Abstract
Babesia spp. are tick-transmitted apicomplexan hemoparasites that infect mammalian red blood cells. Our purpose was to determine the prevalence of Babesia infection in a colony of captive baboons and to evaluate potential experimental routes of the transmission of the hemoparasite. DNA was extracted from the blood of baboons and tested for infection with Babesia by PCR and primers that amplify the 18s rRNA gene of the parasite. The overall prevalence of infection of Babesia in the baboon population was 8.8% (73 of 830). Phylogenetic analysis of the sequenced DNA from 2 baboons revealed that the Babesia isolate found in captive baboons was a novel species most closely related (97% to 99%) to B. leo. Blood from a Babesia-infected donor baboon was inoculated intravenously, intramuscularly, or subcutaneously into 3 naive baboons. The intravenously inoculated baboon was PCR-positive at 7 d after inoculation; the 2 baboons inoculated by other routes became PCR-positive at 10 d after inoculation. All 3 baboons remained PCR-positive for Babesia through day 31. Baboons experimentally inoculated with the new Babesia isolate did not exhibit clinical signs of babesiosis during the experiments. We demonstrated that captive baboons are infected with a novel Babesia isolate. In addition we showed that Babesia can be transmitted in the absence of the organism's definitive host (ticks) by transfer of infected blood through intravenous, intramuscular, and subcutaneous routes to naive baboons.
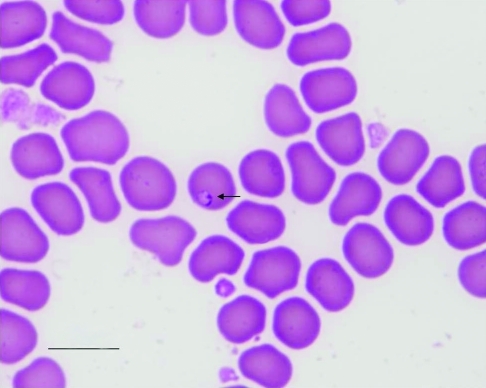
Babesia spp. are apicomplexan hemoprotozoan parasites transmitted to mammals through the bite of an infected ixodid (that is, hard) tick. Sporozoites of Babesia spp. are transferred to an appropriate mammalian host with the saliva of the tick. Once inside their mammalian host, sporozoites enter RBC and undergo asexual reproduction through binary fission. When inside RBC, Babesia spp. often are called piroplasms due to their piriform (that is, pear-shaped) and ‘signet ring’ appearance (Figure 1). Babesiosis can range from subclinical infection to hemolytic anemia, persistent fever, and lethargy in vertebrate hosts. Clinical infections of Babesia spp. have been reported as complications in immunocompromised baboons.1,5 In a xenotransplantation study,5 a baboon (Papio cynocephalus anubis) obtained from the breeding colony at our institution received a pig heart. After a course of immunosuppressive therapy and approximately 5 wk after the transplant, the baboon became lethargic, developed a fever (39.3 °C, 102.9 °F), had a maximum of 39,000 WBC/mm3, and became anemic with hematocrit levels dropping to around 20%. Based on the morphology of the piroplasm and DNA sequencing, the immunosuppressed baboon was diagnosed as being infected with Babesia microti.5
Figure 1.
Blood smear from baboon IVBab at 10 dpi. The signet-shaped piroplasm is indicated by the arrow. Diff-Quik (Dade Behring, Deerfield, IL) stain; bar, 10 µm.
The purpose of the current study was to determine the prevalence of Babesia infection in baboons in our colony, phylogenetically compare the 18s rDNA sequences from infected baboons with orthologous sequences published in GenBank, and ascertain whether Babesia can be transmitted experimentally through the transfer of contaminated blood among baboons without the definitive host (ticks). We found that 8.8% (73 of 830) of baboons in the conventional breeding colony were infected with Babesia. Phylogenetic analyses showed that the Babesia isolate in the colony baboons is novel and most closely related to B. leo. We further demonstrated that the novel Babesia isolate could be transmitted experimentally by the intravenous, intramuscular, and subcutaneous routes.
Materials and Methods
Experimental design.
Captive adult and juvenile olive baboons were used in the present study. PCR using primers that amplify the 18s rRNA gene of Babesia spp. was used to determine the prevalence of Babesia infection within the population of baboons in the conventional breeding colony. The 18s rDNA PCR products from Babesia-infected baboons were sequenced and phylogenetically compared with orthologous sequences of piroplasms published in GenBank. Blood from a baboon confirmed infected with the new Babesia isolate was inoculated into 3 baboons by intramuscular, intravenous, or subcutaneous injection.
Animal housing and husbandry.
All baboons were housed and cared for according to the standards detailed in the Guide for the Care and Use of Laboratory Animals.11 Protocols for maintenance of the baboon colonies were approved by the University of Oklahoma Health Sciences Center Institutional Animal Care and Use Committee. Baboons in the breeding colony were housed in corrals, with approximately 80 animals per corral. Each corral had an open-air outdoor pen as well as an indoor area. Adult baboons housed at the Comparative Medicine annex originated from the breeding colony and were housed separately in approved cages. Cages are designed so feces and urine can pass through the bottom. Rooms where the animals are housed are cleaned twice daily. Baboons were fed Primate Diet 2055 (Harlan, Indianapolis, IN) as well as fresh fruit, vegetables, trail mix, and dry cereal.17 Potable water was available ad libitum from automatic waterers.
Specimens.
Blood samples were collected from individual adult and juvenile olive baboons housed at the conventional colony. Baboons were anesthetized by using ketamine (5 to 10 mg/kg IM; Fort Dodge, Fort Dodge, IA), and blood was drawn from either the femoral region or the forearm. Blood was collected every 6 mo from spring 2007 through spring 2008 at the time of the semiannual health check and tuberculosis test. Samples were transported back to Oklahoma State University on ice, processed within 1 to 2 d, and stored at −20 °C.
DNA extractions.
DNA was extracted from blood samples by using DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to manufacturer's instructions. Briefly, 20 µL proteinase K was added to 200 µL anticoagulated blood, 200 µL Buffer AL was added to each sample, and the tubes were mixed thoroughly by vortexing and incubated at 56 °C for 10 min. After incubation, 200 µL 100% ethanol was added, and samples were vortexed. This mixture was pipetted into DNeasy spin columns placed in 2-mL collection tubes, centrifuged at 6000 × g for 1 min, and collection tubes discarded. Each spin column was placed in a new 2-mL collection tube, 500 µL buffer AW1 was added, and tubes were centrifuged again at 6000 × g for 1 min. The spin column again was placed in a new collection tube, 500 µL of buffer AW2 added, and tubes centrifuged for 3 min at 20,000 × g. The spin columns were placed in clean 1.5-mL microcentrifuge tubes, and 200 µL PCR-grade water warmed to 56 °C was added directly to the membrane. The samples were incubated at 56 °C for 1 min and centrifuged at 6000 × g for 1 min to elute DNA. The elution step was repeated with another 200 µL of warm PCR-grade water for maximal DNA yield. DNA was stored at −20 °C until analyzed by PCR.
PCR assay.
An approximately 1700-bp product of the Babesia 18s rDNA region was amplified by PCR using primers BabAF and BabAR (Table 1). Amplifications were performed in 25-µL volumes containing 0.25 U Taq polymerase (Promega, Madison, WI), 2.4 µL 10× Taq buffer (Promega), 1.5 µL 25 mM MgCl2, 2 µL 10 mM dNTP mixture (Promega), 0.5 µL of a 40-µM solution of each primer, and 5 µL template DNA. For the primary reaction, there was an initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation for 1 min at 94 °C, 1 min for annealing at 56.6 °C, and extension for 2 min at 72 °C. To ensure reactions had gone to completion, a final extension cycle was run at 72 °C for 5 min. A nested PCR reaction was run with 1 µL of the primary PCR product (all other reagents and quantities were the same as used in the primary reaction), and a 460- to 520-bp fragment was amplified by using primers RLBF and RLBR (Table 1). The nested protocol consisted of an initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min, and extension 72 °C for 2 min, followed by the final extension cycle of 72 °C for 5 min. Primers used were previously described.7,14 PCR was carried out in an Eppendorff thermocycler (Eppendorf, Westburg, NY). Amplified products (10 µL) were separated on 1.5% agarose gels stained with ethidium bromide and observed under UV light.
Table 1.
Oligonucleotides used to amplify and sequence the 18s rRNA gene of Babesiafrom baboons.
| Primer | Primer sequence (5′ → 3′) | Reference |
| BABAF | CCG AAT TCG TCG ACA ACC TGG TTG ATC CTG CCA GT | 14 |
| BABAR | CCC GGA TCC AAG CTT GAT CCT TCT GCA GGT TCA CCT AC | 14 |
| RLBF | GAG GTA GTG ACA AGA AAT AAC AAT A | 7 |
| RLBR | TCT TCG ATC CCC TAA CTT TC | 7 |
| BSP1F | TGG CTT ATT CGG ATT CGT CGC TCT | Current study |
| BSP1R | CGC GCA AAT TAC CCA ATC CAG ACA | Current study |
| BSP2F | ATG GCC GTT CTT AGT TGG TGG AGT | Current study |
| BSP2R | CAT CCT TGG CAA ATG CTT TCG CAG | Current study |
| BSP3F | AAG CGC TGT GAA CCC TAT CAC TCT | Current study |
| BSP3R | TGG CTT ATT CGG ATT CGT CGC TCT | Current study |
Purification and sequencing.
Nested PCR products were purified by using the Wizard SV Gel and PCR Clean Up System (Promega) and sequenced at the Oklahoma State University Recombinant DNA–Protein Research Facility (Stillwater, OK) by using an automated DNA sequencer (model 373, Applied Biosystems, Foster City, CA). Three sets of forward and reverse sequencing primers (Table 1) were used to obtain overlapping sequences on both strands of the 18s rDNA. To confirm that each baboon with a positive PCR assay result was infected with Babesia, the nested PCR product was sequenced by using the reverse primer.
Phylogenetics analysis.
Sequences were aligned using ClustalW2 option in Geneious Pro 4.6.1.4,22 For visual inspection and to determine hypervariable regions of the multiple sequence alignment that potentially violated the assumption of positional homology, aligned sequences were imported into MacClade.13 To compare phylogenetic affinities of the Babesia isolate from baboons with other known sequences of Babesia spp. and orthologous sequences of related taxa (Figure 2), we performed phylogenetic analyses under the criteria of maximum parsimony and maximum likelihood by using PAUP21 and Bayesian phylogenetics by using MRBAYES.10 Clades were considered strongly supported if bootstrap values of at least 70% and Bayesian posterior probabilities of at least 0.95 were obtained in at least 2 of the 3 analyses.
Figure 2.
Known sequences of Babesia species and orthologous sequences of related genera from GenBank.
For maximum parsimony, stability of clades was evaluated by performing 1000 bootstrap pseudoreplicates with 25 random additions of input taxa and tree–bisection–reconnection branch-swapping. Prior to maximum likelihood analysis, jModelTest was used to determine the model of DNA sequence evolution that best fit the data.8,16 The GTR + I + Γ model of evolution was chosen with the following parameters: base frequencies = 0.2647, 0.2007, 0.2561, 0.2785; nst = 6; Rmat = 0.9028, 1.8402, 0.9916, 0.4458, 3.8929; rates = Γ with shape parameter (α) = 0.6290, and proportion of invariant sites = 0.5551.8,16 Stability of clades on the resulting tree was evaluated by using a bootstrap analysis with 100 replications and nearest-neighbor interchange branch-swapping. Bayesian analysis was performed by using the GTR + I + Γ model. Four simultaneous Markov chains were run for 5,000,000 generations with random, unconstrained, starting trees. Trees were sampled every 500 generations, with a ‘temperature’ set at 0.02. Finally, percentage sequence divergence calculated by using the GTR + I + Γ model of sequence evolution was computed within and among supported clades.
Inoculation of baboons with Babesia.
Five adult baboons were used for experimental transmission of Babesia. An adult baboon (donor) identified as being infected with the baboon Babesia isolate through PCR and DNA sequencing (as described earlier) was used as the source of infection. Three baboons tested by PCR for infection with Babesia were determined to be negative and used as principals for experimental transmission. Each baboon received 1 mL EDTA-anticoagulated blood from the donor baboon intravenously, intramuscularly, or subcutaneously. A fifth baboon served as an uninfected control and did not receive blood from the Babesia-infected donor baboon. Blood was collected from the principal and control baboons on days 0, 3, 7, 10, 14, 17, 21, 24, and 31 after inoculation for PCR analyses as described earlier.
Statistics.
χ2 tests20 were performed to determine differences in the prevalence of Babesia infection among age groups (younger than 4 y, 5 to 10 y, 11 to 20 y, and 21 y or older), sex, and housing corrals (northeast, northwest, southeast, and southwest). Analyses were performed by using SigmaStat 3.1 statistical software package (Systat Software, Point Richmond, CA). P values of 0.05 or below were considered statistically significant.
Results
Prevalence of Babesia infection.
Overall, the prevalence of infection with Babesia (Figure 1) within the baboon breeding colony was 8.8% (73 of 830). Spring 2007, the first sample period, showed the highest prevalence (12.6%, 34 of 269) of Babesia infection among the breeding population (Table 2), compared with fall 2007 (8.2%, 23 of 281) and spring 2008 (5.7%, 16 of 280; χ2 = 8.31, df = 2, P = 0.01). There was no difference in the prevalence of Babesia infection between male and female baboons during any of the collection periods (Table 2). However, there was a significant difference (χ2 = 23.21, df = 4, P < 0.0001) among age groups for all 3 test periods (Table 3). Adult baboons 11 to 20 y old showed the highest prevalence of infection with Babesia (Table 3). According to housing areas, the northeast corral had the highest prevalence of the 4 different corrals (χ2 = 17.95, 3 df, P < 0.001) in spring 2007, but the northwest corral had the higher prevalence (χ2 = 14.2, 3 df, P = 0.01) in fall 2007; in spring 2008, there was no significant difference among the 4 housing corrals.
Table 2.
Prevalence of the novel Babesiaisolate according to sex and housing corral of baboons
| Sample period | Total no. of baboons | Sex | Northeast corral | Northwest corral | Southeast corral | Southwest corral | Prevalence |
| Spring 2007 | 269 (34) | Male | 15 (4) | 20 (3) | 11 (1) | 16 (0) | 12.60% |
| Female | 58 (11) | 50 (11) | 49 (4) | 50 (0) | |||
| Fall 2007 | 281 (23) | Male | 15 (1) | 20 (4) | 11 (1) | 16 (0) | 8.20% |
| Female | 60 (3) | 52 (8) | 51 (5) | 56 (1) | |||
| Spring 2008 | 280 (16) | Male | 9 (0) | 14 (2) | 6 (0) | 15 (0) | 5.70% |
| Female | 56 (3) | 45 (5) | 54 (6) | 65 (0) |
Numbers in parentheses are the numbers of baboons that tested positive for Babesia.
Table 3.
Numbers of baboons tested and infected with the novel Babesiaisolate within the Oklahoma breeding colony according to age and housing corral
| Sample period | Total no. of baboons | Age (y) | Northeast corral | Northwest corral | Southeast corral | Southwest corral |
| Spring 2007 | 269 (35) | ≤ 4 | 35 (1) | 27 (0) | 26 (1) | 20 (0) |
| 5–10 | 19 (7) | 28 (6) | 18 (1) | 18 (0) | ||
| 11–20 | 5 (3) | 9 (5) | 8 (2) | 22 (0) | ||
| ≥ 21 | 1 (1) | 1 (1) | 2 (0) | 0 | ||
| unknown | 5 (3) | 8 (3) | 8 (1) | 9 (0) | ||
| Fall 2007 | 281 (23) | ≤ 4 | 40 (0) | 28 (1) | 28 (0) | 22 (0) |
| 5–10 | 19 (3) | 26 (3) | 18 (1) | 18 (0) | ||
| 11–20 | 4 (1) | 7 (6) | 10 (4) | 22 (1) | ||
| ≥ 21 | 1 (0) | 1 (1) | 2 (0) | 0 | ||
| unknown | 5 (0) | 8 (2) | 7 (0) | 9 (0) | ||
| Spring 2008 | 280 (16) | ≤ 4 | 34 (0) | 28 (0) | 29(0) | 26 (0) |
| 5–10 | 21 (2) | 25 (3) | 21(2) | 21 (0) | ||
| 11–20 | 12 (1) | 7 (3) | 11(4) | 24 (0) | ||
| ≥ 21 | 1 (0) | 2 (0) | 2(0) | 0 | ||
| unknown | 0 | 4 (1) | 3(0) | 9 (0) |
Numbers in parenthesis are number of baboons positive for Babesia.
Repeat sampling.
Over the 1-y time during which the present study took place, samples were collected 3 times from each animal. There were 7 individual baboons that remained positive for Babesia over the course of the study. Eight baboons that tested positive in the spring of 2007 tested negative in fall 2007 and spring 2008. One baboon was positive for Babesia during the spring 2007 sampling, but that same animal tested negative 6 mo later, in the fall of 2007. Six months later (spring 2008), the baboon again tested positive. When we compared the prevalence of Babesia infection among the 3 sampling periods (Table 2), the prevalence of infection appeared to be decreasing. However, 3 baboons were negative for Babesia infection during the first test sampling but were positive during fall 2007 and spring 2008.
Phylogenetic analysis.
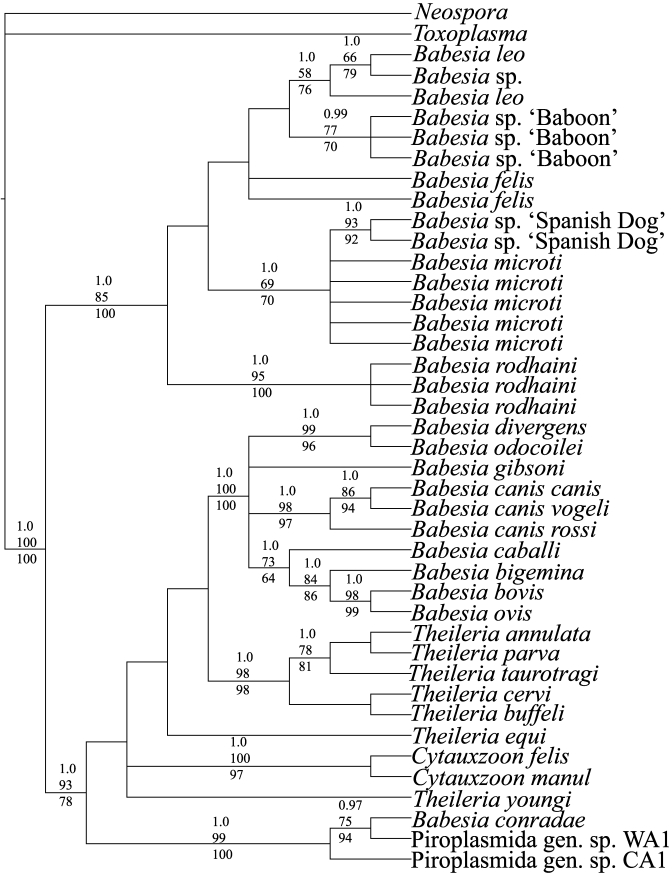
Alignment of 18S rDNA sequences for the 40 ingroup and 2 outgroup taxa resulted in 1745 aligned positions. Of these, 286 occurred in hypervariable regions and were excluded from all phylogenetic analyses because they possibly violated the assumption of positional homology. Maximum parsimony analysis resulted in 32 equally parsimonious trees of 597 steps (consistency index excluding uninformative characters = 0.5796; retention index = 0.8408). Bootstrap analysis revealed 21 clades supported in at least 70% of the bootstrap iterations. Maximum likelihood analysis under the GTR + I + Γ model of nucleotide substitution produced a single optimal tree (score = –5870.278), and bootstrap analysis revealed 18 clades supported in at least 70% of the bootstrap iterations. Bayesian analysis reached stationary at 5000 generations and revealed 21 clades supported with a posterior probability of at least 0.95. Figure 3 shows the results of the Bayesian phylogenetic analysis, along with maximum parsimony and maximum likelihood bootstrap results.
Figure 3.
Phylogenetic relationships among species of Babesia. Numbers above clades are Bayesian posterior support, maximum likelihood, and maximum parsimony bootstrap support values, respectively.
Results of Bayesian, maximum likelihood, and maximum parsimony analyses were concordant in documenting 2 large, strongly supported clades, with each clade consisting of several strongly supported polytomys (Figure 3). In addition to containing the Babesia isolate obtained from baboons, clade I consists of representatives of B. leo, B. felis, B. microti, and B. rhodaini. Clade 2 contained representatives of 8 additional species of Babesia as well as representatives of Cytauxzoon and Theileria.
Although not supported based on bootstrap or Bayesian analyses, all 3 methods of phylogenetic analysis produced a tree topology of Babesia ‘baboon’ that was sister to the clade of B. leo. Further support for the closer phylogenetic affinities of the Babesia-positive baboon sequences to B. leo than B. microti comes from comparisons of corrected percentage sequence divergence. Corrected percentage sequence divergence between Babesia ‘baboon’ and B. leo is 0.94% whereas the comparison of Babesia ‘baboon’ and B. microti is 1.19%. Taken together, these data support the conclusion that the Babesia isolate in captive baboons is most likely not a member of B. microti and likely represents an undescribed species of Babesia most closely related to B. leo. Two sequences of the novel Babesia isolate from baboons in the present study were designated BabBabesiaSeq1 and BabBabesiaSeq2, were submitted to GenBank, and given accession numbers of FJ897741 and GQ225744, respectively.
Experimental transmission.
Baboons inoculated intravenously, intramuscularly, or subcutaneously with 1 mL whole blood from the donor baboon each became infected with Babesia (Table 4). The baboon inoculated intravenously became PCR-positive for Babesia on day 7 after inoculation, whereas those inoculated intramuscularly or subcutaneously were PCR-positive beginning on day 10 after inoculation. All of these baboons remained PCR-positive for Babesia through day 31, when the study ended. None of the experimental baboons exhibited any clinical signs of babesiosis. The control baboon did not become infected with Babesia.
Table 4.
PCR results of baboons inoculated with Babesiaspp.
| Route |
||||
| No. of days after inoculation | IV | IM | SC | Control |
| 0 | – | – | – | – |
| 3 | – | – | – | – |
| 7 | + | – | – | – |
| 10 | + | + | + | – |
| 14 | + | + | + | – |
| 17 | + | + | + | – |
| 21 | + | + | + | – |
| 24 | + | + | + | – |
| 31 | + | + | + | – |
Discussion
Infections of Babesia in captive baboons typically are reported as being complications in immunocompromised animals. In one report,1 2 baboons demonstrated acute hemolytic crises due to infection of a B. microti-like piroplasm after experimental stem cell transplantation. Other authors5 reported anemia, leukocytosis, fever, and anorexia due to infection of baboons with B. microti after heart transplantation and immunosuppressive therapy.
A survey of 65 baboons housed at a Regional Primate Research Center revealed 20 (31%) baboons subclinically infected with Babesia.1 Of the 65 baboons surveyed, 23 originated within the facility's breeding colony, 26 were from an out-of-state breeding facility, and 16 were imported from Africa.1 Through sequencing of a 500-bp 5′ portion of the nuclear single-stranded rDNA and phylogenetic analysis, the authors deduced that a Babesia isolate found in one of their baboons was 97.9% similar to B. microti.1
In the current study, we found that the prevalence of Babesia within our facility's baboon breeding colony averaged 8.8%, with no significant difference between male and female baboons. Prevalence of Babesia infection differed among age groups, with baboons 11 to 20 y old being most likely to be infected with Babesia. Throughout the 3 sampling periods over 1 y, most of the baboons that were infected maintained their infection and were still infected a year later. Eight baboons initially tested positive but later tested negative on the next 2 samplings. These baboons may have cleared their infections, or perhaps they represented false-positive results, which could have been due to errors in sample collection, DNA extraction, or PCR assays. Three baboons that tested positive for Babesia during the second and third sampling periods were negative for Babesia during the first sampling.
Although the phylogenetics of piroplasmids have been studied in depth,2 little is known regarding Babesia spp. found in captive or wild baboons. Previous reports of Babesia spp. of captive baboons indicated that the piroplasms were B. microti5 or most similar1 to B. microti. Here we report a novel species of Babesia in colony-reared baboons. The 18s rDNA Babesia sequences we obtained from 2 captive baboons were most similar to B. leo, which is a piroplasm of African lions.15 Our facility's breeding colony includes wild-caught baboons that were imported from Africa. We speculate that some of these wild-caught baboons were infected with the novel Babesia isolate when they were captured. For example, one of our baboons that yielded a novel Babesia sequence was a wild-caught 14-y-old female from Africa; another animal that yielded sequence from the novel isolate was a 7-y-old male that had been born in our facility's breeding colony.
One previously reported baboon with babesiosis5 was born in our facility's breeding colony and then shipped to the authors’ institution, where the transplantation study occurred. The authors of the cited study5 stated that the baboon was infected with B. microti and that the baboon almost certainly carried the parasite when admitted to their clinic. However, the authors did not publish their sequencing data to allow independent analyses to confirm or refute their taxonomic identification of B. microti. In our study, we found a novel species of Babesia in our baboon colony. The novel species of Babesia in the baboons is more closely related to B. leo than B. microti. In the United States, B. microti is most prevalent in the northeastern and north central regions of the country.23 B. microti is a piroplasm of rodents and can be transmitted to humans by the bite of infected Ixodes scapularis (black-legged tick or deer tick). B. microti is considered an emerging infectious agent of humans because of increased contact with ticks and reservoir hosts.18
Babesia spp. are found throughout the world and usually are transmitted to vertebrate hosts by ixodid (that is, hard) ticks. However, the habitat conditions (for example, sandy ground, no live foliage, concrete floors) under which the baboons are housed at our facility's breeding colony, as well as the social grooming behavior among baboons, are not conducive to maintaining tick populations. For a related piroplasm, B. gibsoni in dogs, it has been postulated that the hemoparasite can be transmitted and maintained in dog populations without an ixodid vector by blood-to-blood contact through fighting among dogs.12,24 Baboons in our breeding colony frequently fight to determine and maintain dominance. In addition in the current study, we demonstrated iatrogenic transmission of the novel Babesia isolate through intravenous, intramuscular, and subcutaneous routes of inoculation. We speculate that Babesia can be transmitted among baboons within the colony through infected blood during fights. Passage of contaminated blood during fighting has also been proposed for the transmission of simian T-lymphotrophic virus 1 among baboons in this same captive breeding colony.3
In addition, vertical transmission of B. gibsoni from an infected dam to offspring may also be possible.6 We found that baboons 11 to 20 y of age had the highest prevalence of infection (21.3%; 30 of 141), whereas only a few (0.9%, 3 of 343) baboons 4 y or younger were infected with Babesia. Because few immature and young baboons were infected with Babesia spp. in our colony, we speculate that vertical transmission does not play a major role in the transmission of the hemoparasite. However, more rigorous investigation with controlled studies will be necessary to determine the role of vertical transmission of Babesia in our colony.
Here we report a novel species of Babesia that infects captive baboons in our facility's breeding colony. The novel Babesia isolate is most closely related to a B. leo from African lions. The prevalence of Babesia within the baboons colony was relatively low, averaging about 8%, with baboons 11 to 20 y old being the most likely to be infected. Because baboons are fastidious groomers and due to the fact that they are housed under conditions that do not support populations of ticks, we speculate that Babesia is being maintained in colony baboons through the transfer of contaminated blood during fights. Due to the physiologic similarities between baboons and humans, these nonhuman primates are becoming important models in biomedical research.19 As the role of baboons in biomedical research continues to expand, so will the need to recognize latent or subclinical infections that could introduce confounding variables in subsequent studies.9
Acknowledgments
We thank animal handlers Aaron Admire, Gail Goodson, and Tammy McKnight. This research project was funded in part through NIH grant P40 RR12317 to Gary L White, Roman F Wolf, Jean M d'Offay, and Mason V Reichard; NIH grant R24 RR016556 to Gary L White and Roman F Wolf; and Morris Animal Foundation grant D07Z0-6444 to Christine M Simecka and Mason V Reichard.
References
- 1.Bronsdon MA, Homer MJ, Magera JMH, Harrison C, Andrews RG, Bielitzki JT, Emerson CL, Persing DH, Fritsche TR. 1999. Detection of enzoonotic babesiosis in baboons (Papio cynoephalus) and phylogenetic evidence supporting synonymy of the genera Entopolypoides and Babesia. J Clin Microbiol 37:1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Criado-Fornelio A, Gonzalez-Del-Rio MA, Buling-Sarana A, Barba-Carretero JC. 2004. The ‘expanding universe’ of piroplasma. Vet Parasitol 119:337–345 [DOI] [PubMed] [Google Scholar]
- 3.d'Offay JM, Eberle R, Sucol Y, Schoelkopf L, White MA, Valentine BD, White GL, Lerche NW. 2007. Transmission dynamics of simian T-lymphotropic virus type 1 (STLV1) in a baboon breeding colony: predominance of female-to-female transmission. Comp Med 57:105–114 [PubMed] [Google Scholar]
- 4.Drummond AJ, Kearse M, Heled J, Moir R, Thierer T, Ashton B, Wilson A, Stones-Havas S.2008. [Internet] Geneious version 4.6.1. [Cited 01 November 2010]. Available at: http://www.geneious.com.
- 5.Ezzelarab M, Yeh P, Wagner R, Cooper DK. 2007. Babesia as a complication of immunosuppression following pig-to-baboon heart transplant. Xenotransplantation 14:162–165 [DOI] [PubMed] [Google Scholar]
- 6.Fukumoto S, Suzuki H, Igarashi I, Xuan X. 2005. Fatal experimental transplacental Babesia gibsoni infections in dogs. Int J Parasitol 35:1031–1035 [DOI] [PubMed] [Google Scholar]
- 7.Gubbels JM, de Vos AP, van der Weide M, Viseras J, Schouls LM, de Vries E, Jongejan F. 1999. Simultaneous detection of bovine Theileria and Babesia species by reverse line-blot hybridization. J Clin Microbiol 37:1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704 [DOI] [PubMed] [Google Scholar]
- 9.Haustein SV, Kolterman AJ, Sundblad JJ, Fechner JH, Knechtie SJ. 2008. Nonhuman primate infections after organ transplantation. ILAR J 49:209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huelsenbeck JP, Ronquist FR. 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 11.Institute for Laboratory Animal Research 1996. Guide for the care and use of laboratory animals. Washington (DC): National Academies Press; [PubMed] [Google Scholar]
- 12.Jefferies R, Ryan UM, Jardine J, Broughton DK, Robertson ID, Irwin PJ. 2007. Blood, bull terriers, and babesiosis: further evidence for direct transmission of Babesia gibsoni in dogs. Aust Vet J 85:459–463 [DOI] [PubMed] [Google Scholar]
- 13.Maddison DR, Maddison WP. 2000. Analysis of phylogeny and character evolution. Sunderland (MA): Sinauer Associates [Google Scholar]
- 14.Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499 [DOI] [PubMed] [Google Scholar]
- 15.Penzhorn BL, Kjemtrup AM, Lopez-Rebollar LM, Conrad PA. 2001. Babesia leo n. sp. from lions in the Kruger National Park, South Africa, and its relation to other small piroplasms. J Parasitol 87:681–685 [DOI] [PubMed] [Google Scholar]
- 16.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 17.Reichard MV, Wolf RF, Carey DW, Garrett JJ, Briscoe HA. 2007. Efficacy of fenbendazole and milbemycin oxime for treating baboons (Papio cynocephalus anubis) infected with Trichuris trichiura. J Am Assoc Lab Anim Sci 46:42–45 [PubMed] [Google Scholar]
- 18.Rodgers SE, Mather TN. 2007. Human Babesia microti incidence and Ixodes scapularis distribution, Rhode Island, 1998–2004. Emerg Infect Dis 13:633–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers J, Hixson JE. 1997. Baboons as an animal model for genetic studies of common human disease. Am J Hum Genet 61:489–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokal RR, Rohlf FJ. 1997. Bimetry, 3rd ed. San Franscisco (CA): WH Freeman and Company. [Google Scholar]
- 21.Swofford DL. 2000. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sunderland (MA): Sinauer Associates [Google Scholar]
- 22.Thompson JD, Gibson TJ, Plewkiak F, Jeanmougin F, Higgins D. 1997. The CLUSTAL X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vannier E, Gewurz BE, Krause PJ. 2008. Human babesiosis. Infect Dis Clin North Am 22:469–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeagley TJ, Reichard MV, Hempstead JE, Allen KE, Parsons LM, White MA, Little SE, Meinkoth JH. 2009. Detection of Babesia gibsoni and the canine small Babesia ‘Spanish isolate’ in blood samples obtained from dogs confiscated from dogfighting operations. J Am Vet Med Assoc 235:535–539 [DOI] [PubMed] [Google Scholar]