Abstract
Accumulating data suggest that the biological responses to high and low doses of radiation are qualitatively different, necessitating the direct study of low-dose responses to better understand potential risks. Most such studies have used two-dimensional culture systems, which may not fully represent responses in three-dimensional tissues. To gain insight into low-dose responses in tissue, we have profiled global gene expression in EPI-200, a three-dimensional tissue model that imitates the structure and function of human epidermis, at 4, 16 and 24 h after exposure to high (2.5 Gy) and low (0.1 Gy) doses of low-LET protons. The most significant gene ontology groups among genes altered in expression were consistent with effects observed at the tissue level, where the low dose was associated with recovery and tissue repair, while the high dose resulted in loss of structural integrity and terminal differentiation. Network analysis of the significantly responding genes suggested that TP53 dominated the response to 2.5 Gy, while HNF4A, a novel transcription factor not previously associated with radiation response, was most prominent in the low-dose response. HNF4A protein levels and phosphorylation were found to increase in tissues and cells after low- but not high-dose irradiation.
Introduction
The annual per capita exposure of the U.S. population to medical radiation has increased almost 600% in the last 25 years (1), mainly due to diagnostic procedures, raising concerns about the potential risks of low doses of radiation to public health. There is also increasing epidemiological evidence at lower doses for the induction of health effects such as cancer (2–4) and possibly cardiovascular and other chronic diseases (5, 6). At the same time, novel low-dose effects such as adaptive response and bystander effects are gaining recognition, meaning that direct linear extrapolation from effects at high doses may not be appropriate for low-dose exposures. Exposure to a low dose of radiation can induce an adaptive response that weakens the effects of a subsequent high dose, reflected as lowered mutation rate (7), reduced tumor growth (8) and protection against cytogenetic damage (9). Bystander effects, in which cells or tissue not directly irradiated respond to a stress signal from nearby irradiated cells, may further complicate responses in the low-dose range. Effects in bystander tissue may be either deleterious, such as chromosome damage (10), mutation induction (11) or carcinogenesis (12), or protective, such as apoptosis (13) or terminal differentiation (14).
Gene expression changes were established as an early indicator of cellular responses to low-dose radiation (15). More recently, microarray analysis and whole genome screening have been applied to investigate the effects of low dose rate (16–18) as well as responses to low radiation doses in several cell culture (19, 20) and in vivo (21–24) models. Such whole genome studies can be used to reverse engineer signaling networks and help to unravel the molecular mechanisms of radiation response (25).
While in vivo studies using human material are limited by clinical protocols, there is increasing awareness that two-dimensional tissue culture models may not fully represent the signaling interactions necessary to understand radiation response (26). Well-characterized reconstructed skin models are commercially available, and because the skin is the largest organ in the body and will always be exposed when there is external radiation exposure, three-dimensional skin models are attractive for the study of low-dose radiation effects. We have used EPI-200, a reconstructed human epidermis model that maintains the microstructure and functions of human skin, and compared the global gene expression response at 4, 16 and 24 h after exposure to a high (2.5 Gy) or low (0.1 Gy) dose of low-LET protons.
The pattern of structural changes observed in the tissues differed after exposure to low and high radiation doses, and these changes were consistent with the most significantly over-represented functional classes of genes responding to the two doses. The gene expression results were also subjected to network analysis, which allowed the identification of TP53 as the most connected regulatory hub after high-dose exposure and HNF4A as the major hub after low-dose exposure. Immunofluorescence staining and Western blot analysis confirmed increased nuclear expression of HNF4A at the protein level after exposure to low-dose radiation and suggests a complex interplay between TP53 and HNF4A. HNF4A is a novel low-dose-responsive transcription factor with no previously described role in radiation response.
Materials and Methods
Tissue Culture and Irradiation
EPI-200 (MatTek, Ashland, MA) tissues were cultured in 6-well tissue culture dishes with NMM-100 medium (0.9 ml/well) according to the manufacturer's instructions as described (27). Samples were exposed to 0, 0.1 or 2.5 Gy 4.5 MeV protons using the track segment irradiation facility of the 5.5-MV Singletron accelerator at the Radiological Research Accelerator Facility (RARAF) of Columbia University. After irradiation, samples were returned to plates with fresh culture medium and cultured for 4, 16 or 24 h. During sample processing, the tissue and its supporting membrane were held in growth medium at room temperature for up to 10 min. From three to six biological repeats were performed for each condition and end point.
HCT116 and HCT116 p53−/− cells (28) were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% FBS in Lab-Tek 2-well chamber slides (Thermo Scientific, Waltham, MA). X-ray exposures were performed using a Coronado 250 kV therapy X-ray machine (Westinghouse, Pittsburgh, PA). After incubation for 4, 16 or 24 h, the cells were fixed in ice-cold methanol:acetic acid (3:1) for 20 min at 4°C, air-dried at room temperature, and stored at −20°C.
RNA Isolation and Characterization
Total RNA was isolated using the RNAqueous Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions and stored at −70°C. Briefly, the procedure included homogenization in a Potter homogenizer, binding to a spin column and on-column treatment with DNase I. The impurities were washed out and RNA was eluted with a commercial elution buffer preheated to 75°C. RNA yield and quality were assessed with the NanoDrop spectrophotometer (Thermo Scientific) and the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). All samples used for microarray analysis had ratio A260/A280 > 1.8 and RIN > 8.0.
Microarray Hybridization, Washing and Scanning
Unless otherwise specified, all microarray reagents and hardware were from Agilent Technologies. RNA (100 ng of each sample) was labeled and amplified with the one-color Quick Amp Labeling Kit. Labeled cRNA was purified with RNeasy (Qiagen, Valencia, CA) following the Agilent protocol and checked to ensure incorporation of >8.0 pmol of Cy3 per μg of cRNA prior to fragmentation and hybridization to Agilent Whole Human Genome Oligo Microarrays (G4112F) for 17 h at 65°C using the Gene Expression Hybridization Kit. Arrays were then washed following Agilent's recommendations, dried by centrifugation (1,500 rpm; 10 min), and immediately scanned at 5 μm resolution with an Agilent DNA Microarray Scanner (G2505B). Scan images were analyzed with default parameters using Feature Extraction Software 9.1 for background correction and flagging of non-uniform features.
Quantitative Real-Time PCR (qPCR)
The SuperScript® III First-Strand Synthesis System (Invitrogen) was used to prepare cDNA from total RNA. TaqMan gene expression assays were purchased from Applied Biosystems (Table 1), with the exception of PTGS2, for which previously designed primer/probe sequences were used (29). The TaqMan Endogenous Control for ACTB (4333762F, Applied Biosystems) was used as a control. Using 100 ng cDNA as input, qPCR reactions were performed with the ABI 7900 Real Time PCR System (Applied Biosystems) using Universal PCR Master Mix (Applied Biosystems) with initial activation at 50°C for 120 s and 94.5°C for 10 min followed by 40 cycles of 97°C for 30 s and 59.7°C for 60 s. All samples were run in triplicate.
TABLE 1. Applied Biosystems Predesigned TaqMan Assays Used for qPCR.
| Gene | Assay number |
|---|---|
| ATF3 | Hs00231069_m1 |
| CDK9 | Hs00176222_m1 |
| ADAMTS1 | Hs01095532_g1 |
| GPRC5A | Hs00173681_m1 |
| CYP1B1 | Hs00164383_m1 |
| ROR1 | Hs00178178_m1 |
| PSCA | Hs00194665_m1 |
| IGF2 | Hs01005964_g1 |
| APOBEC3A | Hs00377444_m1 |
| BCAS3 | Hs00375126_m1 |
Data Analysis
Background-corrected hybridization intensities were imported into BRB-ArrayTools, Version 3.7.1 (30) log2-transformed and median normalized. Features not significantly above background intensity in >25% of the hybridizations and those without a minimum 1.5-fold change in at least 20% of the hybridizations were filtered out, yielding a set of 13,683 features that were used in subsequent analyses. Class comparisons using a random variance t test were employed to identify differentially expressed genes in the various groups. Genes with P values less than 0.001 were considered statistically significant, and the false discovery rate (FDR) was estimated for each gene using the method of Benjamini and Hochberg (31). The microarray data are available through the Gene Expression Omnibus database with accession number GSE16935.
The lists of significantly responding genes were annotated with PANTHER (32, 33), which was also used for gene ontology analysis. The gene lists were imported into IPA (Ingenuity® Systems, http://www.ingenuity.com, Redwood City, CA) for core analysis. The most connected regulatory nodes from this core analysis were explored further using IPA Pathway Explorer to determine how many genes were potentially regulated by each node after exposure to the different doses.
Micronucleus Assay and Immunostaining
Irradiated and control tissues were fixed in 10% formalin for 24 h, then washed in PBS and gradually transferred to 70% ethanol for another 24 h prior to embedding in paraffin and sectioning. Hematoxylin and eosin (H&E) staining was performed according to a standard protocol. Micronuclei were counted in binucleate cells as described previously (34). The proliferation index was defined as the percentage of binucleate cells in proliferating cell layers on the slides used to score micronuclei. The thickness of the cornified layer relative to total tissue thickness was measured on H&E-stained sections using Image J software (NIH, Bethesda, MD).
For immunostaining, slides were deparaffinized in EZ-DeWax™ solution (BioGenex, San Ramon, CA), washed and treated for antigen retrieval in citrate-Tween buffer, pH 6.0, preheated to 95°C. The slides were left in the buffer for 30 min until they cooled to room temperature and then washed briefly. For cell lines, fixed cells were rehydrated in 1× PBS (2 × 5 min) and permeabilized for 5 min in 0.2% Triton X-100 in PBS. Using the antibody concentrations recommended by the manufacturers, the slides were hybridized to anti-HNF4A antibody (HPA004712, Sigma-Aldrich, St. Louis, MO) and detected with Texas Red-labeled secondary antibody (TI-1000, Vector Labs, Burlingame, CA). After washing in PBS, the slides were sealed with nail polish. Results were visualized on a Nikon confocal microscope C1 and merged using Image Pro Plus software (Media Cybernetics, Inc., Bethesda, MD). The fluorescence intensity of at least 300 cells was quantified for each point in repeated experiments.
Western Blotting
Nuclear extracts were isolated from HCT116 cells at 4, 16 and 24 h after irradiation with 0, 0.1 or 2.5 Gy of X rays. Nuclear extracts were prepared using the NucBuster protein extraction kit (Novagen, San Diego, CA). Prior to gel loading, samples were boiled in sample buffer (Invitrogen) for 5 min. Proteins were resolved by denaturing electrophoresis on discontinuous polyacrylamide slab gels (SDS-PAGE) and transferred to PVDF membranes (Invitrogen) according to the manufacturer's protocol. Membranes were blocked with 5% non-fat dried milk in TBS-T for 1 h at room temperature followed by overnight incubation with primary antibodies against HNF4A (HPA004712, Sigma-Aldrich) or HNF4A phospho-Ser142 (ab76649, Abcam, Cambridge, MA) at 4°C, with shaking. After washing in TBS-T, membranes were incubated with horseradish-peroxidase (HRP)-conjugated secondary antibodies (Sigma-Aldrich) for 1 h at room temperature. The signal was visualized with the enhanced chemiluminescence ECL Plus detection system (GE Healthcare, Piscataway, NJ). Then membranes were stripped and reprobed with antibodies specific to TATA-box binding protein (Sigma-Aldrich). The Western blot data were quantified by using an AlphaImager 2200 digital image system (Alpha Innotech, San Leandro, CA) and normalized to the TATA-box binding protein signals.
Results
Physiological End Points
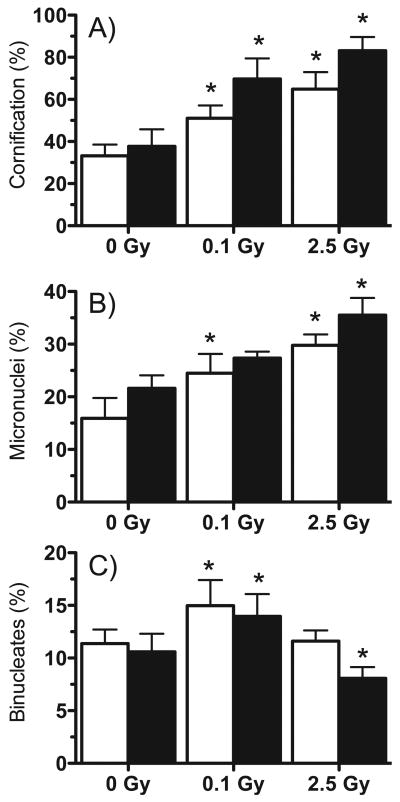
Tissues were examined microscopically in parallel with microarray experiments to monitor structural changes and effects on the tissues' normal differentiation program after radiation exposure. While nonirradiated controls showed normal differentiation and structural integrity, both radiation doses disrupted differentiation, resulting in hypercornification (Figs. 1 and 2A) and impairment of the granular layer (Fig. 1). Furthermore, the high dose resulted in severe disruptions of the basal layer (Fig. 1C). The fraction of micronucleated binucleate cells was also measured as an indicator of DNA damage. Irradiated cells showed a dose- and time-dependent increase in micronucleus frequency after both low and high doses (Fig. 2B). The proliferative index assessed as the ratio of binucleate cells in proliferating cell layers increased at 48 and 72 h after low-dose exposure, in contrast with a decrease in proliferation at 72 h after exposure to the high dose (Fig. 2C). Tissues were also examined at earlier times after irradiation, but no significant differences from controls were observed.
FIG. 1.
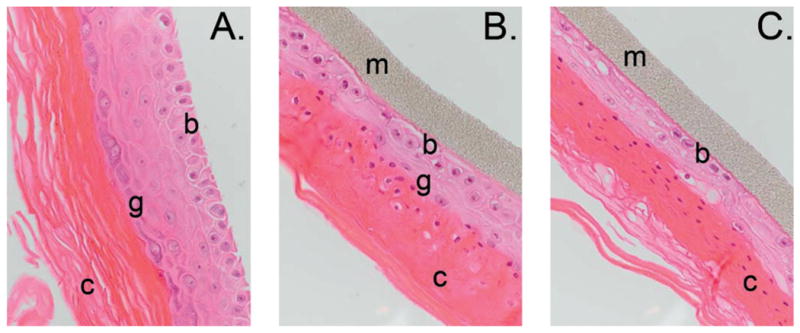
Hematoxylin and eosin (H&E)-stained paraffin-embedded sections. Maximum tissue disruption was evident at 72 h postirradiation, shown here. Panel A: Nonirradiated control; panel B: exposed to 0.1 Gy; panel C: exposed to 2.5 Gy. The supporting membrane (m) and basal (b), granular (g) and cornified (c) layers are indicated. Original magnification 400×.
FIG. 2.
Structural changes in irradiated EPI-200. Tissues were irradiated with 0, 0.1 or 2.5 Gy protons and examined 48 h (white bars) and 72 h (black bars) later. Panel A: Ratio of cornified layer to total tissue thickness. Panel B: Percentage of binucleate cells with micronuclei. Panel C: Percentage of binucleated cells in the basal layers as an indicator of proliferation. All results are means ± SEM (n = 6); (*) indicates significant difference from time-matched controls (P < 0.01).
Gene Expression Profiles in Tissues Irradiated with Low and High Dose
The class comparison feature of BRB-Array Tools (30) was used for each of the six dose-time combinations to identify genes with significantly different expression levels (P < 0.001, FDR < 10%) from their time-matched controls (Table 2; Supplementary Tables 1–6; available online at http://dx.doi.org/10.1667/RR2483.1.S1, http://dx.doi.org/10.1667/RR2483.1.S2, http://dx.doi.org/10.1667/RR2483.1.S3, http://dx.doi.org/10.1667/RR2483.1.S4, http://dx.doi.org/10.1667/RR2483.1.S5, http://dx.doi.org/10.1667/RR2483.1.S6). Irradiation with low and high doses resulted in broadly different patterns of postirradiation response. After exposure to the lower dose, the number of differentially expressed genes reached a maximum at 16 h postirradiation, with fewer genes responding at 4 h and no genes with FDR < 10% at 24 h. Relaxing the FDR requirement for this data set yielded 85 genes with P < 0.001 and FDR between 12 and 16%, however. In contrast, fewer genes responded at 16 h after the higher dose, with the most genes responding at 24 h (Table 2). Comparison of differentially expressed genes at the same times after high- or low-dose exposure revealed relatively few genes in common (98, 32 and 11 genes, at 4, 16 and 24 h, respectively). Only 7 genes were differentially expressed at all times after the higher dose, while none were differentially expressed at all time after the low dose. Of interest, 428 genes were regulated in response to the low dose but were not significantly altered after the high dose at any of the times assayed (Supplementary Table 7; available online at http://dx.doi.org/10.1667/RR2483.1.S7). The complete microarray data are available through the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) using accession number GSE16935.
TABLE 2. Differentially Expressed Genes at Different Times after Irradiation.
| Time | 10 cGy | 2.5 Gy |
|---|---|---|
| 4 h | 184 | 557 |
| 16 h | 352 | 156 |
| 24 h | 85a | 1150 |
FDR for these genes ranged between 12 and 16%.
Validation of Gene Expression with qPCR
To confirm the microarray results, we measured the expression levels of individual genes using TaqMan Quantitative real-time reverse transcription PCR (qPCR). We focused on genes regulated by the low dose that showed different patterns of regulation after the high dose. PTGS2, a well-defined radiation response gene, was included as a positive control, while ACTB was used as an endogenous control for normalization. qPCR was performed on all samples used for microarray hybridization. The pattern of relative gene expression measured by qPCR generally agreed with the microarray results (Table 3).
TABLE 3. Relative Gene Expression Ratios by Microarray and qPCR.
| 4 h | 16 h | 24 h | |||||
|---|---|---|---|---|---|---|---|
| Gene | Method | 10 cGy | 2.5 Gy | 10 cGy | 2.5 Gy | 1 Gy | 2.5 Gy |
| PTGS2 | Microarray | 3.59 | 3.37 | 0.69 | 0.9 | 3.43 | 3.98 |
| qPCR | 2.84 ± 0.21 | 2.57 ± 0.18 | 0.73 ± 0.18 | 1.17 ± 0.32 | 1.64 ± 0.01 | 2.77 ± 0.64 | |
| CDK9 | Microarray | 1.3 | 1.38 | 1.57 | 0.91 | 0.86 | 0.9 |
| qPCR | 2.80 ± 0.40 | 0.98 ± 0.51 | 2.19 ± 0.11 | 1.48 ± 0.19 | 0.95 ± 0.05 | 1.11 ± 0.01 | |
| GPRC5A | Microarray | 2.79 | 3.02 | 2.31 | 1.61 | 1.7 | 1.36 |
| qPCR | 1.47 ± 0.12 | 1.82 ± 0.10 | 1.68 ± 0.10 | 1.51 ± 0.17 | 0.86 ± 0.22 | 1.59 ± 0.05 | |
| ADAMTS1 | Microarray | 1.51 | 1 | 1.98 | 1.47 | 1.98 | 1.43 |
| qPCR | 2.11 ± 0.27 | 1.59 ± 0.04 | 2.25 ± 0.22 | 2.27 ± 0.12 | 1.21 ± 0.47 | 2.04 ± 0.14 | |
| ATF3 | Microarray | 1.67 | 1.44 | 1.72 | 1.41 | 1.85 | 0.94 |
| qPCR | 2.53 ± 0.42 | 2.67 ± 0.11 | 3.94 ± 0.97 | 1.98 ± 0.64 | 1.03 ± 0.21 | 0.84 ± 0.21 | |
| CYP1B1 | Microarray | 4.42 | 2.54 | 1.07 | 0.53 | 1.76 | 0.73 |
| qPCR | 3.98 ± 0.13 | 1.98 ± 0.07 | 1.18 ± 0.04 | 2.00 ± 0.22 | 0.81 ± 0.04 | 0.40 ± 0.04 | |
| ROR1 | Microarray | 0.53 | 0.95 | 11.98 | 0.85 | 0.63 | 0.69 |
| qPCR | 0.55 ± 0.01 | 1.06 ± 0.01 | 4.81 ± 0.07 | 0.49 ± 0.01 | 0.51 ± 0.03 | 0.50 ± 0.02 | |
| PSCA | Microarray | 2.74 | 1.83 | 3.44 | 0.97 | 0.52 | 0.67 |
| qPCR | 3.20 ± 0.22 | 0.72 ± 0.03 | 2.82 ± 0.10 | 1.78 ± 0.08 | 0.34 ± 0.02 | 0.99 ± 0.02 | |
| IGF2 | Microarray | 0.99 | 1.28 | 3.36 | 0.97 | 2.21 | 2.84 |
| qPCR | 0.98 ± 0.03 | 1.33 ± 0.05 | 3.30 ± 0.11 | 0.93 ± 0.01 | 2.18 ± 0.03 | 2.68 ± 0.03 | |
| APOBEC3A | Microarray | 2.27 | 3.34 | 1.79 | 0.91 | 5.37 | 1.46 |
| qPCR | 3.63 ± 0.03 | 5.01 ± 0.05 | 2.15 ± 0.02 | 0.80 ± 0.01 | 2.25 ± 0.05 | 1.23 ± 0.10 | |
| BCAS3 | Microarray | 0.67 | 0.72 | 1.35 | 1.63 | 2.78 | 1.04 |
| qPCR | 0.47 ± 0.02 | 0.79 ± 0.01 | 1.03 ± 0.01 | 1.30 ± 0.01 | 1.80 ± 0.04 | 1.07 ± 0.01 | |
Gene Ontology Analysis
We next analyzed the differentially expressed gene lists from the microarray studies for enrichment of gene groups with PANTHER (32, 33), which uses protein sequence information as well as gene families to assign a gene to an ontology group (Table 4). In samples irradiated with low-dose protons, the most significant biological processes represented among the responding genes were mRNA transcription and nucleotide metabolism (4 h) and cell cycle regulation, as well as blood circulation and gas exchange (16 h). The response to the high dose was dominated by cell cycle processes (4 and 16 h) and later by signal transduction and developmental processes (24 h). Regulation of cell cycle was significantly enriched after both doses, but with different time dependence.
TABLE 4. Gene Ontology Analysis Performed using PANTHER.
| Biological process | P value,a low dose | P value,a high dose |
|---|---|---|
| Time: 4 h | ||
| mRNA transcription | 4.9 × 10−4 | NSb |
| mRNA transcriptional regulation | 6.1 × 10−3 | NS |
| Cell cycle | NS | 7.8 × 10−9 |
| Cell cycle control | NS | 1.6 × 10−2 |
| Cell proliferation and differentiation | NS | 1.1 × 10−6 |
| Cell structure and motility | NS | 1.5 × 10−2 |
| Chromosome segregation | NS | 2.7 × 10−2 |
| Mitosis | NS | 1.7 × 10−2 |
| Nucleoside, nucleotide and nucleic acid metabolism | 4.6 × 10−2 | NS |
| Other receptor mediated signaling pathways | NS | 5.9 × 10−3 |
| Oncogenesis | NS | 1.4 × 10−2 |
| Time: 16 h | ||
| Cell cycle | 2.2 × 10−3 | 1.9 × 10−2 |
| Cell proliferation and differentiation | 2.9 × 10−3 | 2.3 × 10−2 |
| Chromosome segregation | NS | 2.5 × 10−2 |
| Blood circulation and gas exchange | 3.5 × 10−2 | NS |
| Time: 24 h | ||
| Signal transduction | NS | 8.2 × 10−19 |
| Cell surface receptor mediated transduction | NS | 1.9 × 10−12 |
| Developmental processes | NS | 2.5 × 10−11 |
| Transport | NS | 6.8 × 10−9 |
| Cell adhesion | NS | 2.6 × 10−3 |
| Immunity and defense | NS | 1.6 × 10−2 |
| G-protein mediated signaling | NS | 3.9 × 10−7 |
| Neuronal activity | NS | 1.9 × 10−6 |
| Cell communication | NS | 5.0 × 10−5 |
| Receptor protein tyrosine kinase signaling pathway | NS | 1.4 × 10−4 |
| Ion transport | NS | 3.2 × 10−5 |
| Cation transport | NS | 3.6 × 10−4 |
| Ectoderm development | NS | 1.4 × 10−2 |
| Blood circulation and gas exchange | NS | 2.3 × 10−2 |
| Cell structure and motility | NS | 4.9 × 10−2 |
| Cell adhesion-mediated signaling | NS | 2.9 × 10−2 |
| Intracellular signaling cascade | NS | 4.1 × 10−2 |
Bonferroni corrected.
Not statistically significant (P > 0.05).
Network Analysis
Next we performed pathway analysis of the differentially expressed gene sets with the Ingenuity core analysis tool. The top interacting networks among genes responding to low-dose radiation were significantly enriched at all times for functions of cancer (4 h: P = 2.25 × 10−5, 16 h: P = 1.55 × 10−9 and 24 h: P = 1.27 × 10−3), cell cycle (4 h: P = 9.25 × 10−3, 16 h: P = 4.37 × 10−10 and 24 h P = 3.34 × 10−3) and cell death (4 h: P = 1.72 × 10−4, 16 h: P = 2.32 × 10−5, 24 h: P = 1.66 × 10−2). Similarly, following high dose, functions of cell cycle (4 h: P = 3.15 × 10−17, 16 h: P = 4.97 × 10−12, 24 h: P = 1.01 × 10−4) and cancer (4 h: P = 1.05 × 10−11, 16 h: P = 4.97 × 10−12, 24 h: P = 1.01 × 10−4) were also significantly enriched in the top interacting networks. Additional categories significantly enriched at all times only among the high dose responsive genes included protein synthesis (4 h: P = 1.60 × 10−3, 16 h: P = 4.02 × 10−3, 24 h: P = 6.78 × 10−3) and energy production (4 h: P = 3.39 × 10−3, 16 h: P = 5.31 × 10−3, 24 h: P = 8.94 × 10−3).
To explore potential regulatory hubs, we examined the regulatory factors from the IPA core analysis that had the most network connections. Each regulatory factor was analyzed independently using the IPA Pathway Explorer Tool and information from the Ingenuity Knowledge Base to grow a network of all genes known to be regulated by that factor. Differential expression data from high- and low-dose responses at each time were overlaid on these networks to visualize the impact of radiation. Hubs connected to the largest number of radiation-responsive genes after high- and low-dose exposures were identified. Not surprisingly, TP53 emerged as the dominant hub after high-dose exposure (Fig. 3), while HNF4A was the most highly connected hub after low-dose exposure (Fig. 4).
FIG. 3.
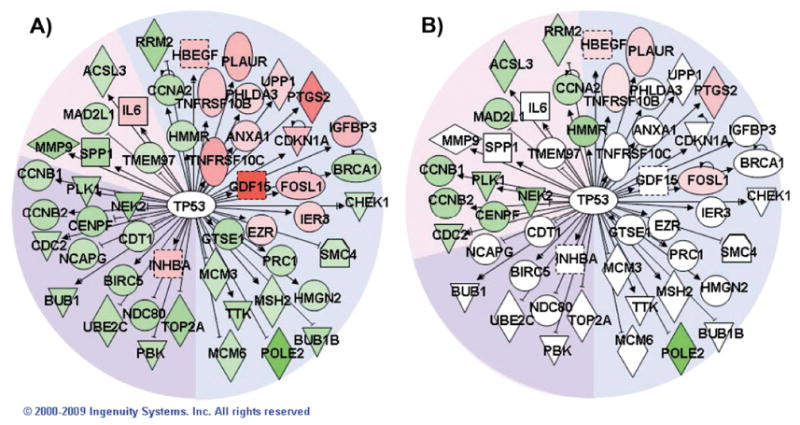
Network of TP53-regulated genes. Pathway Explorer was used to build a network of all transcriptional targets of TP53 from the Ingenuity Knowledge Base (IPA) that were differentially expressed after irradiation with 2.5 Gy. The nodes were colored by overlaying significant expression ratios from the data for 2.5 Gy (panel A) or 0.1 Gy (panel B), with red indicating upregulation and green downregulation. The time after irradiation when a response was observed is indicated by the background shading of the figure (blue: 4 h, pink: 16 h, purple: both 4 and 16 h). Additional TP53 targets also responded to the higher dose at 24 h, but none of those genes responded significantly to the lower dose.
FIG. 4.
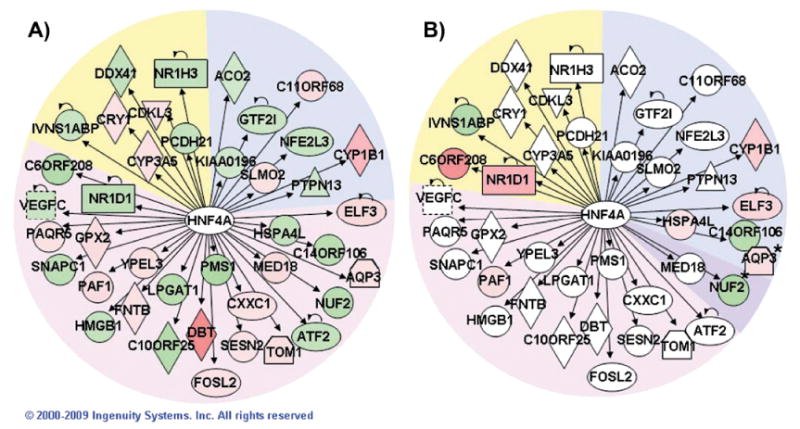
Network of HNF4A-regulated genes. Pathway Explorer was used to build a network of all transcriptional targets of HNF4A from the Ingenuity Knowledge Base (IPA) that were differentially expressed after irradiation with 0.1 Gy (panel A). The nodes were colored by overlaying significant expression ratios from the data for 0.1 Gy (panel A) or 2.5 Gy (panel B), with red indicating upregulation and green downregulation. The time after irradiation when a response was observed is indicated by the background shading of the figure (blue: 4 h, pink: 16 h, purple: both 4 and 16 h, yellow: 24 h).
Immunostaining and Western Blot Analysis
Tissues were stained for HNF4A; expression was found to be confined to the nuclei and increased in low-dose irradiated tissues relative to nonirradiated controls (Fig. 5A) at 16 h postirradiation. This is consistent with a role for HNF4A in the observed low-dose gene expression responses. We also performed Western blot analysis using nuclear extracts from the HCT116 colon cancer cell line, which has wild-type TP53, and its p53−/− knockout derivative (28). The active phosphorylated form of HNF4A was specifically associated with the nuclear fraction in HCT116, and levels increased after irradiation (Fig. 5B). In response to 2.5 Gy of X rays, levels of phosphorylated HNF4A peaked at 4 h after irradiation and then dropped to well below control levels by 24 h. Levels of active HNF4A increased more slowly after the low 0.1-Gy dose, peaking at 16 h postirradiation. In contrast, the TP53 knockout cells derived from HCT116 cells did not show detectable expression of active HNF4A (Fig. 5B) at any time or dose tested, suggesting that p53 may be required for expression of HNF4A.
FIG. 5.
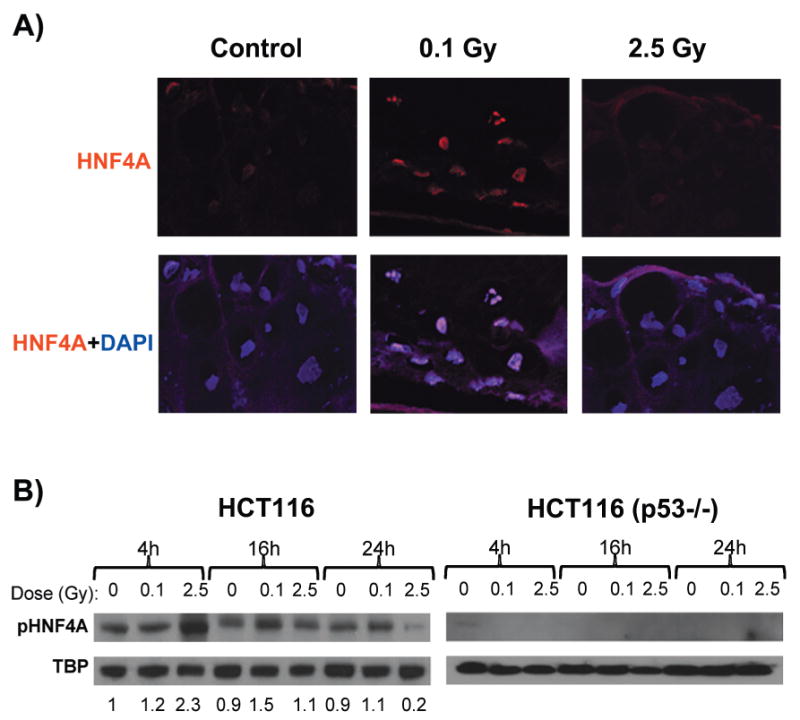
Detection of HNF4A protein. Panel A: Tissues fixed 16 h after exposure (control or 0.1 Gy). HNF4A staining was detected with Texas Red conjugated secondary antibody (red), and nuclei were counterstained (blue) with DAPI (original magnification 600×). Panel B: Western blot analysis of HNF4A phospho-Ser142 (pHNF42A) expression in HCT116 and HCT116 p53−/− nuclear extracts at 4, 16 and 24 h after exposure (control, 0.1 Gy or 2.5 Gy). Results are normalized to TATA-box binding protein (TBP), and relative change ratios compared to the 4-h control are indicated beneath each lane for HCT116. Both HNF4A and pHNF4A were expressed at very low levels in the p53−/− cells.
Discussion
Tissues irradiated with low or high doses of protons exhibited significant structural abnormalities (Figs. 1 and 2). Compared to nonirradiated controls, EPI-200 irradiated with the high dose lacked a continuous basal layer (Fig. 1C). Proliferating cells and postmitotic squamous cells also appeared together in the same layer. Moreover, the granular layer had lost integrity and was represented by rare cells underlying a thickened cornified layer. In contrast, tissues irradiated with the low dose showed elevated cell proliferation (Fig. 2C), suggesting initiation of tissue recovery processes. Cornification was also milder than that occurring after the high dose (Fig. 2A), suggesting a dose-dependent effect for induction of terminal differentiation. Furthermore, all irradiated samples contained numerous nucleated cells in the cornified layer. Such nucleated corneocytes indicate premature cornification in irradiated tissues and suggest failure of nuclear disintegration at the transition from squamous to cornified layers. The cornified layer in irradiated tissues was also sharply delineated and appeared very dense, suggesting dysfunction of the epidermal barrier.
Although there was little overlap of the genes that significantly responded to high and low doses or at the different times, gene ontology analysis can provide insight into common or divergent processes potentially affected by different sets of genes. Gene ontology analysis of the expression profiling data (Table 4) indicated that processes involved in transcriptional regulation are most affected at early times by exposure to the low dose, while cell cycle and proliferation functions dominate the early response to the high dose. Effects on genes related to cell cycle, proliferation and differentiation continued in tissues exposed to the high dose until at least 16 h after exposure and were also significant in the low-dose-exposed tissues at this time. By 24 h after exposure, signal transduction and developmental processes strongly dominated the tissues exposed to high doses, while significant gene expression changes were no longer detected in the tissues exposed to low doses. These processes are generally consistent with the altered structure ultimately observed in the irradiated tissue and suggest a shift from tissue restoration at the lower dose to greater damage followed by terminal differentiation and tissue restructuring at the higher dose.
In earlier studies of gene expression changes after low-dose irradiation, Yin et al. (21) reported the induction in mouse brain of genes involved in protective functions and downregulation of genes involved in specialized functions, such as neural signaling. Although significance of enrichment was not calculated, cell cycle control and signal transduction functions were well represented among genes responding to 10 cG γ rays in that study. In cell culture studies, normal human fibroblasts regulated a large number of genes between 1 and 24 h after exposure to 2 cGy X rays (19), with functions of signal transduction and development being significantly over-represented among responders. Normal human keratinocytes also responded to 1 cGy X rays by regulating expression of many genes, mostly at 48 and 72 h after exposure (20). The genes reported as responding only to 1 cGy and not to 1 Gy were significantly enriched for protein metabolism and modification (P = 1.74 × 10−5 by PANTHER analysis), a category not seen in our study, perhaps due to our focus on earlier times. The category of cell cycle, significant in our low-dose analysis, was the next most significant in the 2D keratinocyte response. Its significance was low (P = 0.1), however, perhaps due to the exclusion from that analysis of genes responding to both high and low doses. The differences with our results may also be due in part to the measurement of only about 7000 genes in the older studies rather than the whole genome, to the later timeframe of the keratinocyte study, or to differences in radiation response and cellular interactions between cells grown in 2- and 3D cultures.
In a whole genome expression study of full-thickness human skin irradiated in vivo with low doses during radiotherapy, variation between donors was found to be too great to allow confident identification of individual responding genes, leading to the development of a unique gene set enrichment analysis approach (35). This approach was used to identify gene groups and pathways responding linearly to exposures of 1 and 10 cGy and 1 Gy. Among the responders, cytokines, MAP kinases and zinc finger proteins were significantly upregulated, while S100 and keratins were downregulated (23, 24). Although our study did not look for linear dose dependence, these responses are consistent with our significant enrichment of signaling and tissue remodeling functions at high dose and transcriptional regulation at low dose.
Perhaps the strongest theme emerging from the gene ontology analyses of the EPI-200 data was that of functions related to cell cycle and proliferation. These functions dominated the high-dose response 4 h after exposure and both high- and low-dose responses 16 h after exposure (Table 4). A more detailed analysis revealed coordinate downregulation of many promitotic genes. Coordinate regulation of such cell cycle genes as CDC2, cyclins, topoisomerase II and centromere-associated proteins has been described previously (36), and the coordinate downregulation of these and other mitosis-related genes represents the most consistent response to ionizing radiation observed across a set of 60 diverse cancer cell lines (37).
In the 3D tissue model, the cyclins CCNA2, CCNB1 and CCNB2 as well as CENPE, CENPF and CEP55 were downregulated after both high- and low-dose irradiation. In contrast, microtubule motor (KIF2C, KIF4A, KIFC1, KNTC1, KIF11, KIF15, KIF20A and KIF22) and non-motor (CKAP5 and PRC1) binding proteins, centromere proteins (CENPA, CENPJ, CENPM and CENPT), and KNTC2, a gene involved in spindle checkpoint signaling (38), were all downregulated only in response to the high dose. Similarly, the G1 checkpoint inhibitors CDKN1A and CDKN1C were upregulated only by the high dose. By 24 h after exposure to the high but not the low dose, MYC expression was strongly elevated (2.6-fold). Overexpression of Myc in keratinocytes has been shown to stimulate their terminal differentiation (39). CABP5 and CALML5 were also upregulated in these samples. These genes are involved in calcium signaling, which is the dominant signaling pathway regulating terminal differentiation of keratinocytes. Taken together, this suggests a period of strong cell cycle arrest followed by increasing terminal differentiation in response to high-dose radiation exposure, consistent with the decrease in proliferating cells (Fig. 2C) and increased cornification (Fig. 2A) observed at later times in these tissues.
In contrast, signals promoting cell cycle arrest after low-dose exposure appear to be less broadly based and shorter lived. In addition, some cell cycle-related genes such as KIF26A and SESN2 were upregulated only by the low dose. SESN2 is of particular interest because it helps regulate cellular protection against oxidative stress (40), which may promote cell survival and help reduce the propensity toward terminal differentiation after radiation exposure.
Genes linked to blood circulation and gas exchange were also significantly enriched in tissues exposed to both the low dose (at 16 h) and high dose (at 24 h). These responses may be explained by the need to maintain homeostasis of the oxygen supply as the differentiation state and integrity of the tissue changes after radiation exposure. Being avascular tissue, epidermis depends on oxygen supplies from external sources. In the normal human epidermis, the upper skin layers to a depth of 0.25–0.40 mm are almost exclusively supplied by external oxygen (41), whereas the bloodstream contributes to oxygen supply at deeper tissue levels. Significant hypercornification observed after irradiation (Figs. 1 and 2A) suggests impairment of the epidermal barrier function, possibly disrupting normal gas exchange.
Among the genes in the gas exchange functional group, three hemoglobin genes (HBA, HBM and CYGB) were upregulated, suggesting a response to an insufficient oxygen supply. Hemoglobins are normally expressed in a variety of tissues including the skin (42). At least one of the represented genes, CYGB, is known to be directly regulated by hypoxia (43). Besides its main function, CYGB also contributes to scavenging NO and reactive oxygen species, contributing a protective function in response to oxidative stress (44). Other upregulated genes involved in gas exchange represent cation channels. SCNN1D, SCNN1G and KCNE1, regulated by the low dose, are involved in regulation of pH in the intracellular milieu (45, 46). Their upregulation may be a response to a pH change in the tissue. P2RX2, the only ion channel gene upregulated by the high dose, is also upregulated by hypoxia (47). Moreover, P2RX2 is an ATP receptor and calcium channel, possibly suggesting another link to calcium signaling, which accelerates differentiation and cornification of keratinocytes, as observed in the tissues exposed to the high dose.
Network analysis of the microarray data indicated TP53 as a major hub involved in the response to high-dose radiation with lesser response after low-dose exposure, consistent with prior studies (15, 29, 48, 49). In addition, network analysis implicated HNF4A as a transcription factor responsive to ionizing radiation for the first time. The increased levels of the active phosphorylated form of HNF4A protein present in the nuclear protein fraction after low-dose radiation exposure (Fig. 5) further supports the involvement of HNF4A in radiation response. A role for HNF4A in stress response is not completely unprecedented, however, because it has been shown to differentially activate specific target genes in response to oxidative stress (50). The acute-phase gene expression response induced in liver by tunicamycin, which causes endoplasmic reticulum stress, was also shown to be partly dependent on HNF4A (51).
Because the transcriptional activity of HNF4A had been shown to be repressed by TP53 (52), this presented an attractive possible mechanism for the observed shift in the pattern of regulated genes as the radiation dose was increased and levels of TP53 rose. We postulated that disruption of the TP53 gene might allow continued increases in HNF4A levels in cells irradiated by higher doses. This was found not to be the case in HCT116, however, because in contrast to the wild-type cells, the HCT116 p53−/− cells showed no detectable levels of HNF4A protein and no increase in activation (Fig. 5B) after either low- or high-dose exposure. Although this finding supports a role for TP53 in the regulation of HNF4A expression, it suggests a more complex relationship, possibly involving interactions with additional regulatory factors. For instance, c-Myc competes with HNF4A for control of the promoters of genes including CDKN1A and APOC3 (53), and MYC mRNA was regulated by exposure to high but not low doses in the present study. Binding of SMAD3 and SMAD4 to the HNF4A transactivation domain also affects HNF4A transcriptional activity (54). Expression of different splice variants has also been implicated in shaping the transactivation profile of HNF4A, with at least 6 adult and 3 prenatal isoforms described (55). In the absence of wild-type p53, it is possible that methylation of the HNF4A promoter may account for the observed lack of HNF4A expression. The overall methylation level of genomic DNA is often higher in cell lines with disrupted or non-functional TP53 (56), and HCT116 p53−/− has specifically been shown to have a 6-fold higher expression of the methylation enzyme DNMT1 than its p53 wild-type parent line (57), suggesting increased methylation and silencing of target genes compared to the parent cell line. HNF4A expression has recently been shown to be regulated by methylation of the promoter (58), making this a plausible potential explanation. Further studies will be required to unravel the complex regulation of HNF4A, which has been shown here for the first time to respond specifically to low-dose ionizing radiation.
Supplementary Material
Acknowledgments
We thank Stephen Marino for assistance with the irradiations and Howard Lieberman for critical reading of the manuscript. Gene expression analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and Amy Peng Lam. This work was supported by the Office of Science (BER), U.S. DOE, Grant No. DE-FG02-07ER46336, and by grant number P41 EB002033-13, Radiological Research Accelerator Facility (RARAF), from the National Institutes of Health/National Institute of Biomedical Imaging and Bioengineering (NIBIB).
Footnotes
Note. The online version of this article (DOI: 10.1667/RR2483.1) contains supplementary information that is available to all authorized users.
References
- 1.Mettler FA, Jr, Thomadsen BR, Bhargavan M, Gilley DB, Gray JE, Lipoti JA, McCrohan J, Yoshizumi TT, Mahesh M. Medical radiation exposure in the U.S. in 2006: preliminary results. Health Phys. 2008;95:502–507. doi: 10.1097/01.HP.0000326333.42287.a2. [DOI] [PubMed] [Google Scholar]
- 2.Hwang SL, Hwang JS, Yang YT, Hsieh WA, Chang TC, Guo HR, Tsai MH, Tang JL, Chang WP. Estimates of relative risks for cancers in a population after prolonged low-dose-rate radiation exposure: a follow-up assessment from 1983 to 2005. Radiat Res. 2008;170:143–148. doi: 10.1667/RR0732.1. [DOI] [PubMed] [Google Scholar]
- 3.Cardis E, Vrijheid M, Blettner M, Gilbert E, Hakama M, Hill C, Howe G, Kaldor J, Muirhead CR, Veress K. The 15-Country Collaborative Study of Cancer Risk among Radiation Workers in the Nuclear Industry: estimates of radiation-related cancer risks. Radiat Res. 2007;167:396–416. doi: 10.1667/RR0553.1. [DOI] [PubMed] [Google Scholar]
- 4.Ronckers CM, Doody MM, Lonstein JE, Stovall M, Land CE. Multiple diagnostic X-rays for spine deformities and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:605–613. doi: 10.1158/1055-9965.EPI-07-2628. [DOI] [PubMed] [Google Scholar]
- 5.Vrijheid M, Cardis E, Ashmore P, Auvinen A, Bae JM, Engels H, Gilbert E, Gulis G, Habib R, Veress K. Mortality from diseases other than cancer following low doses of ionizing radiation: results from the 15-Country Study of nuclear industry workers. Int J Epidemiol. 2007;36:1126–1135. doi: 10.1093/ije/dym138. [DOI] [PubMed] [Google Scholar]
- 6.Zielinski JM, Ashmore PJ, Band PR, Jiang H, Shilnikova NS, Tait VK, Krewski D. Low dose ionizing radiation exposure and cardiovascular disease mortality: cohort study based on Canadian national dose registry of radiation workers. Int J Occup Med Environ Health. 2009;22:27–33. doi: 10.2478/v10001-009-0001-z. [DOI] [PubMed] [Google Scholar]
- 7.Sykes PJ, Day TK, Swinburne SJ, Lane JM, Morley AA, Hooker AM, Bhat M. In vivo mutagenic effect of very low dose radiation. Dose Response. 2006;4:309–316. doi: 10.2203/dose-response.06-004.Sykes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson RE, Tokuda S, Williams WL, Warner NL. Radiation-induced augmentation of the response of A/J mice to SaI tumor cells. Am J Pathol. 1982;108:24–38. [PMC free article] [PubMed] [Google Scholar]
- 9.Stoilov LM, Mullenders LH, Darroudi F, Natarajan AT. Adaptive response to DNA and chromosomal damage induced by X-rays in human blood lymphocytes. Mutagenesis. 2007;22:117–122. doi: 10.1093/mutage/gel061. [DOI] [PubMed] [Google Scholar]
- 10.Nagasawa H, Little JB. Induction of sister chromatid exchanges by extremely low doses of alpha-particles. Cancer Res. 1992;52:6394–6396. [PubMed] [Google Scholar]
- 11.Zhou H, Randers-Pehrson G, Waldren CA, Vannais D, Hall EJ, Hei TK. Induction of a bystander mutagenic effect of alpha particles in mammalian cells. Proc Natl Acad Sci USA. 2000;97:2099–2104. doi: 10.1073/pnas.030420797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mancuso M, Pasquali E, Leonardi S, Tanori M, Rebessi S, Di Majo V, Pazzaglia S, Toni MP, Pimpinella M, Saran A. Oncogenic bystander radiation effects in Patched heterozygous mouse cerebellum. Proc Natl Acad Sci USA. 2008;105:12445–12450. doi: 10.1073/pnas.0804186105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawant SG, Randers-Pehrson G, Geard CR, Brenner DJ, Hall EJ. The bystander effect in radiation oncogenesis: I. Transformation in C3H 10T½ cells in vitro can be initiated in the unirradiated neighbors of irradiated cells. Radiat Res. 2001;155:397–401. doi: 10.1667/0033-7587(2001)155[0397:tbeiro]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Belyakov OV, Folkard M, Mothersill C, Prise KM, Michael BD. Bystander-induced differentiation: a major response to targeted irradiation of a urothelial explant model. Mutat Res. 2006;597:43–49. doi: 10.1016/j.mrfmmm.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Amundson SA, Do KT, Fornace AJ., Jr Induction of stress genes by low doses of gamma rays. Radiat Res. 1999;152:225–231. [PubMed] [Google Scholar]
- 16.Amundson SA, Lee RA, Koch-Paiz CA, Bittner ML, Meltzer P, Trent JM, Fornace AJ., Jr Differential responses of stress genes to low dose-rate gamma irradiation. Mol Cancer Res. 2003;1:445–452. [PubMed] [Google Scholar]
- 17.Sugihara T, Magae J, Wadhwa R, Kaul SC, Kawakami Y, Matsumoto T, Tanaka K. Dose and dose-rate effects of low-dose ionizing radiation on activation of Trp53 in immortalized murine cells. Radiat Res. 2004;162:296–307. doi: 10.1667/rr3223. [DOI] [PubMed] [Google Scholar]
- 18.Sokolov MV, Smirnova NA, Camerini-Otero RD, Neumann RD, Panyutin IG. Microarray analysis of differentially expressed genes after exposure of normal human fibroblasts to ionizing radiation from an external source and from DNA-incorporated iodine-125 radionuclide. Gene. 2006;382:47–56. doi: 10.1016/j.gene.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Ding LH, Shingyoji M, Chen F, Hwang JJ, Burma S, Lee C, Cheng JF, Chen DJ. Gene expression profiles of normal human fibroblasts after exposure to ionizing radiation: a comparative study of low and high doses. Radiat Res. 2005;164:17–26. doi: 10.1667/rr3354. [DOI] [PubMed] [Google Scholar]
- 20.Franco N, Lamartine J, Frouin V, Le Minter P, Petat C, Leplat JJ, Libert F, Gidrol X, Martin MT. Low-dose exposure to gamma rays induces specific gene regulations in normal human keratinocytes. Radiat Res. 2005;163:623–635. doi: 10.1667/rr3391. [DOI] [PubMed] [Google Scholar]
- 21.Yin E, Nelson DO, Coleman MA, Peterson LE, Wyrobek AJ. Gene expression changes in mouse brain after exposure to low-dose ionizing radiation. Int J Radiat Biol. 2003;79:759–775. doi: 10.1080/09553000310001610961. [DOI] [PubMed] [Google Scholar]
- 22.Voy BH, Scharff JA, Perkins AD, Saxton AM, Borate B, Chesler EJ, Branstetter LK, Langston MA. Extracting gene networks for low-dose radiation using graph theoretical algorithms. PLoS Comput Biol. 2006;2:e89. doi: 10.1371/journal.pcbi.0020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg Z, Rocke DM, Schwietert C, Berglund SR, Santana A, Jones A, Lehmann J, Stern R, Lu R, Hartmann Siantar C. Human in vivo dose-response to controlled, low-dose low linear energy transfer ionizing radiation exposure. Clin Cancer Res. 2006;12:3723–3729. doi: 10.1158/1078-0432.CCR-05-2625. [DOI] [PubMed] [Google Scholar]
- 24.Berglund SR, Rocke DM, Dai J, Schwietert CW, Santana A, Stern RL, Lehmann J, Hartmann Siantar CL, Goldberg Z. Transient genome-wide transcriptional response to low-dose ionizing radiation in vivo in humans. Int J Radiat Oncol Biol Phys. 2008;70:229–234. doi: 10.1016/j.ijrobp.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amundson SA. Functional genomics in radiation biology: a gateway to cellular systems-level studies. Radiat Environ Biophys. 2008;47:25–31. doi: 10.1007/s00411-007-0140-1. [DOI] [PubMed] [Google Scholar]
- 26.Barcellos-Hoff MH, Costes SV. A systems biology approach to multicellular and multi-generational radiation responses. Mutat Res. 2006;597:32–38. doi: 10.1016/j.mrfmmm.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Curren RD, Mun GC, Gibson DP, Aardema MJ. Development of a method for assessing micronucleus induction in a 3D human skin model (EpiDerm) Mutat Res. 2006;607:192–204. doi: 10.1016/j.mrgentox.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 28.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 29.Ghandhi SA, Yaghoubian B, Amundson SA. Global gene expression analyses of bystander and alpha particle irradiated normal human lung fibroblasts: Synchronous and differential responses. BMC Med Genomics. 2008;1:63. doi: 10.1186/1755-8794-1-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-Array Tools. Cancer Informatics. 2007;2:11–17. [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 32.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belyakov OV, Mitchell SA, Parikh D, Randers-Pehrson G, Marino SA, Amundson SA, Geard CR, Brenner DJ. Biological effects in unirradiated human tissue induced by radiation damage up to 1 mm away. Proc Natl Acad Sci USA. 2005;102:14203–14208. doi: 10.1073/pnas.0505020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocke DM, Goldberg Z, Schweitert C, Santana A. A method for detection of differential gene expression in the presence of inter-individual variability in response. Bioinformatics. 2005;21:3990–3992. doi: 10.1093/bioinformatics/bti667. [DOI] [PubMed] [Google Scholar]
- 36.Walker MG. Drug target discovery by gene expression analysis: cell cycle genes. Curr Cancer Drug Targets. 2001;1:73–83. doi: 10.2174/1568009013334241. [DOI] [PubMed] [Google Scholar]
- 37.Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, Ahn J, Reimers M, Chen Y, Scudiero DA, Fornace AJ., Jr Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68:415–424. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Riley DJ, Chen PL, Lee WH. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gandarillas A, Watt FM. c-Myc promotes differentiation of human epidermal stem cells. Genes Dev. 1997;11:2869–2882. doi: 10.1101/gad.11.21.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 41.Stucker M, Struk A, Altmeyer P, Herde M, Baumgartl H, Lubbers DW. The cutaneous uptake of atmospheric oxygen contributes significantly to the oxygen supply of human dermis and epidermis. J Physiol. 2002;538:985–994. doi: 10.1113/jphysiol.2001.013067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo X, Philipsen S, Tan-Un KC. Study of the hypoxia-dependent regulation of human CYGB gene. Biochem Biophys Res Commun. 2007;364:145–150. doi: 10.1016/j.bbrc.2007.09.108. [DOI] [PubMed] [Google Scholar]
- 44.Li D, Chen XQ, Li WJ, Yang YH, Wang JZ, Yu AC. Cytoglobin up-regulated by hydrogen peroxide plays a protective role in oxidative stress. Neurochem Res. 2007;32:1375–1380. doi: 10.1007/s11064-007-9317-x. [DOI] [PubMed] [Google Scholar]
- 45.Yamamura H, Ugawa S, Ueda T, Nagao M, Shimada S. Protons activate the delta-subunit of the epithelial Na+ channel in humans. J Biol Chem. 2004;279:12529–12534. doi: 10.1074/jbc.M400274200. [DOI] [PubMed] [Google Scholar]
- 46.Heitzmann D, Koren V, Wagner M, Sterner C, Reichold M, Tegtmeier I, Volk T, Warth R. KCNE beta subunits determine pH sensitivity of KCNQ1 potassium channels. Cell Physiol Biochem. 2007;19:21–32. doi: 10.1159/000099189. [DOI] [PubMed] [Google Scholar]
- 47.Mason HS, Bourke S, Kemp PJ. Selective modulation of ligand-gated P2X purinoceptor channels by acute hypoxia is mediated by reactive oxygen species. Mol Pharmacol. 2004;66:1525–1535. doi: 10.1124/mol.104.000851. [DOI] [PubMed] [Google Scholar]
- 48.Jen KY, Cheung VG. Identification of novel p53 target genes in ionizing radiation response. Cancer Res. 2005;65:7666–7673. doi: 10.1158/0008-5472.CAN-05-1039. [DOI] [PubMed] [Google Scholar]
- 49.Kis E, Szatmari T, Keszei M, Farkas R, Esik O, Lumniczky K, Falus A, Safrany G. Microarray analysis of radiation response genes in primary human fibroblasts. Int J Radiat Oncol Biol Phys. 2006;66:1506–1514. doi: 10.1016/j.ijrobp.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Guo H, Gao C, Mi Z, Wai PY, Kuo PC. Phosphorylation of Ser158 regulates inflammatory redox-dependent hepatocyte nuclear factor-4alpha transcriptional activity. Biochem J. 2006;394:379–387. doi: 10.1042/BJ20051730. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Luebke-Wheeler J, Zhang K, Battle M, Si-Tayeb K, Garrison W, Chhinder S, Li J, Kaufman RJ, Duncan SA. Hepatocyte nuclear factor 4alpha is implicated in endoplasmic reticulum stress-induced acute phase response by regulating expression of cyclic adenosine monophosphate responsive element binding protein H. Hepatology. 2008;48:1242–1250. doi: 10.1002/hep.22439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maeda Y, Seidel SD, Wei G, Liu X, Sladek FM. Repression of hepatocyte nuclear factor 4alpha tumor suppressor p53: involvement of the ligand-binding domain and histone deacetylase activity. Mol Endocrinol. 2002;16:402–410. doi: 10.1210/mend.16.2.0769. [DOI] [PubMed] [Google Scholar]
- 53.Hwang-Verslues WW, Sladek FM. Nuclear receptor hepatocyte nuclear factor 4alpha1 competes with oncoprotein c-Myc for control of the p21/WAF1 promoter. Mol Endocrinol. 2008;22:78–90. doi: 10.1210/me.2007-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chou WC, Prokova V, Shiraishi K, Valcourt U, Moustakas A, Hadzopoulou-Cladaras M, Zannis VI, Kardassis D. Mechanism of a transcriptional cross talk between transforming growth factor-beta-regulated Smad3 and Smad4 proteins and orphan nuclear receptor hepatocyte nuclear factor-4. Mol Biol Cell. 2003;14:1279–1294. doi: 10.1091/mbc.E02-07-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakhei H, Lingott A, Lemm I, Ryffel GU. An alternative splice variant of the tissue specific transcription factor HNF4alpha predominates in undifferentiated murine cell types. Nucleic Acids Res. 1998;26:497–504. doi: 10.1093/nar/26.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 57.Peterson EJ, Bogler O, Taylor SM. p53-mediated repression of DNA methyltransferase 1 expression by specific DNA binding. Cancer Res. 2003;63:6579–6582. [PubMed] [Google Scholar]
- 58.Einstein F, Thompson RF, Bhagat TD, Fazzari MJ, Verma A, Barzilai N, Greally JM. Cytosine methylation dysregulation in neonates following intrauterine growth restriction. PLoS One. 2010;5:e8887. doi: 10.1371/journal.pone.0008887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.