Abstract
Chemical investigation of the Mediterranean sponge Sarcotragus spinosulus led to the isolation of a new hydroxylated nonaprenylhydroquinone, along with two known metabolites, hepta- and octaprenylhydroquinones. The structure of the new metabolite was assigned by extensive 1D and 2D NMR analyses and MS studies. The antileukemic effect of the three compounds towards the chronic myelogenous leukemia (CML) cells line K562 was also evaluated.
Keywords: sponges, Sarcotragus spinosulus, marine natural products, hydroxylated polyprenylhydroquinone, bioactivity
1. Introduction
Several linear and cyclic prenylated hydroquinones and related secondary metabolites have been isolated from sponges [1–6], tunicates [7,8], and algae [9,10]. A large number of linear polyprenylhydroquinones have been isolated from sponges, especially from the Irciniidae family (mainly from the genera Ircinia and Sarcotragus). Biological studies conducted on several polyprenylhydroquinones showed them to have a moderate antibacterial [11,12], antiviral [13], anti-inflammatory [14,15], and phospholipase A2 activity [15]. These linear polyprenylhydroquinones could be further divided in two main groups with polyprenylhydroquinones sulfates [16–20] and hydroxylated polyprenylhydroquinones. To date only four hydroxylated polyprenylhydroquinones [5,6,20,21] have been isolated from Sarcotragus spinosulus (under the name Ircinia spinosula). They have been shown to have an enhanced antibacterial, anti-inflammatory, cytotoxic [12,15], and antioxidant activity [22] potentially associated to the presence of the hydroxyl group on the prenyl side chain.
In the course of our search for bioactive marine natural products, we have investigated the Mediterranean marine sponge Sarcotragus spinosulus (Dictyoceratida, Irciniidae) collected from Callejones, Ceuta. In this paper we report the isolation and structural elucidation of a new hydroxylated nonaprenylhydroquinone, along with two known polyprenylhydroquinones, hepta- and octaprenylhydroquinones. We also report the antileukemic effect of the three compounds towards the chronic myelogenous leukemia (CML) cells line K562.
2. Results and Discussion
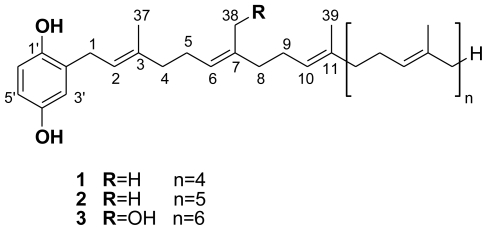
The CH2Cl2/MeOH (1:1, v/v) crude extract of Sarcotragus spinosulus was fractionated by Flash Vacuum Liquid Chromatography, eluting with a gradient of decreasing polarity from H2O to MeOH. The subsequent MeOH fraction was purified by semipreparative reverse-phase HPLC (Phenomenex Luna C6-Pheny, 250 × 10 mm id, 5 μm, gradient H2O/MeCN/Formic Acid, 96:04:0.1 to 0:100:0.1) to afford pure compounds 1 (41.7 mg), 2 (30.6 mg), and 3 (4.7 mg) (Figure 1).
Figure 1.
Structure of compounds 1, 2, and 3.
A preliminary NMR spectral analysis of all isolated compounds showed similarities and strongly supported the presence of a 2-polyprenylhydroquinone skeleton in each. Compounds 1–3 showed the same UV (280 nm) and IR (3350, 1500, 1445, 910, 785, 730 cm−1) spectra, indicating the presence of a monosubstituted hydroquinone structure. This was supported by the occurrence of three aromatic protons in the 1H NMR spectrum, two doublets at δ 6.56 (J = 8.5 Hz) and 6.52 (J = 3.0 Hz), and a double doublet at δ 6.57 (J = 8.5 and 3.0 Hz). An extensive examination of the spectroscopic data (IR, UV, 1D and 2D NMR, and MS spectra) of 1 and 2 led to their identification as hepta- and octaprenylhydroquinones, respectively. All spectral data of 1 and 2 were in agreement with previous published data [2,4].
The 1H- and 13C-NMR spectra of compound 3 were similar to those of compounds 1 and 2, except for the presence of signals at δ 4.06 (2H) and δ 60.0, respectively, in the 1H and 13C NMR spectra (Table 1), assigned to the primary alcohol group of the side chain, and small differences in the chemical shifts around the OH group. Further examinations of the 1H- and 13C-NMR data of 3 led to its identification as a hydroxylated polyprenylhydroquinone, close analogues of which have been previously isolated from S. spinosulus [5,6,20,21]. The molecular formula C51H78O3 of 3 was deduced from the HR-MALDITOF-MS data which showed a pseudomolecular adduct ion at m/z 845.4874 [M + Ag]+ (Calcd. for C51H78107AgO3 845.5001) [23]. Thus, NMR and MS data of 3 led to its identification as a hydroxylated nonaprenylhydroquinone in which one of the methyls has been oxidized to hydroxymethylene. The position of the OH group was unequivocally assigned on the second isoprene unit based on the shift of the olefinic proton H-6 to δ 5.25 (vs. 5.10 in polyprenylhydroquinone), and key H-4/H-5/H-6 COSY correlations. Confirmation was given by the key H-38/C-6, H-38/C-7, and H-38/C-8 HMBC correlations. Thus, the new natural product 3 was identified as 2′-[38-Hydroxy]nonaprenyl-1′,4′-hydroquinone. Finally, the configurations of double bonds were all assigned as E by 1H and 13C NMR chemical shifts (δ 1.68–1.54 and 17.8–16.1 for 1H and 13C, respectively) of the vinyl methyls [24].
Table 1.
1H NMR (500 MHz, CD3OD) and 13C NMR (125 MHz, CD3OD) data of compound 3.
| No. | δC (ppm) | Mult. | δH (ppm) | J (Hz) | Mult. | COSY | HMBC |
|---|---|---|---|---|---|---|---|
| 1′ | 148.9 | qC | |||||
| 2′ | 130.2 | qC | |||||
| 3′ | 117.2 | CH | 6.52 | 3.00 | d | 1 | 1, 1′, 4′, 5′ |
| 4′ | 151.1 | qC | |||||
| 5′ | 113.8 | CH | 6.41 | 8.50, 3.00 | dd | 6′ | 3′, 4′ |
| 6′ | 116.5 | CH | 6.56 | 8.50 | d | 5′ | 1, 1′, 2, 5′ |
| 1 | 29.1 | CH2 | 3.22 | 7.33 | d | 2 | 1′, 2′, 3, 3′ |
| 2 | 124.0 | CH | 5.30 | m | 1 | 1, 4 | |
| 3 | 136.8 | qC | |||||
| 4 | 40.9 | CH2 | 1.98 | m | 5 | ||
| 5 | 27.7 | CH2 | 2.16 | m | 4, 6 | 3, 4, 6, 7 | |
| 6 | 128.8 | CH | 5.25 | m | 5 | ||
| 7 | 139.4 | qC | |||||
| 8 | 35.9 | CH2 | 2.11 | m | 9 | 9, 6, 10, 38 | |
| 9 | 27.7 | CH2 | 2.05 | m | 8, 10 | ||
| 10 | 125.6 | CH | 5.10 | m | 9 | ||
| 11 | 135.8 | qC | |||||
| 12 | 40.9 | CH2 | 1.96 | m | |||
| 13 | 27.7 | CH2 | 2.05 | m | |||
| 14 | 125.6 | CH | 5.10 | m | |||
| 15 | 135.8 | qC | |||||
| 16 | 40.9 | CH2 | 1.96 | m | |||
| 17 | 27.7 | CH2 | 2.05 | m | |||
| 18 | 125.6 | CH | 5.10 | m | |||
| 19 | 135.8 | qC | |||||
| 20 | 40.9 | CH2 | 1.96 | m | |||
| 21 | 27.7 | CH2 | 2.05 | m | |||
| 22 | 125.6 | CH | 5.10 | m | |||
| 23 | 135.8 | qC | |||||
| 24 | 40.9 | CH2 | 1.96 | m | |||
| 25 | 27.7 | CH2 | 2.05 | m | |||
| 26 | 125.6 | CH | 5.10 | m | |||
| 27 | 135.8 | qC | |||||
| 28 | 40.9 | CH2 | 1.96 | m | |||
| 29 | 27.7 | CH2 | 2.05 | m | |||
| 30 | 125.6 | CH | 5.10 | m | |||
| 31 | 135.8 | qC | |||||
| 32 | 40.9 | CH2 | 1.96 | m | |||
| 33 | 27.7 | CH2 | 2.05 | m | |||
| 34 | 125.6 | CH | 5.10 | m | |||
| 35 | 135.8 | qC | |||||
| 36 | 25.9 | CH3 | 1.65 | s | 34,45 | ||
| 37 | 16.2 | CH3 | 1.69 | s | 1,2 | 2,3,4 | |
| 38 | 60.0 | CH2 | 4.06 | s | 6,7,8 | ||
| 39 | 16.2 | CH3 | 1.58 | s | |||
| 40 | 16.2 | CH3 | 1.58 | s | |||
| 41 | 16.2 | CH3 | 1.58 | s | |||
| 42 | 16.2 | CH3 | 1.58 | s | |||
| 43 | 16.2 | CH3 | 1.58 | s | |||
| 44 | 16.2 | CH3 | 1.58 | s | |||
| 45 | 17.8 | CH3 | 1.58 | s |
To the best of our knowledge this is the first report on the isolation and structure identification of a hydroxylated nonaprenylhydroquinone. Until now, four hydroxylated polyprenylhydroquinones have been reported: two heptaprenyl bearing the OH group on the first [20] and fifth prenyl moiety [6]; and two octaprenyl congeners bearing the OH group on the fourth [21] and fifth [5] isoprene unit.
From a chemotaxonomic point of view hydroxylated polyprenylhydroquinones could constitute potential markers for S. spinosulus species.
The metabolites 1–3 were evaluated for their potential antileukemic effect towards the chronic myelogenous leukemia (CML) cells line K562, which is widely used for cytotoxicity assays. The effect of compounds 1–3 was compared with that of Imatinib, the leading compound to treat patients suffering CML. This compound has proven very efficient in killing Bcr-Abl-positive cells in a caspase-dependent manner [25,26]. The IC50 values for Imatinib and compounds 1–3 for loss of cell metabolism (XTT assay) and cell number are given in Table 2.
Table 2.
IC50 values for Imatinib and compounds 1–3 for loss of cell metabolism (XTT assay) and cell number. Cells (15 × 104/mL) were incubated for 48 h at 37 °C with either increasing concentration of compounds 1–3 in the 2.5–250 μM range or with Imatinib in the 0.1–2.5 μM range. Cell metabolism was measured using the XTT assay and cell numbers was assessed by flow cytometry as indicated in the Experimental section. IC50 values are representative of three experiments made in quadruplicate.
| Compounds | Cell metabolism (IC50) | Cell number (IC50) |
|---|---|---|
| 1 | 8 | 7 |
| 2 | 10 | 12 |
| 3 | 193 | 191 |
| Imatinib | 0.4 | 0.5 |
Compounds 1 and 2 inhibited cell metabolism and cell number with very similar IC50 values (around 10 μM). Compound 3 was less efficient that the two former compounds with IC50 values of 193 and 191 μM, respectively. As a control, Imatinib was shown to inhibit cell metabolism and cell number with IC50 values of 0.4 and 0.5 μM, respectively, in agreement with previous results [26]. As compounds 1 and 2 were also found to induce annexin V externalisation in K562 cells (not shown), it is likely that the main mechanism by which both compounds inhibit cells metabolism and cells number is by inducing apoptosis.
3. Experimental Section
3.1. General
All organic solvents used for material extraction were of analytical grade and purchased from Merck (Darmstadt, Germany). Acetonitrile used for HPLC was of HPLC-grade and purchased from Fisher, USA. Formic acid of HPLC grade was purchased from Acros, USA. 2,5-Dihydroxybenzoic acid (DHB, used as the matrix for MALDI-TOF experiments, was of the highest grade available and used without further purification) and Silver trifluoroacetate (AgTFA, used as the cationizing agent) were purchased from Sigma Aldrich Co. The Chromabond C18 preparative column used for flash chromatography was obtained from Merck, USA. Imatinib was kindly provided by Novartis Pharma. UV measurements were performed on a Varian Cary 300 Scan UV-visible spectrometer. IR spectra were obtained with a Perkin-Elmer Paragon 1000 FT-IR spectrometer. Flash chromatography was performed on an Armen Instrument Spot Liquid Chromatography system, the detection wavelength was set at 254 nm. HPLC purifications were carried out on a Waters 600 system equipped with a Waters 717 plus autosampler, a Waters 996 photodiode array detector, and a Sedex 55 evaporative light-scattering detector (SEDERE, France). Detection wavelengths were set at 214, 254 and 280 nm. 1H and 13C NMR spectra were recorded with 500 MHz Bruker Avance NMR spectrometers. Chemical shifts (δ) are recorded in ppm with CD3OD (δH 3.31 ppm and δC 49.0 ppm) as internal standards with multiplicity (s, singlet; d, doublet; t, triplet; m, multiplet). High resolution mass spectra (HRMALDITOFMS) were conducted on a Perseptive Voyager DE-STR MALDI-TOF mass spectrometer (Perseptive Biosystems, Framingham, MA, USA), equipped with a 337 nm pulsed nitrogen laser (20 Hz) and a Acqiris® 2 GHz digitizer board, was used for all experiments. Mass spectra were obtained in reflectron positive ion mode with the following settings: accelerating voltage 20 kV, grid voltage 62% of accelerating voltage, extraction delay time of 100 ns. The laser intensity was set just above the ion generation threshold to obtain peaks with the highest possible signal-to-noise (S/N) ratio without significant peak broadening. All data were processed using the Data Explorer software package (Applied Biosystems).
3.2. Sponge Material
A sponge sample of Sarcotragus spinosulus (Schmidt, 1862) (Figure 2) (Demospongiae, Dictyoceratida, Irciniidae) was collected by hand using scuba at a depth of about 10 m from Callejones, Ceuta in June 2009. Taxonomic examination of the sponge was made by the authors (C.A. and J.V). The sponge shape is massive and subspherical. The color in situ is dark grey exterior, white to beige interior, and in EtOH the flesh color turned slightly to reddish. The texture is difficult to tear and has a firm and compressible consistency. The ectosome is unarmoured, thick with conules (1 to 2 mm height) irregularly scattered over the entire surface. The choanosome is cavernous. Its skeleton consists of laminated primary and secondary fibers and comprises very dense spongin filaments. Primary fibers (90 to 180 μm diameter) are pithed and clear of foreign detritus. Secondary fibers (50 to 100 μm diameter) are uncored, and the spongin filaments are long with a very thin diameter of 0.7 to 2 μm. A sponge sample (090618Ce7-05) is kept in the sponge collection of the Centre d’Océanologie de Marseille (Station Marine d’Endoume, France). The sponge was kept frozen until the extraction process.
Figure 2.
Sarcotragus spinosulus.
3.3. Extraction and Isolation
A portion of S. spinosulus was freeze-dried and ground to obtain a dry powder (20 g), which was exhaustively extracted with a mixture of MeOH/CH2Cl2 (1:1, v/v) to yield 2.2 g of the crude extract after concentration under reduced pressure. The crude extract was fractionated by RP-C18 flash chromatography (elution with a decreasing polarity gradient of H2O/MeOH from 1:0 to 0:1, then MeOH/CH2Cl2 from 1:0 to 0:1) (flow: 10 mL/min). The MeOH fraction (448 mg) was then subjected to semi-preparative HPLC-DAD (Phenomenex Luna C6-Pheny, 250 × 10 mm id, 5 μm) with a gradient of H2O/MeCN/Formic acid 96:4:0.1 to 0:100:0.1) (flow: 3.0 mL/min, injection volume: 100 μL) to afford pure compounds 1–3. All of them were identified by a combination of spectroscopic methods (1D and 2D NMR, MS) and comparison with the literature data [2,4].
3.4. Characterization Data
Compound 1: HR-MALDITOF-MS m/z 693.3808 [M + Ag]+ (Calcd. for C41H62107AgO2, 693.3795); For 1H NMR and 13C NMR data see [2,4].
Compound 2: HR-MALDITOF-MS m/z 761.4389 [M + Ag]+ (Calcd. for C46H70107AgO2, 761.4421); For 1H and 13C NMR data see [2,4].
Compound 3: yellow solid; IR (CHC13) 3350, 1500, 1445, 910, 785, 730 cm−1, UV λmax (MeOH) 280 nm (ɛ 2500); HR-MALDITOF-MS m/z 845.4874 [M + Ag]+ (Calcd. for C51H78107AgO3, 845.5001); For 1H NMR (500 MHz) and 13C NMR (125 MHz) data see Table 1.
3.5. Biological Activity
Cell Line: The human CML K562 cell line was provided by ATCC and were grown at 37 °C under 10% CO2 in RPMI 1640 medium (Gibco BRL, Paisley, UK) supplemented with 5% FCS (Gibco BRL, Paisley, UK) completed with 50 units/mL penicillin, 50 mg/mL streptomycin and 1 mM sodium pyruvate.
Measurement of Cell Metabolism (XTT): Cells (15 × 104/mL) were incubated with 1, 2 or 3 for the times indicated. 50 mL of XTT reagent (sodium 39-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy-6-nitro)benzene sulfonic acid hydrate) was added to each well. Absorbance of the formazan dye produced by metabolically active cells was measured at 490 nm as described earlier [27]. Each assay was performed in quadruplicate.
Cell death measurement: After the indicated treatment, cells were harvested and percentage of viability was measured by propidium iodide (PI) staining (0.5 Ag/mL) and flow cytometry analysis in FL-3.
4. Conclusions
A new hydroxylated nonaprenylhydroquinone, 2′-[38-Hydroxy]nonaprenyl-1′,4′-hydroquinone, along with two known metabolites, hepta- and octaprenyl-1′,4′-hydroquinones, have been isolated from the Mediterranean marine sponge Sarcotragus spinosulus. These compounds exhibited a good activity against K562 cells which will warrant further analysis at the molecular level and offer promising opportunities for the development of new antitumor agents.
Acknowledgments
This work was partially funded by the ECIMAR program (ANR-06-BDIV-001) of the French National Agency for Research. We are grateful to the Lebanese National Council for Scientific Research and EGIDE for financial support (C.A). Finally, we are grateful to T. Perez and all the diver scientists of the ECIMAR program who provided us with the specimens.
Footnotes
Samples Availability: Available from the authors.
References and Notes
- 1.Cimino G, De Stefano S, Minale L. Prenylated quinones in marine sponges Ircinia species. Experientia. 1972;28:1401–1402. [Google Scholar]
- 2.Pouchus YF, Verbist JF, Biard JF, Boukef K. Polyprenylhydroquinones de l’éponge Hippospongia communis: isolement et mise en évidence de formation d’artéfacts sur supports chromatographiques. J. Nat. Prod. 1988;51:188–189. [Google Scholar]
- 3.Venkateswarlu Y, Venkata Rami Reddy M. Three new heptaprenylhydroquinone derivatives from the sponge Ircinia fasciculata. J. Nat. Prod. 1994;57:1286–1289. [Google Scholar]
- 4.Waetjen W, Putz A, Chovolou Y, Kampkoetter A, Totzke F, Kubbutat MHG, Proksch P, Konuklugil B. Hexa-, hepta- and nonaprenylhydroquinones isolated from marine sponges Sarcotragus muscarum and Ircinia fasciculata inhibit NF-κB signalling in H4IIE cells. J. Pharm. Pharmacol. 2009;61:919–924. doi: 10.1211/jpp/61.07.0011. [DOI] [PubMed] [Google Scholar]
- 5.Cimino G, De Stefano S, Minale L. Polyprenyl derivatives from the sponge Ircinia spinosula: 2-Polyprenylbenzoquinones, 2-polyprenylbenzoquinols, prenylated furans and a C-31 difuranoterpene. Tetrahedron. 1972;28:1315–1324. [Google Scholar]
- 6.Tziveleka LA, Abatis D, Paulus K, Bauer R, Vagias C, Roussis V. Marine polyprenylated hydroquinones, quinones, and chromenols with inhibitory effects on leukotriene formation. Chem. Biodivers. 2005;2:901–909. doi: 10.1002/cbdv.200590066. [DOI] [PubMed] [Google Scholar]
- 7.Fenical W, McConnell O. Simple antibiotics from the red seaweed Dasya pedicellata var Stanfordiana. Phytochemistry. 1976;15:435–436. [Google Scholar]
- 8.Sato A, Shindo T, Kasanuki N, Hasegawa K. Antioxidant metabolites from the tunicate Amaroucium multiplicatum. J. Nat. Prod. 1989;52:975–981. doi: 10.1021/np50065a010. [DOI] [PubMed] [Google Scholar]
- 9.Ochi M, Kotsuki H, Inoue S, Taniguchi M, Tokoroyama T. Isolation of 2-(3,7,11-trimethyl-2,6,10-dodecatrienyl)hydroquinone from the brown seaweed Dictyopetris undulate. Chem. Lett. 1979;8:831–832. [Google Scholar]
- 10.Rochfort SJ, Capon RJ. New sesquiterpene/phenol from the Australian marine brown alga Perithalia caudata. J. Nat. Prod. 1994;57:849–851. [Google Scholar]
- 11.De Rosa S, Crispino A, De Guilio A, Iodice C. Biological effects of prenylated hydroquinones: in antimicrobial, brine shrimp, and fish lethality assays structure-activity relationship studies. J. Nat. Prod. 1994;57:1711–1716. doi: 10.1021/np50114a015. [DOI] [PubMed] [Google Scholar]
- 12.Mihopoulos N, Vagias C, Chinou I, Roussakis C, Scoullos M, Harvala C, Roussis VZ. Antibacterial and cytotoxic natural and synthesized hydroquinones from sponge Ircinia spinosula. Z. Naturforsch. 1999;54c:417–423. doi: 10.1515/znc-1999-5-618. [DOI] [PubMed] [Google Scholar]
- 13.Loya S, Rudi A, Kashman Y, Hizi A. Mode of inhibition of HIV reverse transcriptase by 2-hexaprenylhydroquinone, a novel general inhibitor of RNA- and DNA-directed DNA polymerases. Biochem. J. 1997;324:721–727. doi: 10.1042/bj3240721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terencio MC, Ferrandiz ML, Posadas I, Roig E, De Rosa S, De Guilo A, Paya M, Alcaraz MJ. Suppression of leukotriene B4 and tumour necrosis factor α release in acute inflammatory responses by novel prenylated hydroquinone derivatives. Arch. Pharmacol. 1998;357:565–572. doi: 10.1007/pl00005209. [DOI] [PubMed] [Google Scholar]
- 15.Gil B, Sanz MJ, Terencio MC, De Guilio A, De Rosa S, Alcaraz MJ, Paya M. Effects of marine 2-polyprenyl-1,4-hydroquinones on phospholipase A2 activity and some inflammatory responses. Eur. J. Pharmacol. 1995;285:281–288. doi: 10.1016/0014-2999(95)00419-l. [DOI] [PubMed] [Google Scholar]
- 16.Fusetani N, Sugano M, Matsunaga S, Hashimoto K, Shikama H, Ohta A, Nagano H. Isolation of a hexaprenylhydroquinone sulfate from the marine sponge Dysidea sp. as a proton-potassium ATPase inhibitor. Experientia. 1987;43:1233–1234. doi: 10.1007/BF01945539. [DOI] [PubMed] [Google Scholar]
- 17.Stonik VA, Makarieva TN, Dmitrenok AS. Sarcochromenol sulfates A–C and Sarcohydroquinone sulfates A–C, new natural products from the sponge Sarcotragus spinulosus. J. Nat. Prod. 1992;55:1256–1260. doi: 10.1021/np50087a012. [DOI] [PubMed] [Google Scholar]
- 18.De Rosa S, Crispino A, De Guilio A, Iodice C, Milone A. Sulfated polyprenylhydroquinones from the sponge Ircinia spinosula. J. Nat. Prod. 1995;58:1450–1454. [Google Scholar]
- 19.Wakimoto T, Maruyama A, Matsunaga S, Fusetani N. Octa- and Nonaprenylhydroquinone Sulfates, Inhibitors of αl,3-Fucosyltransferase VII, from an Australian Marine Sponge Sarcotragus sp. Bioorg. Med. Chem. Lett. 1999;9:727–730. doi: 10.1016/s0960-894x(99)00059-1. [DOI] [PubMed] [Google Scholar]
- 20.Bifulco G, Bruno I, Minale L, Riccio R, Debitus C, Bourdy G, Vassas A, Lavayre J. Bioactive prenylhydroquinones sulfates and a novel C31 furanoterpene alcohol sulfate from the marine sponge, Ircinia sp. J. Nat. Prod. 1995;58:1444–1449. [Google Scholar]
- 21.Erdogan I, Tanaka J, Higa T, Sener B. Terpenoids from two sponge species of the Aegean Sea. Nat. Prod. Sci. 1999;5:177–180. [Google Scholar]
- 22.Tziveleka LA, Kourounakis AP, Kourounakis PN, Roussis V, Vagias C. Antioxidant potential of natural and synthesised polyprenylated hydroquinones. Bioorg. Med. Chem. 2002;10:935–939. doi: 10.1016/s0968-0896(01)00346-7. [DOI] [PubMed] [Google Scholar]
- 23.Direct analysis of long polyprenylhydroquinones by electrospray ionization mass spectrometry (ESI-MS) or MALDITOF-MS often produces poor results requiring off-line time and sample-consuming derivatization techniques. Thus, silver trifluoroacetate was used as indicated in the experimental section to promote cationization of the isolated compounds by intense formation of [M + Ag]+ adducts.
- 24.Formally, the side-chain OH group gives rise to one (Z)-configured C=C bond, but the parent C=C arrangement remains (all-E).
- 25.Jacquel A, Herrant M, Legros L, Belhacene N, Luciano F, Pages G, Hofman P, Auberger P. Imatinib induces mitochondria-dependent apoptosis of the Bcr-Abl positive K562 cell line and its differentiation towards the erythroid lineage. FASEB J. 2003;17:2160–2162. doi: 10.1096/fj.03-0322. [DOI] [PubMed] [Google Scholar]
- 26.Jacquel A, Colosetti P, Grosso S, Belhacene N, Puissant A, Marchetti S, Breittmayer JP, Auberger P. Apoptosis and erythroid differentiation triggered by Bcr-Abl inhibitors in CML cell lines are fully distinguishable processes that exhibit different sensitivity to caspase inhibition. Oncogene. 2007;26:2445–2458. doi: 10.1038/sj.onc.1210034. [DOI] [PubMed] [Google Scholar]
- 27.Puissant A, Grosso S, Jacquel A, Belhacene N, Colosetti P, Cassuto JP, Auberger P. Imatinib mesylate-resistant human chronic myelogenous leukemia cell lines exhibit high sensitivity to the phytoalexin resveratrol. FASEB J. 2008;22:1894–1904. doi: 10.1096/fj.07-101394. [DOI] [PubMed] [Google Scholar]