Abstract
Herein we report the synthesis of water-soluble polyglycerol-dendronized perylenediimides (PGD-PDIs) with a single reactive group that undergoes high-yielding click reactions. Single-molecule studies and target-specific biolabeling are reported, including the highly specific labeling of proteins on the surface of living bacterial and mammalian cells.
Fluorescent compounds are useful reporters in biology because they signal their environment and probe a wide range of biological processes, including enzyme catalysis and protein folding and trafficking.1 The performance of these probes is enhanced when the following criteria are met: i) high brightness and photostability, ii) water-solubility, iii) biocompatibility, and iv) absorption and emission maxima above 500 nm to minimize the autofluorescence background of cells.2 In this regard, perylene tetracarboxylic acid diimides (PDIs) are an attractive class of fluorophores because of their high thermal and photochemical stability with high fluorescence quantum yields in organic solvents.3 The many successful efforts to prepare soluble and fuctional PDIs have been reviewed by Langhals, Müllen, Würthner, Nagao and others.3,4
Perhaps the largest challenge with PDI chromophores is developing water-soluble, nonaggregating analogs that retain their exceptional properties. One of the most powerful methods, introduced by Müllen and coworkers, is the incorporation of ionic moieties such as sulfonate, pyridinium, and quaternary ammonium groups into the bay region of the PDI chromophores.5 These highly charged PDIs are very effective at staining membranes, have the added advantage that the donor groups in the bay region cause a significant red shift in the absorption maximum and an increased Stokes shift, and recently have been prepared with a single reactive group for protein conjugation.6
Our interest in developing stable fluorophores for bioimaging led us to seek a neutral and biocompatible PDI for targeted imaging.7 While this work was in progress, Haag and coworkers reported neutral, water-soluble PDI chromophores prepared by reaction of amine-cored polyglycerol dendrons (PGs) and perylene tetracarboxylic acid bisanhydride. The dendritic PDIs exhibit high fluorescence quantum yields in water as a result of reduced aggregation.8 By combining the advantages of the approaches pioneered by Müllen and Haag, we prepared neutral PDIs that are bay-substituted with PG dendrons and contain a single azide group with high reactivity in the click reaction, despite the dense, globular PG shell.
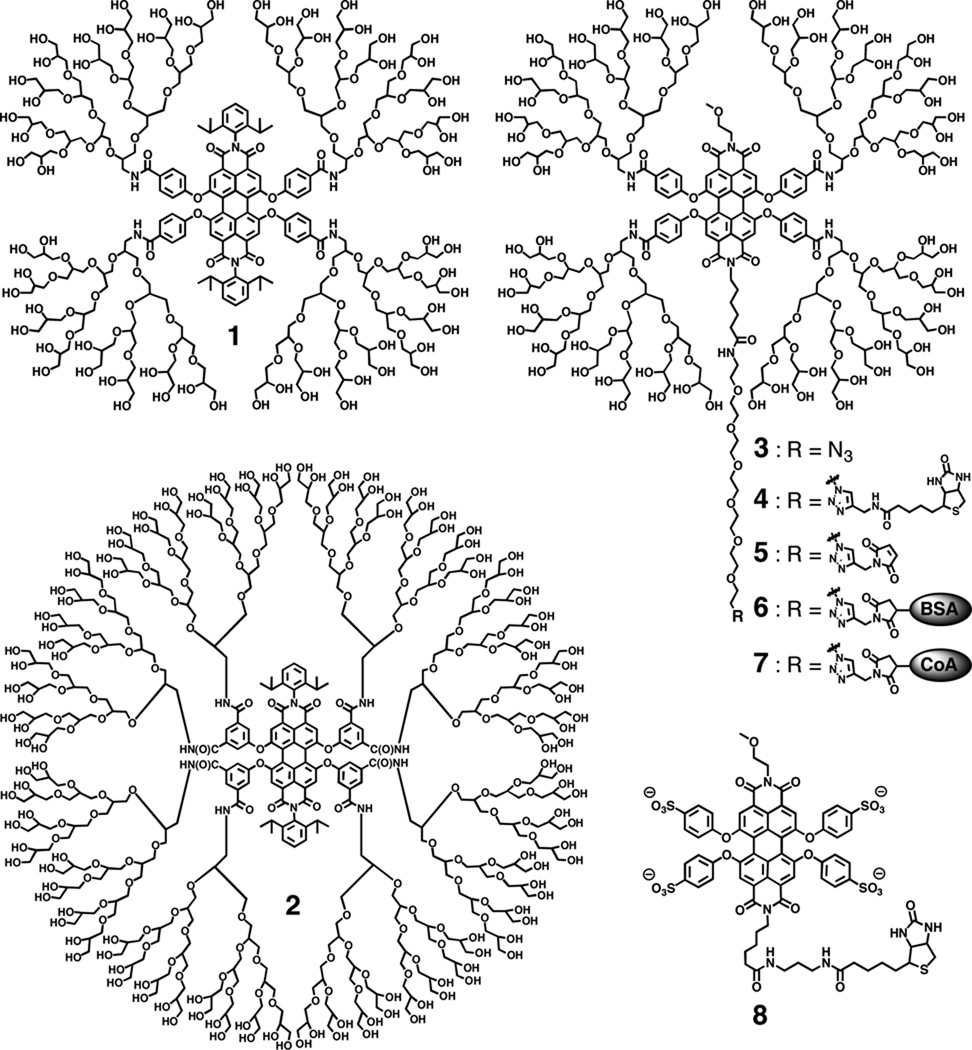
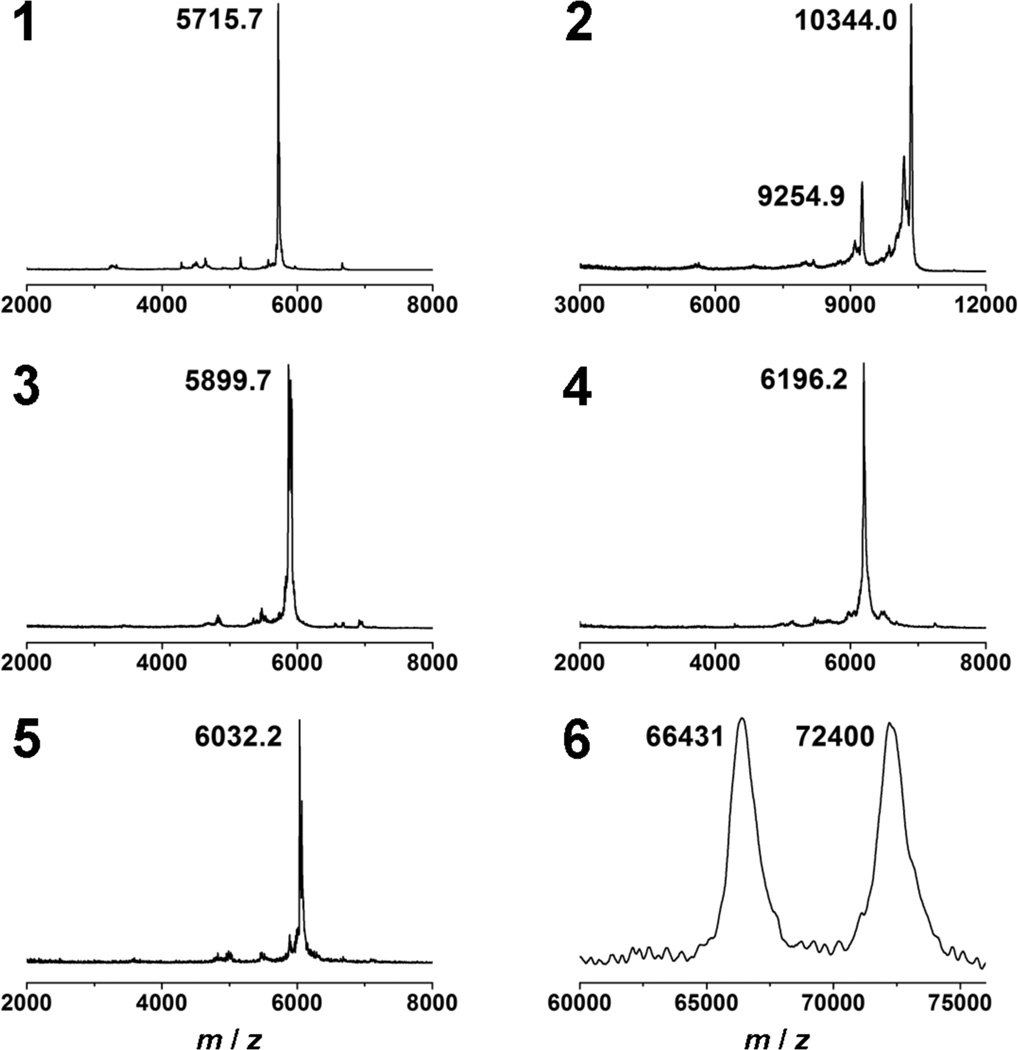
The key structural feature of the PGD-PDIs reported in this study is a fluorescent PDI core that is linked through amides to four (1 and 3) and eight dendrons (2), which give 64 and 128 hydroxyl end-groups, respectively (Figure 1). PGD-PDIs 1–3 were prepared by HATU-mediated amide coupling of the PDI cores with amine-functionalized PGDs containing eight allyl groups followed by dihydroxylation. The synthetic pathway is described in the Supporting Information. The purification was achieved by a combination of column chromatography, SEC and dialysis, and the purity was determined by analytical SEC, 1H NMR spectroscopy, and MALDI-TOF mass spectrometry. PGD-PDIs 1 and 3 appear to be homogeneous with a complete attachment of four dendrons in each, while the MALDI mass spectrum of PGD-PDI 2 reveals a mixture of 7 and 8 attachments, with the latter predominating (Figure 2).
Figure 1.
Structures of PGD-PDIs 1–7 and PDI 8.
Figure 2.
MALDI-TOF mass spectra of PGD-PDIs 1–6.
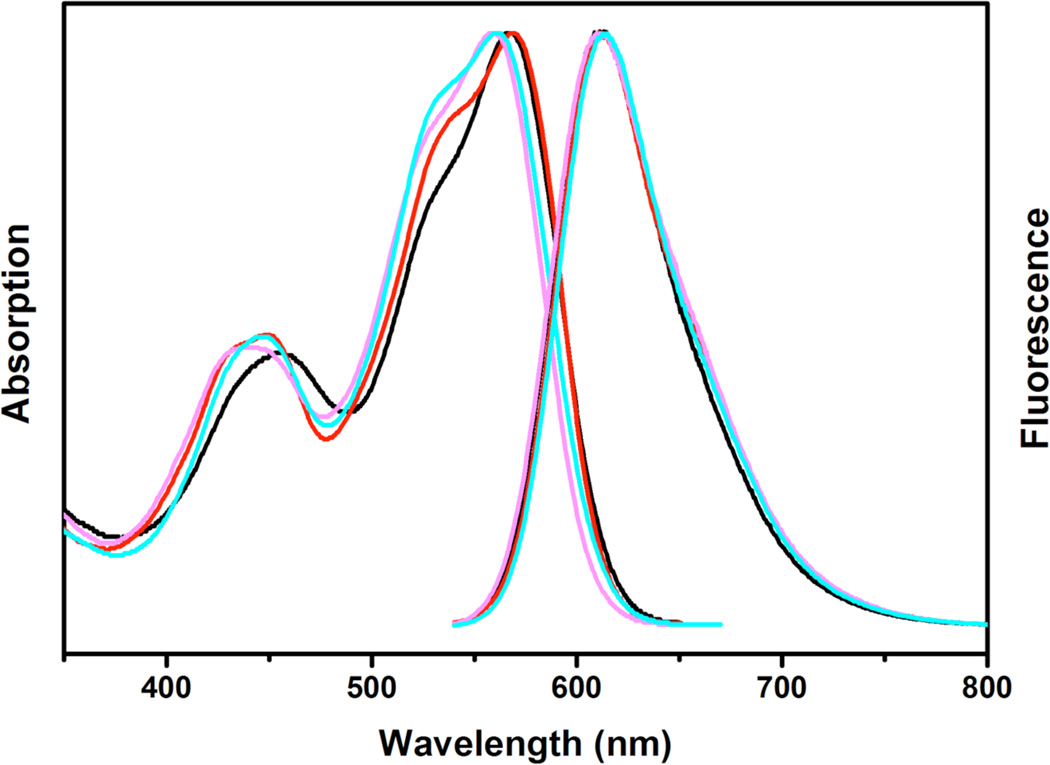
All PGD-PDIs we synthesized are fully water-soluble and highly fluorescent in water with the quantum yields ranging from 0.57 to 0.83. The absorption and emission spectra (Figure 3 and S1) show maxima at 559–569 nm and 612 nm, respectively, consistent with those of the nondendronized PDI analogues5 reported in the literature. These results indicate that the attachment of bulky polyglycerol dendrons not only retains the excellent photophysical properties of original PDI chromophores, but improves the water-solubility with minimal aggregation in water, making them useful in bioapplications.
Figure 3.
Normalized absorption and emission spectra of PGD-PDIs 1–3 and PDI 8 in water. λabs, max (nm), λem, max (nm), Φ (in water) = 567, 612, 0.62 (1, black); 569, 612, 0.83 (2, red); 559, 612, 0.57 (3, pink); 561, 612, 0.39 (8, cyan), respectively.
Furthermore, the reactivity of monofunctional PGD-PDI 3 with alkyne-functionalized biotin and maleimide was investigated using Click chemistry.9 Quantitative conversions to PGD-PDIs 4 and 5 containing a single biotin or maleimide function, respectively, were confirmed by comparing the MALDI mass spectra of 3–5 (Figure 2), demonstrating the potential of our clickable monofunctional PGD-PDIs for conjugation to biomolecules.
The use of copper in the click reaction and the known quenching of fluorescence by metal ions, raised the question of whether the fluorescence of the PGD-PDIs might be affected by copper ion. At concentrations higher than those likely to be present from the click reaction the fluorescence of PGD-PDIs was almost unchanged whereas the nondendronized PDI analogues were significantly quenched (Supporting Information).
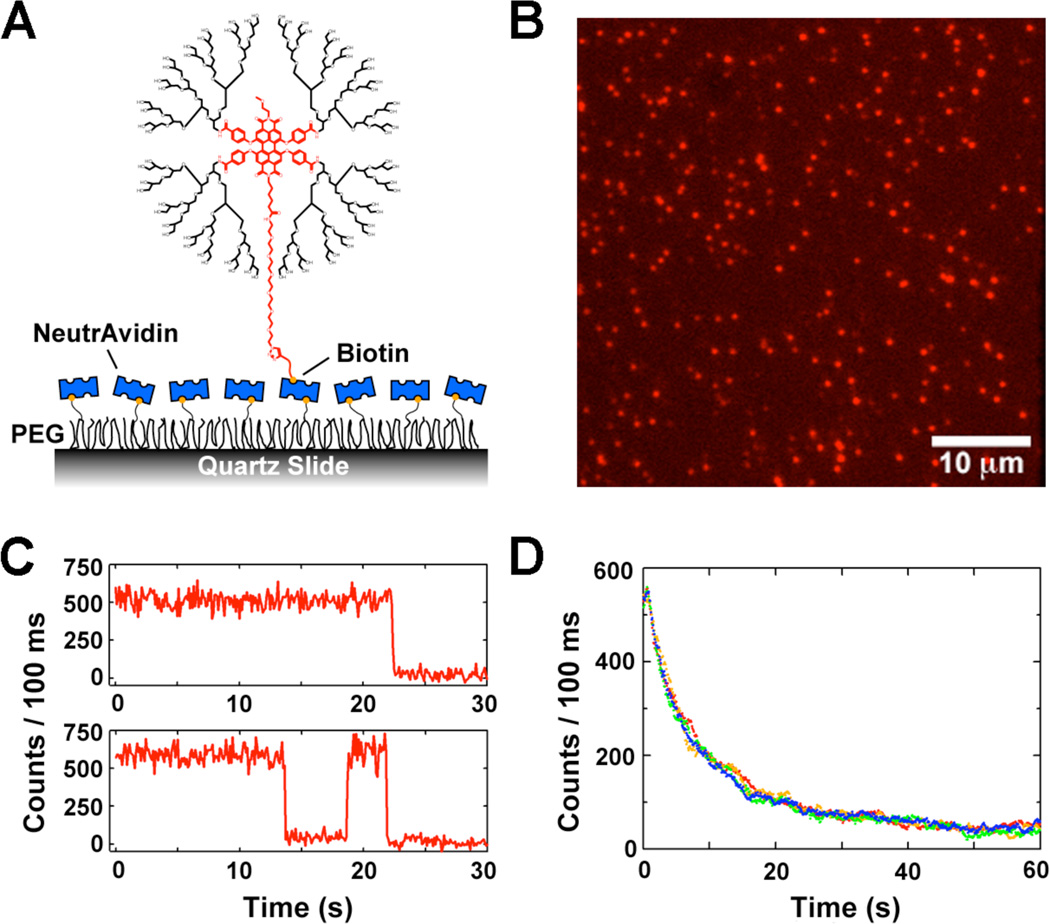
To characterize the photostability of PGD-PDIs for single-molecule applications, biotinylated PGD-PDI 4 was immobilized on a PEG-passivated surface through biotinneutravidin linkages (Figure 4A). This immobilization was indeed highly specific because only negligible binding of 4 was observed in the absence of preincubated neutravidin. The optics used were modified from those that are normally implemented in Cy3/Cy5 FRET experiments,10 by removing the dichroic beamsplitter (see Supporting Information). Moreover, this image clearly shows that the fluorescence intensity of single PGD-PDI molecules can be easily distinguished from the background signal with an integration time of 100 ms (Figure 4B). Thus, individual PGD-PDI molecules are bright enough for single-molecule detection under typical imaging conditions.
Figure 4.
Single-molecule analysis of PGD-PDI 4. (A) Experimental scheme, in which 4 is immobilized on a PEG-coated surface through biotin-neutravidin linkages. (B) Fluorescence image of immobilized molecules with pseudo colors. (C) Typical fluorescence intensity time traces of individual molecules in T50 buffer. (D) Average fluorescence intensity as a function of time in T50 buffer.
The intensity time traces of hundreds of isolated molecules were obtained in T50 buffer,10b containing 10 mM Tris and 50 mM NaCl at pH 8.0 (Figure 4C). As expected, the averaged intensity of these molecules decayed with time (Figure 4D), and this decay was fit to a single exponential, with the fitted lifetime defined as the single-molecule photobleaching time, τphotobleaching. From six repeated measurements in different imaging areas, τphotobleaching was determined as 9.6 ± 2.0 s for 4 while the nondendronized analogue 8 exhibited 7.8 ± 1.2 s under the same conditions (Supporting Information). The fluorescence intensity of a single molecule was also determined by the average number of detected photons: 21000 ± 2000 photons for 4 and 17000 ± 2000 photons for 8. This result indicates that the original photostability of PDIs can be retained despite the bulky dendron substitution on the PDIs.
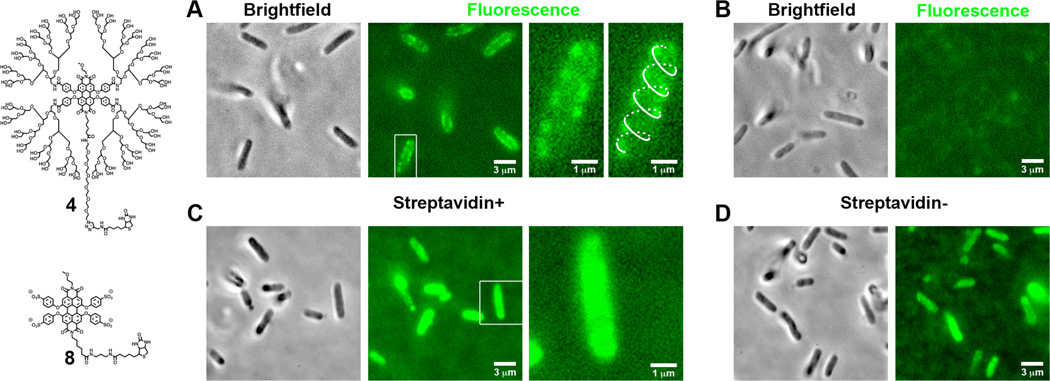
To illustrate the usefulness of PGD-PDI 4 in targeted biological imaging, the fluorescent labeling of λ-receptors on the surface of living bacterial cells using 4 was explored. This receptor protein, also known as LamB or maltoporin, functions as a receptor for bacteriophage λ and also facilitates the transport of maltodextrins across the outer membrane of E. coli.11 The E. coli strain used in this study, called pLO16, produces biotinylated λ-receptors in vivo with the biotin on the extracellular side.12 The low efficiency of biotinylation in this strain allowed for the detection of λ-receptor localization pattern in these cells. As seen in Figure 5A, PGD-PDI 4 produced bright fluorescent spots only at the surface of the cells that were preincubated with streptavidin. Furthermore, in some bacteria a helical pattern of the spots can be discerned along the long axis of the bacterium consistent with the surface arrangement of the proteins.13 This labeling was highly target-specific, as seen in control experiments where labeling with 4 in the absence of streptavidin showed no detectable staining of the bacteria cells, either on the surface or inside the bacterium (Figure 5B).
Figure 5.
Fluorescent labeling of λ-receptors on the surface of living bacteria cells. (A) Brightfield and fluorescence images of E. coli cells labeled with PGD-PDI 4 after preincubation with streptavidin. The helical pattern of λ-receptors is highlighted for one cell (right panel, enlargement of a white box in the middle panel). (B) As a control, cells were labeled with 4 in the absence of streptavidin. In (C) and (D), cells were labeled with nondendronized PDI 8 with and without streptavidin preincubation, respectively. All images were processed using the software ImageJ (NIH) and an intensity range of (10133, 23589) and (28856, 65378) was used for the fluorescence images of 4 and 8, respectively. Note that a scale greater in intensity value was used for 8 because of a higher background and absolute intensity therein.
The importance of the dendronized, neutral character of PDI 4 is seen by comparison with nondendronized analogue 8. Although 8 carries a biotin unit, it exhibited only nonspecific staining of bacteria at the same concentration as used for 4 and in the presence of streptavidin. As seen in Figure 5C, cells stained with 8 exhibited a largely homogeneous distribution in fluorescence signal throughout the entire bacterium that could not be removed even after extensive washes. That ionic interactions dominate the cell staining is further seen in experiments where 8 was used without preincubation with streptavidin. The cells in this case showed a comparable level of staining throughout the bacterium (Figure 5D). These studies demonstrate the importance of the neutral, dendronized PDI in achieving target-specific biolabeling.
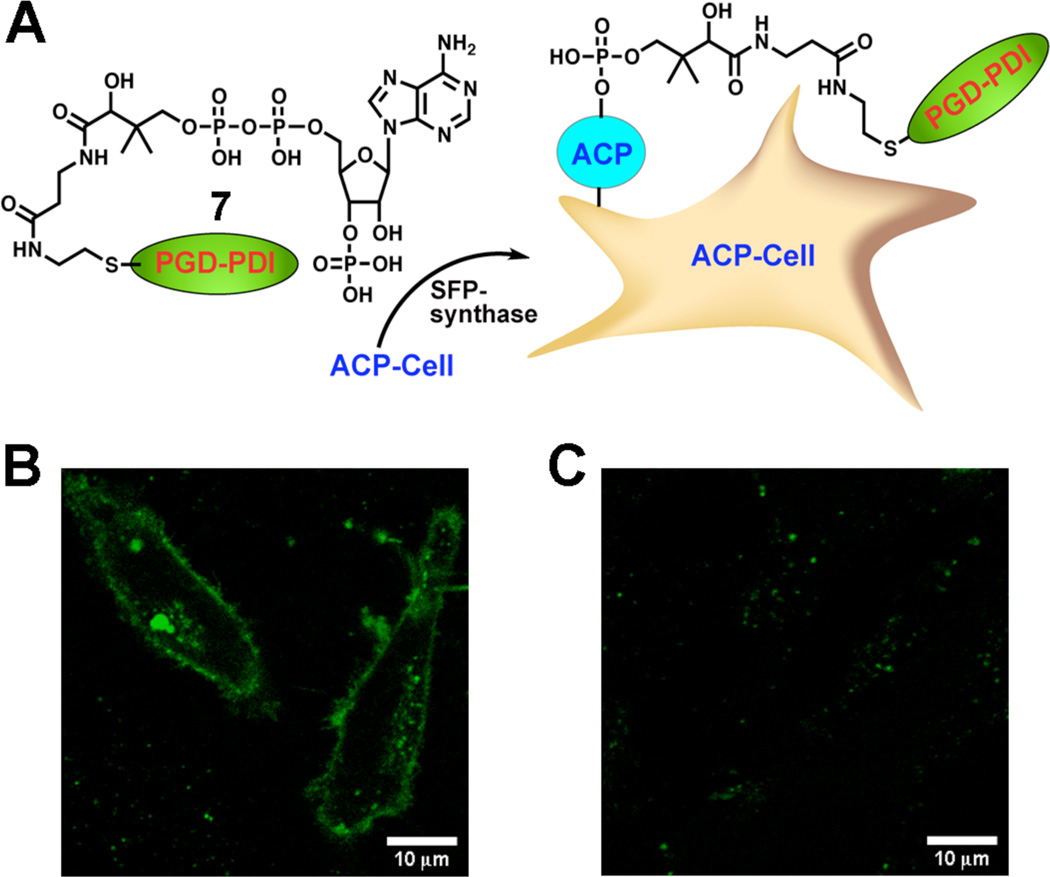
To further illustrate the usefulness of PGD-PDI in targeted biological imaging, fluorescent labeling of ACP-tagged fusion proteins on the surface of living mammalian cells was explored with PGD-PDI 7 synthesized by maleimide-thiol coupling between 5 and coenzyme A (CoA). This fusion protein contains a surface-localized glycosylphosphatidylinositol (GPI) anchor upstream of the ACP tag,14 exposing this tag on the outer surface of the plasma membrane. The ACP tag used here is based on the E. coli acyl carrier protein that can be conjugated to fluorophore derivatives of CoA in the presence of an enzyme named SFP synthase (Figure 6A).15 As seen in Figure 6B, PGD-PDI 7 clearly produced a bright, surface-localized fluorescence signal on live HeLa cells that were transiently transfected and incubated with SFP synthase. This labeling was also target-specific, as seen from the control experiments in which labeling with 7 in the absence of SFP synthase or transfection with the plasmid encoding the abovementioned fusion protein showed no apparent fluorescent labeling on the cell surface (Figure 6C and S10).
Figure 6.
Fluorescent labeling of GPI-ACP fusion proteins on the surface of living mammalian cells. (A) Scheme of enzymatic labeling. (B) Fluorescence image of HeLa cells labeled with PGD-PDI 7 after incubation in the presence of SFP synthase. (C) As a control, cells were labeled without SFP synthase.
The methodology for specific protein labeling can be generalized by employing PGD-PDI 5 functionalized with a single maleimide that enables efficient labeling of cysteine residues in proteins in a specific fashion. In this study, 5 was reacted with bovine serum albumin (BSA) which contains a free surface cysteine at amino acid 34, and the formation of PGD-PDI-BSA conjugates was confirmed by the MALDI mass spectrum of 6 showing two intense peaks corresponding to native BSA and PGD-PDI 6 at 66431 and 72400 m/z, respectively (Figure 2). Unreacted BSA was observed because partial oxidation at Cys-34 results in only ~50% of the cysteine residues being available for reaction.16 Quantification using Ellman’s assay17 revealed 46% and ~0% available cysteine sites on BSA before and after reaction with PGD-PDI 5, respectively, indicating quantitative labeling of BSA with 5. These results demonstrate the versatility of the mono-functional PGD-PDIs for efficient, specific protein labeling.
In conclusion, we have reported the synthesis of a series of highly water-soluble and fluorescent PGD-PDIs and the ability of a clickable monofunctional PGD-PDI to singly link biomolecules in a specific fashion. Single-molecule and live bacteria imaging were performed using a single biotinylated PGD-PDI. The key finding was the ability of uncharged, site-isolated PGD-PDIs for highly specific protein labeling on the surface of living bacteria cells that is not possible with previous ionic nondendronized PDI analogues. This approach using dendrimers to encapsulate dyes clearly enhances the performance of dyes and thereby opens up a new generation of biolabels where the design would couple a monofunctional fluorescent core with a multivalent periphery with cellular receptors, therapeutic agents, targeting groups, or ligands.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health and the National Science Foundation for financial support of this research and Kyung Suk Lee for helpful discussions on single-molecule data processing.
Footnotes
Supporting Information Available: Synthetic details, characterization data, experimental procedures and additional data for single-molecule and live bacteria imaging, and movies S1, S2, and S3 showing the images acquired at z-positions 1–6 in Figure S4, S6, and S8, respectively. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]; (b) Johnsson N, Johnsson K. ACS Chem. Biol. 2007;2:31–38. doi: 10.1021/cb6003977. [DOI] [PubMed] [Google Scholar]; (c) Lavis LD, Raines RT. ACS Chem. Biol. 2008;3:142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Aubin JE. J. Histochem. Cytochem. 1979;27:36–43. doi: 10.1177/27.1.220325. [DOI] [PubMed] [Google Scholar]; (b) Ballou B, Ernst LA, Waggoner AS. Curr. Med. Chem. 2005;12:795–805. doi: 10.2174/0929867053507324. [DOI] [PubMed] [Google Scholar]; (c) Kobayashi H, Ogawa M, Alford R, Choyke PL, Urano Y. Chem. Rev. 2010;110:2620–2640. doi: 10.1021/cr900263j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Langhals H. Chimia. 1994;48:503–505. [Google Scholar]; (b) Pasaogullari N, Icil H, Demuth M. Dyes Pigments. 2006;69:118–127. [Google Scholar]; (c) Weil T, Vosch T, Hofkens J, Peneva K, Müllen K. Angew. Chem., Int. Ed. 2010;49:9068–9093. doi: 10.1002/anie.200902532. [DOI] [PubMed] [Google Scholar]
- 4.(a) Langhals H. Heterocycles. 1995;40:477–500. [Google Scholar]; (b) Nagao Y. Prog. Org. Coatings. 1997;31:43–49. [Google Scholar]; (c) Würthner F. Chem. Commun. 2004:1564–1579. doi: 10.1039/b401630k. [DOI] [PubMed] [Google Scholar]; (d) Langhals H. Helv. Chim. Acta. 2005;88:1309–1343. [Google Scholar]; (e) Würthner F. Pure Appl. Chem. 2006;78:2341–2349. [Google Scholar]; (f) Micheli E, D’Ambrosio D, Franceschin M, Savino M. Mini-Rev. Med. Chem. 2009;9:1622–1632. doi: 10.2174/138955709791012274. [DOI] [PubMed] [Google Scholar]; (g) Avlasevich Y, Li C, Müllen K. J. Mater. Chem. 2010;20:3814–3826. [Google Scholar]
- 5.(a) Margineanu A, Hofkens J, Cotlet M, Habuchi S, Stefan A, Qu J, Kohl C, Müllen K, Vercammen J, Engelborghs Y, Gensch T, De Schryver FC. J. Phys. Chem. B. 2004;108:12242–12251. [Google Scholar]; (b) Qu J, Kohl C, Pottek M, Müllen K. Angew. Chem., Int. Ed. 2004;43:1528–1531. doi: 10.1002/anie.200353208. [DOI] [PubMed] [Google Scholar]; (c) Kohl C, Weil T, Qu J, Müllen K. Chem.-Eur. J. 2004;10:5297–5310. doi: 10.1002/chem.200400291. [DOI] [PubMed] [Google Scholar]; (d) Jung C, Müller BK, Lamb DC, Nolde F, Müllen K, Bräuchle C. J. Am. Chem. Soc. 2006;128:5283–5291. doi: 10.1021/ja0588104. [DOI] [PubMed] [Google Scholar]
- 6.(a) Abdalla MA, Bayer J, Rädler JO, Müllen K. Angew. Chem., Int. Ed. 2004;43:3967–3970. doi: 10.1002/anie.200353621. [DOI] [PubMed] [Google Scholar]; (b) Peneva K, Mihov G, Nolde F, Rocha S, Hotta J, Braeckmans K, Hofkens J, Uji-i H, Herrmann A, Müllen K. Angew. Chem., Int. Ed. 2008;47:3372–3375. doi: 10.1002/anie.200705409. [DOI] [PubMed] [Google Scholar]; (c) Peneva K, Mihov G, Herrmann A, Zarrabi N, Börsch M, Duncan TM, Müllen K. J. Am. Chem. Soc. 2008;130:5398–5399. doi: 10.1021/ja711322g. [DOI] [PubMed] [Google Scholar]; (d) Yin M, Shen J, Pflugfelder GO, Müllen K. J. Am. Chem. Soc. 2008;130:7806–7807. doi: 10.1021/ja8022362. [DOI] [PubMed] [Google Scholar]; (e) Yin M, Shen J, Gropeanu RA, Pflugfelder GO, Weil T, Müllen K. Small. 2008;4:894–898. doi: 10.1002/smll.200701107. [DOI] [PubMed] [Google Scholar]; (f) Cordes T, Vogelsang J, Anaya M, Spagnuolo C, Gietl A, Summerer W, Herrmann A, Müllen K, Tinnefeld P. J. Am. Chem. Soc. 2010;132:2404–2409. doi: 10.1021/ja9099714. [DOI] [PubMed] [Google Scholar]
- 7.Zill A, Rutz AL, Kohman RE, Alkilany AM, Murphy CJ, Kong H, Zimmerman SC. Chem. Commun. 2011;47:1279–1281. doi: 10.1039/c0cc04096g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heek T, Fasting C, Rest C, Zhang X, Würthner F, Haag R. Chem. Commun. 2010;46:1884–1886. doi: 10.1039/b923806a. [DOI] [PubMed] [Google Scholar]
- 9.Kolb HC, Finn MG, Sharpless KB. Angew. Chem., Int. Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 10.(a) Roy R, Hohng S, Ha T. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shi X, Lim J, Ha T. Anal. Chem. 2010;82:6132–6138. doi: 10.1021/ac1008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boos W, Shuman H. Microbiol. Mol. Biol. Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oddershede L, Dreyer JK, Grego S, Brown S, Berg-Sørensen K. Biophys. J. 2002;83:3152–3161. doi: 10.1016/S0006-3495(02)75318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbs KA, Isaac DD, Xu J, Hendrix RW, Silhavy TJ, Theriot JA. Mol. Microbiol. 2004;53:1771–1783. doi: 10.1111/j.1365-2958.2004.04242.x. [DOI] [PubMed] [Google Scholar]
- 14.Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schönle A, Hell SW. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 15.(a) Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. Chem. Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]; (b) George N, Pick H, Vogel H, Johnsson N, Johnsson K. J. Am. Chem. Soc. 2004;126:8896–8897. doi: 10.1021/ja048396s. [DOI] [PubMed] [Google Scholar]
- 16.Janatova J, Fuller JK, Hunter MJ. J. Biol. Chem. 1968;243:3612–3622. [PubMed] [Google Scholar]
- 17.Ellman GL. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.