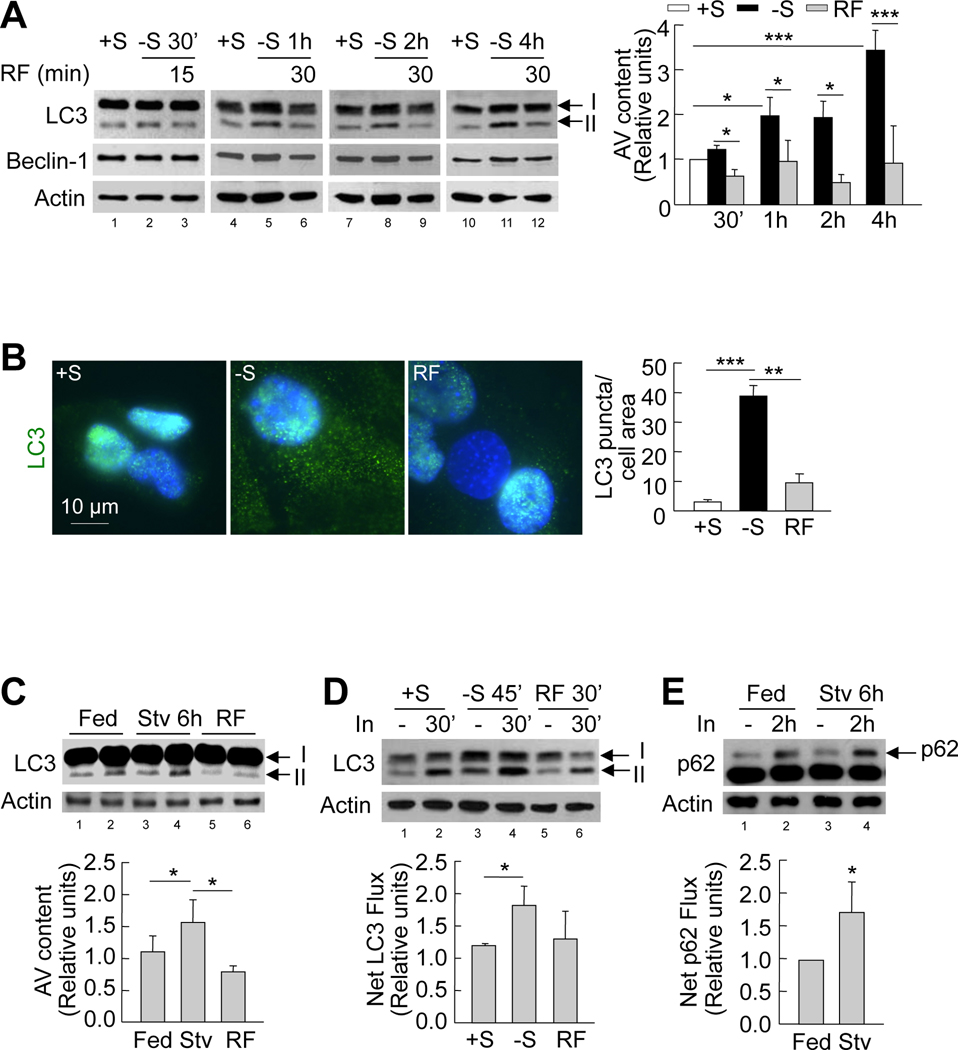
To determine if hypothalamic autophagy is nutritionally regulated, we first examined the effect of serum removal on autophagy in hypothalamic GT1–7 cells in culture. The induction of autophagy was indicated by examining for increased levels of the lipid-conjugated form of the autophagosome marker light chain 3 (LC3)-II, a measure of autophagosome content. In contrast to other neuronal types in which starvation fails to stimulate autophagy (Mizushima et al., 2004), serum removal in GT1–7 cells led to time-dependent increases in LC3-II content, suggesting progressive induction of autophagy in response to nutrient-deprivation (Figure 1A). However, serum removal did not affect levels of Beclin-1, an autophagy-related protein that forms part of the initiation complex involved in autophagosome formation, in GT1–7 cells (Figure 1A). Supporting autophagy induction, indirect immunofluorescence revealed increased membrane-associated LC3-II puncta following 1h of serum removal in GT1–7 cells (Figure 1B). As described in other cell types, activation of autophagy by nutrient-deprivation in these cells associated to phosphorylation of adenosine monophosphate-activated protein kinase (AMPK) (threonine 172), and of its recently elucidated (Egan et al., 2011) downstream substrate unc-51-like kinase (ULK1) (serine 555) a known regulator of autophagy (Figure S1A). To examine whether resupplementing serum reversed the induction of hypothalamic autophagy, we subjected serum-starved GT1–7 cells to serum refeeding. Refeeding serum decreased autophagosome content, as reflected by decreased LC3-II levels (Figure 1A) and LC3-II puncta (Figure 1B), as early as 15min into refeeding (Figure 1A).
Figure 1. Starvation induces autophagy in the hypothalamus.
(A) Immunoblots for LC3 and Beclin-1 from GT1–7 cells in presence (+S) or absence of serum (−S), or serum refed (RF) after serum removal for indicated times. Values for LC3-II represent autophagic vacuole (AV) content relative to +S (n=3). See also Figure S1. (B) Indirect immunofluorescence for LC3 (green) in GT1–7 cells in +S or −S for 1h, or RF for 30min following serum removal. DAPI-stained nuclei are in blue. Values represent LC3 puncta per cell area, and are mean+SE of 20 different cells in each experiment, n=3. (C) Immunoblots for LC3 in mediobasal hypothalamus (MBH) lysates from fed (lanes 1, 2), 6h-starved (Stv) (lanes 3, 4), or starved mice subjected to refeeding for 3h (RF) (lanes 5, 6). Values depict AV content, calculated as in A (n=5). (D) Immunoblots for LC3 from GT1–7 cells in +S or −S or RF following serum removal for times indicated. Cells were treated with (lanes 2, 4, 6) or without lysosomal inhibitors (In) (lanes 1, 3, 5) for 30min. Values represent net LC3 flux determined by subtracting LC3-II densitometric value of untreated samples from corresponding In-treated sample (n=4). (E) Immunoblot for p62 from MBH explants of fed (lanes 1, 2) and 6h-starved (Stv) (lanes 3, 4) mice cultured in presence or absence of lysosomal inhibitors (In) for 2h. Values represent net p62 flux (n=4). Values are mean+SE. *p<0.05, **p<0.01, ***p<0.001.
The observed reduction of autophagy upon nutrient resupplementation does not depend upon serum growth factors, as treatment of serum-starved GT1–7 cultured cells with methylpyruvate (MP), an alternate energy source, inhibited AMPK and ULK1 phosphorylation (Figure S1A), and markedly decreased LC3-II levels (Figure S1B). MP inhibited autophagy by blocking nutrient-responsive AMPK signaling, and not by reducing levels of oxidation in these cells (data not shown). Treatment of GT1–7 cells with another nutrient, methylsuccinate also partially decreased LC3-II levels (Figure S1C). These data support that autophagy is nutritionally regulated in hypothalamic GT1–7 cells.
The crucial cellular nutrient sensor, mammalian target of rapamycin complex 1 (mTORC1) is a known upstream inhibitor of autophagy (Pattingre et al., 2008). To test whether the induction of autophagy in the absence of serum occurred by inhibition of mTOR, we checked for the levels of phosphorylated mTOR (serine 2448) and its substrate p70 S6 kinase in GT1–7 cells cultured in presence or absence of serum. In parallel with the induction of hypothalamic autophagy, serum removal for 30min significantly decreased phosphorylation of both mTOR and p70 S6 kinase in GT1–7 cells (Figure S1D). Consistent with the inhibitory function of mTOR on autophagy, serum refeeding for 15min led to robust phosphorylation and activation of mTOR and p70 S6 kinase (Figure S1D).
To verify whether our observations in GT1–7 cells were reproducible in mediobasal hypothalami (MBH) in vivo, we evaluated C57BL/6 mice that had ad libitum access to food and water or were 6h food-restricted with free access to water. A subset of the starved group was refed for 3h following 6h of food restriction. Consistent with our results in GT1–7 cells, LC3-II content increased in hypothalamic lysates from 6h-starved mice indicating increased autophagosome content, while refeeding of fasted rodents significantly decreased LC3-II content (Figure 1C). Because steady-state LC3-II levels only represent autophagosome content and not turnover of autophagic substrates within lysosomes (autophagic flux), we used inhibitors of lysosomal hydrolases (In) in GT1–7 cells and in hypothalamic explants to determine whether serum removal or food-restriction, respectively, led to increased net autophagic flux. When compared to controls, 30min of serum removal or 6h of food restriction increased lysosomal accumulation of LC3-II and the autophagic substrate p62 in cultured GT1–7 cells (Figure 1D) and in hypothalamic explants (Figure 1E), respectively, demonstrating lysosomal turnover of hypothalamic cargo via autophagy during starvation. These results support that, in contrast to findings in neuronal populations in other brain regions, physiological periods of fasting and refeeding regulate hypothalamic autophagy.
Food intake and energy balance in mice knock-out for atg7 in AgRP neurons
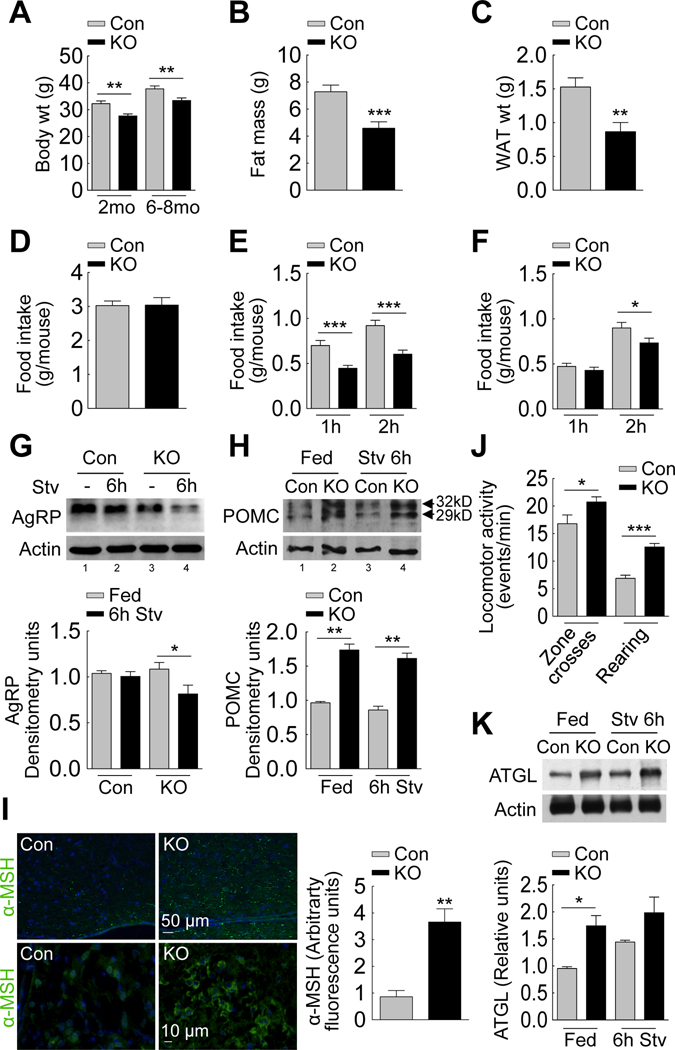
To identify the possible roles of hypothalamic autophagy in regulating energy balance, we modified autophagy in hypothalamic AgRP neurons. To this end, we assessed food intake and body weight in mice that lacked the autophagy gene atg7 specifically within AgRP neurons by crossing Atg7F/F mice with AgRP-Cre mice. AgRP neuron-specific ablation of autophagy was determined by immunohistochemical analysis for the absence of Atg7 in Cre-positive neurons that were distinct from neurons positive for POMC within MBH sections from AgRP neuron autophagy-deficient (Atg7F/F-AgRP-Cre or KO) mice (Figures S2A and S2B). Although AgRP expression in adrenals, lungs and skeletal muscle has been reported, our KO mice did not display changes in levels of Atg7 or LC3-II in any of these organs, supporting that expression levels were not enough to delete atg7 in these tissues (Figures S2C). Inhibition of autophagy within AgRP neurons did not result in higher mortality compared to controls (data not shown). Inhibition of autophagy in AgRP neurons significantly reduced body weight (Figure 2A), total fat mass (Figure 2B) and WAT mass (Figure 2C), without affecting 24h food intake (Figure 2D) or liver mass (Figure S2D). The KO mice also consumed significantly less food than controls when refed for 1h or 2h after a 6h fast (Figure 2E), and when refed for 2h after a 24h fast (Figure 2F). These findings demonstrate a function of autophagy in hypothalamic AgRP neurons in modulating feeding and energy balance.
Figure 2. Food intake and energy balance in mice knock-out for atg7 in AgRP neurons.
(A) Body weights of 2 month (mo) (n=10–12) and 6–8 mo-old control (Con) and Atg7F/F-AgRP-Cre (KO) mice (n=12–15). (B) Total body fat mass of 6–8 month old Con and KO mice (n=10–12). (C) Epididymal fat weights of 6–8 month old Con and KO mice (n=15–16). See also Figure S2D. (D) Food intake over 24h by Con and KO mice (n=27–31). (E) Food intake by 6h- (n=12–24), and (F) 24h-starved Con and KO mice when refed for indicated times (n=15–18). (G) Immunoblot for AgRP from mediobasal hypothalamus (MBH) of Con (lanes 1, 2) and KO (lanes 3, 4) mice fed or starved (Stv) for 6h. Values represent densitometric values for AgRP (n=10). See also Figure S2F–G. (H) Immunoblot for POMC preproprotein from MBH of fed Con and KO (lanes 1, 2) and 6h-starved Con and KO (lanes 3, 4) mice (n=6). (I) Indirect immunofluorescence (at similar exposure times) for α-MSH (green) in MBH sections from Con and KO mice (n=3). (J) Locomotor activity reflecting “zone crosses” and “rearing” by fed Con and KO rodents (n=14–19). (K) Immunoblot for ATGL from adipose tissues of fed Con and KO (lanes 1, 2) and 6h-starved Con and KO (lanes 3, 4) mice. Values represent densitometric values for ATGL (n=3–4). Values are mean+SE. *p<0.05, **p<0.01, ***p<0.001.
Appetite regulation is closely linked to hypothalamic AgRP expression (Kitamura et al., 2006); thus, we asked whether hypothalamic autophagy functions to regulate neuropeptide levels to modulate appetite and energy homeostasis. To explore the possibility that autophagy regulates AgRP expression in vivo, and that decreased refeeding in food-deprived KO mice (Figure 2E and 2F) results from altered AgRP levels, we analyzed the levels of AgRP in MBH from fed and food-deprived control and KO mice. Extended fasting typically increases hypothalamic AgRP mRNA levels, consistent with its orexigenic role in driving feeding after a fast. Accordingly, we found that 24h fasting led to modest increases in hypothalamic AgRP protein levels in control mice (Fig S2E). While no difference in AgRP levels was observed between fed control and KO mice (Figure 2G), 6h or 24h of fasting actually reduced AgRP levels in KO mice (Figure 2G and S2E), consistent with reduced refeeding in these mice (Figure 2E and 2F). AgRP neurons provide inhibitory inputs on to the hypothalamic arcuate POMC neurons (Garfield et al., 2009), consequently, we asked whether loss of autophagy in AgRP neurons affected POMC preproprotein levels in adjacent MBH POMC neurons. Inhibition of autophagy in AgRP neurons significantly increased hypothalamic levels of the 32kDa (glycosylated) and 29kDa (non-glycosylated) forms of POMC preproprotein in fed and 6h fasted animals relative to controls (Figure 2H). Consistent with higher POMC levels, MBH sections from KO mice displayed increased immunostaining for α-melanocyte stimulating hormone (α-MSH) (Figure 2I). To test whether higher MBH POMC/α-MSH levels directly correlated with increased peripheral energy expenditure that contributed to reduced body weight and fat mass in the KO mice, we subjected control and KO rodents to standard measurements of locomotor activity. Mice lacking autophagy in AgRP neurons displayed marked increases in the number of “zone crosses”, and “rearing”, as scored in a manual open field test (Figure 2J). The reduced body weight in KO mice could also be in part attributable to possible increases in lipid mobilization, since immunoblot analysis of adipose tissues from fed or 6h fasted KO mice revealed increased constitutive expression of the adipose triglyceride lipase (ATGL), compared to controls (Figure 2K). Thus, elevated POMC levels in KO mice may partly explain their decreased body weight and adipose mass.
These effects of changes in autophagy on neuropeptide levels were also verified in hypothalamic GT1–7 cells using lentiviral vectors (Piva et al., 2006) that expressed a small hairpin RNA against a second autophagy gene atg5. Efficient ablation of autophagy was confirmed by immunoblotting for autophagy proteins Atg5 and LC3-II in lysates from control and autophagy-deficient (siAtg5) cells (Figure S2F). Serum removal in controls increased AgRP protein (Figure S2G) and mRNA (Figure S2H). In contrast, siAtg5 GT1–7 cells failed to upregulate AgRP (Figures S2G and S2H) in response to serum removal. Decreased AgRP levels in siAtg5 cells following serum removal did not result from increased secretion, as siAtg5 cells exhibited blunted AgRP secretion into the medium by ~50% in response to serum-deprivation (Figure S2I). A modest increase in AgRP protein levels was observed in serum-fed siAtg5 cells compared to serum-fed controls (Figure S2G), which was for the most part due to transcriptional upregulation as we found higher basal AgRP mRNA in these cells (Figure S2H). Interestingly, in clear contrast to controls, siAtg5 cells failed to transcriptionally upregulate AgRP expression in response to starvation (Figure S2H). These results demonstrate that autophagy within hypothalamic neurons regulates neuropeptide levels and in this way may contribute to the control of appetite and energy homeostasis.
Starvation induces hypothalamic autophagy by increasing fatty acid uptake
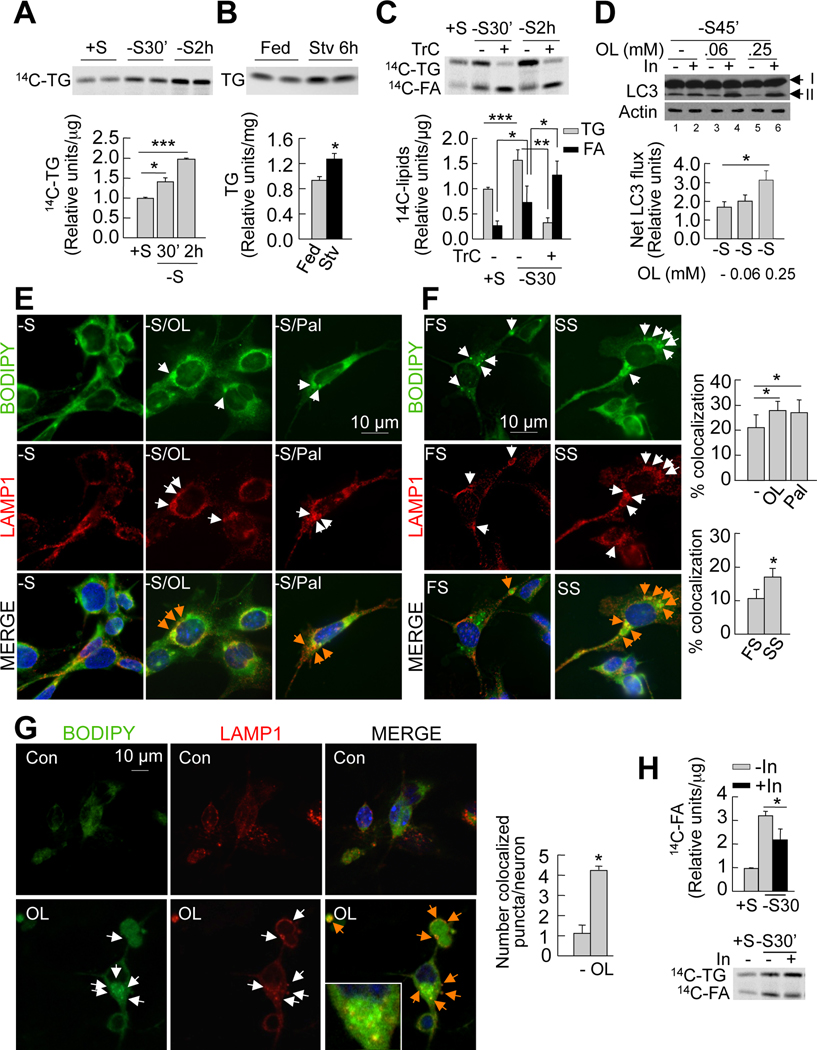
We have shown earlier that addition of a lipid stimulus, such as fatty acid treatment or starvation induces autophagy in cultured hepatocytes and in the liver, respectively (Singh et al., 2009a). Starvation reduces triglycerides in serum (Figure S3A, left), but increases circulating fatty acids (Figure S3A, right) that are rapidly taken up by the liver. We asked whether fasting-induced increases in fatty acid availability increase hypothalamic fatty acid uptake and thereby promote hypothalamic autophagy. To begin to test this, we examined the effect of serum removal on fatty acid uptake and triglyceride synthesis by GT1–7 cells in culture. We maintained 14C-oleic acid-treated GT1–7 cells in presence or absence of serum, following which 14C-labeled intracellular lipids were extracted and resolved by thin layer chromatography. The effect of serum removal in GT1–7 cells for 30min and 2h was a progressive ~1.4-fold and ~2-fold increase in triglyceride accumulation, respectively (Figure 3A). Second, to determine whether elevated serum fatty acids during starvation contribute to hypothalamic lipid accumulation, we examined the effect of culturing GT1–7 cells in serum from fed or 24h food-deprived rats on cellular lipid accumulation. GT1–7 cells incubated in serum from food-deprived rats had higher triglyceride levels (Figure S3B) and more lipid droplets (Figure S3C) than those incubated in serum from fed rats. Furthermore, thin layer chromatograms of extracted lipids from MBH revealed a ~1.5-fold increase in triglyceride levels in 6h-fasted mice as compared to fed littermates (Figure 3B). We confirmed that the observed increase in hypothalamic triglycerides in response to serum from fasted animals results from increased triglyceride synthesis, because addition of triacsin C (inhibitor of triglyceride synthesis) completely abolished triglyceride accumulation (Figure 3C). The decreased triglyceride synthesis in the presence of triacsin C was also associated with ~2-fold increase in intracellular 14C-free fatty acids that failed to form triglycerides (Figure 3C). These results confirm that starvation increases hypothalamic free fatty acid uptake and triglyceride synthesis.
Figure 3. Fatty acid uptake during starvation induces hypothalamic autophagy.
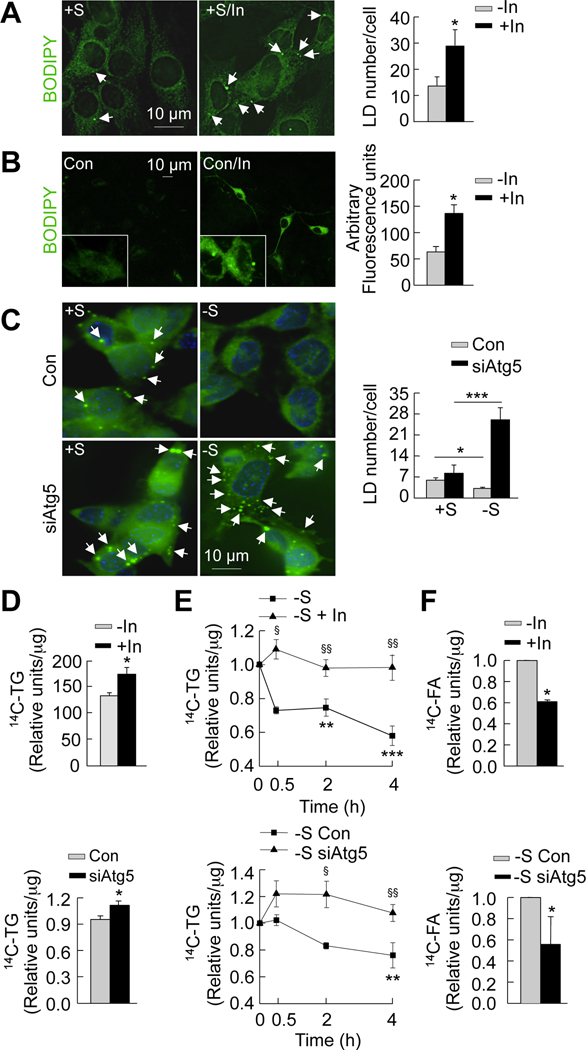
(A) Thin layer chromatogram (TLC) of 14C-triglycerides (14C-TG) from GT1–7 cells in presence (+S) or absence (−S) of serum for the indicated times. Relative densitometric values represent 14C-triglycerides per µg protein (n=3). (B) TLC of TG extracted from mediobasal hypothalamus (MBH) from fed and 6hstarved (Stv) mice. Relative densitometric values represent triglycerides per mg tissue weight (n=4–5). See also Figures S3A and S3B. (C) TLC of 14C-TG and 14C-fatty acids (14C-FA) from GT1–7 cells maintained in +S or −S treated with or without triacsin C (TrC) for indicated times. Relative densitometric values represent corresponding 14C-lipid per µg protein (n=5). (D) Immunoblot for LC3 from GT1–7 cells cultured in −S and treated with 0.06 or 0.25mM oleic acid for indicated times. Cells were treated with or without lysosomal inhibitors (In) for 30min. Values represent net LC3 flux determined by subtracting LC3-II densitometric value of untreated samples from corresponding Intreated samples (n=3). See also Figure S3D. (E) Immunofluorescence for BODIPY/LAMP1 in GT1–7 cells cultured in −S and co-treated with or without 0.25mM oleic (−S/OL) or palmitic acid (−S/Pal) for 30min, and in cells (F) cultured in 1% serum from fed (FS) or 24h-starved (SS) rats for 30min.Values represent %colocalization between BODIPY and LAMP1 (orange arrows), and are mean+SE of 20 different cells in each experiment, n=3. See also Figure S3H. (G) Confocal images for BODIPY/LAMP1 in primary hypothalamic neurons treated with or without 0.25mM oleic (OL). Values represent number of colocalized (orange arrows) puncta, and are mean+SE of more than 20 different cells in each experiment, n=3. (H) TLC of 14C-TG and 14C-FA from GT1–7 cells in +S or −S and treated with lysosomal inhibitors (In) for 30min. Relative densitometric values represent 14C-FA per µg protein (n=3). Values are mean+SE. *p<0.05, **p<0.01, ***p<0.001.
To examine whether fasting-induced increases in hypothalamic fatty acid uptake promote neuronal autophagy, serum-deprived GT1–7 cells were treated with two physiological fatty acids, oleic and palmitic acid or maintained in serum from fed or starved rats and autophagy induction was determined by assessing autophagic flux by immunoblotting for LC3 in presence or absence of lysosomal inhibitors. Treatment with either oleic (Figure 3D) or palmitic acid (Figure S3D) or culture in starved rat serum (data not shown) significantly increased autophagic flux compared to untreated controls or fed rat serum-treated cells. In addition, treating GT1–7 cells with oleic acid for 2h increased rates of lysosomal and macroautophagic (3-methyladenine (3MA)-sensitive) proteolysis by ~25% and ~9% respectively (Figure S3E). Free fatty acid treatment or culturing in serum from starved rats induced autophagy in GT1–7 cells by phosphorylating and activating AMPK, and ULK1 (Figure S3F), and not by increasing mitochondrial β-oxidation, since blockage of β-oxidation with Etomoxir did not modify LC3 flux (Figure S3G).
Hypothalamic availability of free fatty acids has been proposed to regulate food intake (Lam et al., 2005). Thus, we asked whether hypothalamic autophagy during food deprivation functioned to degrade hypothalamic lipids and thereby increase the intracellular availability of free fatty acids. To test this possibility, we performed immunofluorescence microscopy in GT1–7 cells, and confocal microscopy in primary hypothalamic neurons to determine whether fatty acid treatment increased interactions between neuronal lipid stores and autophagic compartments. In serum-starved GT1–7 cells, exposure to oleic or palmitic acid significantly increased colocalization of neutral lipid dye BODIPY 493/503 and the lysosomal marker LAMP1 (Figure 3E). Similarly, serum from 24h fooddeprived rats markedly increased BODIPY/LAMP1 colocalization in GT1–7 cells relative to those cultured in serum from fed rats (Figure 3F). Oleic and palmitic acid treatment also increased the colocalization of lipid droplets with the autophagosome marker LC3 (BODIPY/LC3; Figure S3H), demonstrating that delivery of lipids to lysosomes occurs through autophagosomes. Supporting our data from GT1–7 cells, exposure of primary hypothalamic neurons to oleic acid markedly increased colocalization of BODIPY 493/503 and LAMP1 (Figures 3G). These findings support the idea that fasting-induced increase in hypothalamic uptake of fatty acid rapidly induces autophagy, and may in turn drive lipophagy to deliver cellular lipids to lysosomes for hydrolysis.
To examine whether interactions between autophagic compartments and hypothalamic lipid stores led to their hydrolysis and the subsequent increased availability of endogenous free fatty acids, we treated serum-deprived GT1–7 cells with 14C-oleic acid in presence or absence of lysosomal inhibitors (In). Chromatographic analysis of 14C-labeled lipids revealed that inhibition of lysosomal function significantly reduced endogenous fatty acid generation from decreased breakdown of cellular triglycerides (Figure 3H). In summary, our results demonstrate that starvation increases hypothalamic fatty acid uptake and triglyceride synthesis, and support the idea that one effect of fatty acid uptake is the induction of hypothalamic autophagy to mobilize intracellular lipids.
Autophagy mobilizes hypothalamic lipids to generate endogenous fatty acids
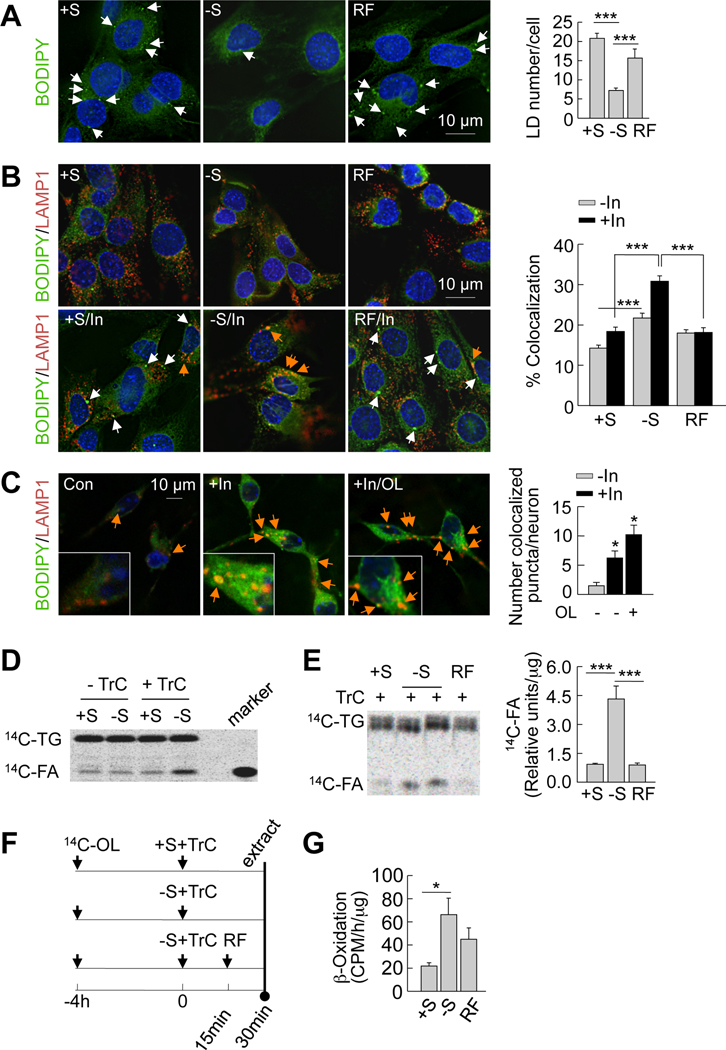
To identify the role of autophagy in the mobilization of endogenous hypothalamic lipids, we first monitored BODIPY staining for lipid stores in GT1–7 cells cultured in the presence or absence of serum or subjected to serum refeeding. Serum removal for 30min led to ~2-fold decrease in lipid droplet number (Figure 4A), consistent with the notion that induction of autophagy mobilizes hypothalamic lipid stores. Serum refeeding in GT1–7 cells restored endogenous lipid droplet content to basal levels (Figure 4A) that occurred in parallel with the decline in autophagy seen during refeeding (Figures 1A and 1B).
Figure 4. Autophagy mobilizes hypothalamic lipids to generate endogenous fatty acids.
(A) BODIPY staining in GT1–7 cells cultured in presence (+S) or absence of serum (−S) for 30min or refed (RF) for 15min following serum removal. Values represents lipid droplet (LD) number per cell, and are mean+SE, for 20 different cells in each experiment, n=3. (B) Immunofluorescence for BODIPY/LAMP1 in GT1–7 cells in +S or −S for 30min or RF for 15min, and treated with or without lysosomal inhibitors (In) for 30min. Values represent % colocalization between BODIPY and LAMP1 (orange arrows), and are mean+SE for 20 different cells in each experiment, n=3. (C) Confocal images for BODIPY/LAMP1 in primary hypothalamic neurons co-treated with or without 0.25mM oleic (OL) or lysosomal inhibitors (In) for 4h. Values represent number of colocalized (orange arrows) puncta, and are mean+SE of more than 20 different cells in each experiment, n=3. See also Figure S4. (D) Thin layer chromatogram (TLC) of 14C-triglycerides (14C-TG) and 14C-fatty acids (14C-FA) from GT1–7 cells maintained in +S or −S and treated with or without triacsin C (TrC) for 30min. (E) TLC of 14C-TG and 14C-FA from TrC-treated GT1–7 cells in +S or −S for 30min or RF for 15min. Relative densitometric values represent 14C-FA per µg protein, and are mean+SE, n=3. (F) Scheme for experiment in E. (G) β-Oxidation assay in GT1–7 cells in +S or −S for 1h or RF for 30min. Values represent 14CO2 release expressed as counts per minute (CPM)/h/µg protein, and are mean+SE, n=3. *p<0.05, ***p<0.001.
To examine if these changes in lipid droplet content were secondary to break down via autophagy, we stained hypothalamic GT1–7 cells, and primary hypothalamic neurons for BODIPY and LAMP1 in the presence or absence of lysosomal inhibitors. These should promote the accumulation of lipids delivered to the lysosomes and thereby increase BODIPY/LAMP1 colocalization. Basal serum-fed conditions resulted in minimal interaction between lipid droplets and lysosomes (Figure 4B), suggesting an inhibitory effect of nutrients on the autophagy of lipids. In contrast, the addition of lysosomal inhibitors during serum-deprivation markedly increased BODIPY/LAMP1 colocalization demonstrating increased endogenous hypothalamic lipid flux to lysosomes for degradation. Remarkably, 15min of serum refeeding dramatically reduced lipid flux to the lysosomes as observed by decreased BODIPY/LAMP1 colocalization that was not affected by addition of lysosomal inhibitors (Figure 4B). In parallel with data from GT1–7 cells, inhibition of lysosomal hydrolysis in primary hypothalamic neurons remarkably increased BODIPY/LAMP1 colocalization (Figure 4C). Addition of oleic acid to primary hypothalamic neurons treated with inhibitors of lysosomal degradation further increased BODIPY/LAMP1 colocalization (Figure 4C). Colocalizations between the lipid droplet marker, perilipin 2 and lysosomal markers, LAMP1 (Figure S4A) and LAMP-2A (Figure S4B) were also observed in MBH sections of fed mice. In totality, these results reveal dynamic interactions between endogenous lipids and the hypothalamic autophagic-lysosomal system in response to the prevailing nutrient status, and support the idea that fasting-induced hypothalamic autophagy mobilizes cellular lipids to generate free fatty acids.
To examine the degree to which starvation-induced autophagy generates endogenous fatty acids, GT1–7 cells were pulsed with 14C-labeled oleic acid for 4h allowing for esterification to 14C-labeled triglycerides, following which excess labeled oleic acid was removed and cells were chased in the presence or absence of serum. Although levels of 14C-labeled oleic acid generated from endogenous 14C-triglycerides appeared to be similar under fed and serum-starved conditions (Figure 4D), the addition of triacsin C during the chase (scheme; Figure 4F) revealed a marked ~4.5-fold increase in generation of free fatty acids (Figures 4D and 4E) in response to serum removal. These data suggest that the fate of autophagy-generated endogenous free fatty acids is consumption for cellular respiration, as reflected by ~3-fold increase in rates of β-oxidation following serum removal (Figure 4G), or re-esterification into triglycerides. The latter is supported by the finding that triacsin C treatment, which blocks triglyceride synthesis, led to accumulation of intracellular free fatty acids in GT1–7 cells during starvation (Figure 4D). Refeeding significantly decreased generation of 14C-oleic acid (Figure 4E), consistent with the inhibitory effect of refeeding on lipophagy. Taken together, these findings demonstrate that starvation increases hypothalamic lipid delivery to the lysosomes via autophagy and promotes the generation of intracellular free fatty acids.
Inhibition of autophagy in hypothalamic cells promotes neuronal lipid accumulation
To provide further support for the idea that free fatty acids are generated through autophagic degradation of endogenous lipids, we examined the effect of inhibition of autophagy on hypothalamic lipid metabolism in vitro. BODIPY staining of GT1–7 cells, and primary hypothalamic neurons cultured in the presence or absence of lysosomal inhibitors revealed that lysosomal inhibition significantly increased lipid droplet number in these cells (Figure 5A and B). Although, autophagy-deficient siAtg5 cells cultured in the presence of serum exhibited a modest increase in lipid droplet number, this difference did not reach statistical significance (Figure 5C). However, in response to serum removal, siAtg5 cells failed to mobilize cellular lipid stores compared to cells with intact autophagy (Figure 5C). These findings are consistent with the significant elevations in 14C-triglycerides in GT1–7 cells subjected to pharmacological (Figure 5D, top) or genetic (Figure 5D, bottom) inhibition of autophagy.
Figure 5. Inhibition of autophagy in hypothalamic cells promotes neuronal lipid accumulation.
(A) BODIPY staining (acquired at similar exposure times) in serum-fed GT1–7 cells (+S) cultured in presence or absence of lysosomal inhibitors (In) for 30min. Values represent lipid droplet (LD) number per cell, and are mean+SE for 20 different cells, n=3. (B) BODIPY staining (acquired at similar exposure times) in primary hypothalamic neurons cultured in presence or absence of lysosomal inhibitors (In) for 4h. Values represent arbitrary fluorescence intensity, and are mean+SE for 20 different cells, n=3. (C) BODIPY staining in control (Con) and autophagy-deficient (siAtg5) GT1–7 cells in +S or −S for 30min. Values represent LD number per cell, and are mean+SE for 20 different cells, n=3. (D) Relative densitometric values representing 14C-triglycerides (14C-TG) per µg protein calculated from thin layer chromatograms from GT1–7 cells cultured in +S for 4h (Con) or subjected to pharmacologic (In; Top) or genetic (siAtg5; Bottom) inhibition of autophagy. Values are mean+SE, n=3. (E) Decay assays for 14C-TG in GT1–7 cells chased in absence of serum for the indicated times (Con) or following pharmacologic (In; Top) or genetic (siAtg5; Bottom) inhibition of autophagy. Relative densitometric values represent residual 14C-TG per µg protein, and are mean+SE, n=4. (F) Relative densitometric values representing 14C-fatty acid (14C-FA) per µg protein calculated from thin layer chromatograms from GT1–7 cells cultured in −S for 30min (Con) or subjected to pharmacologic (In; Top) or genetic (siAtg5; Bottom) inhibition of autophagy. Values relative to −S control are mean+SE, n=3. *p<0.05, **p<0.01, and ***p<0.001. §p<0.05, §§p<0.005 compared to corresponding −S or −S Con.
We then performed 14C-labeled triglyceride decay assays to determine the amount of residual lipids in control and autophagy-deficient cells subjected to serum removal for the indicated times. Both pharmacological (Figure 5E, top) and genetic (Figure 5E, bottom) inhibition of autophagy blocked degradation of 14C-labeled triglycerides when cells were chased in absence of serum. We next directly analyzed the generation of endogenous free fatty acids in response to serum removal by pulsing cells with 14C-oleic acid and chasing in the absence of serum for 30min. We confirmed that starvation-induced generation of endogenous free fatty acids was decreased by ~40% following pharmacological (Figure 5F, top) or genetic (Figure 5F, bottom) inhibition of autophagy. These findings confirm that active hypothalamic autophagy is required to generate free fatty acids in response to starvation.
Autophagy-derived intracellular free fatty acids promote AgRP expression
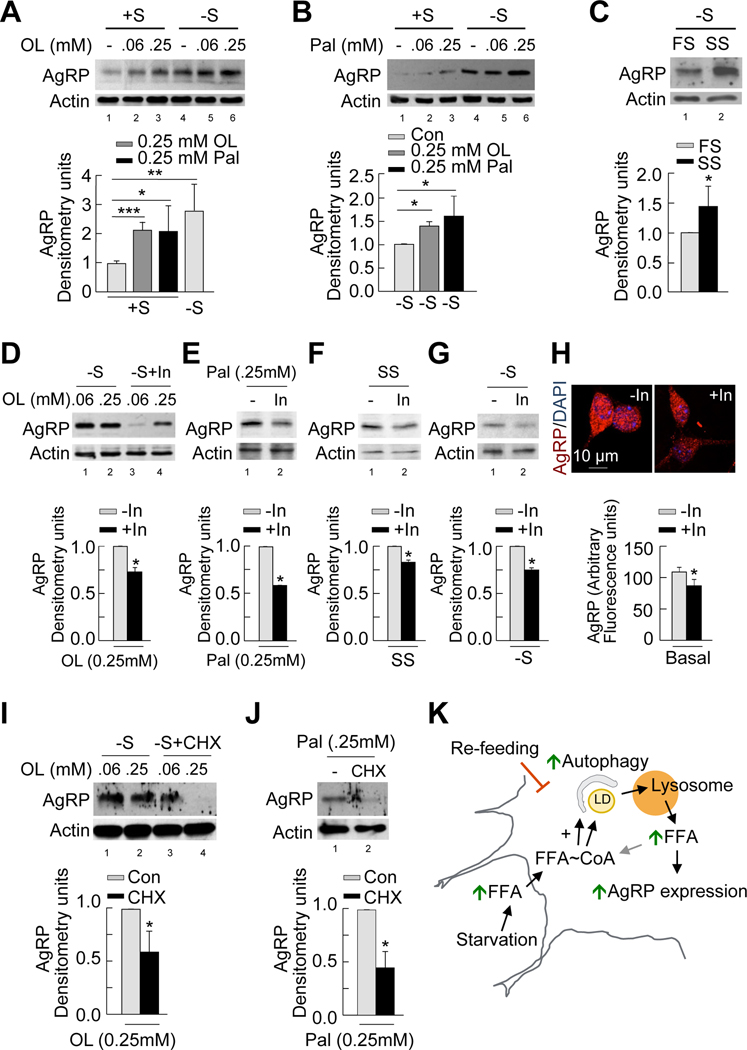
To establish a mechanistic link between free fatty acids generated by autophagy and neuropeptide expression, we first assessed the effect of exogenous administration of oleic or palmitic acid on AgRP levels. GT1–7 cells cultured in the presence or absence of serum were either untreated or treated with oleic or palmitic acid for the indicated times, following which cell lysates were immunoblotted for AgRP. Consistent with data from control cells in Figure S2F, serum removal led to a ~2.5-fold increase in AgRP levels compared to cells cultured in the presence of serum (Figures 6A and 6B, top; Figure 6A, bottom). Addition of oleic (Figure 6A, top) or palmitic acid (Figure 6B, top) to serum-supplemented cells increased AgRP levels comparable to those achieved during serum-deprivation (Figure 6A, bottom). Furthermore, 0.25mM oleic and palmitic acid increased AgRP levels by ~40 and ~60%, respectively, (Figures 6A and 6B, top; Figure 6B, bottom) compared to untreated serum-starved controls. Since fatty acid treatment induced autophagy (Figures 3D, S3D and S3E) and increased AgRP levels (Figures 6A and 6B) in GT1–7 cells, we tested whether culturing GT1–7 cells in serum from starved rats increased AgRP expression. Indeed, AgRP levels were increased ~40% in GT1–7 cells cultured in serum from starved rats as compared to cells maintained in serum from fed rats (Figure 6C). Overall, these findings suggest that increased hypothalamic fatty acid availability provides a functional link between autophagic mobilization of hypothalamic lipids and AgRP expression.
Figure 6. Autophagy-derived intracellular free fatty acids promote AgRP expression.
(A–C) Immunoblot for AgRP from GT1–7 cells in presence (+S) or absence of serum (−S) for 1h and treated without or with 0.06 and 0.25mM oleic acid (OL) (A) or palmitic acid (Pal) (B) or treated with 1% fed (FS) or starved rat serum (SS) (C). Densitometric values of AgRP normalized to untreated +S (lower panel: left); normalized to untreated −S (lower panel: middle); and normalized to FS (lower panel: right). Values are mean+SE, n=3. (D–G) Top: Immunoblot for AgRP from GT1–7 cells in −S in absence or presence of lysosomal inhibitors (In) for 1h and treated with 0.06 and 0.25mM OL (D), 0.25mM Pal (E), 1% SS (F), or untreated (G). Bottom: Densitometric values for AgRP normalized to In-untreated cells. Values are mean+SE, n=3. See also Figure S5. (H) Immunostaining for AgRP (acquired at similar exposure times) in primary hypothalamic neurons cultured in presence or absence of lysosomal inhibitors (In) for 4h. Values represent arbitrary fluorescence intensity for AgRP, and are mean+SE for 20 different cells, n=3. (I–J) Top: Immunoblot for AgRP from serum-deprived GT1–7 cells in absence or presence of the protein synthesis inhibitor, cycloheximide (CHX) for 1h following treatment with 0.06 and 0.25mM OL (I), or 0.25mM Pal (J). Bottom: Densitometric values for AgRP normalized to CHX-untreated cells. Values are mean+SE, n=3. (K) Proposed model depicting role of autophagy in hypothalamic AgRP expression. *p<0.05, **p<0.01, ***p<0.001.
If autophagy functions to increase intracellular availability of fatty acids for AgRP expression, inhibition of lysosomal function should block fatty acid-induced AgRP expression. To test this possibility, we performed similar experiments in the presence of lysosomal inhibitors. Inhibition of lysosomal degradation significantly reduced the increase in AgRP levels that occurred in response to oleic (Figure 6D) and palmitic acid (Figure 6E) treatment, following culture in starved rat serum (Figure 6F), in response to serum-deprivation (Figure 6G), and in primary hypothalamic neurons cultured under basal conditions (Figure 6H). In addition, genetic inhibition of autophagy in GT1–7 cells (siAtg5 cells) decreased AgRP induction in response to culture in starved rat serum (Figure S5A). Decreased AgRP levels following lysosomal inhibition (Figures 6D, 6E, 6F, 6G and 6H) or genetic inhibition (Figures S2F and S5A) in GT1–7 cells are consistent with the observed reduction in AgRP levels in 6h-starved AgRP neuronal autophagy-deficient mice (Figure 2G). Lysosomal inhibition in GT1–7 cells did not affect kisspeptin-1 levels, a neuropeptide unrelated to appetite regulation (Figure S5B). Exposure of GT1–7 cells to rapamycin, which induces autophagy led to increased AgRP levels (Figure S5C). These results establish that active autophagy is tightly linked to AgRP expression. We confirmed that the observed effect was not related to changes in degradation of AgRP itself, but directly to changes in AgRP synthesis. Inhibition of protein synthesis with cycloheximide significantly decreased hypothalamic AgRP levels in response to oleic (Figure 6I) and palmitic acid (Figure 6J). Based on these results, we propose the model linking increased circulating fatty acids observed in fasted conditions to the induction of hypothalamic autophagy, which in turn mobilizes neuronal lipids to generate endogenous free fatty acids that increase AgRP gene expression (Figure 6K).
DISCUSSION
In this study, we present evidence for a role of autophagy in hypothalamic AgRP neurons in the regulation of food intake and energy balance. Specific ablation of autophagy in orexigenic AgRP neurons in mice resulted in decreased body weight, reduced total body fat content, and decreased food intake in response to fasting. Mechanistic studies in AgRP-expressing hypothalamic GT1–7 cells, and in primary hypothalamic neurons revealed that starvation-induced autophagy mobilized hypothalamic lipids to generate endogenous free fatty acids that increased AgRP levels, thus revealing a new role for hypothalamic autophagy in regulating orexigenic AgRP.
A critical response to starvation is the activation of autophagy for the provision of energy (He and Klionsky, 2009). Both in hypothalamic GT1–7 cells and in murine MBH used in our studies, starvation induced autophagy. Prior studies have shown that, unlike other organs, the brain is relatively resistant to the activation of autophagy in response to starvation (Mizushima et al., 2004). Others have shown that neuronal autophagy is constitutively active under basal conditions (Boland et al., 2008) possibly to maintain cellular homeostasis (Hara et al., 2006; Komatsu et al., 2006). The present study demonstrates the unique nature of hypothalamic neurons in its ability to upregulate autophagy in response to starvation that is consistent with the roles of hypothalamic neurons in feeding and energy homeostasis.
We propose that starvation-induced increases in circulating fatty acids drive fasting-induced increases in hypothalamic fatty acid uptake and triglyceride synthesis and we suggest that hypothalamic fatty acid uptake, in turn induces autophagy during fasting. Indeed, exogenous addition of oleic or palmitic acid induced hypothalamic autophagy in GT1–7 cells, in parallel with our earlier finding that an acute lipid stimulus induces autophagy in hepatocytes in culture (Singh et al., 2009a). These results suggest that cellular free fatty acid uptake may be a general mechanism for the induction of autophagy during starvation. In contrast to an acute lipid stimulus that induces autophagy, chronic lipid loading, such as high fat feeding, leads to the inhibition of autophagy, at least as observed in the liver (Singh et al., 2009a). This begs the question whether chronic overnutrition or sustained elevation of free fatty acids as observed in insulin resistance (Lionetti et al., 2009) could alter hypothalamic lipid metabolism thus setting up a vicious circle of overfeeding and altered energy balance.
The mechanisms that regulate neuron-intrinsic availability of free fatty acids are unclear. To this end, we demonstrate in this work that starvation-induced interactions between autophagy and hypothalamic lipids stores increase the availability of hypothalamic free fatty acids. It may be asked, why would autophagy be required for the generation of an endogenous pool of fatty acids despite the large influx of free fatty acids into the hypothalamus during starvation? The immediate fate of starvation-induced fatty acid uptake is rapid triglyceride synthesis, possibly to counteract cellular toxicity arising from a rapid surge of free fatty acids into the cell. That free fatty acids rapidly form triglycerides within lipid droplets points to a crucial role for autophagy or other possible lipolytic pathways to breakdown lipid stores. This would help maintain a controlled availability of an endogenous pool of free fatty acids, functionally distinct from circulating free fatty acids during starvation. Neuronal availability of endogenous free fatty acids (Andrews et al., 2008) has been shown to play an important role in the regulation of feeding (Lam et al., 2005). Indeed, through our work, we have been able to demonstrate that the previously described regulatory role of free fatty acids on feeding originates from the fact that orexigenic AgRP expression displays a critical requirement for availability of free fatty acids. The addition of fatty acids to serum-fed hypothalamic cells in culture increased AgRP levels suggesting that free fatty acids per se can fully reproduce AgRP expression that would normally occur following serum removal.
The functional consequence of the loss of autophagy in GT1–7 cells and in AgRP neuron autophagy-deficient mice is the failure to upregulate AgRP in response to starvation. We cannot discard that other mechanisms may contribute to regulate AgRP levels during the basal fed state, and that hypothalamic autophagy seems to be the predominant mechanism for regulation of AgRP in response to starvation. Chronic inhibition of autophagy in siAtg5 cells may generate compensatory mechanisms thus explaining the higher basal AgRP mRNA expression in siAtg5 GT1–7 cells. An acute pharmacological inhibition of the autophagic-lysosomal system completely blocked the AgRP expression that occurred either in response to treatment of oleic and palmitic acid, or following culture in serum from starved rodents. These data support the role of autophagy in the generation of endogenous free fatty acids to promote AgRP expression.
Hypothalamic POMC neurons receive inhibitory projections from adjacent AgRP neurons (Garfield et al., 2009). We found that deletion of atg7 in AgRP neurons increased MBH levels of POMC preproprotein and α-MSH, suggesting that deficient autophagy may downregulate the inhibitory actions of AgRP projections at POMC neurons, thereby promoting the effect of POMC neurons on energy balance. Centrally administered α-MSH has been shown to drive behavioral and physiological determinants of overall energy balance, including reduced food intake and increased locomotor activity (Semjonous et al., 2009). These energetic consequences of elevated POMC and α-MSH levels in KO mice may contribute to their decreased body weight and adipose mass. Reduced body weight could be partly attributable to increased lipid mobilization from higher ATGL expression.
In summary, this study proposes a new conceptual framework as to how starvation can affect autophagy-regulated hypothalamic lipid metabolism that in turn regulates neuropeptide levels to modulate food intake and energy homeostasis. The ability to regulate hypothalamic autophagy to modulate energy homeostasis may have implications for the development of new therapeutic options for conditions such as obesity and the metabolic syndrome. In addition, as autophagy has been shown to decline with age (Cuervo, 2008), altered food intake and energy homeostasis observed with aging may be a consequence of decreased hypothalamic autophagy.
EXPERIMENTAL PROCEDURES
Animals and cells
We used 2–8 month male mice for our studies. AgRP-neuron specific autophagy-deficient mice (Agouti background) were generated by crossing Atg7F/F (Komatsu et al., 2005) with AgRP-Cre mice. The Atg7F/F mice, and AgRP-Cre mice were gifts from Drs. M. Komatsu and K. Tanaka (Tokyo Metropolitan Institute of Medical Science, Japan), and Dr. B.B. Lowell (Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA), respectively. Studies were performed in Atg7F/F-AgRP-Cre mice and their littermate controls that lacked the cre transgene. Mice were fed regular chow (no. 5058; LabDiet) and maintained in 12h light/dark cycles. Food restrictions were performed at night. Genotyping was performed using established primers (Komatsu et al., 2005). Mice were used under an animal protocol approved by the Institutional Animal Care and Use Committee of the Albert Einstein College of Medicine. Hypothalamic GT1–7 cells were kindly provided by Dr. P. Mellon (University of California, San Diego) and maintained in DMEM (Sigma-Aldrich) with 10% fetal bovine serum (FBS, Invitrogen) and 1% antibiotics (Invitrogen), as described (Morrison et al., 2007). For serum removal, cells were washed with phosphate-buffered saline (PBS) followed by culture in plain DMEM, where indicated. For refeeding, 10% FBS was added. Methylpyruvate, methylsuccinate, triacsin C and cycloheximide were used at 5mM, 5mM, 10µM and 20µg/ml final concentrations, respectively. Lysosomal inhibitors, leupeptin and ammonium chloride were used at 100µM and 20mM, respectively. Serum from fed and 24h-starved rats was de-complemented at 56°C for 30min prior to use.
Mouse hypothalamic primary cultures
Hypothalami were dissected from newborn pups and stored on ice in Hank’s balanced salt solution (HBSS; Gibco). Tissues were washed in 0.1M PBS (pH 7.4), incubated for 5min in a 37°C water bath in 0.25% trypsin (Gibco), and then washed with PBS. Tissues were transferred into 1ml of adult neuronal growth medium consisting of Neurobasal-A medium, B-27 supplement, 0.5mM L-glutamine (Gibco), supplemented with 10% FBS (Hyclone), 5% heat-inactivated horse serum (Gibco), and antibiotics. Tissues were gently triturated until uniform cellular dissociation was achieved, and incubated on ice for 3min to allow debris to settle. Supernatants were diluted 1:16 in adult neuronal medium and distributed into cell culture slides (BD Biosciences) coated with poly-L-lysine (Sigma-Aldrich) for immunostaining.
Immunofluorescence and immunohistochemistry
Cells were grown on coverslips, fixed in 4% paraformaldehyde, blocked, incubated in primary and fluorophore-conjugated secondary antibody (Invitrogen), and mounted in medium containing DAPI (4’,6-diamidino-2-phenylindole) (Invitrogen) for nuclear staining. Neutral lipids were stained with BODIPY 493/503 as previously described (Singh et al., 2009a). Images were acquired on an Axiovert 200 fluorescence microscope (Carl Zeiss, Inc.) with X63 objective and 1.4 numerical aperture and subjected to deconvolution with the manufacturer’s software. All images were prepared using Adobe Photoshop 6.0 software (Adobe Systems Inc.). Quantification was performed in individual frames after appropriate thresholding using ImageJ software (NIH) in a minimum of 20 cells per slide. Colocalization was calculated by JACoP plug-in (Bolte and Cordelieres, 2006) in single Z-stack sections of deconvolved images after appropriate thresholding, as described (Singh et al., 2009a). Images of primary hypothalamic cultures were acquired on a Confocal SP5 AOBS microscope with a 63X 1.4 NA oil-immersion objective (Leica, Mannheim, Germany) using Leica Confocal Software (LCS). Immunohistochemistry was done in mediobasal hypothalamic sections from brains prefixed with paraformaldehyde and incubated in 20% sucrose (Fisher Scientific) overnight at 4°C. Sections were fixed in pre-cooled acetone at −20°C, blocked with 3% fetal bovine serum in PBS containing 0.01% triton X-100. Sections were treated with the appropriate primary and fluorophore-tagged secondary antibody and visualized as above.
Thin layer chromatography of lipids
A 12.7mM sodium oleate-albumin mixture (ratio 7.1:1) was prepared as detailed in Supplemental Experimental Procedures. 14C-labeled cellular lipids and hypothalamic lipids were extracted in organic solvents (Hexanes: isopropanol; 3:2), dried in nitrogen gas, redissolved in chloroform and resolved on silica-based thin layer chromatography plates (Sigma-Aldrich) using two successive resolving systems containing chloroform, acetone, methanol, acetic acid and water (100:40:20:20:10), and hexanes, methanol and acetic acid (80:20:1). Plates were dried and images acquired by Storm analyzers using storage phosphor screens (Amersham). Densitometric analyses of images were done using ImageJ.
Fatty acid β-oxidation
The rate of fatty acid β-oxidation, as described previously (Singh et al., 2009a), was determined by measuring the amount of 14CO2 released by 14C-oleic acid-labeled cultured GT1–7 cells over 1h. The rate of β-oxidation was expressed as CPM per µg protein per hour.
Magnetic resonance spectroscopy
An ECHO (Echo Medical Systems) magnetic resonance spectroscopy instrument was used for body composition determination (Singh et al., 2009b).
Manual open field test
Mice were acclimated into test room for 1h. Tests were performed in a plexiglass black box with clear cover for an open field chamber (18” long, 18” wide, 12” high) with grid lines drawn inside creating 2” × 2” zones. During sessions rodents were scored for zone crosses and rearing. A zone cross was defined as when both hind limbs of the mouse crossed any of the gridlines in the chamber. Rearing was denoted as when the animal extended its body from its hind legs with its fore limbs supported by the sides of the chamber. Each mouse was analyzed for 10min. Locomotor activity was scored as the number of events per minute.
RT-PCR
cDNA was generated from total RNA from GT1–7 cells, and gene expression determined using SYBR Green in a Smart Cycler II thermocycler (Cepheid, Sunnyvale, CA) as described (Hubbard et al., 2010). Expression of agrp was normalized to actin. To determine AgRP levels the following primers were used: forward GTACCGCCACGAACCTCTGT; reverse TCCCCTGCCTTTCCCAAA, using conditions described previously (Morrison et al., 2007).
ELISA
AgRP secretion from GT1–7 cells was assessed using a commercial ELISA kit (Phoenix Pharmaceuticals, Inc, Burlingame, CA) according to the instructions provided.
Statistics
Numerical results are mean±SE and represent data from a minimum of 3 independent experiments. Groups were compared by 2-tailed Student’s t test (Sigma Plot, Jandel Scientific). Statistical significance was defined as p<0.05. Multiple means comparisons, based on number of groups, were done by one-way analysis of variance (ANOVA) followed by Bonferroni post hoc test or the Newman-Keuls multiple comparison test to determine statistical significance.
HIGHLIGHTS.
Starvation induces hypothalamic autophagy
Autophagy in AgRP neurons regulates food intake and energy balance
Starvation-induced hypothalamic autophagy mobilizes lipid droplets
Autophagy-derived free fatty acids increase hypothalamic AgRP levels
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the NIH NIDDK grant DK087776, and an Einstein Nathan Shock Basic Biology of Aging pilot grant to RS, AG031782 to AMC, and grants from Skirball Institute for Nutrient Sensing, NY Obesity Research Center DK026687, and Einstein Diabetes Research Center DK020541. SK and RK are supported by NIH grant T32AG023475 and a Ruth L. Kirschstein fellowship, respectively. EA is supported by Micinn/Fulbright Fellowship 2008-0128. We thank the Biostatistics Division of the Department of Epidemiology and Population Heath for their assistance with data analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors have no competing financial interests to declare.
REFERENCES
- Andrews ZB, Liu ZW, Walllingford N, Erion DM, Borok E, Friedman JM, Tschop MH, Shanabrough M, Cline G, Shulman GI, et al. UCP2 mediates ghrelin's action on NPY/AgRP neurons by lowering free radicals. Nature. 2008;454:846–851. doi: 10.1038/nature07181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgardt BF, Bruning JC. CNS leptin and insulin action in the control of energy homeostasis. Ann N Y Acad Sci. 2010;1212:97–113. doi: 10.1111/j.1749-6632.2010.05799.x. [DOI] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Cota D, Proulx K, Smith KA, Kozma SC, Thomas G, Woods SC, Seeley RJ. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Luca C, Olefsky JM. Inflammation and insulin resistance. FEBS Lett. 2008;582:97–105. doi: 10.1016/j.febslet.2007.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfield AS, Lam DD, Marston OJ, Przydzial MJ, Heisler LK. Role of central melanocortin pathways in energy homeostasis. Trends Endocrinol Metab. 2009;20:203–215. doi: 10.1016/j.tem.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. Journal of immunology. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Feng Y, Kitamura YI, Chua SC, Jr, Xu AW, Barsh GS, Rossetti L, Accili D. Forkhead protein FoxO1 mediates Agrp-dependent effects of leptin on food intake. Nat Med. 2006;12:534–540. doi: 10.1038/nm1392. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TK, Schwartz GJ, Rossetti L. Hypothalamic sensing of fatty acids. Nat Neurosci. 2005;8:579–584. doi: 10.1038/nn1456. [DOI] [PubMed] [Google Scholar]
- Lionetti L, Mollica MP, Lombardi A, Cavaliere G, Gifuni G, Barletta A. From chronic overnutrition to insulin resistance: the role of fat-storing capacity and inflammation. Nutr Metab Cardiovasc Dis. 2009;19:146–152. doi: 10.1016/j.numecd.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DL, Rui L. Recent advances in understanding leptin signaling and leptin resistance. Am J Physiol Endocrinol Metab. 2009;297:E1247–E1259. doi: 10.1152/ajpendo.00274.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison CD, Xi X, White CL, Ye J, Martin RJ. Amino acids inhibit Agrp gene expression via an mTOR-dependent mechanism. Am J Physiol Endocrinol Metab. 2007;293:E165–E171. doi: 10.1152/ajpendo.00675.2006.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountjoy KG. Functions for pro-opiomelanocortin-derived peptides in obesity and diabetes. Biochem J. 2010;428:305–324. doi: 10.1042/BJ20091957. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Piva R, Chiarle R, Manazza AD, Taulli R, Simmons W, Ambrogio C, D'Escamard V, Pellegrino E, Ponzetto C, Palestro G, et al. Ablation of oncogenic ALK is a viable therapeutic approach for anaplastic large-cell lymphomas. Blood. 2006;107:689–697. doi: 10.1182/blood-2005-05-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury A, Zhang L. Role of the arcuate nucleus of the hypothalamus in regulation of body weight during energy deficit. Mol Cell Endocrinol. 2010;316:109–119. doi: 10.1016/j.mce.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Semjonous NM, Smith KL, Parkinson JR, Gunner DJ, Liu YL, Murphy KG, Ghatei MA, Bloom SR, Small CJ. Coordinated changes in energy intake and expenditure following hypothalamic administration of neuropeptides involved in energy balance. Int J Obes (Lond) 2009;33:775–785. doi: 10.1038/ijo.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009a;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009b;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.