Abstract
The chick embryo is a valuable tool in the study of early embryonic development. Its transparency, accessibility and ease of manipulation, make it an ideal tool for studying gene expression in brain, neural tube, somite and heart primordia formation. This video demonstrates the different steps in 2-color whole mount in situ hybridization; First, the embryo is dissected from the egg and fixed in paraformaldehyde. Second, the embryo is processed for prehybridization. The embryo is then hybridized with two different probes, one coupled to DIG, and one coupled to FITC. Following overnight hybridization, the embryo is incubated with DIG coupled antibody. Color reaction for DIG substrate is performed, and the region of interest appears blue. The embryo is then incubated with FITC coupled antibody. The embryo is processed for color reaction with FITC, and the region of interest appears red. Finally, the embryo is fixed and processed for phtograph and sectioning. A troubleshooting guide is also presented.
Keywords: Developmental Biology, Issue 20, whole mount in situ hybridization, gene expression, chick embryo
Protocol
Part 1: Fixing the embryos
Fill dissection dish with ice cold depc-PBS. Keep on ice. Open the egg by tapping the shell with forceps and remove shell pieces.
Remove thick albumin with forceps. Tilt yolk sac with coarse forceps so that embryo faces upwards.
Using scissors, cut a square of yolk sac around the embryo.
Using spoon, remove embryo from yolk and place on ice cold depc-PBS.
Under the microscope, remove membranes and yolk and transfer to fixation dish.
Pin down, remove depc-PBS. Replace with 4% PFA in depc-PBS and fix O/N at 4°C.
Remove fixative. Replace with ice cold depc-PBS. Using a blunt end microcapillary needle or a microdissection knife, perforate the nervous system and the heart cavities, to prevent trapping of probe.
Wash 2x 10 mn depc-PBS with 0.1% Tween-20 (PBT). Dehydrate 10 mn each in 25%, 50%, 75% methanol in PBT. Dehydrate embryos twice in 100% methanol. Store at -20°C in methanol.
Part 2: Prehybridization
Rehydrate embryos 10 mn each in 75%, 50% and 25% methanol in PBT. Wash 2x 10 mn in PBT.
Meanwhile, prepare ice cold fixative (4% paraformaldehyde in PBT). Replace PBT with proteinase K solution (3 ml/vial; final [5?g/ml] in PBT). Make sure the entire vial is exposed to proteinase K solution by gently rolling the vial; see Troubleshooting for exposure times.
Using RNase free Pasteur pipette remove proteinase K and add fixative (2 ml). Immediately place on ice for exactly 20 mn.
All RT washes are performed on nutator. Wash 2x 10 mn in PBT, and 1x 10mn in PBT containing 50% hybridization buffer.
Wash 1x in hybridization buffer. Incubate at 65°C in 2 ml hybridization buffer for 4-5 hr.
Part 3: Hybridize embryos with probes
DIG and FITC coupled probe synthesis is described in Appendix. The probe expected to give a strong signal, should be synhesized with FITC-containing nucleotides; the weaker probe should be synthesized with with DIG-containing nucleotides.
Prewarm probes in hybridization buffer (500 ng/ml each) using a 2 ml vial. Promptly replace the hybridization buffer with the probe solution. Do not let the embryos dry. Incubate for 16 hr at 65°C.
Replace probe with hybridization buffer, 2 washes, 30 mn each at 65°C. Wash 15 mn in hybridization buffer/MABT (1/1 v/v/) at 65°C.
Part 4: DIG antibody incubation and color reaction
Wash 2x20 mn in MABT. Wash 1 hr in 2% BBR/MABT. Wash 5 hr in 2% BBR/20%HISS/MABT (blocking buffer). Incubate in 1:2000 DIG antibody diluted in blocking buffer, O/N at 4°C on nutator.
Wash 3x 10 mn each in MABT, and 3x 1 hr each in MABT.
Wash 2x 20 mn in NTMT. Add 200ul NBT/BCIP substrate to 10 ml NTMT. Immediately remove NTMT from vial and replace with 1-2 ml NBT/BCIP buffer. Place in dark. Monitor color reaction after 20 mn, and at 20 mn intervals thereafter.
Following color reaction (should appear blue), wash in PBS for 10 mn, pin down on fixation dish filled with PBS, and replace solution with 4% PFA in PBS. Fix O/N at 4°C.
Transfer to new scintillation vial. Wash in PBST (PBS with 1% Tween-20) 2x10 mn. Wash in MABT 2x 10 mn.
Incubate in MABT 30 mn at 63°C.
Part 5: FITC antibody incubation and color reaction
Wash 2x 10 mn in MABT, 1 hr in 2%BBR/MABT, 5 hr in blocking buffer
Incubate in alkaline phosphatase-coupled anti-FITC-antibody (1:500) O/N at 4°C.
Wash 3x 10 mn each in MABT, and 3x 1 hr each in MABT.
Wash in 2x 20 mn in TBS pH 8.45 with 0.1% Tween-20 (TBST).
Prepare Vector Red working solution (5 ml) in TBST using reagents 1,2 and 3 provided in kit. Immediately remove TBST from vial and replace with Vector Red buffer. Place in dark. Monitor color reaction after 40 mn, and at 30 mn intervals thereafter.
Following color reaction (should appear red), transfer embryos to PBS.
Part 6: Fixation and processing
Transfer to fixation dish containing PBS, and fix in 4% PFA for 20 mn at RT or O/N at 4°C.
Wash in PBS, 2x 10 mn. To process for photography, transfer to 20% glycerol in PBS (this will make the embryos translucent). To create a chamber, cut 2 strips of PVC tape, superimpose them on slide. Cut a square in the center using a blade. Remove the square using forceps.
To process for wax sectioning, transfer back to PBS, wash 2x 10 mn, dehydrate in graded series of methanol (25%, 50%, 75% and 100% in PBS, 10 mn each).
Replace with propanol, 3mn. Replace with 1,2,3,4-tetrahydronaphthalene, 20 mn followed by Histoclear (2x 1 hr). Replace with histoclear/wax 1/1 (v/v) 60°C for 1 hr. Replace with wax an process for histology.
Part 7: Solutions
Hybridization buffer, store at -20°C in Falcon, 50 ml: 25 ml formamide; 3.25 ml (20xSSC); 0.5 ml (0.5M EDTA); 1 ml (10% Tween-20); 2.5 ml (1% SDS); 125 ul (20mg/ml tRNA); 100ul (50 mg/ml heparin).MABT, prepare 100mM maleic acid; pH to 7.5 with NaOH pellets. Add 150mM NaCl and 0.1% Tween-20.
NTMT (made just prior to use), 50 ml: 1ml (5M NaCl); 2.5 ml (2M Tris-HCl pH9.5); 1.25 ml (2M MgCl2); 5 ml (10% Tween-20).
TBST: 0.1M Tris-HCl, 0.15M NaCl, pH 8.45, 0.1% Tween-20.
Part 8: Troubleshooting
| Problem | Cause | Remedy |
| Embryo is curled up | Insufficient dehydration/rehydration | Maintain 10 mn intervals during successive dehydration/rehydration steps |
| There is no probe signal | Probe is degraded RNase contamination in vial | Run probe on 1% Agarose gel Make sure that the entire vial is exposed to Proteinase K solution in step 2 |
| Probe is labels cavities of heart an neural tube | Probe is trapped | In step 2, make sure that all cavities are perforated using microdissection knife or microcapillary |
| Embryo disintegrated following hybridization (step 3) | Proteinase K step left too long Hybridization temperature is too high | Proteinase K should not be left longer than 1mn (stages 3+ and 4), 2 mn (stage 5 to 7), 3 mn (stage 8-10), 4 mn (stage 11-13) Do not hybridize at To higher than 65 o C |
Part 9: Representative Results
Representative embryos are shown below: (A) Embryo (stage HH7) is labelled with a DIG-Sox probe (blue) and a FITC-delta-1 probe (red). (B) Embryo (stage HH8) is labelled with a DIG-Pax6 probe (blue) and a FITC-Shh probe (red). Nf, neural fold, anp, anterior neural plate, psm, presomitic mesoderm, n, notochord, nt, neural tube). Probe gifts from the laboratories of Drs. C. Tabin, M. Goulding , D. Henrique, and J. Briscoe.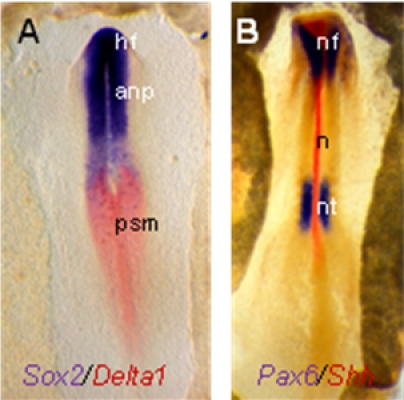
Discussion
The 2-color whole mount in situ hybridization method is used to determine both spatial and temporal patterns of gene expression, using one or two genes of interest. Applications include assessment of gene expression patterns of novel genes (e.g. 1,2, as well as changes in gene expression following insult (embryonic manipulations 3, beads 4, and electroporation of RNA or DNA constructs 5).
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by the Margaret M. Alkek Foundation to RHF.
References
- Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Initiation of neural induction by FGF signaling before gastrulation. Nature. 2000;406:74–78. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- Rodríguez Esteban C, Capdevila J, Economides AN, Pascual J, Ortiz A, Izpisúa Belmonte JC. The novel Cer-like protein Caronte mediates the establishment of embryonic left-right asymmetry. Nature. 1999;401:243–251. doi: 10.1038/45738. [DOI] [PubMed] [Google Scholar]
- Psychoyos D, Stern CD. Restoration of the organizer after radical ablation of Hensen's node and the anterior primitive streak in the chick embryo. Development. 1996;122:3263–3273. doi: 10.1242/dev.122.10.3263. [DOI] [PubMed] [Google Scholar]
- Kawakami M, Nakanishi N. The role of an endogenous PKA inhibitor, PKIalpha, in organizing left-right axis formation. Development. 2001;128:2509–2515. doi: 10.1242/dev.128.13.2509. [DOI] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]


