Abstract
Influenza B virus contains an integral membrane protein, BM2, that oligomerizes in the viral membrane to form pH-activated proton channel. Here we report the solution structures of both the membrane-embedded channel domain and the cytoplasmic domain of BM2. The channel domain forms a left-handed coiled-coil tetramer with a helical packing angle of -37{degree sign} to form a polar pore in the membrane for conducting ions. Mutagenesis and proton flux experiments identified residues involved in proton relay and suggest a mechanism of proton conductance. The cytoplasmic domain of BM2 also forms a coiled-coil tetramer. It has a bipolar charge distribution, in which a negatively charged region interacts specifically with the M1 matrix protein that is involved in packaging the genome in the virion. This interaction suggests another role of BM2 in recruiting the matrix proteins to the cell surface during virus budding. Therefore BM2 is an unusual membrane protein which has the dual functionality of conducting ions and recruiting proteins to the membrane.
Influenza B virus is an important constituent of human seasonal flu that accounts for about 50% of all influenza disease in recent years (see Center for Disease Control website). The virion contains an integral membrane protein, BM2, that is essential for virus replication 1 and oligomerizes in the viral membrane to form pH-activated proton channel 2. The recognized role of the proton channel is to equilibrate pH across the viral membrane during viral entry and across the trans-Golgi membrane of infected cells during viral maturation 3,4.
BM2 protein is a single-span membrane protein of 109 residues; it is a homotetramer in its native state 5. Although BM2 and AM2 (the proton channel from influenza A) both conduct protons and influenza B is the closest relative of influenza A virus, the two proteins are different in amino acid sequence and in channel property. Except for the HXXXW sequence motif in the transmembrane (TM) domain that is essential for pH sensing and channel gating, BM2 shares almost no sequence homology with AM2 (Supplementary Fig. 1). Unlike AM2, the BM2 proton conductance is completely insensitive to amantadine and rimantadine, which were the first effective drugs licensed for influenza treatment 6. BM2 channel activity is also higher than that of AM2 6. A wealth of structural information is available for the channel domain of AM2, including models derived from biochemical and spectroscopic data 7-9, and high resolution NMR 10,11 and X-ray 12 structures. Cysteine scanning mutagenesis suggested a model for the channel domain of BM2 13, but no high resolution structures have been determined. In addition to the channel domain, BM2 and AM2 have relatively large cytoplasmic regions compared to other influenza surface proteins, for which there is no structural information. The cytoplasmic regions of AM2 and BM2 have been suggested to be important for proper virus assembly 14-17.
We previously determined the structure of the AM2 channel bound to the drug rimantadine and proposed allosteric mechanism for drug inhibition and drug resistance 10,18. In an effort to understand how BM2 channel works, we determined the solution structures of the membrane-embedded channel domain and the C-terminal cytoplasmic domain of BM2. The channel assemblies of BM2 and AM2 have many differences that explain the properties unique to the BM2 channel, such as drug resistance and higher proton conductance. We also carried out mutagenesis and liposomal proton flux assays to identify residues involved in proton relay across the channel. The cytoplasmic domain of BM2 is also a coiled-coil tetramer, and its unusually large electrostatic dipole moment suggests a role in molecular recognition. NMR chemical shift perturbation experiments showed that the cytoplasmic domain interacts specifically with the M1 matrix protein. Thus, in addition to the role of conducting protons, BM2 is involved in viral assembly, probably by recruiting the matrix proteins to the cell surface during virus budding.
RESULTS
Solution structure of BM2
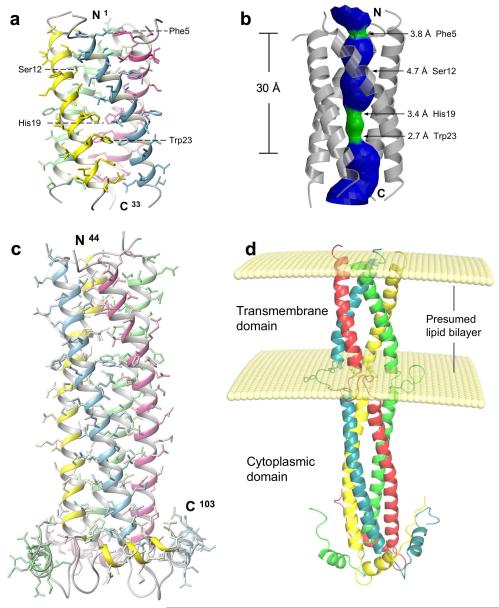
For proteins that contain membrane-embedded and cytoplasmic domains, it is difficult to find a detergent that preserves the structural integrity of both domains. We have screened the intact full-length BM2 in many detergents, but none yielded workable NMR spectra. We then established two protein constructs corresponding to two separate functional domains of BM2 and determined their NMR structures (see NMR Structural statistics in Table 1 and Supplementary Fig. 2). In DHPC (dihexonyl-phosphocholine) micelles, BM2(1-33), a construct encompassing residues 1-33, forms a coiled-coil tetramer with a packing angle of about −37° (Fig. 1a). The tetramer has a well-defined hydrophilic channel that is occluded by Phe5 and Trp23 at the N-, and C-terminal ends, respectively (Fig. 1b). The structure was determined at pH 7.5, and thus corresponds to the closed state. When reconstituted into liposomes made from E. coli lipid extract, BM2(1-33) shows specific proton conductance and that the conductance is completely insensitive to 50 μM of rimantadine (Supplementary Fig. 3). For structural study of the cytoplasmic domain, we used a construct, BM2(26-109), which includes the entire predicted extramembrane region (residues 44-109) as well as six N-terminal, membrane-anchor residues that overlap with BM2(1-33). When bound to the LMPG (14:0 lyso phosphoglycerol) micelles, the structured region of BM2(26-109), for which the atomic coordinates are defined by NMR restraints, commences at Pro44 and ends at Leu103 (Fig. 1c). Residues 45 – 85 form an uninterrupted helix that oligomerizes into a left-handed coiled-coil tetramer. A hairpin-like structure, consisting of residues 86 – 92, connects the coiled-coil structure to a short amphipathic helix that is roughly perpendicular to the coiled-coil helix. This short helix, residues 93 – 103, packs against the hairpin region of the adjacent subunit such that the hydrophobic sides of the amphipathic helix and hairpin are protected from the solvent. Although no inter-subunit nuclear Overhauser effects (NOEs) were observed for residues 34 – 43, the region that connects the TM and cytoplasmic domain, residues 39-45 show NOEs to the glycerol protons of LMPG headgroup (Supplementary Fig. 4). By translating the protein-LMPG NOEs to semi-quantitative distance restraints between protein protons and an imaginary plane that represents the headgroup of the lipid bilayer 19, we modeled the full-length BM2 structure using all NMR restraints measured for the TM and cytoplasmic domains. The model (Fig. 1d) provides a qualitative view of the overall conformation of the BM2 tetramer with respect to the lipid bilayer, although the protein-LMPG interactions observed here may not reflect those in the membrane environment.
Table 1.
NMR and refinement statistics for protein structures
| Residues 2 – 33 | Residues 44 – 103 | |
|---|---|---|
| NMR distance and dihedral constraints | ||
| Distance constraints | ||
| Total NOE | 232 × 4 | 356 × 4 |
| Intra-residue | 57 × 4 | 138 × 4 |
| Inter-residue | 164 × 4 | 202 × 4 |
| Sequential (∣i – j∣ = 1) | 96 × 4 | 118 × 4 |
| Medium-range (∣i – j∣ ≤ 4) | 68 × 4 | 75 × 4 |
| Long-range (∣i – j∣ ≥ 5) | 0 | 9 × 4 |
| Intermolecular | 11 × 4 | 16 × 4 |
| Hydrogen bonds | 38 × 4 | 84 × 4 |
| Total dihedral angle restraints | 51 × 4 | 131 × 4 |
| ϕ (TALOS) | 26 × 4 | 50 × 4 |
| ψ (TALOS) | 25 × 4 | 50 × 4 |
| χ1 (J couplings) | 0 | 31 × 4 |
| Total RDCs | 0 | 56 × 4 |
| Backbone NH | 0 | 56 × 4 |
| Structure statistics | ||
| Violations (mean ± s.d.) | ||
| Distance constraints (Å) | 0.046 ± 0.0021 | 0.068 ± 0.0014 |
| Dihedral angle constraints (°) | 0.247 ± 0.115 | 3.20 ± 0.098 |
| Max. dihedral angle violation (°) | 0.452 | 3.45 |
| Max. distance constraint violation (Å) | 0.049 | 0.070 |
| Q (%) | 0.120 | |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.0035 ± 0.00013 | 0.0029 ± 0.000049 |
| Bond angles (°) | 0.40 ± 0.023 | 0.42 ± 0.0068 |
| Impropers (°) | 0.27 ± 0.011 | 0.36 ± 0.0067 |
| Average pairwise r.m.s. deviation** (Å) | ||
| Heavy | 1.06 | 1.38 |
| Backbone | 0.70 | 0.92 |
Pairwise r.m.s. deviation was calculated among 15 refined structures.
Figure 1. Solution structure of BM2 from influenza B virus.
a, Structure of the channel domain (residues 1-33) in DHPC micelles at pH 7.5. b, The pore surface of the channel domain calculated using the program HOLE 26. The channel is coloured in green for regions wider than 2.7 Å and narrower than 4.6 Å, and coloured in blue for regions wider than 4.6 Å. c, The structured cytoplasmic domain (residues 44-103) of BM2(26-109) anchored to LMPG micelles at pH 6.8. d, A model of the full-length BM2 built with the structures of the channel and cytoplasmic domains. The positioning of the two domains and their localization relative to the lipid bilayer were modeled using intra-subunit and protein-LMPG NOEs, measured for residues 29 – 43 of the BM2(26-109) construct.
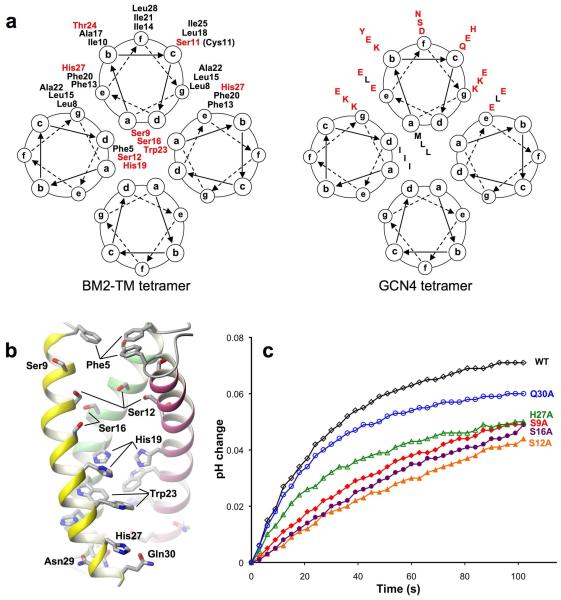
The coiled-coil structure of the channel domain shows at least two heptad repeats: one from Leu8 at position g to Ile14 at position f, and the other from Leu15 at position g to Ile21 at position f (Fig. 2a). Positions a and d, which constitute the core of the coiled-coil tetramer, are occupied mostly by hydrophilic residues such as Ser9, Ser12, and Ser16. His19 at position d and Trp23 at position a are also pore-lining, consistent with their essential roles in pH sensing and channel gating. Positions g and e are occupied by leucines 8 and 15 and phenylalanines 13 and 20, respectively, to allow for peripheral hydrophobic interactions that stabilize helical packing. The above amino acids in positions a, d, g, and e are conserved in all sequenced BM2 variants (Supplementary Fig. 5a). The rest of positions, b, c, and f, of the heptad repeat are occupied by hydrophobic residues (with the exception of Ser11), which form the hydrophobic surface of the tetramer for membrane partition. This arrangement for coiled-coil assembly in membrane is the opposite to that of water-soluble coiled-coil tetramer, in which positions a and d are typically hydrophobic residues and positions g and e are polar residues 20 (Fig. 2a). The structure of the BM2 TM domain thus provide an example for designing membrane-embedded coiled-coil tetramers having a hydrophilic pore, with far reaching implications for ion channel design.
Figure 2. Assembly of the H+ conducting pore.
a, Comparison of helical wheel representations of coiled coil assemblies of the BM2 channel domain (left) and GCN4 (right) 20, illustrating the concept of using the principle of coiled-coil packing to form a polar pore in a hydrophobic environment. Residues with negative or positive hydropathy index are colored in red and black, respectively. b, The pore-lining residues revealed by removing one of the four TM helices. c, Proton conduction traces of BM2(1-33) and its mutants. Protein concentration is the same for all BM2(1-33) variants. Rates of proton flux are reported in Table 2.
Polar residues mediate proton conduction
To investigate the mechanism of proton conductance, we mutated polar pore residues to alanines and quantified the effect of the single mutations on proton conductance using a liposomal proton flux assay. In this assay, we used proton gradient to drive proton conduction because this most directly mimics what happens in the endosome after endocytosis 4. Known quantities of BM2(1-33) variants were reconstituted into liposomes that were made with identical pH and ion concentrations inside and outside, but strongly buffered inside and weakly buffered outside. Proton conduction was initiated under conditions of rapid solution mixing by the addition of concentrated acid to the external solution, and then observed as an increase in pH of the external solution as protons move down the pH gradient into the liposomes 18 (Supplementary Fig. 3a). As a validation of the assay, we verified (i) that nearly no proton translocation is observed in the absence of proton channel (Supplementary Fig. 3b); (ii) that the WT BM2(1-33) has a specific conductance that is completely insensitive to rimantadine (Supplementary Fig. 3c&d); (iii) that the wildtype (WT) AM2 construct containing residues 18 to 60, AM2(18-60), has specific conductance that is inhibited by as little as 10 μM of rimantadine (Supplementary Fig. 3e&f). The oligomeric states of different mutants – S9A, S12A, S16A, H27A, and Q30A – were verified by crosslinking. SDS-PAGE shows no substantial changes upon mutations in these pore lining residues (Supplementary Fig. 6).
We found that the BM2(1-33) proton conductance (~ 14 H+/sec/channel) is about two-fold higher than that of AM2(18-60) (Table 2), consistent with results obtained from whole cell channel recording 6. The BM2 mutants all showed lower conductance than the WT. In particular, S12A and S16A in the middle of the pore between Ser9 and His19 have the largest effect, decreasing conductance by 45% and 44%, respectively. Replacing polar residues near the N- and C-terminal ends of the channel has less effect. The conductance of S9A, H27A, and Q30A are on averaged lower than that of the WT by ~27%. The above results suggest that not one but a combination of polar residues determines the proton conductance of BM2.
Table 2.
Proton conductance ratesa
| Conductance/tetramer | Number of samples |
Standard Deviation |
% of BM2 WT |
|
|---|---|---|---|---|
| BM2 WT | 14.7 | 4 | 1.3 | 100% |
| S9A | 10.5 | 3 | 0.4 | 71% |
| S12A | 8.2 | 4 | 1.5 | 55% |
| S16A | 8.3 | 5 | 1.3 | 56% |
| H27A | 10.8 | 4 | 3.7 | 74% |
| Q30A | 12.7 | 3 | 1.4 | 75% |
| AM2 WT | 7.0 | 2 | 0.3 | 47% |
| Lipids | 0.08 | 4 | 0.3 |
Proton conduction rates were measured as the initial rate of change upon dropping the external pH to 6.05-6.10 (internal pH 7.7). Rates are given in protons/sec/channel.
The cytoplasmic domain interacts with the matrix protein
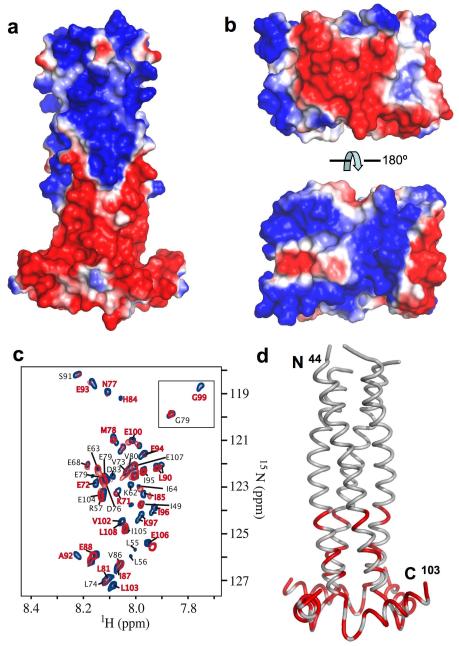
The BM2 cytoplasmic domain shows a striking feature that the surface of the N-terminal half of the domain (residues 44-71) is almost entirely positive, whereas the surface of the C-terminal half (residues 72 – 103) is almost entirely negative (Fig. 3a). This charge separation results in a large electrostatic dipole moment, 4215 Debyes, at neutral pH that is about 4 standard deviations above the mean of all proteins in the database 21. Moreover, the surface charges are highly conserved among different influenza B strains (Supplementary Fig. 5b). Reverse genetics studies suggest that the cytoplasmic domain is involved in the incorporation of ribonucleoprotein (RNP) complex into viral particles, possibly through interaction with the M1 matrix proteins 16. Indeed, the homology model of the N-terminal domain of BM1, derived from the crystal structure of AM1 22 based on 30% sequence identity (Supplementary Fig. 7), also shows a strong bipolar charge distribution (Fig. 3b). To investigate this interaction, we made a water-soluble construct of BM2 encompassing residues 43 – 109 that lacks the membrane-anchor residues. BM2(43-109) was used here instead of BM2(26-109) to avoid complication associated with the detergent required for solubilizing BM2(26-109). Equilibrium sedimentation experiments showed that BM2(43-109) forms a tetramer in water (Supplementary Fig. 8). The BM1 protein used in this experiment was fused at the N-terminus to the maltose binding protein (MBP), which prevented the substantial aggregation that was observed for BM1 protein that had been cleaved from the MBP tag.
Figure 3. Interaction between BM2 cytoplasmic domain and BM1 matrix protein.
a, Surface charge representation of the BM2 cytoplasmic domain, showing the strong segregation of positive (blue) and negative (red) charges. b, Surface charge representation of the N-terminal domain of BM1 (residues 1-156). The homology model of BM1(1-156) was built after the crystal structure of AM1(1-158) 22 based on 30% sequence identity (Supplementary Fig. 7). c, Overlay of 1H-15N TROSY spectra of BM2(43-109) in the absence (blue) and presence (red) of MBP-BM1. The labeled resonances are those whose assignments could be correctly traced. d, Mapping of perturbed resonances onto the cytoplasmic domain structure. The perturbed regions are colored in red.
We performed chemical shift perturbation of 2D 1H-15N correlation spectra of BM2(43-109) by MBP-BM1. At 1:1 molar ratio of BM2(43-109) to MBP-BM1, the majority of resonances of BM2(43-109) are broadened and a specific set of resonances shifted substantially (Fig. 3c), whereas the spectrum is not perturbed by MBP alone (see negative control in Supplementary Fig. 9). The perturbed resonances that could be correctly assigned are mostly from the last amphipathic helix of BM2, of which the solvent exposed surface is negatively charged (Fig. 3d). Therefore, the interaction between BM2 and BM1 is electrostatically driven, possibly between the negatively charged C-terminal end of BM2 (residues 84 – 108) and the positively charged surface of the N-terminal domain of BM1.
DISCUSSION
Although the overall assembly of TM helices of BM2 is similar to that of AM2, e.g., both are left-handed four-helix bundle having a hydrophilic pore, the two channels differ substantially in details. Unlike AM2, the TM domain of BM2 shows strong coiled-coil characteristics with heptad repeats. It is the first of its kind in the known ion channel structures that adopts a coiled-coil assembly to conduct ions. The coiled coil arrangement allows the TM segment of BM2 to form a stable tetramer by itself, but stable channel assembly of AM2 also requires a C-terminal amphipathic helix following the TM domain 10. Indeed, BM2 TM domain (residues 1 – 33) runs as a tetramer in SDS PAGE (Supplementary Fig. 10), but the same could not be achieved with the AM2 TM domain (residues 20-46) 10. Another important difference is that the rimantadine binding observed in the AM2 structure is absent in BM2, which may explain the drug resistance of the BM2 channel. This drug site is the lipid-facing pocket observed in the NMR structure of the AM2 channel, which consists of Trp41, Ile42, and Arg45 from one TM helix and Leu40, Leu43, and Asp44 from the adjacent TM helix. The corresponding residues in BM2 are Trp23, Thr24, and His27 from one TM helix and Ala22, Ile26, and Gly26 from the adjacent TM helix. The above two groups of residues are uncorrelated and thus do not constitute surfaces of similar electrostatic and hydrophobic properties (Supplementary Fig. 11).
The BM2 channel structure suggests a number of polar residues important for hydration of the channel pore and for supporting proton passage during channel activation. Mutating the serines on the N-terminal side of the Trp23 gate to alanine substantially reduced proton conductance, with the largest decrease (~45%) observed for S12A and S16A. Serines 9, 12 and 16 are pore-lining and important for pore hydration. Ser12 sidechain is completely facing the pore, and unlike Ser9 and Ser16, is not involved in helical packing; it is conveniently positioned for relaying protons. It was shown previously that mutating the pH sensor, His19, to alanine dramatically decreases conductance 23. Together, these results suggest that the serines are involved in relaying hydronium ions or coordinating water molecules that may form proton wire to relay protons to His19. The protonated histidines then lead to opening of the Trp23 gate, with a mechanism that is still not understood. Compared to the channel pore of AM2, which is also very hydrophilic 24, the BM2 pore has two more serine hydroxyl groups which can facilitate proton relay. This difference may in part explain the higher conductance of the BM2 channel. Mutating residues on the C-terminal side of the Trp23 gate, H27A and Q30A, also reduced conductance by ~25%. His27 and Asn30 are exposed to the hydrophilic region of the membrane; they probably facilitate proton exit. It is interesting to note that polar residues after the tryptophan gate are also present in AM2 (Asp44 and Arg45), and replacing Asp44 to alanine results in three-fold decrease in conductance 18.
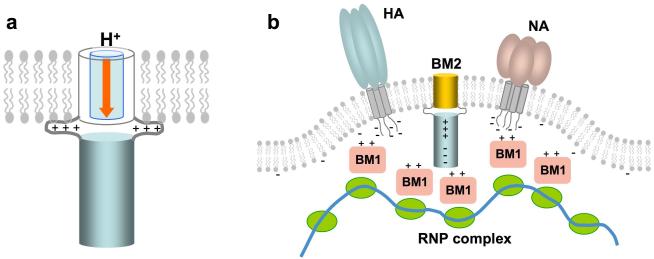
Perturbation of chemical environment of a defined region of the cytoplasmic domain by the M1 matrix protein indicates specific molecular recognition between the two proteins. The perturbed region identified, from residues 84 to 108, is consistent with known deletions and mutations of BM2 that affect virus assembly. Viruses with BM2 deletion of 101-109 contained dramatically reduced RNP complex and affected membrane association of M1 16. Furthermore, alanine-scanning substitution of three consecutive residues showed that the 86-88A, 89-91A, 93-94A, and 95-97A mutants did not grow normally, and contained substantially reduced levels of M1 and nucleoprotein. The data from structural and reverse genetics studies indicate that the interaction between the cytoplasmic regions of the proton channels and matrix proteins play an important role in viral assembly. During virus budding, the matrix proteins and RNPs must coat the plasma membrane such that budding would result in a properly assembled virus. Therefore, the membrane patch that is destined to bud out from the host cell must contain specific sites for recruiting the matrix protein and RNP complexes (Fig. 4). The unusually strong electric dipole moment of BM2 cytoplasmic domain may serve to orient the M1 matrix protein, which also has a strong electric dipole moment, for specific association. Electric dipole facilitated molecular recognition is commonly observed in cellular signaling pathways, e.g., the interactions between the caspase recruitment domains 25. The coating of M1 and RNPs to the virus membrane is likely achieved with cooperative interactions of M1 to the negatively charged membrane, the short cytoplasmic tails of hemagglutinin and neuraminidase, and the cytoplasmic domain of M2.
Figure 4. The dual functionality of BM2.
The channel domain serves to conduct proton across the viral and trans-Golgi membrane (left). The cytoplasmic domain is involved in recruiting the M1 matrix proteins and RNP complexes to the budding membrane (right).
METHODS
Sample preparation
BM2 gene from influenza B (the Maryland/01 strain) with the C11S mutation was E.coli codon optimized and synthesized (Epoch Biolabs). The BM2(1-33) construct was cloned into the pMALc2x vector (New England Biolabs) and expressed as MBP fusion protein. MBP-BM2(1-33) was purified from cell lysis using amylose resin. The fusion protein was cleaved with TEV protease, and BM2(1-33) was isolated by reverse phase HPLC. BM2(1-33) was dissolved in a solution containing 8 M urea and DHPC, followed by dialysis to remove the denaturant. After concentration, the sample typically contains 0.7 mM BM2(1-33), ~300 mM DHPC, 15 mM βME, and 40 mM sodium phosphate (pH 7.5). BM2(26-109) was expressed with C-terminal 6His-tag using the pET21a vector. The over-expressed protein formed inclusion body, which was dissolved in 6 M guanidine HCl and then purified using Ni-resin. After dialysis to remove denaturant, the precipitated BM2(26-109) was reconstituted in LMPG detergent using a dialysis procedure similar to that used for BM2(1-33) above. A typical NMR sample contains 0.7 mM protein, 200 mM LMPG, 15 mM βME, 25 mM NaCl, and 25 mM Tris-HCl (pH 6.8).
Liposomal H+ flux assay
The liposomal H+ flux assay was established based on earlier work described in ref. 18. BM2(1-33) channels were reconstituted into liposomes by mixing 10 mg of E.coli polar lipid extract, 5 nmols of BM2(1-33) peptide, and 0.2 nmol of valinomycin in chloroform/methanol mixture. Thin film was formed and then resuspended in a strong buffer (50 mM phosphate, 50 mM citrate, 122 mM KCl, 122 mM NaCl, 0.01% (w/v) NaN3, pH 7.7), and extruded through 0.2 μM polycarbonate membranes. The external buffer was exchanged to a weak buffer (2 mM phosphate, 2 mM citrate, 122 mM KCl, 122 mM NaCl, 0.01% (w/v) NaN3, pH 7.8). Final eluted volume was 1.5 mL, containing 5 mg mL−1 lipid, 3 μM peptide, and 0.1 μM valinomycin. Proton conductance was initiated by addition of 3.5 μL of 1 M HCl with rapid mixing and terminated by the addition of 5 μM of carbonyl cyanide m-chlorophenylhydrazone (FCCP), which equilibrate pH on both sides of the membrane. Proton flux was monitored by measurement of external pH.
BM2 – BM1 interaction
BM2(43-109) was cloned with N-terminal 6His-tag into the pET21a vector. Protein was expressed in inclusion body and purified using a procedure similar to that for BM2(26-109). The BM1 gene from the same virus strain was cloned into the pMALc2x vector and expressed as MBP fusion protein. MBP-BM1 was purified with amylose resin, followed by Mono-Q ion exchange purification. To obtain MBP protein as a negative control for the BM2-BM1 interaction experiment, MBP-BM1 was cleaved with TEV protease. The released MBP was purified by Mono-Q. The purified BM2(43-109), MBP-BM1, and MBP were dialyzed against a NMR buffer consisting of 25 mM Tris-HCl (pH 6.8), 25 mM NaCl, and 15 mM βME. To investigate specific interaction between BM2 and BM1, 0.3 mM (monomer concentration) of BM2(43-109) was mixed with 0.3 mM of MBP-BM1 or MBP. 2D 1H-15N TROSY spectra of the mixed samples were recorded on a Bruker 750 MHz spectrometer.
NMR spectroscopy
NMR experiments were conducted at 30°C on spectrometers equipped with cryogenic probes (Bruker, Billerica, MA). Sequence specific assignment of backbone and sidechain resonances are described in Supplementary Methods and shown in Supplementary Fig. 10 and 12.
For BM2(1-33), intra-subunit NOEs involving both backbone and sidechain protons were assigned using the 3D 15N-edited and 13C-edited NOESYs recorded with NOE mixing times of 110 and 150 ms, respectively, on a sample containing 15N-, 13C-labeled protein and deuterated DHPC (D35-DHPC) (Avanti Polar Lipids). For identifying contacts between neighboring subunits, intra-subunit NOEs were first assigned to completion. This was possible owing to the low complexity of the NOESY spectra (e.g., see Supplementary Fig. 13). The remaining NOEs, which could not be explained by intra-subunit distances based on the known secondary structures, were identified to be inter-subunit NOEs. For BM2(26-109), the NOESY spectra were collected with deuterated LMPG (D27-LMPG). Similar approach of NOE assignment for BM2(1-33) was used, except for additional 3D NOESY spectra of ILV-labeled protein that were recorded for resolving the increased spectral complexity.
The χ1 rotamers were obtained from measurements of the 3-bond scalar couplings including 3JNCγ and 3JC’Cγ 27. Backbone 1H-15N residual dipolar couplings (RDCs) were measured for the complex of BM2(26-109) and LMPG micelle in the G-Tetrad DNA alignment medium 28.
Structure determination
Structures were calculated using the program XPLOR-NIH 29. The secondary structure of the monomer was first calculated from random coil using intra-subunit NOEs, backbone dihedral restraints derived from chemical shifts (TALOS) 30, and side chain χ1 restraints. A total of 20 monomer structures were calculated using a high-temperature simulated annealing (SA) protocol in which the bath temperature was cooled from 1000 to 200 K. To obtain an initial set of tetramer structures, four copies of the lowest-energy monomer structure calculated above were used. The same SA run was performed in the presence of inter-subunit NOEs and all other intra-subunit restraints. For each experimental inter-subunit NOE between two adjacent subunits, four identical distance restraints were assigned respectively to all pairs of neighboring subunits to satisfy the condition of C4 rotational symmetry. During the annealing run, the bath was cooled from 1000 to 200 K. A total of 100 tetramer structures were calculated. For BM2(1-33), 15 low energy structures were selected as the structural ensemble. Ramachandran plot statistics are as follows: most favored (90.7%), additionally allowed (7.4%), generously allowed (1.9%) and disallowed (0%).
For BM2(26-103), for which RDCs are available, 15 structures of which the individual subunits have on average the best agreement with RDCs (r ~ 0.65 and Q ~ 0.52) were selected for low-temperature refinement (bath cooled from 200 to 20 K) against RDCs in the presence of all other restraints. For each of the 15 structures, 10 RDC-refined structures were generated. From that set, the structure with the lowest total energy was added to the final ensemble to describe the structural diversity of the solution structure. Without violating any NOE restraints, the final subunit structures fit RDCs to r of 0.88 and Q of 0.12, with Da = 9.0 Hz and Rh = 0.62. Ramachandran plot statistics are as follows: most favored (83.3%), additionally allowed (15.3%), generously allowed (1.4%) and disallowed (0%).
Supplementary Material
Acknowledgements
We thank Marcelo Berardi for insightful discussion on the interaction between BM2 and M1 matrix protein. This work was supported by grants from the NIH (AI054520 to JJC) and the Pew Scholars Program in Biomedical Sciences (to JJC).
Footnotes
ACCESSION CODE
The structures have been deposited in the Protein Data Bank under the accession code 2KIX for the channel domain and 2KJ1 for the cytoplasmic domain.
Author Contribution J.W. and J.J.C. determined the BM2 structures; R.M.P. performed proton conductance assays; J.W. and M.A.M. performed the BM2-BM1 interaction experiment; J.J.C., R.M.P. and J.W. wrote the paper; J.J.C. supervised the research.
Author Information The authors declare no competing financial interests.
References
- 1.Hatta M, Goto H, Kawaoka Y. Influenza B virus requires BM2 protein for replication. J Virol. 2004;78:5576–83. doi: 10.1128/JVI.78.11.5576-5583.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 3.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. Embo J. 1985;4:3021–4. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 5.Paterson RG, Takeda M, Ohigashi Y, Pinto LH, Lamb RA. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology. 2003;306:7–17. doi: 10.1016/s0042-6822(02)00083-1. [DOI] [PubMed] [Google Scholar]
- 6.Mould JA, et al. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev Cell. 2003;5:175–84. doi: 10.1016/s1534-5807(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 7.Pinto LH, et al. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci U S A. 1997;94:11301–6. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovacs FA, Denny JK, Song Z, Quine JR, Cross TA. Helix tilt of the M2 transmembrane peptide from influenza A virus: an intrinsic property. J Mol Biol. 2000;295:117–25. doi: 10.1006/jmbi.1999.3322. [DOI] [PubMed] [Google Scholar]
- 9.Kukol A, Adams PD, Rice LM, Brunger AT, Arkin TI. Experimentally based orientational refinement of membrane protein models: A structure for the Influenza A M2 H+ channel. J Mol Biol. 1999;286:951–62. doi: 10.1006/jmbi.1998.2512. [DOI] [PubMed] [Google Scholar]
- 10.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–5. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Kim S, Kovacs F, Cross TA. Structure of the transmembrane region of the M2 protein H(+) channel. Protein Sci. 2001;10:2241–50. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stouffer AL, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–9. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma C, et al. Identification of the pore-lining residues of the BM2 ion channel protein of influenza B virus. J Biol Chem. 2008;283:15921–31. doi: 10.1074/jbc.M710302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol. 2008;82:10059–70. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCown MF, Pekosz A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J Virol. 2006;80:8178–89. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai M, Kawasaki K, Odagiri T. Cytoplasmic domain of influenza B virus BM2 protein plays critical roles in production of infectious virus. J Virol. 2008;82:728–39. doi: 10.1128/JVI.01752-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai M, Watanabe S, Ninomiya A, Obuchi M, Odagiri T. Influenza B virus BM2 protein is a crucial component for incorporation of viral ribonucleoprotein complex into virions during virus assembly. J Virol. 2004;78:11007–15. doi: 10.1128/JVI.78.20.11007-11015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pielak RM, Schnell JR, Chou JJ. Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc Natl Acad Sci U S A. 2009;106:7379–84. doi: 10.1073/pnas.0902548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu C, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell. 2008;135:702–13. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–7. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 21.Felder C, Prilusky J, Silman I, Sussman J. A server and database for dipole moments of proteins. Nucleic Acids Research. 2007;35(special Web Servers Issue) doi: 10.1093/nar/gkm307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sha B, Luo M. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat Struct Biol. 1997;4:239–44. doi: 10.1038/nsb0397-239. [DOI] [PubMed] [Google Scholar]
- 23.Otomo K, Toyama A, Miura T, Takeuchi H. Interactions between histidine and tryptophan residues in the BM2 proton channel from influenza B virus. J Biochem. 2009;145:543–54. doi: 10.1093/jb/mvp009. [DOI] [PubMed] [Google Scholar]
- 24.Du QS, Huang RB, Wang CH, Li XM, Chou KC. Energetic analysis of the two controversial drug binding sites of the M2 proton channel in influenza A virus. J Theor Biol. 2009;259:159–64. doi: 10.1016/j.jtbi.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Chou JJ, Matsuo H, Duan H, Wagner G. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell. 1998;94:171–80. doi: 10.1016/s0092-8674(00)81417-8. [DOI] [PubMed] [Google Scholar]
- 26.Smart OS, Goodfellow JM, Wallace BA. The pore dimensions of gramicidin A. Biophys J. 1993;65:2455–60. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bax A, et al. Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzymol. 1994;239:79–105. doi: 10.1016/s0076-6879(94)39004-5. [DOI] [PubMed] [Google Scholar]
- 28.Lorieau J, Yao L, Bax A. Liquid Crystalline Phase of G-Tetrad DNA for NMR Study of Detergent-Solubilized Proteins. J. Am. Chem. Soc. 2008;130:7536–7. doi: 10.1021/ja801729f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwieters CD, Kuszewski J, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2002;160:66–74. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 30.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.