Abstract
Therapeutic treatments based on the injection of living cells are in clinical use and preclinical development for diseases ranging from cancer to cardiovascular disease to diabetes. To enhance the function of therapeutic cells, a variety of chemical and materials science strategies are being developed that engineer the surface of therapeutic cells with new molecules, artificial receptors, and multifunctional nanomaterials, synthetically endowing donor cells with new properties and functions. These approaches offer a powerful complement to traditional genetic engineering strategies for enhancing the function of living cells.
Keywords: Cell therapy, cell surface bioengineering, adoptive T cell therapy, nanoparticles
Introduction
Cell therapies are clinical procedures involving the direct transplantation, injection, or infusion of live cells to treat disease. These donor cells can be autologous (from the same patient) or allogeneic (from an unrelated donor). Cell therapy is a rapidly expanding field in translational medicine, encompassing both well-established procedures as well as advanced therapies still in early preclinical testing. While hematopoietic stem cell transplants (bone marrow transplants) have essentially become standard-of-care for the treatment of leukemia and related bone and blood cancers with nearly 40,000 procedures per year worldwide[1], a plethora of non-hematopoietic adult stem cell therapies are also undergoing clinical evaluation. In particular, mesenchymal stem cells (MSCs) have emerged as cornerstones of regenerative medicine strategies, based on their diverse spectrum of differentiation into cartilage, bone, cardiomyocytes or neurons, and the ease of culturing quantities appropriate for clinical applications[2–4]. Moreover, MSCs can suppress lymphocyte proliferation, which is being explored clinically to treat T cell-dependent pathologies, such as multiple sclerosis[5], amylotrophic lateral sclerosis[6], transplant rejection[7] or acute graft-versus-host disease[8]. Beyond stem cells, a manifold of differentiated functional cells are clinically exploited. For example, a cell technology poised to have a major impact on the treatment of end-stage heart failure is the intra-coronary infusion of skeletal myoblasts[9]. A related strategy is employed when infusing functional beta-islets cells into the portal vein of type I diabetic patients to restore natural insulin production, thereby eliminating the need for repeated insulin injections[10]. Autologous ex-vivo expanded tumor- or virus-specific effector T lymphocytes have shown tremendous therapeutic potential when adoptively transferred into patients with advanced malignancies[11] or chronic infections, such as HIV[12]. Also in the field of cellular tumor immunotherapy, injections of autologous ex-vivo matured dendritic cells loaded with tumor antigen represent a promising treatment modality to stimulate anti-tumor immunity[13]. Earlier this year, this methodology received FDA approval as the first cellular immunotherapy product (Provenge®) for the treatment of metastatic cancer[13]. Ultimately, the current cell therapy armamentarium will be critically expanded by the addition of induced pluripotent stem cells (iPSCs), which not only offer exciting prospects for regenerative medicine, but also the correction of genetic defects, such as beta-thalassemia or sickle cell anemia, at the stem cell level[14,15]. Thus, the field of cell therapy includes diverse therapeutic applications employing donor cells ranging from the earliest progenitor cells to terminally differentiated tissue cells.
The efficacy of cell therapies hinges on appropriate control of the fate and function of the therapeutic cells, and engineering donor cells for enhanced survival, proliferation, or differentiated function is a topic of central interest. Cell function has traditionally been engineered in three distinct ways, either by (1) preconditioning cells for enhanced in vivo effector function, self renewal or longevity by ex vivo exposure to soluble cytokines[16–18], pharmacological agents[19,20]or stimulatory ligands[21,22] (2) providing supporting adjuvant drug treatment by systemic bolus injections in parallel with cell therapies[23,24] ((modulating cell function via exogenous, systemic external cues) or (3) by genetic engineering, employing the tools of molecular biology to modify the genetic programming of cells[25,26]. Recently, a fourth approach has begun to gain attention, based on using synthetic materials or chemical biology approaches to alter cell surfaces. This complementary strategy lies somewhere between the two extremes of chemotherapy and genetic engineering, allowing exogenous cues to be provided to cells in a novel manner by directly remodeling the cell itself, employing the tools of chemistry and materials science in addition to molecular biology. Success in this arena depends on the recognition of the plasma membrane itself as a complex nanostructured surface, composed of organized lipids, proteins, and polysaccharide assemblies whose nanoscale organization can impact cell function. Synthetic nanomaterials have a significant role to play in cell surface engineering, due to their unique properties and ability to provide functionality beyond that achievable by single molecules. In this review, we will first summarize key methodologies used to manipulate the surface of living mammalian cells with synthetic material. We will then discuss the main challenges to overcome to stably modify cell surfaces and we will highlight how these cell modifications can be applied to enhance the therapeutic potential of cell products in clinic. Finally, we will outline future trends and perspectives of this relatively new and fast-developing discipline.
Motivations for exploiting cell surface bioengineering in cell therapy
Rapid progress in basic biology has defined a host of signaling pathways that could be modulated to endow therapeutic cells with enhanced functionality. The characterization of cell surface receptors, their respective ligands, and downstream signaling pathways that integrate external cues to regulate cell survival, proliferation, and differentiation, have provided motivation for the rational remodeling of cell surfaces for therapeutic purposes. In addition, because the cell surface controls all interactions of the cell with its environment, cell functions such as adhesion, migration, tissue homing, and cell-cell interactions can all be redefined via engineering of the molecular landscape of the plasma membrane.
Genetic engineering is well established as a robust and highly versatile methodology for introducing or knocking down specific surface proteins to modulate cell functionality[27,28]. However, technical challenges of efficient gene transfer, laborious gene transduction protocols and the safety concerns of unintentionally activating oncogenes through the random insertion of transgenes into the target cell genome hinder gene therapy in the clinical setting[29]. Genetic cell engineering is further constrained by limits on the size of genes that can be carried into cells using the most efficient current clinically-useful gene delivery vehicles, recombinant viruses. Thus, it is technically challenging to engineer cells with multiple genes[27]. Finally, gene therapy only allows cells to be engineered within the rule set of cellular biochemistry, and cannot be used to modify cell surfaces with non-biological therapeutic materials (e.g., non-biological small molecules, fluorescent tags, clinical imaging contrast agents, reactive polymers, complex nanocarriers, etc.) or to introduce biomolecules on the cell surface that do not have a biological mechanism for placement there (e.g., DNA or RNA). These limitations have fueled interest in devising novel bioconjugation, protein engineering, chemistry, and material science approaches with the goal of rationally modifying the molecular landscape on the cell surface.
What are the goals of cell surface engineering? Key applications include engineering cell adhesion[30] and in vivo cell migration by the introduction of exogenous targeting ligands into the cell membrane[31–33] or by manipulating cell surface glycosylation[34,35]. Likewise, stimulatory biomolecules or biological components that mimic a supportive microenvironment can be surface-coupled to transplanted cells to enhance their longevity, proliferative reserves and therapeutic potential[36]. Prolonged in vivo persistence and functionality of grafted cells is of particular importance in cell xeno-transplants, which can be rapidly destroyed by the recipient’s immune system[37]. To safeguard against immune rejection during early cell engraftment, transplanted cells can be camouflaged by polymer coatings[38]. Alternatively, the coupling of immune-inhibitory ligands or small molecules to the surface of transplanted cells could be envisioned as a promising strategy to locally blunt immune attacks. With next-generation biomaterials and synthetic nanocarriers emerging at rapid pace, cell bioengineering methodologies are ultimately poised to extend far beyond enhancing the efficacy of established cell-based therapies. One new area of intense research, which is based on the necessity to deliver drugs in a more directed and controlled manner to disease-relevant sites, is to decorate isolated cells, which display a defined in vivo migration pattern, with synthetic drug nanocarriers. Following adoptive transfer, cells actively transmigrate endothelial and stromal barriers and accumulate surface-attached cargo at pathological sites[39,40]. This is in contrast to conventional static drug targeting ligands, like antibodies, which rely on passive mechanisms, such as the enhanced permeation and retention (EPR)[41] to cross vascular endothelium and initially accumulate near target cells. As further outlined below, dynamic cell-nanoparticle hybrid vehicles could be applied to deliver a wide range of therapeutic agents, including small molecules, siRNA, contrast agents or vaccines to therapeutically desired anatomical compartments.
Challenges of cell surface engineering
Criteria for developing clinically-viable cell surface bioengineering strategies
Any cell surface bioengineering strategy developed in a preclinical setting should ideally meet several criteria to be of translational relevance for clinical cell therapy applications. First, to avoid loss of cell viability, conventional bioconjugation/bioengineering-protocols need to be carefully adapted for the purpose of engineering primary cells. Small changes in pH or osmolarity, exposure of cells to organic solvents, heat, excessive agitation, or serum starvation can have profound effects on cell viability and/or differentiated function. In most instances, surface bioengineering should further minimize alterations in membrane fluidity, i.e. the viscosity of the lipid bilayer, or changes to the bending elasticity of the cell membrane, since cell functions, such as adhesion, migration, proliferation and cell signaling critically depend on these physical parameters[42–44]. Considering the complexity and myriad functions of cell membrane surface proteins, glycolipids and polysaccharides, the main challenge of cell surface engineering remains to rationally modify select cell-surface molecules without physically blocking or functionally compromising others.
Cell membrane turnover and internalization as limiting factors
Cell surface engineering is complicated by the fact that the plasma membrane is not a static structure, but rather is always in a dynamic state[45]. In addition to constant redistribution and compartmentation in the lateral plane of the cell membrane, both lipid and protein components of the plasma membrane are continuously internalized, degraded, and replaced by de novo synthesis[46]. This is a highly selective process with specific turnover rates for different proteins and lipids ranging from hours to several weeks[47]. Internalization of lipids and protein from the cell surface occurs through processes of endocytosis, pinocytosis, or phagocytosis. In endocytosis[48], binding of ligands to cell surface receptors triggers a specific response where small pockets (e.g., ~50 nm in diameter) of the plasma membrane around the engaged receptor invaginate and pinch off to form closed vesicles. These budded vesicles are trafficked into the cell, and the internalized materials are sorted for intracellular transport or recycling to the membrane. While endocytosis is cargo-specific, non-specific uptake is dominated by pinocytosis[49]. This process involves continual invagination and budding of vesicles from the cell surface, allowing fluid and extracellular material to enter the cell. Conversely, intracellular transport vesicles fuse with the plasma membrane to release proteins into the extracellular space during exocytosis. Endocytosis and pinocytosis traffic small quantities of membrane and adjacent fluid into the cell. By contrast, large membrane areas are internalized during phagocytosis, a process used by specialized cells such as macrophages, neutrophils, and dendritic cells to remove pathogens, cell debris or dying cells from the extracellular environment[50]. The dynamic nature of the cell surface is a major challenge to cell surface engineering, which may lead to premature internalization and ultimately degradation of surface modifications or cell-conjugated nanocarriers. Therefore, numerous approaches, which are discussed in detail below, have been developed to prolong retention of engineered membrane components on the surfaces of cells. In recent years, significant progress has been made in understanding how the geometry and charge of biomaterials, their surface chemistry, and the strategy chosen to hybridize biomaterials with cell surfaces affect their internalization rate into cells.
Mechanical and biochemical challenges of the in vivo environment
The third key challenge to cell surface engineering is to introduce synthetic modifications that are compatible with the complex mechanical and biochemical environment the cell is exposed to in vivo. Following cell transplantation or adoptive transfer, cells will be exposed to in vivo shear stress and hemodynamic forces, and circulating cells such as leukocytes or stem cells undergo extensive reshaping during endothelial transmigration and migration through tissues[51,52]. Uncompromised elasticity is also prerequisite for a cell to repeatedly squeeze through the dense network of fenestrated sinusoidal vessels in the spleen, which efficiently filters damaged or aged cells from the blood circulation[53]. Once biomaterial-modified cells come into contact with human blood, they are also exposed to soluble and cellular blood components of the immune system and the blood coagulation cascade, which may adversely affect in vivo cell persistence and therapeutic function[54,55]. For instance, cell surface-conjugated particulate biomaterials become prone to phagocytosis by monocytes and macrophages[56]. Immunogenic xenoproteins, such as streptavidin, a common linker for bioconjugation, elicit neutralizing antibodies, which can opsonize and clear the engineered therapeutic cell product. Aggregation of surface-engineered cells with blood platelets or the activation of blood clotting factors could lead to thrombus formation and serious complications[57,58]. In particular negatively charged materials, which to some extent mimic extracellular matrix and bacterial lipopolysaccharides, could render intrinsically athrombogenic cell surfaces thrombogenic and trigger vessel blockade[59].
Efficient “masking” of synthetic modifications on the cell surface from soluble and cellular blood components can be achieved by covalent attachment of a reactive derivative of polyethylene glycol (PEG)[60,61]. The concept of linking one or more highly flexible PEG chains to small molecules, proteins, peptides or whole cells to prolong their body-residence has reached widespread applications in modern pharmaceutical technology, which have been reviewed extensively[62–64]. As a tool to safeguard modified cell surfaces from adsorption of opsonic proteins or unspecific phagocytosis, PEGylation has been implemented in a variety of protocols, detailed further below.
Cell surface engineering with exogenous materials
As described above, materials of interest for modifying the surface of cells range from recombinant proteins to imaging agents to drug-loaded nanoparticles. The starting point for any cell engineering endeavor is to determine how to interface the cell with these exogenous materials, given the constraints to clinical cell engineering just discussed. Reflecting the complex structure of the plasma membrane and its various constituents, a diverse toolbox of bioengineering methodologies has been developed over recent years to rationally modify the surface of therapeutic cells (Fig. 1). The strategy perhaps most straightforward, but which provides the least control over the resulting cell surface remodeling, is chemical conjugation of molecules or nanomaterial cargos to the cell via pre-existing functional groups on cell surface proteins, polysaccharides, or lipids (Fig. 1a, b). Alternatively, cargo materials conjugated with lipophilic molecules can spontaneously associate with the plasma membrane via spontaneous insertion of these hydrophobic anchors into the bilayer, allowing the membrane to be decorated with novel structures (Fig. 1c, d). Finally, cell surface receptors can themselves act as binding sites to interface materials with the cell surface, by building ligands for target receptors into the nanomaterials of interest for cell surface engineering[65] (Fig. 1e). Utilization of antibodies, aptamers, or other engineered binding molecules to attach nanomaterials to the cell surface is a variation on this approach, which in theory could allow any desired molecule on the cell surface to serve as an anchor for attachment of materials to the cell. Each of these approaches finds utility in different situations.
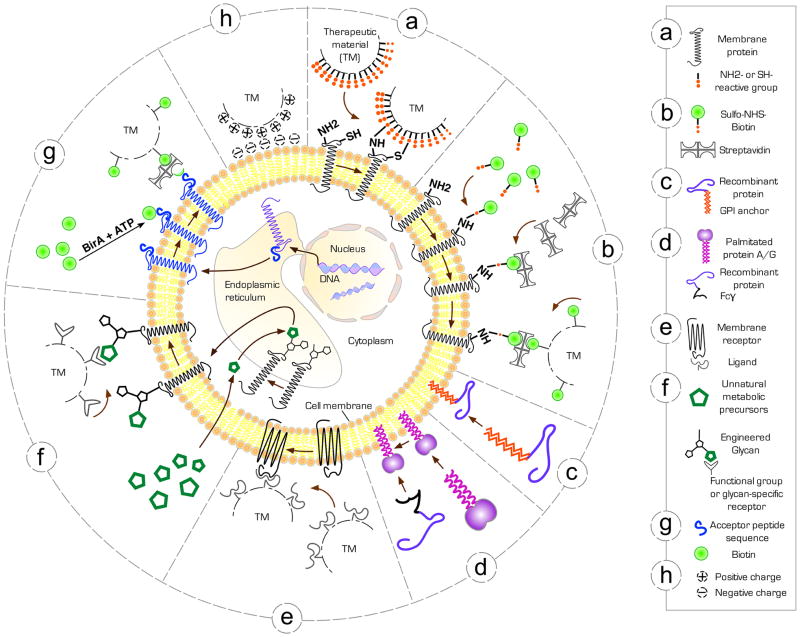
Figure 1. The cell surface bioengineering tool box.
Schematic of key bioengineering methodologies to therapeutically modify the surface of live mammalian cells. (a) Direct conjugation of therapeutic materials (TM) functionalized with reactive groups which covalently bond to amine (−NH2) or thiol (−SH) groups, intrinsic to cell membrane proteins. (b) Covalent attachment of biotin anchors to membrane proteins by reacting N-Hydroxysuccinimide (NHS)-activated biotin with primary amine groups in membrane proteins, followed by streptavidin-biotin linkage of TM. (c) Exogenous insertion of recombinant GPI-anchored proteins into the outer membrane leaflet (d) Exogenous insertion of palmitate-conjugated protein A or G into the cell membrane to subsequently immobilize antibodies or recombinant Fcγ-fusion proteins. (e) Anchoring ligand-functionalized TM to membrane receptors naturally present on the cell surface. (f) Metabolic labeling of surface glycans by the biosynthetic introduction of unnatural sugars containing unique functional groups, which serve as reactive sites for the attachment of TM. (g) Targeting TM site-specifically to surface proteins genetically fused to the BirA biotinylation enzyme acceptor peptide (AP) sequence. (h) Nonspecific electrostatic adsorption of cationic TM to the negatively charged cell membrane.
Chemical/enzymatic cell surface modification exploiting functional groups or molecules naturally present on the cell surface
Functional groups naturally present on the cell surface as part of proteins or carbohydrates are appealing docking sites for the covalent conjugation of therapeutic materials to cells since they do not require any chemical preconditioning of the cell. The most straightforward approach involves direct chemical reaction of an amino (lysine – NH2) or thiol (cysteine – SH) group presented on a cell membrane protein with a compatible reactive group on exogenous small molecules, proteins, polymers, or nanoparticles (Fig. 1a, b). This strategy is exemplified by the direct coupling of succinmidyl ester-functionalized polyethylene glycol (PEG) to cell surface amines of pancreatic islets to camouflage potentially immunogenic surface antigens[66]. Using similar chemistry, Hsiao et al. attached amine-reactive ssDNA conjugates to cells through reaction with cell-surface lysines as a method to array cells on DNA-functionalized surfaces in defined patterns[67]. Biotinylation of cell surfaces is also most commonly achieved through reaction of primary amine groups present on the cell membrane with amine-reactive biotin, such as N-hydroxy-succinimide biotin derivatives; once a cell is labeled with biotin, it can be readily functionalized with a wide range of biotinylated molecules or particles through a streptavidin bridge[31,68,69] (Fig. 1b). To reduce the number of incubation/washing steps protocols have been reported that either genetically fuse streptavidin to recombinant proteins[70,71] or chemically pre-conjugate streptavidin to nanoparticles before incubation with biotinylated cell surfaces[72]. A second approach is to make use of free thiols present in cysteine residues of proteins on the surface of mammalian cells. Reduced thiols at the cell surface play a role in protecting cell surfaces against oxygen radicals[73–75] and fine tune cell signaling and differentiation[76,77]. Thiol-reactive functional groups such as maleimide can thus be employed to link molecules or nanoparticles to the surface of cells (Fig. 1a). Notably, it has been shown that lipid or polymer particles in the 100–300 nm size range can be linked to cells via maleimide-thiol conjugation to live cells without compromising cell function[36].
In addition to amino or thiol groups, aldehydes and ketones have been used successfully to covalently attach surface-modifying molecules. The main drawback of using this approach, however, is that these reactive groups must be generated through chemical or enzymatic treatment of existing cell surface carbohydrates. For example, Yang and coworkers treated macrophages with sodium periodate to generate aldehyde groups on sialic acid residues[78]. These aldehydes were in turn reacted with amino-derivatized PEG-quantum dots to form transient Schiff base linkages. Finally, reduction with NaCNBH3 created a stable amine bond. For a narrow range of applications, enzymatic transformation of existing cell surface molecules to directly produce the desired modification is an efficient option. To this end, McEver pioneered a simple method to improve HSC homing and engraftment by ex vivo fucosylation of cord blood with guanosine diphosphate fucose and exogenous α1–3 fucosyltransferase to increase HSC tethering to P-selectin and E-selectin on activated endothelium[79]. Building on this approach, Sackstein and co-workers fucosylated surface CD44 on human MSCs to promote their adhesion with bone marrow vasculature following intravenous administration[35,80]. This innovative tool to custom engineer therapeutically desirable cell surface glycoforms will be discussed in more detail below.
Metabolic or genetic introduction of reactive functional groups on the cell surface
The concept of metabolically introducing chemical functional groups that are absent from the native plasma membrane as molecular handles for remodeling the cell surface was pioneered by Reutter and colleagues in 1992[81]. Their studies first demonstrated that nonphysiological amino sugar analogues could be metabolically incorporated into membrane glycoconjugates through natural carbohydrate biosynthetic pathways. In particular, sialic acids are an appealing target of metabolic oligosaccharide engineering. As the most abundant terminal components of membrane glycolipids, they are positioned on the outer periphery of the cell and, therefore, situated in ideal proximity for subsequent attachment of externally delivered reagents (Fig. 1f). In a series of studies, Bertozzi and coworkers metabolically introduced ketone- or azide-groups into sialic acid[82–84], thereby enabling surface conjugation of cells with various compounds[85,86] or in tissue engineering applications to covalently attach cells to synthetic scaffolds decorated with complementary functional groups[87]. The same concept can be applied to metabolically install reactive thiol groups into the outer periphery of the glycocalyx where they are most accessible to maleimide-derivatized materials[88].
Genetic engineering can also be used to introduce “bio-orthogonal” reactive groups into cell surface proteins, creating sites for selective modification of cells via a combination of traditional molecular biology and exogenous materials science/chemical methods. For example, Ting and colleagues developed a robust methodology to incorporate biotin groups site-specifically into desired cell surface proteins, exploiting the Escherichia coli enzyme biotin ligase (BirA)[89]: This enzyme biotinylates a lysine side chain within a 15-amino acid acceptor peptide (AP) sequence. As an initial step of this strategy, the AP tag is genetically fused to the N terminus or C terminus of a protein of interest. Following cell transfection, BirA enzyme, which is added to the culture medium with biotin and ATP, biotinylates AP-tags of surface-expressed proteins. These biotin groups can then be targeted with streptavidin conjugates (Fig. 1g). BirA-catalyzed ligations also permit the derivatization of membrane proteins with ketone groups, if ketone analogues of biotin are exogenously supplied, which further extends the spectrum of possible conjugates[90]. In a variation of the BirA ligation approach, a 22 amino acid peptide tag can be genetically engineered into proteins, to serve as a substrate for enzymatic ligation of an unnatural alkyl azide substrate by E. Coli lipoic acid ligase (LplA)[91]. Although these approaches require genetic manipulation of the substrate cell, they allow the location and identity of the reactive site on the cell surface (which protein, in what location) to be specified, providing greater control over the nature of subsequent cell surface remodeling than linkages introduced through naturally-occurring reactive groups described above.
Hydrophobic insertion into the cell membrane
Integral membrane proteins are anchored into the cell membrane through hydrophobic residues in their transmembrane helices, which make complementary interactions with the hydrophobic lipid bilayer[92]. This hydrophobic effect that governs the incorporation and orientation transmembrane proteins can also be put to use for cell surface engineering. When molecules or nanoparticles conjugated with an appropriate hydrophobic anchor are admixed with cultured cells, the hydrophobic moiety can spontaneously insert into the lipid bilayer, anchoring the conjugated cargo on the surface (Fig. 1c, d). One example of this general approach, termed “protein painting” uses hydrophobic glycoinositol phospholipids (GPIs) to anchor proteins to the outer cell membrane[93]. During physiological protein synthesis, GPIs are attached post-translationally to the C-terminus of select proteins, which direct them into lipid rafts of the outer cell surface membrane[94]. Recombinant GPI-anchored proteins form micelles in solution and subsequently can be exogenously re-incorporated into the plasma membrane of any target cell, where they retain their natural function[93,95]. Furthermore, recombinant DNA technologies make it possible to introduce new properties into GPI-linked proteins. For example, Nelson et al. genetically rendered the potent T cell activator RANTES immune-inhibitory and introduced a GPI-anchored version of this construct into the external membrane of endothelial cells to protect vasculature from acute immune rejection in xenotransplanted organs[96]. To immobilize a wide range of therapeutically relevant antibodies on cell surfaces, without the need to individually express them as GPI-tagged fusion proteins, target cells can first be coated with chemically-palmitated protein A or protein G (Fig. 1d)[97–99]. These proteins bind immunoglobulins through their Fc region and anchor antibodies to cell membranes without compromising their affinity or functionality. In theory, this method can be applied to any given protein by genetic fusion with an Fc domain, as demonstrated by Tykocinski and coworkers[100]. Using a two-step procedure, this group first precoated antigen presenting cells with palmitated protein A and subsequently “painted” cells with costimulatory B7.1-Fcγ1 fusion protein.
As a new group of therapeutics, “prosthetic” surface receptors that mimic the architecture or function of physiological receptor proteins can be chemically synthesized[101]. These compounds seamlessly incorporate into the surface membrane of cells and, for instance, enable the delivery of cell-impermeable molecules. The Peterson group has synthesized a range of cell surface receptor mimics composed of a ligand-binding small molecule or peptide linked to an N-alkyl derivative of 3β-cholesterylamine, which acts as membrane anchor[102,103]. In a related study, Bertozzi and coworkers functionalized synthetic glycopolymers, designed to mimic cell surface mucins with a hydrophobic anchor. Following incubation with live cells, these mucin-mimic polymers spontaneously incorporated into the cell surface membrane and retained the ability to recognize glycan-binding proteins[34].
An interesting alternative to unimolecular insertion of therapeutic proteins or molecules into cell membranes was recently reported by Sarkar et al[104]. To surface-functionalize MSCs with the targeting ligand sialyl Lewis X (SLeX), unilamellar lipid vesicles composed of biotinylated lipid were fused with MSCs. Subsequently, biotinylated SLeX could be immobilized on the cell surface through a biotin-streptavidin bridge.
Adsorption
The surface of mammalian cells carries a net negative charge, as a result of phosphate groups of phospholipids, carboxylate groups on proteins, and sialic acids terminating glycoproteins sugar chains[105]. The high ionic strength of physiological solutions makes monovalent electrostatic interactions with cells very weak, but polymers or nanoparticles with many cationic sites can bind stably to cells via multivalent electrostatic interactions (Fig. 1h). To this end, Wilson et al. functionalized PEG polymer with cationic poly-L-lysine for electrostatic adsorption onto the surface of pancreatic islets[38]. Beyond their applications in organ transplantation as a physical barrier between allograft tissue and the host immune system, functionalized multilayer films coated on cells have also shown great promise for protecting damaged blood vessels from platelet adhesion and vessel stenosis[106]. Furthermore, by alternate layering of cells and polyelectrolyte films, multilayer cellular constructs can be engineered which mimic the 3-D structure and cellular composition of functional organs[107]. Presumably due to a combination of van der Waals, electrostatic, hydrogen bonding, and hydrophobic interactions, certain nanoparticle formulations can also nonspecifically adsorb to cell membranes[108]. As further discussed below, such attachment to a cellular chaperone can drastically reduce systemic clearance of nanocarriers[108,109].
Interaction of a ligand with a receptor naturally present on the cell surface
Physiologically-expressed transmembrane receptors are tempting targets to conjugate biomaterials functionalized with their respective ligands to cells. However, receptor-ligand interactions are transient in nature, determined by intrinsic binding and dissociation kinetics[110], which restricts their utility for stable coupling of material to cell surfaces[111]. However, multivalent binding of ligand-decorated materials with cell surface receptors can lead to stable cell surface binding. For example, work by Swiston et al. demonstrated that the multivalent interaction between cell surface CD44 receptors and the “flat” face of polyelectrolyte multilayer disks (with diameters of 5–10 μm but thicknesses of a few hundred nm) displaying hyaluronic acid (the natural ligand of CD44) is strong enough to anchor HA-coated multilayer thin films to T cells[65] (Fig. 2a). To tether single-walled carbon nanotubes (CNTs) to cell surface glycoprotein ligands, Bertozzi and coworkers first coated CNTs with a biopolymer designed to mimic cell surface glycoproteins. Using a hexavalent lectin (polysaccharide-binding protein) as a crosslinker, surface-modified CNTs were efficiently attached to live cells[112]. The unique hollow monolithic structure comprising an outer and inner core, which can be independently functionalized or loaded with therapeutics[113] are especially valuable features of CNTs as cell surface engineering tools. More recently, this approach was extended to surface-functionalize cells with less cytotoxic boron nitride nanotubes[114]. In principle, any ligand-, aptamer-, or antibody-targeted compound or nanoparticle can site-specifically attach to cell surfaces. However cell surface engineering is predicated on retaining the material at the cell surface, and many targeting agents trigger internalization of their receptor on binding, which will preclude stable surface modification (discussed further below).
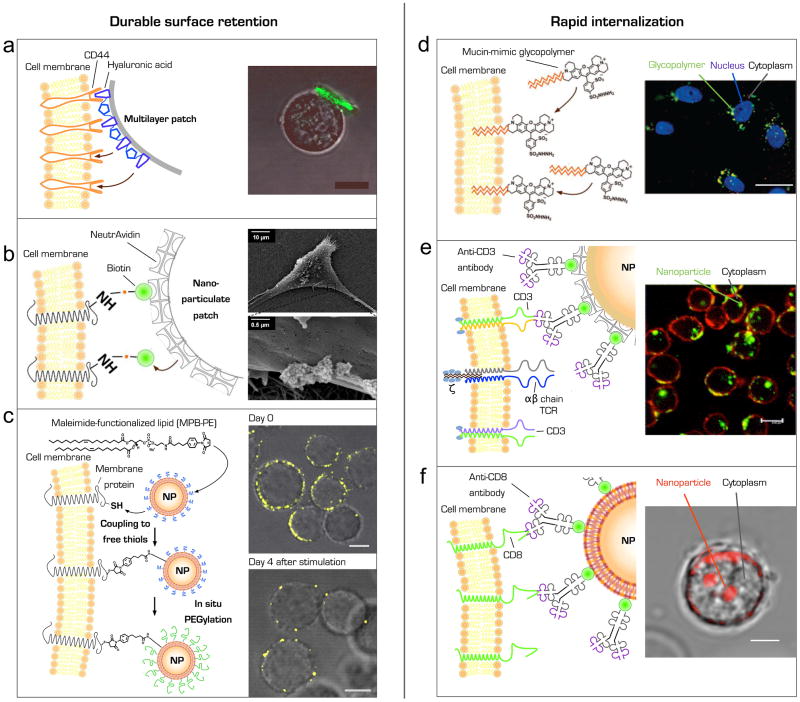
Figure 2. Durable surface coupling (a–c) versus rapid internalization (d–f) of therapeutic material following different cell surface engineering strategies. (a) Attachment of micrometer scale polymer layers to cell membranes.
Schematic of a multilayer hyaluronic acid-functionalized polymer patch attached to the surface of a T lymphocyte through intrinsic CD44 membrane receptors (left panel). Confocal microscopy image of a patch- (green fluorescence) functionalized T-cell. Scale bar, 10 μm (right panel). Adapted with permission from Swiston et al. (2008). (b) Tethering nanostructures to biotinylated cell membrane proteins. Schematic illustration of NeutrAvidin-coated nanoparticulate patches anchored onto a biotinylated plasma membrane (left panel). Scanning electron microscopy images of nanoparticle cluster on a human mesenchymal stem cell membrane (right panel). Adapted with permission from Cheng et al. (2010). (c) Covalent coupling maleimide-functionlized nanocarriers to free thiol groups on membrane proteins. Schematic of maleimide-based conjugation of synthetic lipid-coated nanoparticles (NP) to cell surface thiols and subsequent quenching of residual maleimide headgroups on nanoparticles by in situ conjugation to thiol-terminated polyethylene glycol (PEGylation) (left panel). Confocal microscopy images of CD8+ effector T cells immediately after conjugation with fluorescent multilamellar lipid nanoparticles and after 4-d in vitro T cell expansion (right panel). Scale bar, 2 μm. Adapted with permission from Stephan et al. (2010). (d) Incorporation of bioactive synthetic glycopolymers into cellular membranes. Schematic of synthetic mucin-mimic glycopolymers exogenously inserted into the cellular membrane through hydrophobic anchors (left panel). Fluorescent microscopy image of ldlD CHO cells incubated for 1 h with synthetic glycopolymer (green). Nuclei are stained with Hoechst 33342 in blue (right panel). Scale bar, 10 μm. Colocalization of glycopolymers and early endosomes was confirmed. Adapted with permission from Rabuka et al. (2008). (e) Selective targeting of antibody-conjugated nanoparticles to T lymphocyte T cell receptors. Schematic illustration of an anti-CD3 antibody-functionalized nanoparticle (NP) coupled to the T cell receptor CD3 complex. To immobilize biotinylated anti-CD3 antibodies nanoparticles were modified with NeutrAvidin (left panel). Confocal microscopy image of CD3+ Jurkat cells cultured in the presence of FITC-conjugated anti-CD3-NP for 4 h. To visualize cellular membranes cells were counterstained with Alexa fluor 594-conjugated concanavalin A (right panel). Scale bar, 8 μm. Adapted with permission from Balthasar et al. (2005). (f) Tethering particulate carriers to CD8 antigen on human effector T lymphocytes. Illustration of an antibody-nanoparticle (NP) conjugate targeted to the CD8 antigen on the surface of a human CD8+ T cell (left panel). Confocal microscopy of a human CD8+ effector T lymphocyte incubated with anti-CD8-decorated fluorescent nanoparticles for 45 minutes (right panel). Scale bar, 3 μm (M.T. Stephan, unpublished data).
The role of nanomaterials in achieving stable cell surface modification
The approaches reviewed above for linking exogenous molecules, particles, and polymer films to cells do not necessarily address the larger issue of stable cell surface modification. In general, exogenous molecules linked monovalently to cell surface proteins/lipids/polysaccharides will be retained on the cell surface only as long as the membrane molecule to which they are bound, which may range from minutes to hours. In fact, the rapid and efficient internalization of ligand-, antibody-, or aptamer-conjugated drugs or nanoparticles following binding to target proteins on the cell surface is a critical determinant of efficacy for many targeted drug delivery strategies[115,116]. Notably, multivalent crosslinking of receptors by ligand-functionalized nanoparticles binding to the cell surface does not prevent internalization in many diverse systems and cell types[117,118]. However, studies aiming to create long-lived cell surface modifications using micro- and nano-structured materials have begun to identify some strategies to physically or chemically induce long-lived cell surface association of exogenous materials.
Engineering stable cell surface association via engineered particle shape
One emerging strategy to regulate the internalization of synthetic materials following cell contact is via the physical shape of the exogenous material: In general, materials with large aspect ratios, such as disc- or worm-like shapes, are internalized at significantly decreased rates compared to spherical material[119,120]. In line with these observations, Swiston and co-workers reported that micrometer-diameter polymer patches with nanoscale thickness stably attached to and remain on the surface of T cells[65] and even phagocytic B lymphocytes[121] for several days in cell culture (Fig. 2a). In a related study, Cheng et al. demonstrated that mesenchymal stem cells, which typically internalize nanostructures within minutes or hours[122], retain linked nanoparticulate patches on the cell surface for up to 2 days[72] (Fig. 2b). Notably, to understand the fate of nanomaterials following association with the cell surface, physical geometry and the mechanisms of cell binding must be considered together to understand the fate of exogenous materials binding to the cell surface, as highlighted by Jiang and colleagues[123]: By synthesizing nanoparticles of various sizes coated with high densities of growth factor receptor-binding antibodies, this study demonstrated that cells most efficiently internalized nanocarriers within the 25–50 nm size range. In contrast, larger 70 nm-diam. particles bound to cells but mostly remained localized on the cell surface over a period of hours. Given that the cellular uptake of nanocarriers may depend on receptor-mediated wrapping of the cell membrane around the particle[124], the authors reasoned that larger nanostructures with a high density of receptor binding sites occupy all the receptors available in the local vicinity of the membrane on a single face of the particle. This reduction of receptors in the immediate neighborhood of the adhesion region then limits additional ligand-receptor interactions, which would be required to completely coat nanoparticles and trigger endocytic uptake.
Mechanisms of cell association control cell surface stability of exogenous materials
In addition to size and geometry aspects, the mechanism of cell attachment plays a key role in the stability of cell surface modifications; certain strategies for the attachment of nanomaterials or molecules to cell surfaces lead to rapid internalization, while others appear capable of stably modifying the cell surface (Fig. 2a–c). As stated above, turnover rates of macromolecules which are exogenously inserted into the membrane of live cells, such as biomimetic receptors, glycopolymers, surface glycans or GPI-anchored proteins, tend to mirror those of natural membrane-associated biomolecules[34,35,101]. Rapid membrane dynamics, however, bear the risk of premature loss of therapeutic membrane-embedded macromolecules owing to endosomal internalization (Fig. 2d). A positive surface charge of biomaterials in contact with inherently negatively charged cell membranes can cause local membrane depolarization and also trigger subsequent intracellular uptake[125,126]. To immobilize and maintain nanostructures on the outer membrane of cells for days, even following cell proliferation, covalent linkage strategies to reactive groups inherently present on their exterior cell surface have proven most promising. For example, conjugation of lipid or polymer nanoparticles in the 100–300 nm diam. size range to free thiols on the surface of primary T cells or hematopoietic stem cells led to stable cell surface localization of these nanomaterials, even during/after cell division (Fig. 2c)[36]. Importantly, coupling of up to ~100 particles/cell was nontoxic and was not found to affect key cellular functions, such as in vivo migration, proliferation or cell fate. The mechanistic underpinnings of this durable surface-coupling of nanocarriers to cells remains to be determined. In agreement with previous work (Fig. 2e)[118], our group has observed some degree of internalization of nanoparticles coated with antibodies to CD3 or CD8 by primary T lymphocytes as a result of receptor-induced endocytosis (Fig. 2f and unpublished results). These results suggest that it is not size, shape or surface charge of the nanoparticles that explains the prolonged surface retention seen in our adoptive T-cell therapy studies, but rather the maleimide-thiol coupling strategy itself. Therefore, a key to understanding and further optimizing this promising surface modification approach will be identification of the surface proteins maleimide-functionalized nanocarriers commonly bind, and, ultimately, to determine whether the covalent coupling of nanomaterial to surface proteins delays or even prevents inherent protein turnover and membrane recycling.
Key therapeutic gains achieved by cell surface engineering
Retargeting systemic cell homing
Direct injection of therapeutic cell products into target tissue sites is practicable only for a limited number of applications, such as intracoronary infusions of myoblasts into the damaged heart muscle, pancreatic beta islets transplants through the portal vein into the liver or subcutaneous injections of cellular vaccines[9,10,13]. For most cellular therapies, which target systemic multifocal sites of injured, inflamed or cancerous tissue, intravenous delivery is the preferred mode of cell administration. Hence, efficient homing of donor cells to the tissue or organ of intended action is crucial to the therapeutic success.
Given that only a subset of intravenously injected cells may engraft in the tissue of interest due to the absence of the key homing receptors on infused cells[127], the lack of sufficient chemoattractants, or the presence of suppressive stromal barriers at target locations[128], methods of improving therapeutic cell trafficking are a high priority. A wide variety of simple yet versatile cell engineering methodologies have been reported to program the in vivo trafficking of systemically delivered cells. Sackstein et al. demonstrated that mesenchymal stem cells, which inherently display limited bone marrow tropism, more efficiently engraft to this site when enzymatically surface-engineered ex-vivo with an E-selectin binding motif that is responsible for bone marrow-homing of hematopoietic stem cells[35,80]. This ex vivo glycan engineering strategy was also applied to enhance the engraftment of human umbilical cord blood cells[129]. Traditionally, cord blood has been the standard source of cells for hematopoietic cell transplantation in pediatric patients only, owing to the limited number of stem cells available in a typical cord blood unit[130]. By enzymatically optimizing the bone marrow tropism of infused cord blood cells and thereby significantly reducing the required number of stem cells, adult recipients could potentially be transplanted using cord blood.
Beyond rationally designing cell surface glycoforms, cell homing responses can be induced by functionalizing cell membranes with recombinant targeting ligands. As an alternative to directing regenerative cells to tissue antigens exposed within injury sites, vascular addressins, which are ubiquitously displayed on inflamed endothelial surfaces to mediate rapid deceleration of circulating immune cells, can be directly targeted. To promote attachment to activated endothelium, Dennis and coworkers coated mesenchymal stem cells with antibodies to ICAM-1 through membrane-inserted palmitated protein G as docking points[97] (e.g., method of Fig. 1d). In a related study, the Karp group immobilized the carbohydrate molecule Sialyl Lewis X (SLeX) as high-affinity P-selectin ligand on the surface of mesenchymal stem cells using biotin-streptavidin bridges[31]. Like leukocytes adhering to the luminal surface of inflamed vasculature to subsequently migrate into underlying damaged tissue, SLeX surface-engineered mesenchymal stem cells decelerate and roll on P-selectin-coated substrates.
Providing transplanted cells with autocrine sources of growth factors
Once therapeutic cytoreagents reach their desired location, cell viability, function and expansion critically rely on a sustained supply of oxygen, nutrients and growth factors. Access to these factors for transplanted cells can be limited as a result of competition with endogenous host cells[131] or due to a hostile microenvironment characterized by tissue necrosis, hypoxia or acidosis[132]. In addition, adoptively transferred tumor-targeted T lymphocytes need to overcome an immune-evasive tumor microenvironment with suppressive molecules and inhibitory cells[133]. To promote in vivo longevity and function of cell transplants, adjuvant growth factors, such as cytokines[24] antibodies[134,135] or small molecule drugs[136,137] are administered intravenously. However, systemic adjuvant injections generally do not selectively target the transplanted cell population, and high systemic drug levels need to be maintained through repeated bolus injections. This often results in dose-limiting toxicities, and ultimately precludes the clinical use of many potentially potent adjuvants drugs[23,138]. To focus adjuvant drug action on the transferred cell, thereby minimizing systemic side effects, nanoparticles loaded with growth factors can be directly conjugated onto the surface of donor cells (Fig. 3)[36]. In this strategy, supporting drug molecules are slowly released from cell-bound nanoparticles and primarily recaptured by particle-carrying cells in autocrine signaling loops. Using this approach to enhance the survival and function of anti-tumor T-cells, the cytokines interleukin (IL)-15 and IL-21 were attached to T-cells at minimal doses that had no therapeutic effect when given systemically. When these same cytokine doses were encapsulated in lipid nanoparticles (which released the drugs over the course of ~1 week) and directly attached to the donor T-cells, massive in vivo T cell expansion was induced, leading to complete clearance of systemic melanoma tumor burdens. This simple approach for donor cell modification also enhanced the engraftment and repopulation kinetics of hematopoietic stem cells, by locally releasing a glycogen synthase kinase-3β inhibitor (known to enhance HSC expansion) from cell surface-linked nanoparticles carrying minute quantities of drug[36]. While the clinical introduction of this HSC surface engineering strategy is still distant, the key benefits of this technology compared to conventional HSC transplant regimens are highly appealing. As part of the ongoing clinical effort to enhance HSC proliferation and in vivo homing, a growing number of small molecule drugs, such as 16,16-dimethyl Prostaglandin E2 (FT1050), have entered phase 1 testing in human subjects[20,139]. However, to avert possible systemic toxicities in response to intravenous bolus injections, stem cell modulators need to be administered ex vivo prior to infusion. By slowly releasing the HSC-stimulating agent from cell surface-coupled nanoparticles instead, cells could be infused directly into the recipient without overnight in vitro culture. Furthermore, adjuvant nanoparticles stably conjugated to HSCs follow the characteristic in vivo migration patterns of their cellular vehicles and can promote early stages of engraftment through sustained in vivo drug release.
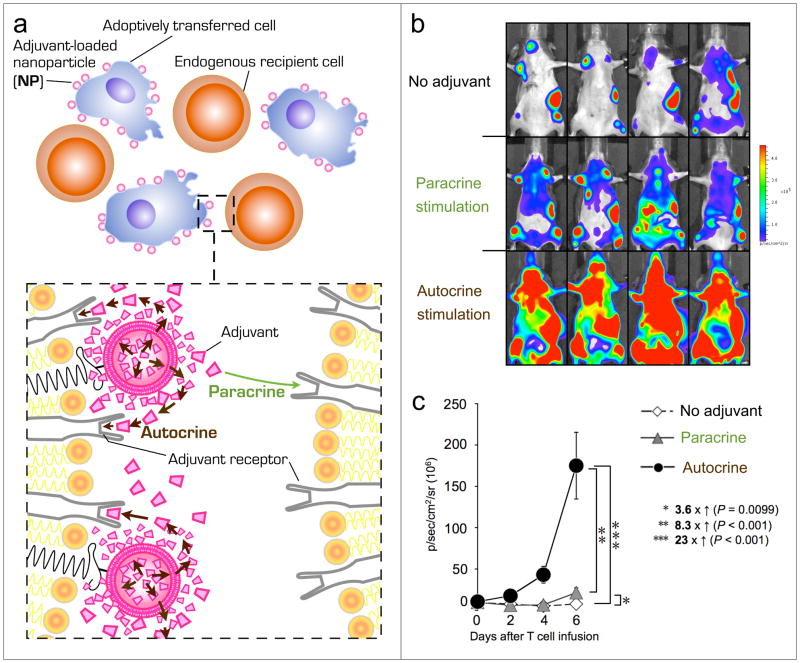
Figure 3. Stable chemical conjugation of adjuvant-loaded nanoparticles (NP) to the surfaces of transplanted cells permits pseudo-autocrine self-stimulation while limiting potentially harmful paracrine stimulation of bystander cells.
(a) Schematic of adoptively transferred therapeutic cells in the presence of recipient bystander cells. To provide sustained autocrine stimulation, adjuvant growth factor-loaded nanocarriers are conjugated to the surfaces of donor cells just prior to cell infusion. A magnified view of nanoparticle-modified versus unmodified surface membranes, which further illustrates autocrine versus paracrine cell stimulation, is shown in the lower inset. (b) T cell-linked adjuvant NPs activate primarily their own cellular carrier with minimal stimulation of bystander cells. B16F10-OVA lung tumor-bearing mice were treated by i.v. co-transfer of luciferase-expressing pmel-1 reporter T-cells together with luciferase-negative OT-1 T cells. In the “no adjuvant” group mice received unmodified T lymphocytes. To quantify “paracrine stimulation” of cell-conjugated adjuvant nanoparticles, luciferase-expressing pmel-1 T cells were co-transferred with luciferase-negative OT-1 T cells decorated with interleukin 15/21-loaded nanoparticles prior to adoptive transfer. “Autocrine stimulation” of surface-tethered adjuvant nanoparticles was determined by adoptive transfer of luciferase-transgenic pmel-1 T cells surface-modified with adjuvant releasing nanoparticles in the presence of unmodified OT-1 T cells. Shown are representative in vivo bioluminescent images. Respective whole body bioluminescent pmel-1 T cell photon counts, quantified every 2 d during initial T cell expansion, are shown in (c). The much greater pmel-1 T-cell expansion seen in the “autocrine stimulation” case shows that most of the drug released from cell-bound particles acts on the carrier cell itself, rather than bystander cells in the local microenvironment. Adapted with permission from Stephan et al. (2010).
Whether nanoparticles loaded with small molecule drugs that freely cross cell membranes or that target cytoplasmic receptors need to be surface-displayed to exhibit their optimal adjuvant effects in vivo is subject to ongoing experiments. Recent in vitro studies by Riley and colleagues demonstrated that microparticles loaded with the differentiation factor Dexamethasone still efficiently induced osteoblastic differentiation following their uptake by human mesenchymal stem cells[140]. Interestingly, in addition to triggering intracrine differentiation pathways, internalized drug particles also controlled the cell fate of unmodified bystander cells through paracrine-like signaling.
Targeting drugs to therapeutically relevant tissue sites using cellular carriers
Following conventional drug administration via oral ingestion or intravenous injection, therapeutic agents are distributed throughout the body through the systemic circulation, with only a small fraction of the injected dose actually reaching the intended target cells[141]. In addition, the temporal window over which drugs are present in the tissues at therapeutically meaningful doses can be extremely short, causing a need for frequent repeat dosing. To overcome these limitations, drug-loaded synthetic nanocarriers are being implemented as a means to deliver therapeutic agents over sustained periods in the bloodstream or to target high concentrations of drug cargos to relevant tissue sites, thus reducing drug toxicity while increasing treatment efficacy[142]. These approaches are rapidly progressing in the field of cancer therapy, where passive accumulation of nanocarriers at tumor sites via the enhanced permeation and retention effect[143] (highly disorganized, leaky tumor vasculature in combination with compromised lymphatic drainage) is often observed. Nonetheless, only a small percentage (typically, a few percent) of the total injected dose is delivered into the tumor microenvironment due to rapid particle clearance by immune cells in the liver and the spleen (collectively referred to as the reticuloendothelial system, or RES)[144]. Particle clearance from the blood also limits the time window over which nanoparticle carriers can provide systemically-available drug. To further improve on these problems, cell surface engineering has been applied to therapeutic drug delivery.
Inspired by the observation that some pathogens efficiently evade immune clearance by attaching themselves to the surface of red blood cells (RBCs)[145,146], protocols were developed to test the hypothesis that RBC-bound drugs or drug carrier nanoparticles would be shielded from rapid scavenging by the RES following systemic administration. Initially, autologous erythrocytes were the prime candidates for use as drug delivery vehicles because of their abundance (~5.4 million cells/mm3 blood), their considerably uniform size and shape, and their long life span in circulation (110–120 days)[147]. Murciano et al. successfully coupled tissue type-plasminogen activator (tPA), which is used clinically to dissolve arterial thrombi, to the surface of isolated erythrocytes, to extend the inherently short circulation half-life of this fibrinolytic drug[69]. Following intravenous injection, tPA-decorated RBCs could durably protect animals from induced thrombi, whereas an equal dose of soluble tPA had no prophylactic effect. Despite significantly increasing the circulation lifetime of a therapeutic agent, the methodology of anchoring drugs to the surface of cellular carriers is inherently limited to agents that are effective while still attached to the cell membrane. To combine the benefits of synthetic nanoparticles as slow-release depots of a wide range of therapeutic cargos with the use of cellular carriers, which might prevent premature clearance of the particles from the blood, cell-nanoparticle hybrid vectors have been developed. In a proof of concept study, Chambers and colleagues reported that the non-covalent attachment of polystyrene beads as large as 450 nm to the surface of RBCs dramatically increased their circulation time compared to particles injected freely into the blood[109]. These highlighted studies only exemplify a multitude of therapeutic applications of erythrocytes as cellular carriers of biopharmaceuticals explored through more than five-decades of intensive development[148,149]. However, the implementation of erythrocytes as widespread drug delivery systems in clinical therapies is hindered by major drawbacks. Most lipophilic drugs with limited water solubility can only be entrapped into red blood cells at concentrations below therapeutic levels, and exhibit a burst release of the loaded drug during early cell preparation stages or in vitro cell storage[150,151]. Moreover, most methods developed to chemically modify red blood cells induce irreversible destructive changes in the cell membrane. This predisposes carrier erythrocytes to premature clearance from the circulation by the reticulo-endothelial system in the liver or spleen[152]. Finally, compared to other cellular blood components, such as monocytes or lymphocytes, erythrocytes lack inherent migratory properties, which preclude their use for active drug targeting to defined tissue sites. Thus, despite much effort, clinical products based on engineered red blood cells have not yet translated to the clinic.
In parallel to the pursuit of red blood cell engineering, recent advances in our understanding of in vivo migration patterns of immune or stem cells to inflamed, hypoxic or cancerous tissue combined with technical advances of cell isolation and in vitro expansion have expanded the field of cellular drug carriers to cell types beyond erythrocytes. For instance, the inherent ability of monocytes/macrophages and mesenchymal stem cells to migrate to hypoxic, necrotic, and inflamed tissue[127], makes them ideal candidates as cellular carriers of cancer therapeutics or tumor imaging agents[153]. Most commonly, isolated monocytes or mesenchymal stem cells are “fed” and loaded in vitro with nanocarriers, which contain membrane-permeable therapeutics[40,154]. Following their systemic injection, tumors actively recruit these cells, which subsequently release their toxic payload in a highly controlled and directed manner. Although the intracellular delivery strategy of therapeutic nanoparticles may seem obvious for phagocytic carrier cells, coupling the therapeutic cargo to the cell surface could reduce the toxicity of the therapeutic agent on the carrier cells. To this end, Chen and colleagues chemically anchored nanoparticle “patches” to the membrane of mesenchymal stem cells without compromising key intrinsic cell functions[72] (Fig. 2b). Using Schiff base linkage chemistry, Holden et al. demonstrated durable coupling of PEGylated dendrimers to the surface of macrophages[78], which sets the stage for the future development of macrophage-nanoparticle hybrid vectors as tumoritropic drug delivery vehicles.
Effector T-cells have also been tested as chaperones of surface-attached therapeutic cargo, motivated by their the clinical track record of adoptive T-cell therapy in cancer and the ease in harvesting, genetically modifying and expanding these cells to clinical scale[155]. Further, T cells can efficiently infiltrate systemic tumor lesions across endothelial and stromal barriers[156]. Based on the unexpected finding that viral particles adsorbed to the membrane of T lymphocytes can be released and taken up by tumor cells in vivo at tumor site[157], several groups have successfully used adoptively-transferred tumor antigen-specific T cells to carry oncolytic viruses to tumor deposits[39,158,159]. By “hitchhiking” on tumoritropic cells, therapeutic viral particles aggregate in poorly accessible compartments and lyse tumor cells in synergy with their cytotoxic carrier T-cells. Extension of this approach to delivery of synthetic drug-loaded nanoparticles will be an attractive goal, since many relevant cancer diagnostics and therapeutics cannot be delivered by viruses, such as small molecule drugs, antibody-drug-conjugates, aptamers, or magnetic imaging agents[160–162]. In vivo biodistribution assays in tumor-bearing animals have confirmed that T-cell-coupled nanoparticles with sizes of 200–300 nm diam. follow the inherent migration pattern of their cellular vehicles and efficiently accumulate at tumor sites, while the same particles injected systemically were rapidly scavenged by the liver and the spleen and showed negligible tumor uptake[36]. This result highlights a key area for further research: Accumulation of freely injected nanoparticles in tumor sites is highly dependent on precise tuning of particle size, with an optimum near 100 nm diam. In contrast, the same size limitations do not apply to cell-carried nanomaterials, and defining the upper limit for particle sizes that are efficiently trafficked into tissues by chaperone cells will be an important parameter to define, since a modest change in particle size will have a tremendous impact on the payload of drug carried per particle. The profoundly altered biodistribution of cell-nanoparticles hybrid vectors and potential to utilize particles with sizes outside the narrow range optimal for “free” particle tumor accumulation strongly motivates the continued translational development of such “pharmacytes” as actively targeting cell products, which deliver therapeutic or diagnostic agents to desired anatomical compartments[163,164].
Tracking adoptively transferred cells in vivo
To optimally evaluate the efficacy of cell-based therapies in clinical studies it is fundamental to monitor cell homing to target tissue and in vivo persistence by noninvasive cell imaging. Currently, two distinct technologies are employed to track cell location in patients: (1) Loading cells ex vivo with probes, including radioisotope-labeled chemicals for Positron Emission Tomography (PET) imaging[165] or contrast agents for Magnetic Resonance Imaging (MRI)[166], and (2) genetically engineering cells ex vivo to constitutively express a reporter gene, such as the widely used PET reporter gene Herpes Simplex Virus 1 thymidine kinase (HSV-tk)[167].
The approach of ex vivo cell labeling is primarily designed to determine the short-term fate of transplanted cells, given that tagged cells dilute out the imaging probe with every cell division following their adoptive transfer. Unfortunately, most imaging agents developed to enhance the resolution in MRI, such as superparamagnetic iron oxide nanoparticles (SPION), are known to induce cellular stress and perturb cell functionalities and gene expression once inside the cytosol[168]. This raises many safety concerns, including the risk of aberrant in vivo cell responses, the risk of induced mutagenesis or the lack of therapeutic potency of a cell product due to premature apoptosis. On the other hand, genetically tagging therapeutic cells, while allowing for long-term longitudinal cell imaging, permanently alters their genetic composition and adds significant costs and regulatory roadblocks[169].
In the light of these shortcomings, the direct coupling of imaging agents or nanoparticulate carrier systems loaded with imaging reagents to the external membrane of cells prior to infusion could provide significant advantages for in vivo cell imaging. Based on the wealth of available nanoparticle formulations and protocols to chemically modify cell surfaces (Fig. 1) the range of imaging tracers that can be linked to therapeutic cells without compromising cell functions extends far beyond the abovementioned magnetic contrast agents. New cell surface engineering strategies could encompass (1) coupling high-affinity antibody fragments, which are widely used reagents to sequester radionucleotides in pretarget radioimmunotherapy[170], to the surface of cells (2) tethering synthetic nanoparticles, which are loaded with recombinant reporter enzymes, such as HSV-tk for PET imaging, or (3) surface-labeling cells with colloidal quantum dots[171] for near-infrared cell imaging. Since many cell types explored therapeutically in clinic, such as myoblasts, neuronal stem cells, or pancreatic islet beta-cells exhibit low in vivo proliferative rates[172], cell surface coupling strategies are not restricted to monitoring the short-term fate of transplanted cells.
Future Directions
Currently, over 500 companies are involved in the development and commercialization of cell-based products[173] to treat a range of diseases, including tissue degeneration, chronic inflammation, autoimmunity, genetic disorders, cancer, and infections. In retrospect, cell therapy has undergone a tremendous metamorphosis from a discipline traditionally defined by blood transfusions and bone marrow transplantations into a nascent healthcare industry. The strong clinical presence of an expanding array of cell therapy products has also catalyzed the field of cell engineering with the goal of maximizing the therapeutic performance of cytoreagents in patients. In this review we have highlighted strategies developed to rationally design the microarchitecture of cell surfaces with synthetic nanomaterials. Cell products are not replenishable “off-the-shelf” reagents, but rather live therapeutics, that are costly to harvest, purify and expand. With this in mind, most cell bioengineering work discussed in this review has been geared towards minimizing cell damage or loss by avoiding non-physiological cell culture conditions or excessive cell handling. However, to successfully transition cell surface bioengineering technologies into clinically viable tools, future research efforts will be needed to optimize additional parameters.
New materials for cell surface engineering
The advent of advanced biomaterial discovery tools, such as combinatorial synthesis, high-throughput experimentation or computational modeling[174], has led to the development of a substantial number of next-generation biomaterials with superior biocompatibility and functionality. These compounds hold the promise to further diversify and accelerate the clinical implementation of cell surface bioengineering approaches.
One emerging class of materials are DNA/RNA molecules as building units of self-assembling secondary and tertiary structures[175,176]. Branched oligonucleotide sequences in combination with “sticky” ends create a powerful molecular assembly kit to rationally modify surfaces of therapeutic cells. Francis and co-workers have shown proof of concept that synthetic DNA strands can be hybridized to the surface plasma membrane of living cells[67]. Since aptamers, which are oligonucleotide molecules that bind to specific targets, have emerged as a class of molecules that rival antibodies in both therapeutic and diagnostic applications[161], their use as cell targeting ligands in the clinic may provide major advantages compared to conventional protein ligands. In contrast to recombinant proteins, aptamers are comparably compact, they can be rapidly developed in vitro against virtually any class of target molecules, including protein antigen, fatty acids, carbohydrates and even synthetic compounds. Moreover, aptamers can be engineered to avoid in vivo immunogenicity[177]. Another novel technology with tantalizing therapeutic potential for cell engineering is the field of bio-responsive or bio-interactive materials[178–181]. This methodology exploits biological events or stimuli, such as cell-secreted enzymes, pH, temperature, photon flux or ligand binding events, as triggers to induce macroscopic transitions in materials. As a result, “smart” or “sensing” compounds can be designed to perform sophisticated functions. In the context of cell surface engineering one could envisage cellular “Trojan horse vehicles” tuned to release their drug payload only once reaching tumor targets with acidic pH environments. Such on-demand drug release would further minimize premature metabolism and excessive background levels of drugs with high toxicity. Disease-specific enzymes could also trigger the display of cell membrane-inserted bioactive ligands to instruct cell behavior on-site only without off-target stimulation of irrelevant bystander cells while circulating in the blood stream.
Regulatory issues and cost-effective scale-up
Personalized cell therapies have high production costs (currently, ~$25,000 per treatment for adoptive T cell therapy[182]) based on labor-intensive cell harvesting, large-scale cell expansion and the requirement for extensively trained personnel and specialized manufacturing facilities. Due to high research and development costs, commercialized cytoreagents are even more costly, exemplified most recently by Dendreon’s $93,000/patient autologous dendritic cell cancer vaccine Provenge® [183]. The large-scale synthesis of therapeutic nanomaterials with GMP- (good manufacturing practices) compliant reagents, especially with recombinant proteins involved, will significantly add to the already high price tag of established cellular therapies.
From a regulatory perspective, the most likely scenario for initial commercialization of cell engineering would be the development of drug-loaded nanomaterials that are themselves off-the-shelf reagents that could be combined with cells just before transfer into patients. This approach would allow GMP-compliant processing and characterization of the materials (e.g., cell-binding nanoparticles) to be independently established and “plugged in” to existing clinical cell therapy protocols such as MSC, islet cell, or stem cell transplants. In this setting, many of the methodologies employed for establishing the safety, pharmacokinetics, and efficacy of existing clinical nanomaterials (e.g., chemotherapy-loaded liposomes) might be applied. Clearly, drug-carrying nanomaterials designed for cell conjugation would face a lower barrier to approval if initially based on existing, clinically-used drugs. Modification of commercial cell therapies a la Provenge® would pose additional hurdles for FDA approval.
Notwithstanding these technical and regulatory issues, cellular engineering tools become economically justified if highly effective. Given that a majority of cell therapies are currently non-curative and fall short of desired clinical efficacy[184], a one-time treatment with a cell product engineered with curative properties is most cost effective since it cuts down on costly palliative care to manage disease relapse. Furthermore, the adoptive transfer of a significantly reduced number of cells demonstrating superior clinical efficacy, compared to the infusion of large quantities of short-lived/low potency cells will reduce the cost and complexity of cell manufacturing. It is likely that the most attractive setting for these concepts in the clinic will be delivery of highly potent yet inexpensive small molecule compounds, which might replace expensive adjuvant drug treatments and produce dramatic changes in the efficacy of cell therapy while avoiding substantial increased costs to these therapies.
In vivo cell surface bioengineering?
In an ideal scenario one could envision directly engineering surface properties of defined therapeutic cell populations in their physiological environment within the patient without the need for ex vivo cell isolation and expansion. Besides being costly and laborious, large-scale cell expansion often renders cells functionally exhausted and significantly reduces their subsequent proliferative potential[185]. It would, therefore, be highly desirable to develop in vivo cell bioengineering methodologies that expand, re-target or functionally enhance endogenous pools of pathophysiologically relevant cell types, such as mesenchymal stem cells, hematopoietic stem cells or tumor antigen-specific T lymphocytes. As a step in this direction, the Fahmy group has recently developed tolerogenic nanoparticles targeted to specifically bind to CD4+ T lymphocyte populations via surface-attached anti-CD4 antibodies. A brief mixing of these targeted particles with unpurified splenocytes ex vivo led to CD4+ T-cell labeling with the drug-loaded particles, and on infusion into animals, these cells differentiated into immune-suppressive regulatory CD4+ T lymphocytes[186]. Fahmy and Saltzman have also shown that dendrimers carrying doxorubicin can be targeted onto antigen-specific T-cells directly in vivo by injecting dendrimers carrying peptide-MHC ligands (which bind to specific T cell receptors of their target cells[187]). Blood-circulating cells such as lymphocytes are the ideal targets to attempt in situ cell engineering, as these cells are in theory fully accessible to nanomaterials injected intravenously and the opportunity exists for cell binding to scavenging by the RES. Beyond nanoparticulate biomaterial, recent studies by Mooney and colleagues demonstrate that bioplymer scaffolds can mimic three-dimensional immune cell niches and in situ instruct endogenous immune cells to efficiently target tumors[188]. Following subcutaneous implantation, this biomaterial vaccine platform, termed Cellarium™, slowly releases GM-CSF and the danger signal CpG-oligodeoxynucleotide, which in combination attract and mature circulating dendritic cells into the porous scaffold. Subsequently, dendritic cells take up tumor antigen incorporated into the matrix, mature and emigrate into regional lymph nodes where they can prime tumor antigen specific CD8 T lymphocytes. Although important questions pertaining to the clinical value of this vaccine device in cancer patients with tumor-induced dendritic cell dysfunction and immune-suppressive dendritic cell populations await detailed clinic testing, Cellarium™ could provide major therapeutic benefits compared to conventional cancer vaccines.
As proof-of-principle that in situ cell surface engineering protocols can be developed to chemically modify selective cell membrane biomolecules in a living organism, Bertozzi and colleagues employed Copper-free click chemistry to metabolically label surface glycans in live mice[189,190]. While encouraging, these in vivo cell engineering efforts warrant further development and refinement to safely perform chemical reactions in living subjects. Key tasks in this regard are the identification and validation of high-affinity and high-specificity agents which target only defined cell populations in vivo, thereby preventing potentially harmful side effects.
Despite the challenges, ample opportunities for the further advancement of cell surface bioengineering as a clinical tool to endow cells with enhanced therapeutic properties lie ahead of us. Many of the robust methodologies described in this review are primed for translation across widespread clinical indications in the near future, while others are still in their infancy and face significant biological or safety uncertainties. To avoid being stalled in preclinical studies cell engineering strategies will need to remain focused on therapeutic efficacy, but also take into consideration factors such as labor intensity and cost which will govern the clinical availability and ultimately the commercial success of cell therapy products.
Acknowledgments
This work was supported in part by the National Science Foundation through the MIT Center for Materials Science and Engineering (DMR - 0819762), the Dept. of Defense Prostate Cancer Research Program (W81XWH-10-1-0290), and the NIH (CA140476 to DJI and EB012352 to MTS). DJI is an investigator of the Howard Hughes Medical Institute.
Biographies
 Matthias T. Stephan received his medical degree in 2002 from the University of Luebeck/Germany before pursuing a Ph.D. in Immunology at Cornell University under the supervision of Prof. Michel Sadelain. During his graduate studies he genetically engineered prostate cancer specificity into human T cells with a T-body specific for prostate-specific membrane antigen (PSMA). To optimally stimulate adoptively transferred T cells in an immune-suppressive tumor microenvironment Dr. Stephan developed a genetic strategy to provide T cells with “auto-costimulation”, thereby triggering robust T cell expansion and in vivo persistence. His expertise in therapeutic cell engineering was further broadened as a postdoctoral fellow in Prof. Darrell Irvine’s research group at the Massachusetts Institute of Technology. By combining the field of synthetic nanocarrier drug delivery with adoptive cell therapy, Dr. Stephan pioneered a methodology to enhance the persistence and functionality of adoptively transferred cells, which is based on the stable chemical conjugation of drug-loaded nanoparticles to the surfaces of cell products just prior to reinfusion. His work was recognized with the 2010 “Future Leader in Translational Medicine Award” by the American Academy for Cancer Research (AACR) and a NIH Ruth L. Kirschstein Research National Research Service Award.
Matthias T. Stephan received his medical degree in 2002 from the University of Luebeck/Germany before pursuing a Ph.D. in Immunology at Cornell University under the supervision of Prof. Michel Sadelain. During his graduate studies he genetically engineered prostate cancer specificity into human T cells with a T-body specific for prostate-specific membrane antigen (PSMA). To optimally stimulate adoptively transferred T cells in an immune-suppressive tumor microenvironment Dr. Stephan developed a genetic strategy to provide T cells with “auto-costimulation”, thereby triggering robust T cell expansion and in vivo persistence. His expertise in therapeutic cell engineering was further broadened as a postdoctoral fellow in Prof. Darrell Irvine’s research group at the Massachusetts Institute of Technology. By combining the field of synthetic nanocarrier drug delivery with adoptive cell therapy, Dr. Stephan pioneered a methodology to enhance the persistence and functionality of adoptively transferred cells, which is based on the stable chemical conjugation of drug-loaded nanoparticles to the surfaces of cell products just prior to reinfusion. His work was recognized with the 2010 “Future Leader in Translational Medicine Award” by the American Academy for Cancer Research (AACR) and a NIH Ruth L. Kirschstein Research National Research Service Award.
 Darrell J. Irvine, Ph.D., is an Associate Professor at the Massachusetts Institute of Technology and an Investigator of the Howard Hughes Medical Institute. He holds appointments in the MIT Department of Materials Science & Engineering and the Department of Biological Engineering, the Koch Institute for Integrative Cancer Research, and is a member of the steering committee for the Ragon Institute of MGH, MIT, and Harvard. His research is focused on the application of engineering tools to problems in cellular immunology and the development of new materials for vaccine and drug delivery. These efforts largely focus on cellular immunology and vaccine development for HIV and immunotherapy of cancer.
Darrell J. Irvine, Ph.D., is an Associate Professor at the Massachusetts Institute of Technology and an Investigator of the Howard Hughes Medical Institute. He holds appointments in the MIT Department of Materials Science & Engineering and the Department of Biological Engineering, the Koch Institute for Integrative Cancer Research, and is a member of the steering committee for the Ragon Institute of MGH, MIT, and Harvard. His research is focused on the application of engineering tools to problems in cellular immunology and the development of new materials for vaccine and drug delivery. These efforts largely focus on cellular immunology and vaccine development for HIV and immunotherapy of cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jenq RR, van den Brink MR. Nat Rev Cancer. 2010;10:213–221. doi: 10.1038/nrc2804. [DOI] [PubMed] [Google Scholar]
- 2.Uccelli A, Moretta L, Pistoia V. Nat Rev Immunol. 2008 doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 3.Martens TP, See F, Schuster MD, Sondermeijer HP, Hefti MM, Zannettino A, et al. Nat Clin Pract Cardiovasc Med. 2006;3(Suppl 1):S18–22. doi: 10.1038/ncpcardio0404. [DOI] [PubMed] [Google Scholar]
- 4.Uccelli A, Pistoia V, Moretta L. Trends Immunol. 2007;28:219–226. doi: 10.1016/j.it.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Karussis D, Karageorgiou C, Vaknin-Dembinsky A, Gowda-Kurkalli B, Gomori JM, Kassis I, et al. Arch Neurol. 2010;67:1187–1194. doi: 10.1001/archneurol.2010.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazzini L, Ferrero I, Luparello V, Rustichelli D, Gunetti M, Mareschi K, et al. Exp Neurol. 2010;223:229–237. doi: 10.1016/j.expneurol.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Perico N, Casiraghi F, Introna M, Gotti E, Todeschini M, Cavinato RA, et al. Clin J Am Soc Nephrol. 2010 doi: 10.2215/CJN.04950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato K, Ozaki K, Mori M, Muroi K, Ozawa K. J Clin Exp Hematop. 2010;50:79–89. doi: 10.3960/jslrt.50.79. [DOI] [PubMed] [Google Scholar]
- 9.Rangappa S, Makkar R, Forrester J. J Cardiovasc Pharmacol Ther. 2010;15:338–343. doi: 10.1177/1074248410376382. [DOI] [PubMed] [Google Scholar]
- 10.Noguchi H. Rev Diabet Stud. 2010;7:105–111. doi: 10.1900/RDS.2010.7.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, et al. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varela-Rohena A, Molloy PE, Dunn SM, Li Y, Suhoski MM, Carroll RG, et al. Nat Med. 2008;14:1390–1395. doi: 10.1038/nm.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jahnisch H, Fussel S, Kiessling A, Wehner R, Zastrow S, Bachmann M, et al. Clin Dev Immunol. 2010;2010:517493. doi: 10.1155/2010/517493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye L, Chang JC, Lin C, Sun X, Yu J, Kan YW. Proc Natl Acad Sci U S A. 2009;106:9826–9830. doi: 10.1073/pnas.0904689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, et al. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 16.Hinrichs CS, Spolski R, Paulos CM, Gattinoni L, Kerstann KW, Palmer DC, et al. Blood. 2008;111:5326–5333. doi: 10.1182/blood-2007-09-113050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brentjens RJ, Latouche JB, Santos E, Marti F, Gong MC, Lyddane C, et al. Nat Med. 2003;9:279–286. doi: 10.1038/nm827. [DOI] [PubMed] [Google Scholar]
- 18.Varnum-Finney B, Brashem-Stein C, Bernstein ID. Blood. 2003;101:1784–1789. doi: 10.1182/blood-2002-06-1862. [DOI] [PubMed] [Google Scholar]
- 19.Gattinoni L, Zhong XS, Palmer DC, Ji Y, Hinrichs CS, Yu Z, et al. Nat Med. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goessling W, Allen RS, Guan X, Jin P, Uchida N, Dovey M, et al. Cell Stem Cell. 2011;8:445–458. doi: 10.1016/j.stem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suhoski MM, Golovina TN, Aqui NA, Tai VC, Varela-Rohena A, Milone MC, et al. Mol Ther. 2007;15:981–988. doi: 10.1038/mt.sj.6300134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaney C, Heimfeld S, Brashem-Stein C, Voorhies H, Manger RL, Bernstein ID. Nat Med. 2010;16:232–236. doi: 10.1038/nm.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger C, Berger M, Hackman RC, Gough M, Elliott C, Jensen MC, et al. Blood. 2009;114:2417–2426. doi: 10.1182/blood-2008-12-189266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberg GL, Zakrzewski JL, Perales MA, van den Brink MR. Semin Immunol. 2007;19:289–296. doi: 10.1016/j.smim.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmqvist L, Pineault N, Wasslavik C, Humphries RK. PLoS One. 2007;2:e768. doi: 10.1371/journal.pone.0000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quintarelli C, Savoldo B, Dotti G. Methods Mol Biol. 2010;651:119–130. doi: 10.1007/978-1-60761-786-0_8. [DOI] [PubMed] [Google Scholar]
- 27.Verma IM, Weitzman MD. Annu Rev Biochem. 2005;74:711–738. doi: 10.1146/annurev.biochem.74.050304.091637. [DOI] [PubMed] [Google Scholar]
- 28.Singer O, Verma IM. Curr Gene Ther. 2008;8:483–488. doi: 10.2174/156652308786848067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohn DB, Sadelain M, Glorioso JC. Nat Rev Cancer. 2003;3:477–488. doi: 10.1038/nrc1122. [DOI] [PubMed] [Google Scholar]
- 30.Kato M, Mrksich M. Journal of the American Chemical Society. 2004;126:6504–6505. doi: 10.1021/ja039058e. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar D, Vemula PK, Teo GSL, Spelke D, Karnik R, Wee LY, et al. Bioconjugate Chemistry. 2008;19:2105–2109. doi: 10.1021/bc800345q. [DOI] [PubMed] [Google Scholar]
- 32.Lee RJ, Fang QZ, Davol PA, Gu YP, Sievers RE, Grabert RC, et al. Stem Cells. 2007;25:712–717. doi: 10.1634/stemcells.2005-0602. [DOI] [PubMed] [Google Scholar]
- 33.Zhao W, Teo GS, Kumar N, Karp JM. Materials Today. 2010;13:14–21. doi: 10.1016/S1369-7021(10)70056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabuka D, Forstner MB, Groves JT, Bertozzi CR. Journal of the American Chemical Society. 2008;130:5947–5953. doi: 10.1021/ja710644g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sackstein R, Merzaban JS, Cain DW, Dagia NM, Spencer JA, Lin CP, et al. Nature Medicine. 2008;14:181–187. doi: 10.1038/nm1703. [DOI] [PubMed] [Google Scholar]
- 36.Stephan MT, Moon JJ, Um SH, Bershteyn A, Irvine DJ. Nature Medicine. 2010;16:1035–1041. doi: 10.1038/nm.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Sanctis JB, Garmendia JV, Radzioch A, Radzioch D. Curr Pharm Des. 2006;12:2383–2395. doi: 10.2174/138161206777698990. [DOI] [PubMed] [Google Scholar]
- 38.Wilson JT, Krishnamurthy VR, Cui WX, Qu Z, Chaikof EL. Journal of the American Chemical Society. 2009;131:18228–18229. doi: 10.1021/ja908887v. [DOI] [PubMed] [Google Scholar]
- 39.Power AT, Bell JC. Gene Therapy. 2008;15:772–779. doi: 10.1038/gt.2008.40. [DOI] [PubMed] [Google Scholar]
- 40.Choi MR, Stanton-Maxey KJ, Stanley JK, Levin CS, Bardhan R, Akin D, et al. Nano Letters. 2007;7:3759–3765. doi: 10.1021/nl072209h. [DOI] [PubMed] [Google Scholar]
- 41.Torchilin VP. Eur J Pharm Sci. 2000;11(Suppl 2):S81–91. doi: 10.1016/s0928-0987(00)00166-4. [DOI] [PubMed] [Google Scholar]
- 42.Lautenschlager F, Paschke S, Schinkinger S, Bruel A, Beil M, Guck J. Proc Natl Acad Sci U S A. 2009;106:15696–15701. doi: 10.1073/pnas.0811261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curtis AS, Chandler C, Picton N. J Cell Sci. 1975;18:375–384. doi: 10.1242/jcs.18.3.375. [DOI] [PubMed] [Google Scholar]
- 44.Novo C, Fonseca E, Freitas AA. Cell Immunol. 1987;106:387–396. doi: 10.1016/0008-8749(87)90181-x. [DOI] [PubMed] [Google Scholar]
- 45.Lingwood D, Simons K. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 46.Cone RE, Marchalonis JJ, Rolley RT. J Exp Med. 1971;134:1373–1384. doi: 10.1084/jem.134.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vellai T, Takacs-Vellai K. Adv Exp Med Biol. 2010;694:69–80. doi: 10.1007/978-1-4419-7002-2_7. [DOI] [PubMed] [Google Scholar]
- 48.Kumari S, Mg S, Mayor S. Cell Res. 2010;20:256–275. doi: 10.1038/cr.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doherty GJ, McMahon HT. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 50.Wynn TA, Barron L. Semin Liver Dis. 2010;30:245–257. doi: 10.1055/s-0030-1255354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Nat Rev Immunol. 2007;7:678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- 52.Katritsis D, Kaiktsis L, Chaniotis A, Pantos J, Efstathopoulos EP, Marmarelis V. Prog Cardiovasc Dis. 2007;49:307–329. doi: 10.1016/j.pcad.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 53.Kierszenbaum A. Mosby. 2002 [Google Scholar]
- 54.Nilsson B, Korsgren O, Lambris JD, Ekdahl KN. Trends Immunol. 2010;31:32–38. doi: 10.1016/j.it.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Mol Pharm. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Sui M, Fan W. Curr Drug Metab. 2010;11:129–141. doi: 10.2174/138920010791110827. [DOI] [PubMed] [Google Scholar]
- 57.Sperling C, Fischer M, Maitz MF, Werner C. Biomaterials. 2009;30:4447–4456. doi: 10.1016/j.biomaterials.2009.05.044. [DOI] [PubMed] [Google Scholar]
- 58.Gorbet MB, Sefton MV. Biomaterials. 2004;25:5681–5703. doi: 10.1016/j.biomaterials.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 59.Ramot Y, Steiner M, Morad V, Leibovitch S, Amouyal N, Cesta MF, et al. Nanotoxicology. 2010;4:98–105. doi: 10.3109/17435390903470093. [DOI] [PubMed] [Google Scholar]
- 60.Wattendorf U, Merkle HP. J Pharm Sci. 2008;97:4655–4669. doi: 10.1002/jps.21350. [DOI] [PubMed] [Google Scholar]
- 61.Veronese FM, Mero A. BioDrugs. 2008;22:315–329. doi: 10.2165/00063030-200822050-00004. [DOI] [PubMed] [Google Scholar]
- 62.Pasut G, Veronese FM. Drugs Today (Barc) 2009;45:687–695. doi: 10.1358/dot.2009.45.9.1416421. [DOI] [PubMed] [Google Scholar]
- 63.Joralemon MJ, McRae S, Emrick T. Chem Commun (Camb) 2010;46:1377–1393. doi: 10.1039/b920570p. [DOI] [PubMed] [Google Scholar]
- 64.Knop K, Hoogenboom R, Fischer D, Schubert US. Angew Chem Int Ed Engl. 2010;49:6288–6308. doi: 10.1002/anie.200902672. [DOI] [PubMed] [Google Scholar]
- 65.Swiston AJ, Cheng C, Um SH, Irvine DJ, Cohen RE, Rubner MF. Nano Letters. 2008;8:4446–4453. doi: 10.1021/nl802404h. [DOI] [PubMed] [Google Scholar]
- 66.Lee DY, Park SJ, Nam JH, Byun Y. Tissue Engineering. 2006;12:615–623. doi: 10.1089/ten.2006.12.615. [DOI] [PubMed] [Google Scholar]
- 67.Hsiao SC, Shum BJ, Onoe H, Douglas ES, Gartner ZJ, Mathies RA, et al. Langmuir. 2009;25:6985–6991. doi: 10.1021/la900150n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krishnamachari Y, Pearce ME, Salem AK. Advanced Materials. 2008;20:989–993. [Google Scholar]
- 69.Murciano JC, Medinilla S, Eslin D, Atochina E, Cines DB, Muzykantov VR. Nature Biotechnology. 2003;21:891–896. doi: 10.1038/nbt846. [DOI] [PubMed] [Google Scholar]
- 70.Yolcu ES, Askenasy N, Singh NP, Cherradi SEL, Shirwan H. Immunity. 2002;17:795–808. doi: 10.1016/s1074-7613(02)00482-x. [DOI] [PubMed] [Google Scholar]
- 71.Singh NP, Yolcu ES, Taylor DD, Gercel-Taylor C, Metzinger DS, Dreisbach SK, et al. Cancer Research. 2003;63:4067–4073. [PubMed] [Google Scholar]
- 72.Cheng H, Kastrup CJ, Ramanathan R, Siegwart DJ, Ma ML, Bogatyrev SR, et al. ACS Nano. 2010;4:625–631. doi: 10.1021/nn901319y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lawrence DA, Song R, Weber P. J Leukoc Biol. 1996;60:611–618. doi: 10.1002/jlb.60.5.611. [DOI] [PubMed] [Google Scholar]
- 74.Sahaf B, Heydari K, Herzenberg LA, Herzenberg LA. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4001–4005. doi: 10.1073/pnas.2628032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thoren FB, Romero AI, Hermodsson S, Hellstrand K. J Immunol. 2007;179:781–785. doi: 10.4049/jimmunol.179.2.781. [DOI] [PubMed] [Google Scholar]
- 76.Hirota M, Suzuki M, Hagino S, Kagatani S, Sasaki Y, Aiba S, et al. J Toxicol Sci. 2009;34:139–150. doi: 10.2131/jts.34.139. [DOI] [PubMed] [Google Scholar]
- 77.Hirota M, Motoyama A, Suzuki M, Yanagi M, Kitagaki M, Kouzuki H, et al. J Toxicol Sci. 2010;35:871–879. doi: 10.2131/jts.35.871. [DOI] [PubMed] [Google Scholar]
- 78.Holden CA, Yuan Q, Yeudall WA, Lebman DA, Yang H. International Journal of Nanomedicine. 2010;5:25–36. [PMC free article] [PubMed] [Google Scholar]
- 79.Xia L, McDaniel JM, Yago T, Doeden A, McEver RP. Blood. 2004;104:3091–3096. doi: 10.1182/blood-2004-02-0650. [DOI] [PubMed] [Google Scholar]
- 80.Sackstein R. Methods Enzymol. 2010;479:93–105. doi: 10.1016/S0076-6879(10)79005-4. [DOI] [PubMed] [Google Scholar]
- 81.Kayser H, Zeitler R, Kannicht C, Grunow D, Nuck R, Reutter W. J Biol Chem. 1992;267:16934–16938. [PubMed] [Google Scholar]
- 82.Prescher JA, Dube DH, Bertozzi CR. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 83.Dube DH, Bertozzi CR. Current Opinion in Chemical Biology. 2003;7:616–625. doi: 10.1016/j.cbpa.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 84.Mahal LK, Yarema KJ, Bertozzi CR. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 85.Lee JH, Baker TJ, Mahal LK, Zabner J, Bertozzi CR, Wiemer DF, et al. J Biol Chem. 1999;274:21878–21884. doi: 10.1074/jbc.274.31.21878. [DOI] [PubMed] [Google Scholar]
- 86.Nauman DA, Bertozzi CR. Biochim Biophys Acta. 2001;1568:147–154. doi: 10.1016/s0304-4165(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 87.Iwasaki Y, Tabata E, Kurita K, Akiyoshi K. Bioconjug Chem. 2005;16:567–575. doi: 10.1021/bc049707r. [DOI] [PubMed] [Google Scholar]
- 88.Sampathkumar SG, Li AV, Jones MB, Sun ZH, Yarema KJ. Nature Chemical Biology. 2006;2:149–152. doi: 10.1038/nchembio770. [DOI] [PubMed] [Google Scholar]
- 89.Howarth M, Takao K, Hayashi Y, Ting AY. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen I, Howarth M, Lin WY, Ting AY. Nature Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 91.Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, et al. Nature Biotechnology. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.White SH, von Heijne G. Curr Opin Struct Biol. 2005;15:378–386. doi: 10.1016/j.sbi.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 93.Medof ME, Nagarajan S, Tykocinski ML. FASEB Journal. 1996;10:574–586. doi: 10.1096/fasebj.10.5.8621057. [DOI] [PubMed] [Google Scholar]
- 94.Paulick MG, Bertozzi CR. Biochemistry. 2008;47:6991–7000. doi: 10.1021/bi8006324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nagarajan S, Selvaraj P. Cancer Res. 2002;62:2869–2874. [PubMed] [Google Scholar]
- 96.Notohamiprodjo M, Djafarzadeh R, Mojaat A, von Luttichau I, Grone HJ, Nelson PJ. Protein Eng Des Sel. 2006;19:27–35. doi: 10.1093/protein/gzi072. [DOI] [PubMed] [Google Scholar]
- 97.Ko IK, Kean TJ, Dennis JE. Biomaterials. 2009;30:3702–3710. doi: 10.1016/j.biomaterials.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dennis JE, Cohen N, Goldberg VM, Caplan AI. Journal of Orthopaedic Research. 2004;22:735–741. doi: 10.1016/j.orthres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 99.Kim SA, Peacock JS. Journal of Immunological Methods. 1993;158:57–65. doi: 10.1016/0022-1759(93)90258-9. [DOI] [PubMed] [Google Scholar]
- 100.Chen AS, Zheng GX, Tykocinski ML. Journal of Immunology. 2000;164:705–711. doi: 10.4049/jimmunol.164.2.705. [DOI] [PubMed] [Google Scholar]
- 101.Peterson BR. Org Biomol Chem. 2005;3:3607–3612. doi: 10.1039/b509866a. [DOI] [PubMed] [Google Scholar]
- 102.Boonyarattanakalin S, Athavankar S, Sun Q, Peterson BR. Journal of the American Chemical Society. 2006;128:386–387. doi: 10.1021/ja056126j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Boonyarattanakalin S, Martin SE, Sun Q, Peterson BR. Journal of the American Chemical Society. 2006;128:11463–11470. doi: 10.1021/ja062377w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sarkar D, Vemula PK, Zhao WA, Gupta A, Karnik R, Karp JM. Biomaterials. 2010;31:5266–5274. doi: 10.1016/j.biomaterials.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.David CMM, Metzler EI. 1978;409 [Google Scholar]
- 106.Thierry B, Winnik FM, Merhi Y, Tabrizian M. J Am Chem Soc. 2003;125:7494–7495. doi: 10.1021/ja034321x. [DOI] [PubMed] [Google Scholar]
- 107.Rajagopalan P, Shen CJ, Berthiaume F, Tilles AW, Toner M, Yarmush ML. Tissue Eng. 2006;12:1553–1563. doi: 10.1089/ten.2006.12.1553. [DOI] [PubMed] [Google Scholar]
- 108.Chambers E, Mitragotri S. Journal of Controlled Release. 2004;100:111–119. doi: 10.1016/j.jconrel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 109.Chambers E, Mitragotri S. Experimental Biology and Medicine. 2007;232:958–966. [PubMed] [Google Scholar]
- 110.Chen W, Zarnitsyna VI, Sarangapani KK, Huang J, Zhu C. Cell Mol Bioeng. 2008;1:276–288. doi: 10.1007/s12195-008-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kickhoefer VA, Han M, Raval-Fernandes S, Poderycki MJ, Moniz RJ, Vaccari D, et al. Acs Nano. 2009;3:27–36. doi: 10.1021/nn800638x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen X, Tam UC, Czlapinski JL, Lee GS, Rabuka D, Zettl A, et al. Journal of the American Chemical Society. 2006;128:6292–6293. doi: 10.1021/ja060276s. [DOI] [PubMed] [Google Scholar]
- 113.Beg S, Rizwan M, Sheikh AM, Hasnain MS, Anwer K, Kohli K. J Pharm Pharmacol. 2011;63:141–163. doi: 10.1111/j.2042-7158.2010.01167.x. [DOI] [PubMed] [Google Scholar]
- 114.Chen X, Wu P, Rousseas M, Okawa D, Gartner Z, Zettl A, et al. J Am Chem Soc. 2009;131:890–891. doi: 10.1021/ja807334b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kirpotin DB, Drummond DC, Shao Y, Shalaby MR, Hong K, Nielsen UB, et al. Cancer Res. 2006;66:6732–6740. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 116.Sapra P, Allen TM. Cancer Res. 2002;62:7190–7194. [PubMed] [Google Scholar]
- 117.Poon Z, Chen S, Engler AC, Lee HI, Atas E, von Maltzahn G, et al. Angew Chem Int Ed Engl. 2010;49:7266–7270. doi: 10.1002/anie.201003445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Balthasar S, Michaelis K, Dinauer N, von Briesen H, Kreuter J, Langer K. Biomaterials. 2005;26:2723–2732. doi: 10.1016/j.biomaterials.2004.07.047. [DOI] [PubMed] [Google Scholar]
- 119.Muro S, Garnacho C, Champion JA, Leferovich J, Gajewski C, Schuchman EH, et al. Molecular Therapy. 2008;16:1450–1458. doi: 10.1038/mt.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Champion JA, Mitragotri S. Proc Natl Acad Sci U S A. 2006;103:4930–4934. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vasconcellos FC, Swiston AJ, Beppu MM, Cohen RE, Rubner MF. Biomacromolecules. 2010;11:2407–2414. doi: 10.1021/bm100570r. [DOI] [PubMed] [Google Scholar]
- 122.Greulich C, Diendorf J, Simon T, Eggeler G, Epple M, Koller M. Acta Biomater. 2010;7:347–354. doi: 10.1016/j.actbio.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 123.Jiang W, Kim BY, Rutka JT, Chan WC. Nat Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 124.Gao H, Shi W, Freund LB. Proc Natl Acad Sci U S A. 2005;102:9469–9474. doi: 10.1073/pnas.0503879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu ZJ, Carboni R, Quercio MJ, Yan B, Miranda OR, Anderton DL, et al. Small. 2010;6:2261–2265. doi: 10.1002/smll.201000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Arvizo RR, Miranda OR, Thompson MA, Pabelick CM, Bhattacharya R, Robertson JD, et al. Nano Letters. 2010;10:2543–2548. doi: 10.1021/nl101140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Karp JM, Teol GSL. Cell Stem Cell. 2009;4:206–216. doi: 10.1016/j.stem.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 128.Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, Jones JO, et al. Science. 2010;330:827–830. doi: 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- 129.Xia LJ, McDaniel JM, Yago T, Doeden A, McEver RP. Blood. 2004;104:3091–3096. doi: 10.1182/blood-2004-02-0650. [DOI] [PubMed] [Google Scholar]
- 130.Zhong XY, Zhang B, Asadollahi R, Low SH, Holzgreve W. Ann N Y Acad Sci. 2010;1205:17–22. doi: 10.1111/j.1749-6632.2010.05659.x. [DOI] [PubMed] [Google Scholar]
- 131.Klebanoff CA, Khong HT, Antony PA, Palmer DC, Restifo NP. Trends Immunol. 2005;26:111–117. doi: 10.1016/j.it.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fukumura D, Jain RK. J Cell Biochem. 2007;101:937–949. doi: 10.1002/jcb.21187. [DOI] [PubMed] [Google Scholar]
- 133.Schreiber H, Rowley DA. Science. 2010;330:761–762. doi: 10.1126/science.1198345. [DOI] [PubMed] [Google Scholar]
- 134.Wolchok JD, Yang AS, Weber JS. Cancer J. 2010;16:311–317. doi: 10.1097/PPO.0b013e3181eb3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Thompson RH, Kwon ED, Allison JP. Immunotherapy. 2009;1:129–139. doi: 10.2217/1750743X.1.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wallace A, Kapoor V, Sun J, Mrass P, Weninger W, Heitjan DF, et al. Clin Cancer Res. 2008;14:3966–3974. doi: 10.1158/1078-0432.CCR-08-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ohta A, Gorelik E, Prasad SJ, Ronchese F, Lukashev D, Wong MK, et al. Proc Natl Acad Sci U S A. 2006;103:13132–13137. doi: 10.1073/pnas.0605251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Thompson JA, Lee DJ, Cox WW, Lindgren CG, Collins C, Neraas KA, et al. Cancer Res. 1987;47:4202–4207. [PubMed] [Google Scholar]
- 139.Durand EM, Zon LI. Curr Opin Hematol. 2010;17:308–312. doi: 10.1097/MOH.0b013e32833a888c. [DOI] [PubMed] [Google Scholar]
- 140.Sarkar D, Ankrum JA, Teo GS, Carman CV, Karp JM. Biomaterials. 2011;32:3053–3061. doi: 10.1016/j.biomaterials.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cai Z, Wang Y, Zhu LJ, Liu ZQ. Curr Drug Metab. 2010;11:197–207. doi: 10.2174/138920010791110836. [DOI] [PubMed] [Google Scholar]
- 142.Davis ME, Chen ZG, Shin DM. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 143.Fang J, Nakamura H, Maeda H. Adv Drug Deliv Rev. 2010 [Google Scholar]
- 144.Owens DE, 3rd, Peppas NA. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 145.Beck Z, Brown BK, Wieczorek L, Peachman KK, Matyas GR, Polonis VR, et al. PLoS One. 2009;4:e8297. doi: 10.1371/journal.pone.0008297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jain KK. Veterinary Hematology. 1993 [Google Scholar]
- 147.Hamidi M, Tajerzadeh H. Drug Deliv. 2003;10:9–20. doi: 10.1080/713840329. [DOI] [PubMed] [Google Scholar]
- 148.Rossi L, Serafini S, Pierige F, Antonelli A, Cerasi A, Fraternale A, et al. Expert Opin Drug Deliv. 2005;2:311–322. doi: 10.1517/17425247.2.2.311. [DOI] [PubMed] [Google Scholar]
- 149.Hamidi M, Zarrin A, Foroozesh M, Mohammadi-Samani S. J Control Release. 2007;118:145–160. doi: 10.1016/j.jconrel.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 150.Eichler HC, Raffesber W, Gasic S, Korn A. Res Exp Med. 1985;185:341–344. doi: 10.1007/BF01851959. [DOI] [PubMed] [Google Scholar]
- 151.Eichler HC, Gasic S, Daum B, Bacher S, Steger G. Adv Biosci (series) 1987;67:11–15. [Google Scholar]
- 152.Hamidi M, Zarrin A, Foroozesh M, Mohammadi-Samani S. Journal of Controlled Release. 2007;118:145–160. doi: 10.1016/j.jconrel.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 153.Tang C, Russell PJ, Martiniello-Wilks R, Rasko JEJ, Khatri A. Stem Cells. 2010;28:1686–1702. doi: 10.1002/stem.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dou H, Destache CJ, Morehead JR, Mosley RL, Boska MD, Kingsley J, et al. Blood. 2006;108:2827–2835. doi: 10.1182/blood-2006-03-012534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.June CH. J Clin Invest. 2007;117:1466–1476. doi: 10.1172/JCI32446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Harlin H, Meng Y, Peterson AC, Zha Y, Tretiakova M, Slingluff C, et al. Cancer Res. 2009;69:3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cole C, Qiao J, Kottke T, Diaz RM, Ahmed A, Sanchez-Perez L, et al. Nature Medicine. 2005;11:1073–1081. doi: 10.1038/nm1297. [DOI] [PubMed] [Google Scholar]
- 158.Russell SJ, Peng KW. Curr Opin Mol Ther. 2008;10:380–386. [PubMed] [Google Scholar]
- 159.Ilett EJ, Prestwich RJ, Kottke T, Errington F, Thompson JM, Harrington KJ, et al. Gene Ther. 2009;16:689–699. doi: 10.1038/gt.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Banerjee R, Katsenovich Y, Lagos L, McIintosh M, Zhang X, Li CZ. Curr Med Chem. 2010;17:3120–3141. doi: 10.2174/092986710791959765. [DOI] [PubMed] [Google Scholar]
- 161.Barbas AS, Mi J, Clary BM, White RR. Future Oncol. 2010;6:1117–1126. doi: 10.2217/fon.10.67. [DOI] [PubMed] [Google Scholar]
- 162.Alley SC, Okeley NM, Senter PD. Curr Opin Chem Biol. 2010;14:529–537. doi: 10.1016/j.cbpa.2010.06.170. [DOI] [PubMed] [Google Scholar]
- 163.Mortensen MW, Kahns L, Hansen T, Sorensen PG, Bjorkdahl O, Jensen MR, et al. J Nanosci Nanotechnol. 2007;7:4575–4580. [PubMed] [Google Scholar]
- 164.Freitas RA., Jr J Nanosci Nanotechnol. 2006;6:2769–2775. doi: 10.1166/jnn.2006.413. [DOI] [PubMed] [Google Scholar]
- 165.Saudek F, Brogren CH, Manohar S. Rev Diabet Stud. 2008;5:6–12. doi: 10.1900/RDS.2008.5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tang C, Russell PJ, Martiniello-Wilks R, Rasko JE, Khatri A. Stem Cells. 2010;28:1686–1702. doi: 10.1002/stem.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yaghoubi SS, Jensen MC, Satyamurthy N, Budhiraja S, Paik D, Czernin J, et al. Nat Clin Pract Oncol. 2009;6:53–58. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Singh N, Jenkins JS, Asadi R, Doak SH. Nano Reviews. 2010;1:5358. doi: 10.3402/nano.v1i0.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bhaumik S. Curr Pharm Biotechnol. 2011 doi: 10.2174/138920111795163896. [DOI] [PubMed] [Google Scholar]
- 170.Orcutt KD, Slusarczyk AL, Cieslewicz M, Ruiz-Yi B, Bhushan KR, Frangioni JV, et al. Nucl Med Biol. 2011;38:223–233. doi: 10.1016/j.nucmedbio.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Medintz IL, Mattoussi H, Clapp AR. Int J Nanomedicine. 2008;3:151–167. doi: 10.2147/ijn.s614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Teta M, Long SY, Wartschow LM, Rankin MM, Kushner JA. Diabetes. 2005;54:2557–2567. doi: 10.2337/diabetes.54.9.2557. [DOI] [PubMed] [Google Scholar]
- 173.Kirouac DC, Zandstra PW. Cell Stem Cell. 2008;3:369–381. doi: 10.1016/j.stem.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 174.Petersen LK, Narasimhan B. Expert Opin Drug Deliv. 2008;5:837–846. doi: 10.1517/17425247.5.8.837. [DOI] [PubMed] [Google Scholar]
- 175.Stadler A, Chi C, van der Lelie D, Gang O. Nanomedicine (Lond) 2010;5:319–334. doi: 10.2217/nnm.10.2. [DOI] [PubMed] [Google Scholar]
- 176.Lee JB, Roh YH, Um SH, Funabashi H, Cheng W, Cha JJ, et al. Nat Nanotechnol. 2009;4:430–436. doi: 10.1038/nnano.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pendergrast PS, Marsh HN, Grate D, Healy JM, Stanton M. J Biomol Tech. 2005;16:224–234. [PMC free article] [PubMed] [Google Scholar]
- 178.Guo M, Que C, Wang C, Liu X, Yan H, Liu K. Biomaterials. 2011;32:185–194. doi: 10.1016/j.biomaterials.2010.09.077. [DOI] [PubMed] [Google Scholar]
- 179.Azagarsamy MA, Sokkalingam P, Thayumanavan S. J Am Chem Soc. 2009;131:14184–14185. doi: 10.1021/ja906162u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.You JO, Almeda D, Ye GJ, Auguste DT. J Biol Eng. 2010;4:15. doi: 10.1186/1754-1611-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lai TC, Bae Y, Yoshida T, Kataoka K, Kwon GS. Pharm Res. 2010;27:2260–2273. doi: 10.1007/s11095-010-0092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Laszlo Radvanyi MACC. International Society for Cellular Therapy. Cell Therapy FDA Liaison Meeting; Bethesda MD. Oct 17; 2008. [Google Scholar]
- 183.Nabhan C. N Engl J Med. 2010;363:1966–1967. doi: 10.1056/NEJMc1009982. author reply 1968. [DOI] [PubMed] [Google Scholar]
- 184.McAllister TN, Dusserre N, Maruszewski M, L’Heureux N. Regen Med. 2008;3:925–937. doi: 10.2217/17460751.3.6.925. [DOI] [PubMed] [Google Scholar]
- 185.Mumprecht S, Schurch C, Schwaller J, Solenthaler M, Ochsenbein AF. Blood. 2009;114:1528–1536. doi: 10.1182/blood-2008-09-179697. [DOI] [PubMed] [Google Scholar]
- 186.Park J, Gao W, Whiston R, Strom TB, Metcalfe S, Fahmy TM. Mol Pharm. 2010 doi: 10.1021/mp100203a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fahmy TM, Schneck JP, Saltzman WM. Nanomedicine. 2007;3:75–85. doi: 10.1016/j.nano.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 188.Ali OA, Emerich D, Dranoff G, Mooney DJ. Sci Transl Med. 2009;1:8ra19. doi: 10.1126/scitranslmed.3000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, et al. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]