Abstract
Background
The planktonic microcrustacean Daphnia pulex is among the best-studied animals in ecological, toxicological and evolutionary research. One aspect that has sustained interest in the study system is the ability of D. pulex to develop inducible defence structures when exposed to predators, such as the phantom midge larvae Chaoborus. The available draft genome sequence for D. pulex is accelerating research to identify genes that confer plastic phenotypes that are regularly cued by environmental stimuli. Yet for quantifying gene expression levels, no experimentally validated set of internal control genes exists for the accurate normalization of qRT-PCR data.
Results
In this study, we tested six candidate reference genes for normalizing transcription levels of D. pulex genes; alpha tubulin (aTub), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), TATA box binding protein (Tbp) syntaxin 16 (Stx16), X-box binding protein 1 (Xbp1) and CAPON, a protein associated with the neuronal nitric oxide synthase, were selected on the basis of an earlier study and from microarray studies. One additional gene, a matrix metalloproteinase (MMP), was tested to validate its transcriptional response to Chaoborus, which was earlier observed in a microarray study. The transcription profiles of these seven genes were assessed by qRT-PCR from RNA of juvenile D. pulex that showed induced defences in comparison to untreated control animals. We tested the individual suitability of genes for expression normalization using the programs geNorm, NormFinder and BestKeeper. Intriguingly, Xbp1, Tbp, CAPON and Stx16 were selected as ideal reference genes. Analyses on the relative expression level using the software REST showed that both classical housekeeping candidate genes (aTub and GAPDH) were significantly downregulated, whereas the MMP gene was shown to be significantly upregulated, as predicted. aTub is a particularly ill suited reference gene because five copies are found in the D. pulex genome sequence. When applying aTub for expression normalization Xbp1 and Tbp are falsely reported as significantly upregulated.
Conclusions
Our results suggest that the genes Xbp1, Tbp, CAPON and Stx16 are suitable reference genes for accurate normalization in qRT-PCR studies using Chaoborus-induced D. pulex specimens. Furthermore, our study underscores the importance of verifying the expression stability of putative reference genes for normalization of expression levels.
Background
Gene-expression studies provide insights into the regulatory processes of genes that modulate phenotypes of organisms. The two most reliable techniques to date that directly measure and compare the differential response in gene expression are microarray studies and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analyses. Microarrays provide a high-throughput measurement of the transcriptional changes for thousands of genes, within short time, from limited number of RNA samples. However, quality of expression data can vary substantially and is often validated by a second method. In contrast to microarrays, qRT-PCR provides precise quantification over a wider dynamic range. Because of the higher sensitivity, qRT-PCR is often used to validate microarray data. Several variables can still introduce biases in qRT-PCR studies. For example, the amount and quality of template RNA, the enzymatic efficiencies and other in vitro artefacts can add technical variability to the data y[1-4]. Hence, experiments are typically standardized and expression levels need to be normalized [5,6]. Normalization is accomplished in several ways; the expression values from target genes of interest can be balanced against the total amount of RNA in the reactions, balanced against synthetic RNA that is "spiked" in each reaction, or measured against internal reference genes. This last method is generally considered reliable and is frequently applied for quantifying relative gene expression [7]. Yet, several studies have shown that this approach can introduce large errors when the expression of such "housekeeping genes" varies under different treatments and in different tissues [e.g. [8]]. To improve robustness of the experiment, it is recommended to use more than one reference gene [7,9] and to verify that their transcriptional activity are stable across conditions and tissue types. Because it is difficult to assess the expression stability of a reference gene by itself, current approaches aim to analyze the expression levels of several candidate reference genes with respect to each other. In samples with different amounts of input RNA the ratio of two ideal reference genes remains constant.
Once suitable reference genes for a certain experiment are selected, a normalization factor (NF), which is the geometric mean of the crossing point (CP) values of the reference genes, is calculated for normalization of the genes under investigation, i.e. to remove nonspecific variation in the data.
Selection of reference genes for Daphnia pulex
So far, no systematic validation of reference genes in D. pulex has been published. In a recent study, Schwarzenberger et al. [10] tested the expression stability of seven genes in a different species, D. magna, and found glyceraldehyde-3-phosphate dehydrogenase (GAPDH), TATA box binding protein (Tbp), and succinate dehydrogenase (sucDH) suitable for normalization of gene expression in predator experiments (fish and phantom midge larvae Chaoborus). Under low-food quality conditions (microcystin-producing strain of the cyanobacterium Microcystis), Tbp, 18S, and alpha tubulin (aTub) were suitable reference genes and GAPDH and ubiquitin conjugating enzyme (UBC) significantly upregulated [10]. Heckmann et al. [11] investigated the expression stability of several genes also in D. magna, when exposed to ibuprofen. They selected GAPDH, an actin gene (similar to actin isoform 3 in D. pulex) and UBC as the most stable reference genes and showed that aTub was differentially regulated. Rider and LeBlanc [12] and Zeis et al. [13] used beta-actin as a single reference gene for D. magna without prior validation of its suitability for expression normalization. The candidate reference genes chosen for our analysis include three frequently applied housekeeping genes, aTub, GAPDH, Tbp, and three genes with microarray support for stable expression: Syntaxin 16 (Stx16), X-box binding protein 1 (Xbp1) and CAPON (see Table 1). Xbp1 is a transcription factor which is activated through differential splicing. It plays a major role in unfolded protein response in eukaryotes [14] and is critical for larval development of Drosophila [15].
Table 1.
Candidate reference and differentially expressed genes with putative function and gene ID from the Dappu V1.1 draft genome annotation, primer sequences, amplicon characteristics.
| Gene symbol | Gene name | (putative) Function | Gene ID | P | Primer sequences [5'→3'] | L (bp) | Localization in gene | E (%) |
|---|---|---|---|---|---|---|---|---|
| aTub | alpha Tubulin | Cytoskeletal protein | Dappu-301837 | 5 | GCATGTTGTCCAACACTACTGC | 135 | 3' exon | 91 |
| CCTCAGAGAACTCTCCCTCCTC | ||||||||
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Glycolytic enzyme | Dappu-302823 | 0 | TGGGATGAGTCACTGGCATAC | 136 | 3' exon | 93 |
| GAAAGGACGACCAACAACAAAC | ||||||||
| Tbp | TATA binding protein | Transcription initiation | Dappu-194512 | 0 | CTACGATGCATTCGATAACATATACC | 144 | 3' exon | 90 |
| AGAACCAGCAATGAGTTAAACAAAG | ||||||||
| Stx16 | Syntaxin 16 | Protein involved in exocytosis | Dappu-194044 | 0 | CACATTGGTCGTCCTTAGTCTTG | 148 | 3' exon | 93 |
| TGCTATACGTTACGCTTGTCCTTAC | ||||||||
| Xbp1 | X-box binding protein 1 | Transcription factor | Dappu-314438 | 0 | CCGATATTCGAGACTGCAATG | 131 | 3' exon | 93 |
| AAAGATGGGTGAGCCAGAAATAC | ||||||||
| MMP | Matrix metallo-proteinase | Degradation of extracellular proteins | Dappu-303491 | 0 | CGAAACATGGACGCATAACTC | 80 | spanning penultimate 3' intron | 92 |
| GTCCCAAAGTGTGACCGAAC | ||||||||
| CAPON | C-terminal pdz ligand of neuronal nitric oxide synthase | Location of neuronal nitric oxide synthase | Dappu-100564 | 0 | TAACGAGTCGGGAGGAAGTG | 140 | 3' exon | 94 |
| GCTGGACTTGAGCCAGTATCTC |
Abbreviations: E: amplification efficiency; P: number of paralogs in D. pulex genome; L: length of the amplicon
CAPON is a protein which targets the neuronal nitric-oxide synthase to the presynaptic nerve terminal in mice [16]. To test the performance of candidate reference genes in an actual experiment, one gene with expected differential expression levels encoding for a matrix metalloproteinase (MMP) was selected on the basis microarray data (manuscript in preparation). MMPs are a family of evolutionary conserved extracellular proteases that play important roles in cell-cell signaling processes in most animal species [17-19]. Most importantly, they process and degrade extra- and pericellular proteins. A precise function of MMP in D. pulex has not yet been described.
The expression levels of these seven genes were measured by qRT-PCR using D. pulex juveniles that were exposed to water-borne chemical cues (kairomones) released by Chaoborus larvae and manifested the characteristic defence-against-predator phenotype called neckteeth ('induced' animals) [20]. These recorded gene transcript levels were compared to expression levels in unexposed juveniles without neckteeth (control) that have not been exposed to Chaoborus larvae. To date, no single best strategy for the selection of reference genes exists. Therefore, the suitability of the genes as reference markers for normalization was assessed using three different algorithms implemented in the programs BestKeeper [9], geNorm [7] and NormFinder [21] and evaluated by normalizing the expression level of a regulated gene against different sets of the candidate reference genes.
Results and discussion
RNA Quality
RNA concentration and purity was measured with the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). The mean (± SD) A260/280 ratio of the samples was 2.02 ± 0.05, indicating pure (protein free) RNA quality. RNA integrity of samples was further checked by capillary gel electrophoresis on the StdSens chip of the Experion RNA StdSens Analysis Kit (Bio-Rad).
qRT-PCR efficiencies and intra-assay variation
The expression levels of the seven candidate reference genes were measured in triplicates with qRT-PCR in 12 biological replicates of each induced and non-induced daphnids (50 individuals per replicate). The CP values, which negatively correlate with the concentration of target sequence present at the very beginning of the amplification reaction [22], ranged from 19.72 cycles for the gene with the highest expression (GAPDH) to 28.95 cycles for MMP, which showed the lowest expression (Table 2). The standard deviation (SD) within triplicates ranged from 0.044 to 0.532 cycles with a mean of 0.242 cycles. Mean PCR efficiencies varied from 90% to 94% (Table 1).
Table 2.
Descriptive statistic analysis with BestKeeper
| Gene | aTub | GAPDH | Tbp | Stx16 | Xbp1 | MMP | CAPON |
|---|---|---|---|---|---|---|---|
| N | 24 | 24 | 24 | 24 | 24 | 24 | 24 |
| GM [CP] | 20.83 | 19.7 | 27.66 | 26.88 | 26.41 | 28.89 | 27.73 |
| AM [CP] | 20.87 | 19.72 | 27.68 | 26.9 | 26.44 | 28.95 | 27.74 |
| Min [CP] | 19.19 | 18.67 | 26.74 | 25.49 | 24.81 | 24.63 | 26.43 |
| Max [CP] | 26.84 | 23.58 | 30.16 | 30.71 | 29.14 | 33.67 | 30.21 |
| SD [CP] | 0.82 | 0.65 | 0.65 | 0.77 | 0.86 | 1.56 | 0.71 |
| CV [% CP] | 3.92 | 3.29 | 2.37 | 2.86 | 3.24 | 5.37 | 2.54 |
| Min [x-fold] | -2.68 | -1.89 | -1.73 | -2.36 | -2.69 | -13.75 | -2.26 |
| Max [x-fold] | 36.98 | 10.96 | 4.4 | 10.6 | 5.43 | 18.92 | 4.72 |
| SD [± x-fold] | 1.62 | 1.47 | 1.47 | 1.58 | 1.66 | 2.51 | 1.52 |
| BK Corr [r] | 0.875 | 0.84 | 0.952 | 0.952 | 0.978 | 0.751 | 0.884 |
| p-value | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| BK Corr -MMP [r] | 0.943 | 0.917 | 0.922 | 0.982 | 0.944 | 0.803 | |
| p-value | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
Abbreviations: N: number of samples; GM [CP]: the geometric mean of CP; AM [CP]: the arithmetic mean of CP; Min [CP] and Max [CP]: the extreme values of CP; SD [CP]: the standard deviation of the CP; CV [% CP]: the coefficient of variance expressed as a percentage on the CP level; Min [x-fold] and Max [x-fold]: the extreme values of expression levels expressed as an absolute x-fold over- or under-regulation coefficient; SD [± x-fold]: standard deviation of the absolute regulation coefficients; BK CorrC [r]: Pearson correlation coefficient, correlation between the BestKeeper index and the contributing gene. BK Corr -MMP [r]: Pearson correlation coefficient between BestKeeper index without MMP in the analysis and the contributing gene.
Descriptive analysis of the reference genes
The expression variation of each candidate reference gene was assessed using BestKeeper v. 1.0 [9]. Pfaffl et al. [9] recommend to exclude genes with a SD of the mean CP of > 1 from the NF (here called BestKeeper Index), which corresponds to a starting template variation by the factor two. MMP had a SD [CP] of 1.56 and was therefore excluded for further analyses (Table 2). The remaining genes showed minor fluctuations in expression levels (0.65 < SD [CP] < 0.86) and a strong correlation with the BestKeeper index (coefficient of correlation r between 0.803 and 0.982) after the exclusion of MMP, which indicates expression stability. The expression of all genes highly correlated with the NF, which is supported by p-values < 0.001.
Ranking the candidate reference genes
The candidate reference genes were ranked with respect to their suitability as reference genes using the programs geNorm v. 3.5 [7] and NormFinder v.0.953 [21]. geNorm utilizes a pairwise comparison approach and calculates a gene-stability measure M, which is the arithmetic mean of the pairwise variations between a particular gene and all other candidate control genes. The least stable genes have the highest M values and are successively excluded. The program also indicates the minimum number of reference genes that should be included in the NF by calculating the pairwise variation V reflecting the effect of the inclusion of an additional control gene on the NF. The authors of geNorm suggest the analysis of at least 8 samples per group and 5 to 10 candidate genes. Figure 1A shows the stepwise exclusion of the least stable genes and the average expression stability measure M of the remaining genes. The last two genes could not be further ranked because the calculation involves the ratio of expression levels. Also in this calculation, MMP was the first gene to be excluded because of the high value for M. The results of geNorm suggest Tbp and CAPON as the most stably expressed reference genes. However, Vandesompele et al. [7] highly recommended using at least three reference genes and a cut-off value of pairwise variation of 0.15. Starting with Tbp and CAPON we included Xbp1 as a third reference gene (Figure 1B). The pairwise variation for the inclusion of Xbp1 was 0.135 (V2/3) and thus lies below the suggested cut-off. Pairwise variation even further decreased with the inclusion of Stx16 (V3/4: 0.122), GAPDH (V4/5: 0.114) and aTub (V5/6: 0.108) and exceeds the 0.15 pairwise variation cut-off only after the inclusion of MMP (V6/7: 0.197).
Figure 1.
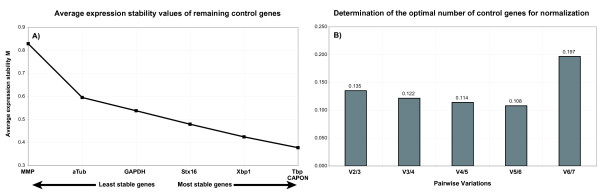
Stability of the investigated candidate reference genes (A) and pairwise variations (B) calculated with geNorm.
The second program utilized was NormFinder. It applies a model-based approach, which in contrast to geNorm allows the assignment of groups to the samples (treatment vs. control). Inter- and intragroup variations are used for the calculation of a stability value i.e., candidates with minimal combined intra- and intergroup variation are ranked as the most stable genes. This approach has advantages over the pairwise comparison approach of geNorm if coregulated genes, inappropriate as reference genes, could bias the results [21].
In a first analysis with NormFinder, all seven genes were tested for intra- and intergroup variation by assigning the samples to the two groups (induced and control).
The intergroup variation was very high for MMP (± 0.476) and lowest for those of CAPON, Tbp and Xbp1, which lay between ± 0.008 and ± 0.037 (Table 3). The program indicated Xbp1 as the best reference gene with a stability value of 0.079 and Tbp and Xbp1 as the best combination of two genes with an even better combined stability value of 0.075.
Table 3.
Candidate reference gene inter- and intragroup variation (conrol vs. induced specimens).
| Gene name | Stability value | Intergroup variation | Intragroup variation | ||
|---|---|---|---|---|---|
| Induced | Control | Induced | Control | ||
| Xbp1 | 0.079 | 0.037 | -0.037 | 0.02 | 0.005 |
| Tbp | 0.092 | 0.019 | -0.019 | 0.06 | 0.014 |
| CAPON | 0.101 | 0.008 | -0.008 | 0.163 | 0.004 |
| Stx16 | 0.175 | -0.121 | 0.121 | 0.062 | 0.001 |
| aTub | 0.337 | -0.202 | 0.202 | 0.409 | 0.046 |
| GAPDH | 0.338 | -0.217 | 0.217 | 0.128 | 0.124 |
| MMP | 0.602 | 0.476 | -0.476 | 0.742 | 0.468 |
| Best gene | Xbp1 | ||||
| Stability value | 0.079 | ||||
| Best combination of two genes | Tbp and Xbp1 | ||||
| Stability value for best combination of two genes | 0.075 | ||||
Ranking according to expression stability calculated with NormFinder (NormFinder I analysis)
The NormFinder approach attempts to compensate for expression differences between treatment and control by selecting combinations of genes with opposite expression and as little intra- and intergroup variation as possible. Therefore, in our second NormFinder analysis, those genes with a high intergroup variation (aTub, GAPDH and MMP) - and thus a high bias on the selection of the best genes for normalization - were excluded (Table 4, NormFinder II analysis). The analyses of these genes in this study revealed that Stx16 has the lowest, i.e. best stability value in combination with Xbp1 (0.076), although Stx16 alone had the highest, i.e. worst stability value (0.146). The variations in expression levels are opposite and thus compensate for each other.
Table 4.
NormFinder analysis of the four genes with the lowest (best) stability value in the previous analysis (NormFinder II analysis)
| Intergroup variation | Intragroup variation | ||||
|---|---|---|---|---|---|
| Stability value | Induced | Control | Induced | Control | |
| Xbp1 | 0.088 | 0.051 | -0.051 | 0.024 | 0.006 |
| Tbp | 0.093 | 0.033 | -0.033 | 0.079 | 0.013 |
| CAPON | 0.102 | 0.022 | -0.022 | 0.02 | 0.044 |
| Stx16 | 0.146 | -0.107 | 0.107 | 0.288 | 0.04 |
| Best gene | Xbp1 | ||||
| Stability value | 0.088 | ||||
| Best combination of two genes | Xbp1 and Stx16 | ||||
| Stability value for best combination of two genes | 0.076 | ||||
The three different algorithmic approaches applied in this study yielded mostly concordant results. All programs clearly identified MMP as an unstably expressed gene. BestKeeper, however, did not provide information, which of the remaining genes should best be used to normalize qRT-PCR results. The programs geNorm and NormFinder both ranked Xbp1, Tbp, CAPON and Stx16 as the genes with the highest expression stability, albeit in a different order. As briefly mentioned above, the use and comparison of both programs is highly recommended, because in principle, results can be strongly biased by the analytical approach selected [23].
Because the results of geNorm and NormFinder are largely concordant, we conclude that there are no significantly coregulated genes in the seven genes studied. Thus, there is no need to choose genes with opposite regulation in induced and control samples. It is largely accepted that at least three reference genes should be used for normalization, as every additional gene increases the robustness of the NF. Based on the analyses using geNorm and NormFinder, we consider Tbp, CAPON and Xbp1 as a good set of internal reference genes for expression analysis of Chaoborus-treated daphnids (Table 5). The results of the NormFinder II analysis (Table 4) showed that the three highest ranked genes (Xbp1, Tbp and CAPON) have a slight tendency towards higher expression in the induced compared to the non-induced samples. It might therefore be advisable to include Stx16 as a fourth reference gene, because of its opposing expression, as advocated by Andersen et al. [21].
Table 5.
Most stable reference genes and optimum number of reference genes calculated by geNorm and NormFinder
| geNorm | NormFinder | ||
| 1 st analysis | 2 nd analysis | ||
| Tbp/CAPON | Xbp1 | Xbp1 | |
| Xbp1 | Tbp | Tbp | |
| Stx16 | CAPON | CAPON | |
| GAPDH | Stx16 | Stx16 | |
| aTub | aTub | ||
| MMP | GAPDH | ||
| MMP | |||
| Optimum number | 3 | 2 | 2 |
Genes are ranked according to Figure 1A (geNorm) and to their stability values in Table 3 and 4, respectively (NormFinder). In bold letters: The recommended optimum combination of genes.
The genes Xbp1 and CAPON have not yet been used as normalization genes in any study. However, the systematic validation in this study provides evidence that they are suitable reference genes under these experimental conditions despite comparatively low transcription levels (CP > 25). Most importantly, they are much more stably expressed between experimental groups than the classical housekeeping genes aTub and GAPDH. In general, variation is expected to be inversely proportional to the amplified target amount but low variation despite high CP (Ct) values has been observed in other studies as well [e.g. [24]].
Evaluation of the selected reference genes
The choice of reference genes can have a strong impact on the results in relative expression studies [25]. To test the impact of reference gene selection and to evaluate the suitability of the reference genes selected in this study, the differential expression of MMP between Chaoborus-induced and non-induced daphnids was assessed using three different sets of reference genes - (1) the most stable genes identified by geNorm and NormFinder, (2) the classical housekeeping genes (aTub, GAPDH and Tbp) and (3) all six candidate reference genes (Table 6, Figure 2). Furthermore, we tested aTub and GAPDH, which were deemed unstable with high intergroup variation, to determine if this was due to unspecific fluctuations or to a significant differential expression of both genes (Table 6, Figure 2). In addition, we assessed the effects of using single traditional housekeeping genes (aTub and GAPDH) for normalization of expression levels only (additional file 1).
Table 6.
Results of the gene expression analysis with REST for aTub, GAPDH and MMP with different normalization strategies
| Normalized against | Target gene | x-fold change in gene expression* | SE range | P |
|---|---|---|---|---|
| geNorm: Tbp, Xbp1, Capon | aTub | 0.62 | 0.50 - 1.02 | 0.009 |
| GAPDH | 0.60 | 0.32 - 1.06 | 0.007 | |
| MMP | 2.64 | 1.06 - 7.18 | 0.003 | |
| NormFinder I: Tbp, Xbp1 | aTub | 0.61 | 0.51 - 0.97 | 0.001 |
| GAPDH | 0.59 | 0.33 - 0.97 | 0.002 | |
| MMP | 2.60 | 0.90 - 7.68 | 0.005 | |
| NormFinder II: Xbp1, Stx16 | aTub | 0.71 | 0.51 - 1.13 | 0.034 |
| GAPDH | 0.69 | 0.44 - 1.08 | 0.013 | |
| MMP | 3.02 | 1.05 - 9.80 | 0.003 | |
| Classical HKGs: Tbp, aTub, GAPDH | MMP | 3.66 | 1.00 - 13.16 | 0.003 |
| All genes | MMP | 3.27 | 1.11 - 11.07 | 0.002 |
*x-fold change in gene expression in the induced samples compared to the control group.
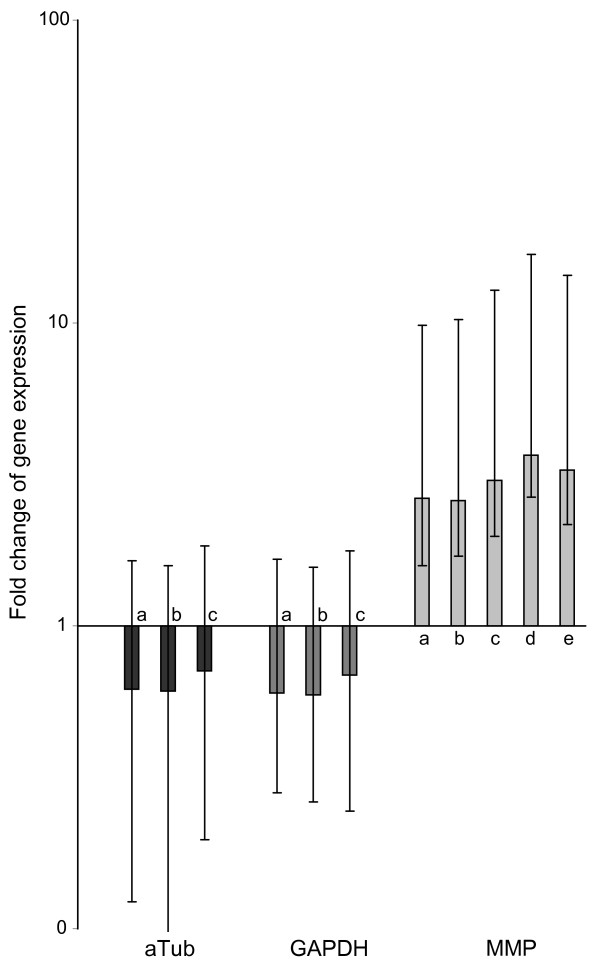
Figure 2.
Differential expression of aTub, GAPDH and MMP evaluated with REST using different normalization strategies. Program and genes used as reference genes for normalization: a) geNorm: Tbp/Xbp1/CAPON; b) NormFinder I: Tbp/Xbp1; c) NormFinder II: Xbp1/Stx16; d) the classical house-keeping genes Tbp/aTub/GAPDH; e) all genes, i.e.: Tbp/aTub/GAPDH/Stx16/Xbp1/CAPON. Boxes represent the interquartile range, or the middle 50% of observations. The dotted line represents the median gene expression. Whiskers represent the minimum and maximum observations.
Expression analyses were done using the relative expression software tool REST v. 2.0.7 [26]. REST applies the efficiency-corrected comparative CP method [27] and performs randomization tests to estimate a sample's expression ratio and the likelihood of up or down regulation, taking into account several reference genes and the individual amplification efficiency of each gene. The P values obtained from the randomization tests in REST indicate the likelihood of observing differences between sample and control groups due to chance alone. They were calculated by 10,000 random reallocations of samples and controls between the groups and counting the number of times the relative expression on the randomly assigned group is greater than the sample data (see REST manual http://www.gmo-qpcr-analysis.com/REST2008_Manual_v207.pdf).
Differential expression measurements of MMP indicated a significant upregulation irrespective of the normalization strategy (Table 6, Figure 2). Similarly, both traditional 'housekeeping' genes aTub and GAPDH were reported significantly downregulated in induced animals, which supports the finding of a significant downregulation on protein level for aTub [28] and differs from the results of Schwarzenberger et al. [10] for D. magna.
Differential expression levels reported by the REST program depended on the reference genes selected for normalization (Figure 2). For MMP they ranged from 2.64 to 3.66 in the induced samples, whereas for aTub (0.61 to 0.71) and GAPDH (0.59 to 0.69) fluctuations were less prominent. Since REST uses expression ratios rather than CP raw values for randomization tests and the data visualized in the Whisker box plots (Figure 2, additional file 1) often portray lopsided distributions with standard errors.
Because aTub and GAPDH show variations in expression, they are not suitable reference genes for such experiments with D. pulex. Even though no strong difference on the regulation of MMP is observed the impact reference genes have on the results become evident when using classical housekeeping genes aTub and GAPDH as reference genes. When selecting aTub as a single reference gene for normalization, REST reports a significant upregulation not only for MMP but also for Xbp1 and Tbp (1.66 and 1.61; see additional file 1a). No differential regulation is then reported for GAPDH due to similar (low) expression. These results highlight the importance of testing for expression stability of reference genes. When selecting GAPDH as the only reference (additional file 1b), MMP is the only gene with significant differential expression, aTub is reported to underlie no significant differential expression. Altogether, the false positive report of differential expression for Xbp1 and Tbp and the false negative report of differential expression for GAPDH when using aTub as reference gene for expression normalization highlights the importance of careful reference gene selection according to the approach outlined in this study.
Alpha tubulin paralogs in Daphnia pulex
Of clear importance for gene expression studies using D. pulex is the large number of duplicated gene families, many comprised of recently derived paralogs. In the genome of D. pulex, at least five aTub genes were identified (Dappu-318433, Dappu-306726, Dappu-301837, Dappu-315806 and Dappu-315805) http://wfleabase.org. Interestingly, in microarray experiments, members of this gene family have opposing directionality in their expression. This has been observed also for other genes in other taxon groups [e.g. [29]]. This issue might bias the results of the qRT-PCR when primers interrogate more than one aTub gene. In any case, such qRT-PCR markers are avoidable by querying the genome sequence.
Functional role of MMP
Our finding that MMP is significantly upregulated by qRT-PCR was expected; this gene is noted in a microarray study (DGC manuscript in preparation) to be upregulated in juvenile daphnids exposed to Chaoborus kairomone. According to the most recent JGI annotation, the MMP gene found is a Meprin A metalloprotease. NCBI BLAST matches it to genes that are members of the protein family Astacin (peptidase family M12A; PF01400). MMPs are known to play an important role in development, in particular in degrading and processing proteins, and are associated with cell-cell signalling pathways [17-19]. Although our data clearly identified MMP as a differentially expressed candidate gene, subsequent analyses should now focus on analyzing the function of this gene product in D. pulex to understand its relevance in the context of predator-induced defences.
Conclusions
Our results suggest that Xbp1, Tbp, CAPON and Stx16 are suitable internal reference genes for studying relative gene expression levels in D. pulex that are challenged by Chaoborus predation. Two traditional housekeeping genes, GAPDH and aTub, were studied with qRT-PCR and found to have a strong expression variation and were significantly downregulated. One candidate gene with assumed differential expression, MMP, was found to be significantly upregulated. Using aTub as reference gene leads to a strong bias in reported expression levels emphasizing the importance of thorough reference gene evaluation prior to target gene expression profiling. In particular, for further studies investigating transcriptional responses of Daphnia to other treatments we recommend to experimentally verify stable expression of reference genes prior to data acquisition in order to improve accuracy and reliability of qRT-PCR data.
Methods
Test species
Daphnia pulex (Clone R9) were used for our study. Daphnia medium consisted of charcoal filtered tab water. All Daphnia cultures and experiments were conducted at 20°C with a 16/8-h light/dark cycle. Culture daphnids were maintained at a density of ~50 animals per litre and fed daily with the unicellular green algae Scenedesmus spp. which was cultured in the laboratory.
Predator assay
Induction of D. pulex was carried out by incubating age- synchronized adult females in 1 l glass beakers with a nylon net cage containing 20 Chaoborus sp. larvae so that the adult females and their offspring had contact with Chaoborus kairomones but would not be preyed upon. Chaoborus larvae were fed daily with 40 juvenile daphnids. The control group was reared in similar glass beakers with nylon net cages that did not contain Chaoborus larvae.
Twelve biological replicates of induction and control, respectively, were conducted.
Neonate daphnids were separated daily from the mothers. They were reared under the same conditions until they reached the second juvenile instar, and batches of 50 animals were preserved in 20 μl RNAlater (Qiagen) and stored at 4°C until RNA extraction. A representative amount of induced and non-induced juveniles were checked for the presence and absence of neckteeth, respectively. Batches that did not show the appropriate phenotypic expression were discarded.
RNA isolation and cDNA synthesis
RNAlater was decanted from the specimens and RNA was extracted using the MasterPure Complete DNA and RNA Purification Kit (Epicentre) according to the manufacturer's protocol. The purified nucleic acids were resuspended in 30 μl of RNAse free water. The integrity of the RNA samples was checked with Experion RNA StdSens Analysis Kit (Bio-Rad) and concentration and purity with the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). An amount of 1 μg of each extraction was reverse-transcribed with QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's protocol, which included a 20 min DNAse I digestion prior to reverse transcription. For reverse transcription oligo(dT) primers (1 μM) were used. The cDNA was diluted ten-fold with RNAse free water.
Samples were checked with PCR (GAPDH primer pair) for genomic DNA contamination after the DNAse I digestion. An additional positive control was the single product of 80 bp in the qRT-PCR reactions with the MMP primer pair which was designed to span an intron.
Identification of candidate reference genes
The sequences of putative genes with microarray and EST support and a gene prediction (data available from http://wfleabase.org) were aligned using tBLASTx against NCBI (National Center for Biotechnology Information) nucleotide database sequences to find homologue genes and assign a putative function. Unpublished microarray data (manuscript in preparation) were then taken as basis to estimate expression stability.
Design and validation of qRT-PCR primers
For primer design, the Primer3 v. 0.4.0 software [30] was used with the following settings differing from the default parameters:
Primer size of 20-27 bp, amplicon size 130-150 bp, melting temperature 60-61°C; maximum temperature difference of 0.5°C, maximum length of a polynucleotide repeat 3, and a number of consecutive Gs and Cs at the 3' end of 1.
Primers were designed preferentially for the 3'-exon. The primers for MMP span the last 3'-intron. To check for mispriming primers were blasted (BLASTn) against the genome of D. pulex. Primers with a binding energy ΔG of less than -3 kcal/mole and -2 kcal/mole for internal hairpins and hairpins at the 3'-end, respectively, as well as those with less than -6 kcal/mole and -5 kcal/mole for internal or 3' self and cross dimers, respectively, calculated with Beacon Designer Free Edition of Premier Biosoft International, were excluded.
Real-time quantitative PCR
The PCR mix consisted of 2 μl cDNA (equivalent to approximately 10 ng cDNA), 10 μl of the DyNAmo Flash SYBR Green qPCR Kit (Finnzymes), primer concentrations of 300 nM of forward and reverse primers each (Stx16, Tbp, MMP, CAPON), 100 nM each (aTub, GAPDH), 400 nM each (Xbp), and PCR-grade water up to a total volume of 20 μl.
Reactions were performed in triplicates and a no-template control was included. Every gene was tested for all biological replicates on a separate 96-well plate.
PCR reactions were performed using the DNA Engine Opticon 2 Two-Color Real-Time PCR Detection System (Bio-Rad) and the following conditions: 10 min at 95°C and 40 cycles of 95°C for 15 s, followed by 60°C for 1 min; finally 1 min at 55°C. Amplification specificity was verified based on the melting curve which was obtained by heating in steps of 0.3°C from 60°C to 95°C.
Analysis of candidate reference genes
Optical raw data (not baseline corrected) were exported from the Opticon Monitor software v. 3.1 (Bio-Rad) into MS Excel (Microsoft) and processed with the program LinRegPCR v. 11.0 [31,32]. LinRegPCR determines CP values for each reaction and a mean PCR efficiency corresponding to a primer pair by a linear regression fit to the data in the exponential phase of a reaction.
The BestKeeper descriptive statistical method was applied [9] on the CP values determined by LinRegPCR.
For subsequent analysis with the programs geNorm v. 3.5 [7] and NormFinder v. 0.953 [21], CP values were converted to linear values.
Gene expression analysis and reference gene evaluation
Gene expression analysis of MMP in the induced versus the non-induced samples was carried out with the relative expression software tool REST v. 2.0.7 [26] REST analyzes gene expression data (based on CP values) with particular emphasis on describing and visualizing uncertainty in expression ratios by introducing a randomization test, calculating confidence intervals and standard errors [see [26]]. Whisker box plots provide a visual representation of variation for each gene.
In this study CP values and mean efficiencies determined with LinRegPCR were used for analysis and randomization tests were performed with 10,000 iterations to assess the significance. The MMP expression levels were normalized using five different strategies. Furthermore, gene expression analysis was carried out for Tbp, Stx16, Xbp1, CAPON and MMP using the classical 'housekeeping genes aTub and GAPDH as reference.
Abbreviations
Genes: aTub: alpha tubulin; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; MMP: matrix metalloproteinase; Stx16: syntaxin 16; sucDH: succinate dehydrogenase; Tbp: TATA binding protein; UBC: ubiquitin conjugating enzyme; Xbp1: X-box binding protein 1.
Other: BLAST: basic local alignment search tool; CP: crossing point; NCBI: National Center for Biotechnology Information; NF: normalization factor; SD: standard deviation; qRT-PCR: quantitative real-time reverse transcription polymerase chain reaction.
Authors' contributions
KIS, FL and RT designed the research. KS and FL carried out the molecular genetic and statistic analyses, made the data interpretation and drafted the manuscript. CM helped with the data interpretation and provided bioinformatic support. MEP, DG and JKC provided the microarray data. RT, MEP, DG and JKC revised critically the manuscript. RT provided financial support to the study. All authors read and approved the final manuscript.
Supplementary Material
Results of the gene expression analysis with REST when using aTub (1a) and GAPDH (1b) as reference genes for expression normalization.
Contributor Information
Katina I Spanier, Email: katina.spanier@uni-wh.de.
Florian Leese, Email: florian.leese@rub.de.
Christoph Mayer, Email: cm@tp4.rub.de.
John K Colbourne, Email: jcolbour@cgb.indiana.edu.
Don Gilbert, Email: gilbertd@cricket.bio.indiana.edu.
Michael E Pfrender, Email: mpfrende@nd.edu.
Ralph Tollrian, Email: tollrian@rub.de.
Acknowledgements
We thank Carsten Balczun (Ruhr-University Bochum) who gave valuable comments on RNA extraction, real-time PCR setup and analysis, Linda Weiss (Ruhr-University Bochum) for helpful comments on the manuscript and the laboratory rearing of Daphnia, and two anonymous reviewers for helpful suggestions. This work was supported by a grant of the Ruhr-University Bochum (Rektoratsprogramm) to FL and CM. MEP was partly supported by NIH grant 5R24GM078274.
The sequencing and portions of the analyses were performed at the DOE Joint Genome Institute under the auspices of the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under Contract No. DE-AC02-05CH11231, Los Alamos National Laboratory under Contract No. W-7405-ENG-36 and in collaboration with the Daphnia Genomics Consortium (DGC) http://daphnia.cgb.indiana.edu. Additional analyses were performed by wFleaBase, developed at the Genome Informatics Lab of Indiana University with support to Don Gilbert from the National Science Foundation and the National Institutes of Health. Coordination infrastructure for the DGC is provided by The Center for Genomics and Bioinformatics at Indiana University, which is supported in part by the METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. Our work benefits from, and contributes to the Daphnia Genomics Consortium.
References
- Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;11:155–566. [PMC free article] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;11:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Pérez-Novo CA, Claeys C, Speleman F, Van Cauwenberge P, Bachert C, Vandesompele J. Impact of RNA quality on reference gene expression stability. Biotechniques. 2005;11:52–56. doi: 10.2144/05391BM05. [DOI] [PubMed] [Google Scholar]
- Suslov O, Steindler DA. PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency. Nucleic Acids Res. 2005;11:e181. doi: 10.1093/nar/gni176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;11:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;11:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;11 doi: 10.1186/gb-2002-3-7-research0034. 0034.1-0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. Journal of Biochemical and Biophysical Methods. 2000;11:69–81. doi: 10.1016/S0165-022X(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;11:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Schwarzenberger A, Courts C, von Elert E. Target gene approaches: Gene expression in Daphnia magna exposed to predator-borne kairomones or to microcystin-producing and microcystin-free Microcystis aeruginosa. BMC Genomics. 2009;11:527. doi: 10.1186/1471-2164-10-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann LH, Connon R, Hutchinson TH, Maund SJ, Sibly RM, Callaghan A. Expression of target and reference genes in Daphnia magna exposed to ibuprofen. BMC Genomics. 2006;11:175. doi: 10.1186/1471-2164-7-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider CV, LeBlanc GA. Atrazine stimulates hemoglobin accumulation in Daphnia magna: is it hormonal or hypoxic? Toxicol Sci. 2006;11:443–449. doi: 10.1093/toxsci/kfl067. [DOI] [PubMed] [Google Scholar]
- Zeis B, Becher B, Goldmann T, Clark R, Vollmer E, Bölke B, Bredebusch I, Lamkemeyer T, Pinkhaus O, Pirow R, Paul RJ. Differential haemoglobin gene expression in the crustacean Daphnia magna exposed to different oxygen partial pressures. Biol Chem. 2003;11:1133–1145. doi: 10.1515/BC.2003.126. [DOI] [PubMed] [Google Scholar]
- Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;11:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Souid S, Lepesant JA, Yanicostas C. The xbp-1 gene is essential for development in Drosophila. Dev Genes Evol. 2007;11:159–167. doi: 10.1007/s00427-006-0124-1. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Benfenati F, Snowman AM, Czernik AJ, Snyder SH. Neuronal nitric-oxide synthase localization mediated by a ternary complex with synapsin and CAPON. Proc Natl Acad Sci USA. 2002;11:3199–3204. doi: 10.1073/pnas.261705799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-McCaw A, Serano J, Santé JM, Rubin GM. Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Dev Cell. 2003;11:95–106. doi: 10.1016/S1534-5807(02)00400-8. [DOI] [PubMed] [Google Scholar]
- Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nature Reviews Molecular Cell Biology. 2007;11:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behaviour. Annu Rev Cell Dev Biol. 2001;11:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollrian R. Chaoborus crystallinus predation on Daphnia pulex: can induced morphological changes balance effects of body size on vulnerability? Oecologia. 1995;11:151–155. doi: 10.1007/BF00317278. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;11:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;11:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Skern R, Frost P, Nilsen F. Relative transcript quantification by quantitative PCR: roughly right or precisely wrong? BMC Mol Biol. 2005;11:10. doi: 10.1186/1471-2199-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;11:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- Dheda K, Huggett JF, Chang JS, Kim LU, Bustin SA, Johnson MA, Rook GA, Zumla A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem. 2005;11:141–143. doi: 10.1016/j.ab.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;11:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;11 doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijanowska J, Kloc M. Daphnia response to predation threat involves heat-shock proteins and the actin and tubulin cytoskeleton. Genesis. 2004;11:81–86. doi: 10.1002/gene.20000. [DOI] [PubMed] [Google Scholar]
- Infante C, Matsuoka MP, Asensio E, Cañavate JP, Reith M, Manchado M. Selection of housekeeping genes for gene expression studies in larvae from flatfish using real-time PCR. BMC Mol Biol. 2008;11:28. doi: 10.1186/1471-2199-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S Totowa, editor. NJ: Humana Press; 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;11:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;11:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of the gene expression analysis with REST when using aTub (1a) and GAPDH (1b) as reference genes for expression normalization.