Abstract
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third leading cause of cancer death worldwide. Despite recent advances in the diagnosis and treatment of HCC, its prognosis remains dismal. Infection with hepatitis B virus (HBV) and hepatitis C virus (HCV) are the major risk factors for HCC. Although both are hepatotropic viral infections, there are important differences between the oncogenic mechanisms of these two viruses. In addition to the oncogenic potential of its viral proteins, HBV, as a DNA virus, can integrate into host DNA and directly transform hepatocytes. In contrast, HCV, an RNA virus, is unable to integrate into the host genome, and viral protein expression has a more critical function in hepatocarcinogenesis. Both HBV and HCV proteins have been implicated in disrupting cellular signal transduction pathways that lead to unchecked cell growth. Most HCC develops in the cirrhotic liver, but the linkage between cirrhosis and HCC is likely multifactorial. In this review, we summarize current knowledge regarding the pathogenetic mechanisms of viral HCC.
Keywords: hepatocellular carcinoma, hepatitis B virus, hepatitis C virus, genomics, signaling pathways pathogenesis
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third leading cause of cancer death worldwide (El-Serag and Rudolph, 2007). In the year 1990, it was estimated that there were 4 37 408 cases of HCC worldwide (Parkin et al., 1999), but by the year 2000, new cases of HCC had increased to 5 64 300 (Parkin et al., 2001). More than 80% of these cases occur in developing countries, especially Southeast Asia and sub-Saharan Africa. However, the incidence of HCC has been rising in Western countries in recent years. In fact, HCC has become the fastest growing cause of cancer death in men in the United States, and age-adjusted HCC incidence rates increased more than twofold between 1985 and 2002 (El-Serag and Rudolph, 2007). The incidence and mortality rates of HCC in the United States are also expected to double over the next 10–20 years (El-Serag, 2002). Surgical resection or liver transplantation remains the most effective treatment options for HCC, but few patients are deemed suitable for operative intervention. Despite recent advances in the diagnosis and treatment of HCC, its prognosis remains dismal.
Infection with hepatitis B virus (HBV) and hepatitis C virus (HCV) is the major risk factor for HCC worldwide. Globally, up to 80% of HCC is attributable to HBV or HCV (Perz et al., 2006). The risk of HCC is increased 5- to 15-fold in chronic HBV carriers (El-Serag and Rudolph, 2007) and 11.5- to 17-fold in HCV-infected patients (Donato et al., 1998, 2002). Antiviral therapy is effective in preventing HCC in only a proportion of patients (Sorrell et al., 2009; Tai and Chung, 2009). Moreover, sustained clearance of HBV or HCV may be difficult to accomplish, particularly among cirrhotic patients. Thus, it is important to understand the molecular mechanisms underlying viral hepatocarcinogenesis, so as to maximize options to interrupt these pathways.
Molecular pathways in hepatocarcinogenesis
HCC is a highly heterogeneous tumor. Hepatocarcino-genesis is a complex multistep process involving a number of genetic and epigenetic alterations, the activation of cellular oncogenes and/or the inactivation of tumor suppressor genes, and dysregulation of multiple signal transduction pathways. These pathways include Wnt/β-catenin, p53, pRb, Ras, mitogen-activated protein kinase (MAPK), Janus kinase (JAK)/signal transducer and activator of transcription (STAT), phosphatidylinositol 3-kinase (PI3K)/Akt, Hedgehog and growth factors such as epidermal growth factor, and transforming growth factor-β (TGF-β) pathways (Branda and Wands 2006; Aravalli et al., 2008; Llovet and Bruix, 2008a).
Fibrosis, cirrhosis and HCC
Of all HCCs, 80–90% develop in a cirrhotic liver (Caldwell and Park, 2009). After 20–30 years of chronic infection, 20–30% of patients develop liver cirrhosis. HCC develops at an annual rate of 1–7% in HCV-infected cirrhotic patients (El-Serag and Rudolph, 2007) and 3–8% in HBV-infected cirrhotic patients (Fattovich, 2003).
During the progression of liver injury, hepatic stellate cells (HSCs) become activated, losing retinoid-containing lipid droplets and transforming into myofibroblast-like cells, which produce extracellular matrix (Okuno et al., 2002), the first step in hepatic fibrosis. Unchecked progression of fibrosis ultimately eventuates in irreversible cirrhosis. The activated HSCs become responsive to both proliferative (platelet-derived growth factor (PDGF)]) (Friedman and Arthur, 1989; Pinzani et al., 1996) and fibrogenic (TGF-β) (Matsuzaki, 2009) cytokines, which are upregulated in fibrogenesis and modulate inflammatory signaling from infiltrating immune cells (Parsons et al., 2007). PDGF can activate both MAPK and PI3K/Akt signaling cascades (Parsons et al., 2007). In PDGF-C transgenic mice, activation and proliferation of HSCs precedes development of fibrosis, which in turn is followed by the occurrence of HCC. This progression is analogous to that seen in human HCC (Campbell et al., 2005). The cirrhotic liver is also associated with telomere shortening, which may in turn lead to chromosomal instability and deletion of checkpoints (Satyanarayana et al., 2004). The decreased liver reserve in cirrhosis may also increase accumulation of toxic metabolites, which could also possibly increase the risk of HCC. Increased survival factors that prevented apoptosis of DNA-damaged hepatocytes and activated stellate cells (for example, Gas6215) and reduced tumor surveillance function due to decreased natural killer cell function are all possible factors related to HCC development in cirrhosis (Friedman, 2008). Recent studies have found that stellate cells express stem cell markers such as CD133, nestin, c-kit and p75 neurotrophin receptor (Fujio et al., 1994; Niki et al., 1999; Cassiman et al., 2001; Kordes et al., 2007), and activated stellate cells appear to contribute to the stem cell niche (Roskams 2006; Gaudio et al., 2009). Hedgehog and Wnt signaling pathways involved in stem cell differentiation and cancer formation are also found in stellate cells (Myung et al., 2007; Yang et al., 2008). These lines of evidences suggest that stellate cells may harbor the potential to transdifferentiate into progenitor cells and possibly be linked to the development of HCC (Friedman, 2008).
HBV hepatocarcinogenesis
Virology
HBV belongs to the Hepadnavirus family. The HBV virion, which is 42 nm in diameter, is composed of an outer envelope formed by hepatitis B surface antigen. This envelope surrounds an inner nucleocapsid, the hepatitis B core antigen, that packages the viral genome and polymerase. The HBV genome is a partially double-stranded relaxed circular DNA molecule about 3200 nt in length. The minus strand contains four partially overlapping open-reading frames encoding the envelope (pre-S/S), core (pre-C/C), polymerase (P) and X proteins (X). After attachment to the hepatocyte, the HBV genome moves to the nucleus and assumes a highly stable conformation, called covalently closed circular DNA. Covalently closed circular DNA serves as a template for transcription of viral RNA, which is translated into viral proteins. The HBV polymerase, which reverse transcribes and replicates HBV DNA, lacks proofreading ability, and is therefore prone to generate mutations. Common mutations include the precore (G1896A) mutation, basal core promoter mutations (A1762T/G1764A) and deletion mutations of pre-S/S genes (Locarnini et al., 2003). On the basis of genomic sequence divergence, HBV can be classified into eight genotypes, A to H (Schaefer, 2005).
HBV DNA integration
HBV genomic integration is present in over 85–90% of HBV-related HCCs and usually precedes the development of HCC. The integration of HBV DNA is not restricted to HCC but also is found in non-tumor tissue of patients with chronic HBV (Brechot et al., 1980; Shafritz et al., 1981). Exposure to oxidative stress or agents that cause DNA damage increases the frequency of HBV integration (Dandri et al., 2002). During chronic inflammation, enhanced DNA replication and DNA damage also promote the process of viral integration (Guerrero and Roberts, 2005).
HBV integration induces a wide range of genetic alterations within the host genome, including chromosomal deletions, translocations, production of fusion transcripts, amplification of cellular DNA and generalized genomic instability (Guerrero and Roberts, 2005; Feitelson and Lee, 2007). Many integrated events occur near or within fragile sites or other cancer-associated regions of the human genome that are prone to instability in tumor development and progression. Genetic instability associated with integration may alter the expression of oncogenes, tumor suppressor genes and microRNAs (Feitelson and Lee, 2007). A recent large-scale analysis of HBV DNA integration sites in cellular DNA found a preference at sites regulating cell signaling, proliferation and viability (Murakami et al., 2005). Common gene targets of integration include the human cyclin A2 gene (Wang et al., 1992), the retinoic acid receptor β gene (Yaginuma et al., 1987), human telomerase reverse transcriptase (Horikawa and Barrett, 2001), PDGF receptor, calcium signaling-related genes, mixed lineage leukemia and 60S ribosomal protein genes (Murakami et al., 2005). A large proportion of HCCs have integrated HBV sequences encoding HBV X (HBx) and/or truncated envelope pre-S2/S proteins, which both contribute to hepatocarcinogenesis.
HBV proteins
HBx protein
HBx is a protein of 154 amino acids that increases virus gene expression and replication by transactivating cellular promoter and enhancers important for persistent viral infection (Keasler et al., 2007; Feitelson et al., 2009). Integration of the HBx sequence into host DNA is a common event in HCC. After integration, HBx promotes genetic instability through a variety of mechanisms. HBx compromises nucleotide excision repair by inactivating the UV-damaged DNA binding protein, inactivating p53 and disrupting the association of p53 to excision repair cross complementing factor 3 and the transcription factor IIH transcription factors, XPB and XPD (Wang et al., 1994; Jia et al., 1999).
HBx transactivates several cytoplasmic signaling pathways, including protein kinase C (PKC), JAK/STAT, PI3K, stress-activated protein kinase (SAPK)/c-Jun NH2-terminal kinase (JNK), ras-raf-MAPK, activator protein-1 (AP-1), AP-2, nuclear factor-κB (NF-κB), Smad and Wnt, and by binding to nuclear transcription factors, including cAMP response element binding (CREB), activating transcription factor 2 (ATF-2), Oct-1 and TATA box binding protein (TBP) (Feitelson and Lee, 2007; Feitelson et al., 2009) (Figure 1).
Figure 1.
Cellular signaling pathways implicated in hepatitis B virus (HBV) X protein-related hepatocarcinogenesis. Bolded boxes indicate key driving forces for carcinogenesis.
HBx expression also increases cytosolic calcium levels and then activates the Ca2+-dependent proline-rich tyrosine kinase 2 (Bouchard et al., 2001; Oh et al., 2003), which in turn activates Src kinase (Lev et al., 1995). Src increases association of Shc-Grb2-Sos, then activates Ras and the Ras-Raf-ERK and Ras-MEKK-JNK cascades and induces transcription of genes of the AP-1 family, such as c-fos and c-jun (Benn and Schneider, 1994; Oh et al., 2003). HBx also activates NF-κBina Ras-dependent manner (Su and Schneider, 1996).
HBx also decreases proteasomal degradation of β-catenin; there is a significant correlation between HBx expression and β-catenin accumulation in HCC tissue (Ding et al., 2005). HBx also increases the downstream targets of β-catenin, c-myc and cyclin D-1. Pin1, a Wnt signal regulator, can interact with a specific serine-proline motif of HBx protein and overexpression of Pin1 increases the protein stability of HBx. HBx-mediated transactivation is also enhanced by co-expression of Pin1 (Pang et al., 2007).
HBx causes transcriptional repression of the p53 gene (Lee and Rho, 2000) and interacts with p53 to inactivate several important p53-dependent activities, including apoptosis, cell-cycle regulation, DNA repair and tumor suppression (Kremsdorf et al., 2006).
TGF-β is an important mediator of fibrosis and apoptosis. TGF-β expression is upregulated by HBx in HCC tissue (Yoo et al., 1996). TGF-β signals through phosphorylation of Smad3 at its middle linker and/or C-terminal regions. Reversible shifting of Smad3-dependent signaling between tumor suppression and oncogenesis indicates that Smad3 phosphorylated at the C-terminal region (pSmad3C) transmits a tumor-suppressive TGF-β signal, whereas oncogenic activities such as cell proliferation and invasion are promoted by Smad3 phosphorylated at the linker region (pSmad3L) (Matsuzaki, 2009). HBx shifts hepatocytic TGF-β signaling from the tumor-suppressive pSmad3C pathway to the oncogenic pSmad3L pathway in the early carcinogenic process (Murata et al., 2009). A recent study found that the HBx protein induces paracrine activation of human HSCs and anti-TGF-β blocks these paracrine effects (Martıń-Vıĺchez et al., 2008).
Steatosis is also an important cofactor for chronic liver disease and HCC. A recent study found that HBx induces hepatic steatosis by transcriptional activation of SREBP1 and PPAR-γ (Kim et al., 2007). Liver X receptor was found to mediate HBx protein-induced lipogenesis in HCC (Kim et al., 2008; Na et al., 2009).
Other potential mechanisms of HBx hepatocarcinogenesis include inhibition of Fas-mediated apoptosis (Diao et al., 2001), induction of genetic instability by binding to damaged DNA binding protein 1 (Martin-Lluesma et al., 2008), modulation of the degradation of cellular and viral proteins through inhibition of serine protease inhibitors and proteasome complexes (Sirma et al., 1998), induction of oxidative stress and mitochondrial translocation of Raf-1 kinase (Chen and Siddiqui, 2007), overexpression of insulin receptor substrate-1 (Longato et al., 2009), upregulation of beclin 1 and a corresponding increase in starvation induced autophagy (Tang et al., 2006, 2009), interaction with DNA methyltransferase (DNMT) with aberrant epigenetic modifications (Park et al., 2007; Zheng et al., 2009) and survival benefit under oxidative stress (Severi et al., 2007).
Pre-S/S protein
The truncated form of the pre-S2/S gene is commonly found from deleted integrated viral genomes (Tai et al., 2002). The pre-S2/S product also has transactivational abilities. These proteins transactivate cellular genes, including c-myc, c-fos and c-Haras (Schluter et al., 1994; Luber et al., 1996). A recent study found that pre-S2 may increase malignant transformation of human HCC cell lines by upregulating human telomerase reverse transcriptase and inducing telomerase activation (Liu et al., 2007). The pre-S2 activator activates PKC and causes phosphorylation of the pre-S domain, then activates the c-Raf-1/MEK/ERK signal transduction cascades (Hildt et al., 2002).
The pre-S mutant large surface antigens can activate endoplasmic reticulum (ER) stress to induce oxidative DNA damage and genomic instability (Wang et al., 2006). The pre-S mutant also can upregulate cyclooxygenase-2 and cyclin A to induce cell-cycle progression and proliferation of hepatocytes (Wang et al., 2006). A recent study found that vascular endothelial growth factor-A (VEGF-A) is upregulated by pre-S mutants and that pre-S mutant-expressed Huh-7 cells exhibited activation of Akt/mTOR (mammalian target of rapamycin) signaling and increased growth advantage, which could be inhibited by VEGF-A neutralization (Yang et al., 2009).
HBV genotype, mutations and viral load
HBV genotypes have distinct geographical and ethnic distributions (Kidd-Ljunggren et al., 2002; Liu et al., 2005). Genotype C infection is associated with delayed HBe seroconversion, more severe liver damage and liver cirrhosis than genotype B (Kao et al., 2000; Chu et al., 2002). Longitudinal and cross-sectional studies have shown that genotype C is significantly more common than genotype B in HCCs (Fujie et al., 2001; Chan et al., 2004; Yu et al., 2005; Yang et al., 2008). A recent prospective study disclosed that HBV genotype C, particularly subgenotype Ce, increased the risk of HCC in chronic hepatitis B (Chan et al., 2008). In a study from India, the prevalence of genotype D was higher in patients with HCC younger than 40 years of age (Thakur et al., 2002). In Alaska, genotype F is associated with the development of HCC (Livingston et al., 2007).
HBV basal core promoter mutations (A1762T/G1764A) are found to be associated with the occurrence of HCC in several cross-sectional (Baptista et al., 1999; Kao et al., 2003) and longitudinal studies (Chou et al., 2008; Fang et al., 2008; Wu et al., 2008; Yuan et al., 2009). The REVEAL-HBV study from Taiwan found that the multivariable-adjusted hazard ratio of developing HCC was 1.73 for basal core promoter mutations and that the risk was highest among participants infected with genotype C HBV and who harbored the precore 1896 variant and mutations for the basal core promoter (Yang et al., 2008).
Several recent studies suggested that HBV viral load is associated with HCC (Yu et al., 2005; Chen et al., 2006, 2009; Chan et al., 2008; Wu et al., 2008). The REVEAL-HBV study found that after adjusting for age, sex, smoking, alcohol use, HBeAg status, ALT level and cirrhosis, the risk of HCC was approximately six times higher for persons with viral load above 105 copies per ml compared with persons with undetectable viral load (Chen et al., 2006). HBV viral load is also closely related to the development of cirrhosis (Iloeje et al., 2006).
HBV animal models
The eastern woodchuck harbors the woodchuck hepatitis virus, which is similar to HBV in structure and life cycle. Woodchuck hepatitis virus can infect the liver, cause acute or chronic hepatitis and lead to HCC development after 2–4 years (Tennant et al., 2004). However, when ducks are infected with duck hepatitis B virus, the risk of HCC development is low. This may be because duck hepatitis B virus does not have an HBx gene, supporting a function for HBx in hepatocarcinogenesis.
In HBV transgenic mouse models, only the large envelope and HBx proteins are closely related to HCC development (Leenders et al., 2008). Chisari et al. (1989) developed a transgenic mice model carrying the integrated HBV large envelope polypeptides on a C57BL/6 genetic background. Toxic quantities of hepatitis B surface antigen accumulate within the hepatocyte and develop hepatocellular injury characterized by inflammation, regenerative hyperplasia, transcriptional deregulation and aneuploidy, which eventually leads to HCC development. Kim et al. (1991) developed an HBx transgenic mouse model using microinjection of HBV DNA containing the HBx gene into single-cell embryos from CD1 mice. On autopsy, 80–91% of male and 60–67% of female mice developed HCC. However, outcomes in other HBx transgenic mice have been variable. This may be attributable to differences in mouse strain, transgene sequences and integration site (Leenders et al., 2008) (Table 1).
Table 1.
Common viral transgenic mouse models of hepatocellular carcinoma
| Viral protein | Promoter/mouse strain | Phenotype | References |
|---|---|---|---|
| HBV large envelope protein | Albumin/C57BL/6 | Inflammation, regenerative hyperplasia, transcriptional deregulation, aneuploidy, HCC in 72% male and 31% female, 72% of male HCC occurred at 16–21 months | Chisari et al., 1989 |
| HBV X protein | HBV X promoter/CD1 | No obvious cell death or regeneration, DNA synthesis induced, HCC in 84% at mean age of 16.7 months | Koike et al., 1994 |
| HBV X protein | HBV X promoter/C57BL/6 | No inflammation, no cirrhotic change, HCC in 64% at 11–18 months | Yu et al., 1999 |
| Core | HBV/C57BL/6 | Steatosis, oxidative stress, HCC in 25.9–38% at 16–19 months | Moriya et al., 1998, 2001; Tanaka et al., 2008a,b |
| Core-E1–E2 | Albumin/C57BL/6 | Steatosis, HCC in 2.3% at age > 13 months | Lerat et al., 2002 |
| Polyprotein | Albumin/C57BL/6 | Steatosis, HCC in 13.2% at age > 13 months | Lerat et al., 2002 |
| Core-E1–E2 | Albumin/FVB × C57BL/6 | Anti-apoptotic, DEN treated: 100% HCC at 32 weeks; HCV core-E1–E2 with largest tumors | Kamegaya et al., 2005 |
| Core-E1–E2 | CMV/C57BL/6 | Steatosis, HCC in 3.7% after 1 year of age | Naas et al., 2005 |
Abbreviations: HBV, hepatitis B virus; HBV X, hepatitis B virus X; HCC, hepatocellular carcinoma; DEN, diethylnitrosamine.
HCV hepatocarcinogenesis
Virology
HCV belongs to the Flaviviridae family. Its genome is a 9.6-kb uncapped linear positive-stranded RNA. It contains 5′ and 3′ untranslated regions including control elements required for translation and replication. The untranslated regions flank an uninterrupted open-reading frame encoding a single polyprotein of 3010 or 3011 amino acids, which is processed into structural (core, envelope glycoproteins E1 and E2) and nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, NS5B) proteins by host and viral proteases (Chisari 2005; Lindenbach and Rice, 2005).
Enveloped virus particles interact with specific surface receptors and are probably internalized. Fusion of the viral and cellular membranes leads to the release of a single-stranded, positive-sense RNA genome into the cytoplasm. This genome serves as a messenger RNA for translation of the viral proteins, as a template for RNA replication, and as a nascent genome packaged into new virus particles (Lindenbach and Rice, 2005; Moradpour et al., 2007).
HCV proteins
HCV is an RNA virus unable to reverse transcribe its genome and thus to integrate it into the host genome. Instead, viral proteins and their evoked host responses contribute mostly to the viral oncogenic processes.
Core protein
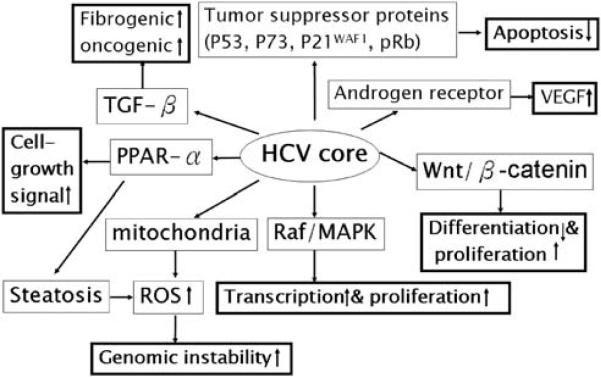
HCV core protein appears to have a diverse range of functions and interacts with many cellular proteins. HCV core protein has been proposed to be involved in apoptosis, signal transduction, reactive oxygen species (ROS) formation, lipid metabolism, transcriptional activation, transformation and immune modulation (Anzola, 2004; Irshad and Dhar, 2006) (Figure 2).
Figure 2.
Cellular signaling pathways implicated in hepatitis C virus (HCV) core protein-related hepatocarcinogenesis. Bolded boxes indicate key driving forces for carcinogenesis.
HCV core protein binds to several tumor suppressor proteins, including p53, p73 and pRb (Ray et al., 1997; Cho et al., 2001). HCV core interacts with p73, causes nuclear translocation of core protein and prevents p73-α-dependent cell growth arrest in a p53-dependent manner (Alisi et al., 2003; Benard et al., 2003). HCV core can also modulate the expression of the cyclin-dependent inhibitor p21WAF1, which is a major target of p53 and regulates the activities of cyclin/cyclin-dependent kinase complexes involved in cell-cycle control and tumor formation (Yamanaka et al., 2002; Kwun and Jang, 2003).
A microarray analysis study revealed threefold or more transcriptional changes in 372 of 12 500 known human genes in core-expressing Huh-7 cells, with most genes involved in cell growth or oncogenic signaling (Fukutomi et al., 2005). Of particular interest is the marked upregulation of both Wnt-1 and its downstream target gene WISP-2. Small-interfering RNA against Wnt-1 blunted growth stimulation by HCV core and conditioned medium from Wnt-1 transfected cells accelerated cell growth. HCV core downregulates E-cadherin expression at the transcriptional level. This effect is strongly correlated with hypermethylation of CpG islands of the E-cadherin promoter through concerted action of both DNMT1 and 3b and is abolished by a specific inhibitor of DNMT (Arora et al., 2008).
HCV core also upregulates the expression of TGF-β (Taniguchi et al., 2004; Shin et al., 2005). As HCV-infected livers progress from chronic hepatitis through cirrhosis to HCC, hepatocytic pSmad3L/PAI-1 increases with fibrotic stage and necroinflammatory grade, and pSmad3C/p21WAF1 decreases. These results indicate that chronic inflammation associated with HCV infection shifts hepatocytic TGF-β signaling from tumor suppression to fibrogenesis, accelerating liver fibrosis and increasing the risk of HCC (Matsuzaki et al., 2007). Another study showed that different thresholds of Smad3 activation dictate TGF-β responses in hepatocytes and that liver cancer-derived HCV core protein, by decreasing Smad3 activation, switches TGF-β growth inhibitory effects to tumor-promoting responses (Battaglia et al., 2009). A recent study found that HCV core triggers the production of both TGF-β2 and VEGF proteins through multiple pathways, including PKC, RB/E2F1, ASK1-JNK/p38 and ERK (Hassan et al., 2009). HCV core protein also behaves as a positive regulator in androgen receptor signaling and enhances the expression of VEGF in hepatocytes (Kanda et al., 2008).
HCV core protein can activate the Raf/MAPK signal pathway (Hayashi et al., 2000; Erhardt et al., 2002), has immunosuppressive activities through its interaction with the complement receptor C1qR on T cells (Kittlesen et al., 2000) and can interact with numerous transcription factors, including heterogeneous nuclear ribonucleoprotein K, leucine zipper transcription factor, 14-3-3 protein and RNA helicase CAP-Rf (Hsieh et al., 1998; You et al., 1999; Aoki et al., 2000; Jin et al., 2000). HCV core proteins also contribute to oxidative and ER stress and steatosis (see below).
HCV envelope protein
The HCV envelope protein E2 exerts an inhibitory effect on natural killer cells through engagement of CD81 (Crotta et al., 2002). E2 also activates the MAPK/ERK pathway, including the downstream transcription factor ATF-2, and maintains survival and growth of target cells (Zhao et al., 2005).
HCV nonstructural proteins
HCV NS3 protein
HCV NS3 protein may exert its hepatocarcinogenic effect on host cells in early stages (Sakamuro et al., 1995; Zemel et al., 2001). NS3 inhibits the activity of the p21WAF1 promoter in a dose-dependent manner and is synergistic with core in this regard (Kwun et al., 2001). NS3 inhibits the function of p53 in an NS3 sequence in an NS3 sequence-dependent manner (Deng et al., 2006).
The expression of NS3 enhances cell growth, JNK activation and DNA-binding activities of the transcription factors AP-1 and ATF-2 (Hassan et al., 2005). NS3 also induces TNF-α production by activation of AP-1 and NF-κB (Hassan et al., 2007).
HCV NS5A protein
NS5A is involved in a large number of cellular functions, including apoptosis, signal transduction, transcription, transformation and ROS production. NS5A binds directly to p53, inhibits transcriptional transactivation by p53 in a dose-dependent manner and inhibits gene transcription of p21WAF1 (Majumder et al., 2001; Lan et al., 2002). NS5A interacts with and partially sequesters p53 and hTAF(II)32, a component of TFIID and an essential coactivator of p53, and suppresses p53-mediated transcriptional transactivation and apoptosis during HCV infection (Lan et al., 2002). NS5A protein forms complexes with the TBP and p53 and inhibits the binding of both p53 and TBP to their DNA consensus binding sequences in vitro. It also inhibits the p53-TBP and p53-excision repair cross complementing factor 3 protein–protein complex formation (Qadri et al., 2002). NS5A also interacts with Bax as a Bcl-2 homologue and prevents apoptosis in a p53-independent manner (Chung et al., 2003).
NS5A prevents TNF-α-mediated apoptotic cell death by blocking the activation of caspase-3 and inhibiting proteolytic cleavage of the death substrate, poly (ADP-ribose) polymerase (Ghosh et al., 2000). NS5A can also activate NF-κB and lead to anti-apoptotic activity (Bonte et al., 2004).
NS5A interacts with growth-factor-receptor-bound protein 2 to inhibit mitogenic signaling while simultaneously promoting the PI3K/Akt cell survival pathway by interaction with p85 PI3K (He et al., 2002). NS5A expression in the context of HCV polyprotein inhibits the Akt substrate Forkhead transcription factor and stimulates the phosphorylation of glycogen synthase kinase-3β, leading to stabilization of cellular β-catenin and stimulation of β-catenin-responsive transcription (Street et al., 2005).
NS5A can also modulate TGF-β signaling through interaction with TGF-β receptor I (Choi and Hwang, 2006). Another recent study found that NS5A protein downregulates the expression of the mitotic spindle protein ASPM through the PKR-p38 signaling pathway and induces aberrant mitoses, chromosome instability and HCC (Wu et al., 2008).
HCV, ER and oxidative stress
Chronic HCV infection is characterized by increased oxidative stress. Lipid peroxidation and oxidative DNA damage are enhanced in serum and liver specimens of patients with HCV infection (Sumida et al., 2000; Mahmood et al., 2004). Mitochondrial dysfunction, ER stress and immune-cell-mediated oxidative bursts contribute to HCV-associated oxidative stress (Wang and Weinman, 2006). The increased oxidative stress may be accounted for by NAD(P)H oxidase, especially Nox-2 in PMNs and Kupffer cells in the liver (Takeya and Sumimoto, 2006). Both structural and nonstructural proteins of HCV can contribute to the generation of ROS. HCV NS5A protein induces ER stress and Ca2+ release, resulting in increased Ca2+ uptake and ROS generation by mitochondria, which has been linked to the activation of NF-κB and STAT-3 (Gong et al., 2001; Tardif et al., 2002). Cellular responses to oxidative stress in NS5A-expressing cells are regulated by both p38 MAPK and JNK by AP-1 (Qadri et al., 2004). NS3 has been shown to activate Nox2, which generates ROS (Bureau et al., 2001). Expression of HCV core uniformly increases ROS and also increases lipid peroxidation products and induces antioxidant gene expression (Okuda et al., 2002). HCV core gene expression decreases intracellular/mitochondrial GSH levels and mitochondrial NADPH levels, which are accompanied by increased Ca2+ uptake as well as ROS generation at complex I in mitochondria (Korenaga et al., 2005). Core protein has also been shown to induce ER stress and modulate the production of cytokines and host enzymes that can increase ROS, such as inducible nitric oxide synthase and cyclooxygenase-2 (de Lucas et al., 2003; Nunez et al., 2004; Benali-Furet et al., 2005). A recent study found that HCV triggers the mitochondrial permeability transition with production of ROS, leading to induction of double-stranded DNA breaks and STAT-3 activation (Machida et al., 2006). Autophagy may also function as a survival mechanism in liver cancer cells (Longo et al., 2008), and HCV also induces an incomplete autophagic response. Autophagy also appears to be important in HCV pathogenesis (Sir et al., 2008).
By altering or participating in different signaling pathways, ROS may modulate gene expression, cell adhesion, cell metabolism, cell cycle and cell death and induce oxidative DNA damage, which in turn increases chromosomal aberrations associated with cell transformation (Choi and Ou, 2006). ROS may also activate cellular signal pathways including the MAPK, NF-κB, PI3K, p53, β-catenin/Wnt and angiogenesis signaling pathways (Martindale and Holbrook, 2002; Tien Kuo and Savaraj, 2006; Czaja, 2007). We have found that HCV increases ROS production and TGF-β1 expression in the JFH1 HCV infection model and that HCV-mediated TGF-β1 enhancement occurs through an ROS-regulated and p38 MAPK-, JNK-, MEK- and NF-κB-dependent pathway (Lin et al., unpublished data).
HCV and steatosis
The prevalence of steatosis in patients with chronic HCV infection is 30–70% (Lonardo et al., 2004). Steatosis is associated with worsening fibrosis in chronic HCV infection (Hourigan et al., 1999; Adinolfi et al., 2001). In patients with HCV-related cirrhosis, steatosis is independently associated with the development of HCC (Ohata et al., 2003; Pekow et al., 2007).
In HCV transgenic mice that develop HCC, carcinogenesis is preceded by development of steatosis (Moriya et al., 1998). In addition, steatosis may induce chronic hepatic inflammation, ROS and DNA damage in animal models (Moriya et al., 2001; Okuda et al., 2002; Arkan et al., 2005; Cai et al., 2005; Koike, 2007). Core protein has been found to interact with retinoid X receptor-α (Tsutsumi et al., 2002). A dominant-negative retinoic acid receptor causes steatohepatitis and liver tumors in transgenic mice (Yanagitani et al., 2004). HCV core protein induces spontaneous and persistent activation of PPAR-αa in transgenic mice (Tanaka et al., 2008a). A recent transgenic mouse study using PPAR-α-homozygous, heterozygous and null mice with liver-specific transgenic expression of the core protein gene found that severe steatosis was observed only in PPAR +/+:HCVcoreTg mice and HCC developed in 35% of 24-month-old PPAR +/+:HCVcoreTg mice, but not in the other genotypes (Tanaka et al., 2008b). Long-term treatment of these mice with clofibrate, a PPAR-α activator, induced HCC with mitochondrial abnormalities and hepatic steatosis. The core protein induces steatosis by several pathways, leading to `fatty acid spiral' in the presence of the mitochondrial complex 1 dysfunction and PPAR-α activation. These intracellular alterations collectively contribute to hepatocarcinogenesis by inducing oxidative stress overproduction and cell growth signal activation (Koike, 2009).
HCV and iron
Elevated serum iron and increased hepatic iron deposition are relatively common in patients with chronic HCV (Kato et al., 2007). However, recent studies have shown that excess hepatic iron accumulation in patients with HCV contributes to liver injury and fibrosis progression (Bassett et al., 1999; Kato et al., 2007; Missiha et al., 2008). Free iron increases oxidative stress and leads to lipid peroxidation. Cytotoxic by-products of lipid peroxidation can impair cellular function, protein synthesis and cause DNA damage (Kew, 2009). Transgenic mice expressing the HCV polyprotein fed an excess-iron diet had greater hepatic content of lipid peroxidation products and 8-hydroxy-2′-deoxyguanosine and increased the risk of HCC development (Furutani et al., 2006). A recent study found that retinoids can suppress iron-induced oxidative stress of the liver by regulating hemojuvelin expression (Tsuchiya et al., 2009).
HCV animal models
Various HCV gene products have been expressed either alone or together in transgenic mice using different liver-specific promoters (Barth et al., 2008; Newell et al., 2008) (Table 1). Among these lines, only the transgenic mice expressing the core gene developed HCC (Moriya et al., 1998). Moriya et al. described a transgenic mouse strain expressing HCV core protein under control of the HBV promoter. These mice developed steatosis without inflammation after 3 months and HCC in 26–31% of male transgenic mice after 16–19 months (Moriya et al., 1998). The same study found no carcinoma in transgenic mice overexpressing HCV envelope genes. Our recent study disclosed that transgenic mice expressing core, E1, E2 proteins treated with diethylnitrosamine were found to have an accelerated tumor growth than mice expressing core protein only and the accelerated tumor phenotype is attributable to suppression of apoptosis rather than enhanced proliferation (Kamegaya et al., 2005). The transgenic mice expressing the complete HCV polyprotein were found to have an increased risk of liver tumor development than mice expressing structural proteins only (Lerat et al., 2002). However, the transgenic mice expressing HCV nonstructural proteins only do not cause any spontaneous liver pathology (Majumder et al., 2003). Core protein continues to have the most important function in the hepatocarcinogenesis of HCV.
HBV, HCV and host genomics
Several studies have reported genomic alterations in HBV-related HCC (Villanueva et al., 2007). Gain of 11q and 2q and loss of 1p, 16p, 16q and 17p were found to be associated with HBV-related HCC (Lin et al., 1999; Zondervan et al., 2000; Kusano et al., 2002; Katoh et al., 2005) (Table 2). A summary of 428 different comparative genomic hybridization analyses of human HCCs found that losses at 4q (43.4 vs 19.6%), 16q (41.8 vs 18.5%), 13q (31.1 vs 19.6%) and 8p (40.6 vs 29.3%) were correlated with HBV etiology (Moinzadeh et al., 2005). Another comparative genomic hybridization study of 158 HBV-related HCCs found that gains of 1q21–23 and 8q22–24 were associated with the early development of HCC; gain of 3q22–24 was associated with tumor recurrence and poor survival (Poon et al., 2006). A recent genome-wide analysis of gene expression in human HCC found that expression of UGT1A1, UGT2B10 and GPX2 was preferentially repressed in HBV-HCCs, but were unchanged in most other HCVHCCs (Okabe et al., 2001) (Table 3). Viral factors are also involved in the methylation of some genes. A recent study disclosed that E-cadherin and GSTP1 were preferentially methylated in HBV-HCC compared with HCV-HCC (Su et al., 2007). TNF-α-863A genotype, IL-1B-511C allele, C-A haplotype of IL-10 gene and XRCC1 Gln allele at codon 399, −509C→T polymorphism in the TGF-β1 gene promoter are all associated with HCC in chronic HBV carriers (Shin et al., 2003; Yu et al., 2003; Hirankarn et al., 2006; Kummee et al., 2007; Qi et al., 2009).
Table 2.
Chromosomal aberrations in viral-hepatocellular carcinoma
| Gain | Loss | References | |
|---|---|---|---|
| HBV vs non-HBV | 2q (50 vs 9.5%) | 16q (57 vs 8.3%) | Lin et al., 1999 |
| 4p (P = 0.007), 16q (P = 0.04), 17p (P = 0.04), 18q (P = 0.03), 1P (50 vs 9.5%) | Zondervan et al., 2000 | ||
| 16p (45 vs 4.8%), 16q (75 vs 23.8%), 17p (70 vs 23.8%), 4q (43.4 vs 19.6%) | Katoh et al., 2005 | ||
| 16q (41.8 vs 18.5%), 13q (31.1 vs 19.6%), 8p (40.6 vs 29.3%) | Moinzadeh et al., 2005 | ||
| LOH on 13q and 16q (p <.001) | Okabe et al., 2000 | ||
| HBV vs HCV | 11q13 (36 vs 3%) | 10q (26 vs 0%) | Kusano et al., 1999 |
| HBV (early vs advanced tumors) | 1q21–q22 (61.6 vs 77.8%), 3q22–q28 (6.8 vs 29.6%), 7q21–q22 (26.0 vs 48.1%), 7q34–q36 (16.4 vs 40.7%) | NR | Sy et al., 2005 |
| HBV (early development of HCC) | 1q21–23 (P <0.001), 8q22–24 (P <0.001) | NR | Poon et al., 2006 |
| HBV (tumor recurrence and poor survival) | 3q22–24(P = 0.039) | NR | Poon et al., 2006 |
| Non-HCV vs HCV | NR | 8p (36 vs 20%) | Moinzadeh et al., 2005 |
Abbreviations: HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; NR, not reported.
Table 3.
Gene expression analysis in HBV- and HCV-related hepatocellular carcinoma
| Gene expression analysis | HBV | HCV | References |
|---|---|---|---|
| UGT1A1, UGT2B10 and GPX2 | Repressed | No change | Okabe et al., 2001 |
| Genes encoding phase I enzymes CYP2E, AKR1C4, EPHX1 and FMO3 | NR | Exclusively in HCV | Okabe et al., 2001 |
| Imprinted genes (H19 and IGF-2) | Upregulated compared to HCV | Iizuka et al., 2002 | |
| Genes related to signal transduction, transcription and metastasis | Upregulated compared to HCV | Iizuka et al., 2002 | |
| Genes for detoxification and immune response | Upregulated compared to HBV | Iizuka et al., 2002 | |
| Inflammatory and anti-apoptotic phenotypes | Predominant compared to HBV | Honda et al., 2006 | |
| Pro-apoptotic and DNA repair response | Predominant compared to HCV | Honda et al., 2006 | |
| Notch and Toll-like receptor pathways in cirrhosis | NR | Deregulation in cirrhosis | Wurmbach et al., 2007 |
| JAK/STAT pathway | NR | Deregulation in early carcinogenesis | Wurmbach et al., 2007 |
| Genes involved in DNA replication and repair and cell cycle | NR | Upregulation in late carcinogenesis | Wurmbach et al., 2007 |
| miR-122, miR-100, miR-10a | NR | Upregulated | Varnholt et al., 2008 |
| miR-198, niR-145 | NR | Downregulated | Varnholt et al., 2008 |
| Pathways related to cell death, DNA damage, recombination and signal transduction | Activated | Not activated | Ura et al., 2009 |
| Pathways related to immune response, antigen presentation, cell cycle, proteasome and lipid metabolism | Not activated | Activated | Ura et al., 2009 |
| Genes involved in the regulation of transcription and DNA repair | NR | Upregulated | Mas et al., 2009 |
| Coagulation factors and apoptosis genes | NR | Downregulated | Mas et al., 2009 |
Abbreviations: HBV, hepatitis B virus; HCV, hepatitis C virus; NR, not reported.
A previous study found that gain of 10q was detected exclusively in cases of HCC with HCV infection (Kusano et al., 1999). A review of 338 different comparative genomic hybridization analyses of human HCCs, of which 100 were HCV positive, found that only losses of 8p were more frequent in HCV-negative cases (Moinzadeh et al., 2005) (Table 2). A genome-wide analysis study of gene expression found increased expression of genes encoding phase I enzymes CYP2E, AKR1C4, EPHX1 and FMO3 exclusively in HCV-positive HCCs (Okabe et al., 2001) (Table 3). Another study found that genes involved in the regulation of transcription and DNA repair were upregulated, whereas coagulation factors and apoptosis genes were downregulated in HCV-HCC samples when compared with cirrhotic tissues from patients with HCV-HCC (Mas et al., 2009). When comparing cirrhotic tissues with and without HCC, the top canonical pathways associated with HCV cirrhosis in patients with HCC included p53 signaling, acute-phase response signaling, xenobiotic metabolism signaling, IL-6 signaling and NFR2-mediated oxidative stress response. From normal liver to HCV cirrhosis to HCV-HCC, a positive trend was found in MHC class-I receptor activity, DNA damage checkpoint cell division and ubiquitin cycle genes (Mas et al., 2009). Recently, a three-gene set signature (GPC3, LYVE1 and survivin) that can distinguish dysplastic nodules from early HCC in HCV cirrhosis with a discriminative accuracy of 94% was discovered (Llovet et al., 2006). Another recent study that compared the gene expression profiles of HCV tissue samples representing the stepwise carcinogenic process from preneoplastic lesions (cirrhosis and dysplasia) to HCC disclosed the dysregulation of pathways, including the Notch and Toll-like receptor pathways in cirrhosis, followed by dysregulation of several components of the JAK/STAT pathway in early carcinogenesis, then upregulation of genes involved in DNA replication and repair and cell cycle in late cancerous stages (Wurmbach et al., 2007). A microarray study found that inflammatory and anti-apoptotic phenotypes were predominant in HCV but pro-apoptotic and DNA repair response were predominant in HBV with p53 and 14-3-3 interacting genes having an important function (Honda et al., 2006). Another study found that imprinted genes (H19 and IGF2) and the genes relating to signal transduction, transcription and metastasis are upregulated in HBV-HCC and the genes responsible for detoxification and immune response are upregulated in HCV-HCC (Iizuka et al., 2002). Another study revealed differential microRNA expression between HBV and HCV: the pathways related to cell death, DNA damage, recombination and signal transduction were activated in HBV-HCC and those related to immune response, antigen presentation, cell cycle, proteasome and lipid metabolism were activated in HCV-HCC (Ura et al., 2009). These differences suggested different oncogenic factors in HBV and HCV. Another microRNA gene expression study disclosed that miR-122, miR-100 and miR-10a were overexpressed whereas miR-198 and miR-145 were up to fivefold downregulated in HCV-HCC tumor tissues compared to normal liver tissues (Varnholt et al., 2008). Aberrant DNA methylation is an important epigenetic alteration in HCC. This process was found to be associated with viral infection and is more prominent in HCV than HBV in a recent study (Nishida et al., 2008). Taken together, although there are suggestions, there is no clear `signature' for HBV- or HCV-related hepatocarcinogenesis.
Chemical carcinogens, hormones, micronutrients and viral hepatitis
The combination of HBV and aflatoxin B1 exposure may underlie the high frequency of TP53 249ser mutations in HCC (Ming et al., 2002). Aflatoxin exposure may also be associated with advanced liver disease in patients with chronic hepatitis C (Chen et al., 2007).
An intronic polymorphism in the corticotropin-releasing hormone receptor 2 gene increases susceptibility to HBV-related HCC (Gu et al., 2009). Pathways involving androgen signaling may affect the risk of HBV-related HCC among men (Yu et al., 2001). In addition, HCV core protein behaves as a positive regulator in androgen receptor signaling (Kanda et al., 2008).
In HBV carriers, a high serum level of retinol is associated with a decreased risk of HCC (Yuan et al., 2006). In patients with chronic HCV, zinc concentration is lower in patients with HCC (Moriyama et al., 2006) and hepatic copper also may contribute to the development of HCC (Ebara et al., 2003). In patients with chronic viral hepatitis, selenium levels were significantly lower in patients with HCC (Yu et al., 1999).
Conclusions
HCC is a highly complex and heterogenous tumor. Hepatocarcinogenesis is a multistep process and involves multiple cellular signaling pathways. Although HBV and HCV are the major risk factors leading to the development of HCC, the precise pathogenetic mechanisms linking viral infection and HCC remain uncertain. A number of transgenic mouse model studies have found that the expression of various viral proteins either alone or in combination lead to the development of HCC. Viral proteins also have been implicated in disrupting several cellular signal transduction pathways that affect cell survival, proliferation, migration and transformation (Figure 3). HBV and HCV also appear to have distinct paths to cancer (Table 4). Current advances in the understanding of gene expression profile with high-throughput technologies and selective messenger RNA analysis have revolutionized our approach to the pathogenesis of HCC. Recently a multikinase inhibitor, sorafenib, has been shown to provide survival benefit in patients with advanced HCC (Llovet et al., 2008b; Cheng et al., 2009). This represents a breakthrough in the understanding of the mechanisms and treatment of HCC and proves that molecular therapies can be effective in HCC. However, this is only the first step toward cure. Because there are still likely several complex pathogenetic mechanisms that require targeting, it will therefore be of great importance to fully understand the molecular mechanisms underlying viral hepatocarcinogenesis so as to maximize our options to interrupt these pathways.
Figure 3.
A unified model of viral hepatocarcinogenesis.
Table 4.
Comparison of the pathogenetic mechanisms between HBV and HCV hepatocarcinogenesis
| HBV | HCV | References | |
|---|---|---|---|
| Similar pathways | |||
| Inflammation/fibrosis/cirrhosis | Chronic hepatitis/immune response | Chronic hepatitis/immune response | El-Serag and Rudolph, 2007 |
| Ca2+ signaling | HBx, HBV integration | HCV core, NS5A | Chami et al., 2006; Feitelson and Lee, 2007 |
| Raf/MAPK pathway | HBx | HCV core | Benn and Schneider, 1994; Hayashi et al., 2000; Erhardt et al., 2002; Oh et al., 2003 |
| TGF-β pathway | HBx | HCV core | Matsuzaki et al., 2007; Matsuzaki, 2009 |
| Oxidative stress | HBx, pre-S mutant | HCV core, NS3, NS5A | Bureau et al., 2001; Gong et al., 2001; Okuda et al., 2002; Wang et al., 2006; Chen and Siddiqui, 2007 |
| Endoplasmic reticulum stress | HBx, pre-S mutant | HCV core, NS5A | Tardif et al., 2002; Benali-Furet et al., 2005; Wang et al., 2006; Li et al., 2007 |
| p53 pathway | HBx | HCV core, NS3, NS5A | Ray et al., 1997; Lee and Rho, 2000; Lan et al., 2002; Kremsdorf et al., 2006; Deng et al., 2006 |
| Androgen signaling pathway | HBx | HCV core | Chiu et al., 2007; Kanda et al., 2008 |
| Wnt/β-catenin pathway | HBx | HCV core, NS5A | Ding et al., 2005; Wong and Ng, 2008 |
| Different pathways | |||
| Genomic instability | Insertional mutagenesis | HCV core, NS3, NS5A | Gong et al., 2001; Okuda et al., 2002; Guerrero and Roberts, 2005; Machida et al., 2006; Feitelson and Lee, 2007 |
| Oncogene activation | Insertional mutagenesis | HCV proteins | Fukutomi et al., 2005; Guerrero and Roberts, 2005; Feitelson and Lee, 2007 |
| Iron deposition | Less common | HCV polyprotein | Furutani et al., 2006 |
| Chromosomal aberrations | monLess common | Less common | Zondervan et al., 2000 |
| Methylation | Less common | More common | Nishida et al., 2008 |
Abbreviations: HBV, hepatitis B virus; HBx, HBV X; HCV, hepatitis C virus; TGF-β, transforming growth factor-β.
Acknowledgements
This work was supported by NIH Grants DK078772 and AI069939 (both to RTC).
Footnotes
Conflict of interest The authors declare no conflict of interest.
References
- Adinolfi LE, Gambardella M, Andreana A, Tripodi MF, Utili R, Ruggiero G. Steatosis accelerates the progression of liver damage of chronic hepatitis C patients and correlates with specific HCV genotype and visceral obesity. Hepatology. 2001;33:1358–1364. doi: 10.1053/jhep.2001.24432. [DOI] [PubMed] [Google Scholar]
- Alisi A, Giambartolomei S, Cupelli F, Merlo P, Fontemaggi G, Spaziani A, et al. Physical and functional interaction between HCV core protein and the different p73 isoforms. Oncogene. 2003;22:2573–2580. doi: 10.1038/sj.onc.1206333. [DOI] [PubMed] [Google Scholar]
- Anzola M. Hepatocellular carcinoma: role of hepatitis B and hepaititis C viruses proteins in hepatocarcinogenesis. J Viral Hepat. 2004;11:383–393. doi: 10.1111/j.1365-2893.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- Aoki H, Hayashi J, Moriyama M, Arkawa Y, Hino O. Hepatitis C virus core protein interacts with 14-3-3 protein and activates the kinase Raf-1. J Virol. 2000;74:1736–1741. doi: 10.1128/jvi.74.4.1736-1741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravalli RN, Steer CJ, Cressman ENK. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, Long JM, et al. IKK-beta links inflammation to obesity-induced insulin resistance. Nat Med. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- Arora P, Kim EO, Jung JK, Jang KL. Hepatitis C virus core protein downregulates E-cadherin expression via activation of DNA methyltransferase 1 and 3b. Cancer Lett. 2008;261:244–252. doi: 10.1016/j.canlet.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Baptista M, Kramvis A, Kew MC. High prevalence of 1762(T) 1764(A) mutations in the basic core promoter of hepatitis B virus isolated from black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology. 1999;29:946–953. doi: 10.1002/hep.510290336. [DOI] [PubMed] [Google Scholar]
- Barth H, Robinet E, Liang TJ, Baumert TF. Mouse models for the study of HCV infection and virus-host interactions. J Hepatol. 2008;49:134–142. doi: 10.1016/j.jhep.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett SE, Di Bisceglie AM, Bacon BR, Sharp RM, Govin darajan S, Hubbard GB, et al. Effects of iron loading on pathogenicity in hepatitis C virus-infected chimpanzees. Hepatology. 1999;29:1884–1892. doi: 10.1002/hep.510290623. [DOI] [PubMed] [Google Scholar]
- Battaglia S, Benzoubir N, Nobilet S, Charneau P, Samuel D, Zignego AL, et al. Liver cancer-derived hepatitis C virus core proteins shift TGF-beta responses from tumor suppression to epithelialmesenchymal transition. PLoS ONE. 2009;42:e4355. doi: 10.1371/journal.pone.0004355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benali-Furet NL, Chami M, Houel L, De Giorgi F, Vernejoul F, Lagorce D, et al. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene. 2005;24:4921–4933. doi: 10.1038/sj.onc.1208673. [DOI] [PubMed] [Google Scholar]
- Benard J, Douc-Rasy S, Ahomadegbe JC. TP 53 family members and human cancers. Hum Mutat. 2003;21:182–191. doi: 10.1002/humu.10172. [DOI] [PubMed] [Google Scholar]
- Benn J, Schneider RJ. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonte D, François C, Castelain S, Wychowski C, Dubuisson J, Meurs EF, et al. Positive effect of the hepatitis C virus nonstructural 5A protein on viral multiplication. Arch Virol. 2004;149:1353–1371. doi: 10.1007/s00705-003-0291-6. [DOI] [PubMed] [Google Scholar]
- Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- Branda M, Wands JR. Signal transduction cascades and hepatitis B and C related hepatocellular carcinoma. Hepatology. 2006;43:891–902. doi: 10.1002/hep.21196. [DOI] [PubMed] [Google Scholar]
- Brechot C, Pourcel C, Louise A, Rain B, Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286:533–535. doi: 10.1038/286533a0. [DOI] [PubMed] [Google Scholar]
- Bureau C, Bernad J, Chaouche N, Orfila C, Beraud M, Gonindard C, et al. Nonstructural 3 protein of hepatitis C virus triggers an oxidative burst in human monocytes via activation of NADPH oxidase. J Biol Chem. 2001;276:23077–23083. doi: 10.1074/jbc.M100698200. [DOI] [PubMed] [Google Scholar]
- Cai D, Yuan M, Frantz DF, Melendez PA, Hansen L, Lee J, et al. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med. 2005;11:183–190. doi: 10.1038/nm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell S, Park SH. The epidemiology of hepatocellular cancer: from the perspectives of public health problem to tumor biology. J Gastroenterol. 2009;44(Suppl XIX):96–101. doi: 10.1007/s00535-008-2258-6. [DOI] [PubMed] [Google Scholar]
- Campbell JS, Hughes SD, Gilbertson DG, Palmer TE, Holdren MS, Haran AC, et al. Platelet-derived growth factor C induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc Natl Acad Sci USA. 2005;102:3389–3394. doi: 10.1073/pnas.0409722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassiman D, Denef C, Desmet VJ, Roskams T. Human and rat hepatic stellate cells express neurotrophins and neurotrophin receptors. Hepatology. 2001;33:148–158. doi: 10.1053/jhep.2001.20793. [DOI] [PubMed] [Google Scholar]
- Chami M, Oulés B, Paterlini-Brećhot P. Cytobiological consequences of calcium-signaling alterations induced by human viral proteins. Biochim Biophys Acta. 2006;1763:1344–1362. doi: 10.1016/j.bbamcr.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Chan HL, Hui AY, Wong ML, Tse AM, Hung LC, Wong VW, et al. Genotype C hepatitis B virus infection is associated with increased risk of hepatocellular carcinoma. Gut. 2004;53:1494–1498. doi: 10.1136/gut.2003.033324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HL, Tse CH, Mo F, Koh J, Wong VW, Wong GL, et al. High viral load and hepatitis B virus subgenotype ce are associated with increased risk of hepatocellular carcinoma. J Clin Oncol. 2008;26:177–182. doi: 10.1200/JCO.2007.13.2043. [DOI] [PubMed] [Google Scholar]
- Chen CH, Wang MH, Wang JH, Hung CH, Hu TH, Lee SC, et al. Aflatoxin exposure and hepatitis C virus in advanced liver disease in a hepatitis C virus endemic area in Taiwan. Am J Trop Med Hyg. 2007;77:747–752. [PubMed] [Google Scholar]
- Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, et al. REVEAL-HBV Study Group Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Yang HI, Iloeje UH, REVEAL-HBV Study Group Hepatitis B Virus DNA levels and outcomes in chronic Hepatitis B. Hepatology. 2009;49:S72–S84. doi: 10.1002/hep.22884. [DOI] [PubMed] [Google Scholar]
- Chen J, Siddiqui A. Hepatitis B virus X protein stimulates the mitochondrial translocation of Raf-1 via oxidative stress. J Virol. 2007;81:6757–6760. doi: 10.1128/JVI.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- Chisari FV. Unscrambling hepatitis C virus-host interactions. Nature. 2005;436:930–932. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- Chisari FV, Klopchin K, Moriyama T, Pasquinelli C, Dunsford HA, Sell S, et al. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145–1156. doi: 10.1016/0092-8674(89)90770-8. [DOI] [PubMed] [Google Scholar]
- Chiu CM, Yeh SH, Chen PJ, et al. Hepatitis B virus X protein enhances androgen receptor-responsive gene expression depending on androgen level. Proc Natl Acad Sci USA. 2007;104:2571–2578. doi: 10.1073/pnas.0609498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Baek W, Yang S, Chang J, Sung YC, Suh M. HCV core protein modulates Rb pathway through pRb down-regulation and E2F-1 up-regulation. Biochim Biophys Acta. 2001;1538:59–66. doi: 10.1016/s0167-4889(00)00137-3. [DOI] [PubMed] [Google Scholar]
- Choi J, Ou JHJ. Mechanisms of liver injury III oxidative stress in the pathogenesis of hepatitis C virus. Am J Physiol Gastrointest Liver Physiol. 2006;290:G847–G851. doi: 10.1152/ajpgi.00522.2005. [DOI] [PubMed] [Google Scholar]
- Choi SH, Hwang SB. Modulation of the transforming growth factor-beta signal transduction pathway by hepatitis C virus nonstructural 5A protein. J Biol Chem. 2006;281:7468–7478. doi: 10.1074/jbc.M512438200. [DOI] [PubMed] [Google Scholar]
- Chou YC, Yu MW, Wu CF, Yang SY, Lin CL, Liu CJ, et al. Temporal relationship between hepatitis B virus enhancer II/basal core promoter sequence variation and risk of hepatocellular carcinoma. Gut. 2008;57:91–97. doi: 10.1136/gut.2006.114066. [DOI] [PubMed] [Google Scholar]
- Chu CJ, Hussain M, Lok ASF. Hepatitis B virus genotype is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology. 2002;122:1756–1762. doi: 10.1053/gast.2002.33588. [DOI] [PubMed] [Google Scholar]
- Chung YL, Sheu ML, Yen SH. Hepatitis C virus NS5A as a potential viral Bcl-2 homologue interacts with Bax and inhibits apoptosis in hepatocellular carcinoma. Int J Cancer. 2003;107:65–73. doi: 10.1002/ijc.11303. [DOI] [PubMed] [Google Scholar]
- Crotta S, Stilla A, Wack A, D'Andrea A, Nuti S, D'Oro U, et al. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J Exp Med. 2002;195:35–41. doi: 10.1084/jem.20011124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja MJ. Cell signaling in oxidative stress-induced liver injury. Semin Liver dis. 2007;27:378–389. doi: 10.1055/s-2007-991514. [DOI] [PubMed] [Google Scholar]
- Dandri M, Burda MR, Burkle A, Zuckerman DM, Will H, Rogler CE, et al. Increase in de novo HBV DNA integrations in response to oxidative DNA damage or inhibition of poly(ADPribosyl)ation. Hepatology. 2002;35:217–223. doi: 10.1053/jhep.2002.30203. [DOI] [PubMed] [Google Scholar]
- de Lucas S, Bartolome J, Amaro MJ, Carreno V. Hepatitis C virus core protein transactivates the inducible nitric oxide synthase promoter via NF-kappaB activation. Antiviral Res. 2003;60:117–124. doi: 10.1016/j.antiviral.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Deng L, Nagano-Fujii M, Tanaka M, Nomura-Takigawa Y, Ikeda M, Kato N, et al. NS3 protein of hepatitis C virus associates with the tumour suppressor p53 and inhibits its function in an NS3 sequence-dependent manner. J Gen Virol. 2006;87:1703–1713. doi: 10.1099/vir.0.81735-0. [DOI] [PubMed] [Google Scholar]
- Diao J, Khine AA, Sarangi F, Hsu E, Iorio C, Tibbles LA, et al. X protein of hepatitis B virus inhibits Fas-mediated apoptosis and is associated with up-regulation of the SAPK/JNK pathway. J Biol Chem. 2001;16:8328–8340. doi: 10.1074/jbc.M006026200. [DOI] [PubMed] [Google Scholar]
- Ding Q, Xia W, Liu JC, Yang JY, Lee DF, Xia J, et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell. 2005;19:159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Donato F, Boffetta P, Puoti M. A meta-analysis of epidemiological studies on the combined effect of hepatitis B and C virus infections in causing hepatocellular carcinoma. Int J Cancer. 1998;75:347–354. doi: 10.1002/(sici)1097-0215(19980130)75:3<347::aid-ijc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Donato F, Tagger A, Gelatti U, Parrinello G, Boffetta P, Albertini A, et al. Alcohol and hepatocellular carcinoma: the effect of lifetime intake and hepatitis virus infections in men and women. Am J Epidemiol. 2002;155:323–331. doi: 10.1093/aje/155.4.323. [DOI] [PubMed] [Google Scholar]
- Ebara M, Fukuda H, Hatano R, Yoshikawa M, Sugiura N, Saisho H, et al. Metal contents in the liver of patients with chronic liver disease caused by hepatitis C virus. Reference to hepatocellular carcinoma. Oncology. 2003;65:323–330. doi: 10.1159/000074645. [DOI] [PubMed] [Google Scholar]
- El-Serag HB. Hepatocellular carcinoma and hepatitis C in the United States. Hepatology. 2002;36:S74–S83. doi: 10.1053/jhep.2002.36807. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- Erhardt A, Hassan M, Heintges T, Haussinger D. Hepatitis C virus core protein induces cell proliferation and activates ERK, JNK, and p38 MAP kinases together with the MAP kinase phosphatase MKP-1 in a HepG2 Tet-Off cell line. Virology. 2002;292:272–284. doi: 10.1006/viro.2001.1227. [DOI] [PubMed] [Google Scholar]
- Fang ZL, Sabin CA, Dong BQ, Ge LY, Wei SC, Chen QY, et al. HBV A1762T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am J Gastroenterol. 2008;103:2254–2262. doi: 10.1111/j.1572-0241.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattovich G. Natural history and prognosis of hepatitis B. Sem Liver Diseas. 2003;23:47–58. doi: 10.1055/s-2003-37590. [DOI] [PubMed] [Google Scholar]
- Feitelson MA, Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007;252:157–170. doi: 10.1016/j.canlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Feitelson MA, Reis HM, Lale Tufan N, Sun B, Pan J, Lian Z. Putative roles of hepatitis B x antigen in the pathogenesis of chronic liver disease. Cancer Lett. 2009;286:69–79. doi: 10.1016/j.canlet.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman SL. Mechanism of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman SL, Arthur MJ. Activation of cultured rat hepatic lipocytes by Kupffer cell conditioned medium. Direct enhancement of matrix synthesis and stimulation of cell proliferation via induction of platelet-derived growth factor receptors. J Clin Invest. 1989;84:1780–1785. doi: 10.1172/JCI114362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujie H, Moriya K, Shintani Y, Yotsuyanagi H. Hepatitis B virus genotypes and hepatocellular carcinoma in Japan. Gastroenterology. 2001;120:1564–1565. doi: 10.1053/gast.2001.24501. [DOI] [PubMed] [Google Scholar]
- Fujio K, Evarts RP, Hu Z, Marsden ER, Thorgeirsson SS. Expression of stem cell factor and its receptor, c-kit, during liver regeneration from putative stem cells in adult rat. Lab Invest. 1994;70:511–516. [PubMed] [Google Scholar]
- Fukutomi T, Zhou Y, Kawai S, Eguchi H, Wands JR, Li J. Hepatitis C virus core protein stimulates hepatocyte growth: correlation with upregulation of wnt-1 expression. Hepatology. 2005;41:1096–1105. doi: 10.1002/hep.20668. [DOI] [PubMed] [Google Scholar]
- Furutani T, Hino K, Okuda M, Gondo T, Nishina S, Kitase A, et al. Hepatic iron overload induces hepatocellular carcinoma in transgenic mice expressing the hepatitis C virus polyprotein. Gastroenterology. 2006;130:2087–2098. doi: 10.1053/j.gastro.2006.02.060. [DOI] [PubMed] [Google Scholar]
- Gaudio E, Carpino G, Cardinale V, Franchitto A, Onori P, Alvaro D. New insights into liver stem cells. Dig Liver Dis. 2009;41:455–462. doi: 10.1016/j.dld.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Majumder M, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein protects against TNFalpha mediated apoptotic cell death. Virus Res. 2000;67:173–178. doi: 10.1016/s0168-1702(00)00141-6. [DOI] [PubMed] [Google Scholar]
- Gong G, Waris G, Tanveer R, Siddiqui A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc Natl Acad Sci USA. 2001;98:9599–9604. doi: 10.1073/pnas.171311298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Qi P, Zhou F, Ji Q, Wang H, Dou T, et al. An intronic polymorphism in the corticotropin-releasing hormone receptor 2 gene increases susceptibility to HBV-related hepatocellular carcinoma in Chinese population. Hum Genet. 2009;127:75–81. doi: 10.1007/s00439-009-0750-6. [DOI] [PubMed] [Google Scholar]
- Guerrero RB, Roberts LR. The role of hepatitis B virus integrations in the pathogenesis of human hepatocellular carcinoma. J Hepatol. 2005;42:760–777. doi: 10.1016/j.jhep.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Hassan M, Ghozlan H, Abdel-Kader O. Activation of c-Jun NH2-terminal kinase (JNK) signalling pathway is essential for the stimulation of hepatitis C virus (HCV) nonstructural protein 3 (NS3)-mediated cell growth. Virology. 2005;333:324–336. doi: 10.1016/j.virol.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Hassan M, Selimovic D, Ghozlan H, Abdel-Kader O. Induction of high-molecular-weight (HMW) tumor necrosis factor(TNF) alpha by hepatitis C virus (HCV) non-structural protein 3 (NS3) in liver cells is AP-1 and NF-kappaB-dependent activation. Cell Signal. 2007;19:301–311. doi: 10.1016/j.cellsig.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Hassan M, Selimovic D, Ghozlan H, Abdel-kader O. Hepatitis C virus core protein triggers hepatic angiogenesis by a mechanism including multiple pathways. Hepatology. 2009;49:1469–1482. doi: 10.1002/hep.22849. [DOI] [PubMed] [Google Scholar]
- Hayashi J, Aoki H, Kajino K, Moriyama M, Arakawa Y, Hino O. Hepatitis C virus core protein activates the MAPK/ERK cascade synergistically with tumor promoter TPA, but not with epidermal growth factor or transforming growth factor alpha. Hepatology. 2000;32:958–961. doi: 10.1053/jhep.2000.19343. [DOI] [PubMed] [Google Scholar]
- He Y, Nakao H, Tan SL, Polyak SJ, Neddermann P, Vijaysri S, et al. Subversion of cell signaling pathways by hepatitis C virus nonstructural 5A protein via interaction with Grb2 and P85 phosphatidylinositol 3-kinase. J Virol. 2002;76:9207–9217. doi: 10.1128/JVI.76.18.9207-9217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildt E, Munz B, Saher G, Reifenberg K, Hofschneider PH. The PreS2 activator MHBs(t) of hepatitis B virus activates c-raf-1/Erk2 signaling in transgenic mice. EMBO J. 2002;21:525–535. doi: 10.1093/emboj/21.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirankarn N, Kimkong I, Kummee P, Tangkijvanich P, Poovorawan Y. Interleukin-1beta gene polymorphism associated with hepatocellular carcinoma in hepatitis B virus infection. World J Gastroenterol. 2006;12:776–779. doi: 10.3748/wjg.v12.i5.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Kaneko S. Different signaling pathways in the livers of patients with chronic hepatitis B or chronic hepatitis C. Hepatology. 2006;44:1122–1138. doi: 10.1002/hep.21383. [DOI] [PubMed] [Google Scholar]
- Horikawa I, Barrett JC. Cis-activation of the human telomerase gene (hTERT) by the hepatitis B virus genome. J Natl Cancer Inst. 2001;93:1171–1173. doi: 10.1093/jnci/93.15.1171. [DOI] [PubMed] [Google Scholar]
- Hourigan LF, Macdonald GA, Purdie D, Whitehall VH, Shorthouse C, Clouston A, et al. Fibrosis in chronic hepatitis C correlates significantly with body mass index and steatosis. Hepatology. 1999;29:1215–1219. doi: 10.1002/hep.510290401. [DOI] [PubMed] [Google Scholar]
- Hsieh TY, Matsumoto M, Chou HC, Schneider R, Hwang SB, Lee AS, et al. Hepatitis C virus core protein interacts with heterogenous nuclear ribonucleoprotein K. J Biol Chem. 1998;273:17651–17659. doi: 10.1074/jbc.273.28.17651. [DOI] [PubMed] [Google Scholar]
- Iizuka N, Oka M, Yamada-Okabe H, Mori N, Tamesa T, Okada T, et al. Comparison of gene expression profiles between hepatitis B virus- and hepatitis C virus-infected hepatocellular carcinoma by oligonucleotide microarray data on the basis of a supervised learning method. Cancer Res. 2002;62:3939–3944. [PubMed] [Google Scholar]
- Iloeje UH, Yang HI, Su J, Jen CL, You SL, Chen CJ. Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology. 2006;130:678–686. doi: 10.1053/j.gastro.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Irshad M, Dhar I. Hepatitis C virus core protein: an update on its molecular biology, cellular function and clinical implications. Med Princ Pract. 2006;15:405–416. doi: 10.1159/000095485. [DOI] [PubMed] [Google Scholar]
- Jia L, Wang XW, Harris C. Hepatitis B virus X protein inhibits nucleotide excision repair. Int J Cancer. 1999;80:875–879. doi: 10.1002/(sici)1097-0215(19990315)80:6<875::aid-ijc13>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Jin DY, Wang HL, Zhou Y, Chun AC, Kibler KV, Hou YD, et al. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 2000;19:729–740. doi: 10.1093/emboj/19.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamegaya Y, Hiasa Y, Zukerberg L, Fowler N, Blackard JT, Lin W, et al. Hepatitis C virus acts as a tumor accelerator by blocking apoptosis in a mouse model of hepatocarcinogenesis. Hepatology. 2005;41:660–667. doi: 10.1002/hep.20621. [DOI] [PubMed] [Google Scholar]
- Kanda T, Steele R, Ray R, Ray RB. Hepatitis C virus core protein augments androgen receptor-mediated signaling. J Virol. 2008;82:11066–11072. doi: 10.1128/JVI.01300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554–559. doi: 10.1016/s0016-5085(00)70261-7. [DOI] [PubMed] [Google Scholar]
- Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124:327–334. doi: 10.1053/gast.2003.50053. [DOI] [PubMed] [Google Scholar]
- Kato J, Miyanishi K, Kobune M. Iron, hepatitis C virus, and hepatocellular carcinoma: iron reduction preaches the gospel for chronic hepatitis C. J Gastroenterol. 2007;42:923–926. doi: 10.1007/s00535-007-2110-4. [DOI] [PubMed] [Google Scholar]
- Katoh H, Shibata T, Kokubu A, Ojima H, Loukopoulos P, Kanai Y, et al. Genetic profile of hepatocellular carcinoma revealed by array-based comparative genomic hybridization: identification of genetic indicators to predict patient outcome. J Hepatol. 2005;43:863–874. doi: 10.1016/j.jhep.2005.05.033. [DOI] [PubMed] [Google Scholar]
- Keasler VV, Hodgson AJ, Madden CR, Slagle BL. Enhancement of hepatitis B virus replication by the regulatory X protein in vitro and in vivo. J Virol. 2007;81:2656–2662. doi: 10.1128/JVI.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew MC. Hepatic iron overload and hepatocellular carcinoma. Cancer Lett. 2009;286:38–43. doi: 10.1016/j.canlet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Kidd-Ljunggren K, Miyakawa Y, Kidd AH. Genetic variability in hepatitis B viruses. J Gen Virol. 2002;83:1267–1280. doi: 10.1099/0022-1317-83-6-1267. [DOI] [PubMed] [Google Scholar]
- Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- Kim K, Kim KH, Kim HH, Cheong J. Hepatitis B virus X protein induces lipogenic transcription factor SREBP1 and fatty acid synthase through the activation of nuclear receptor LXRalpha. Biochem J. 2008;416:219–230. doi: 10.1042/BJ20081336. [DOI] [PubMed] [Google Scholar]
- Kim KH, Shin HJ, Kim K, Choi HM, Rhee SH, Moon HB, et al. Hepatitis B virus X protein induces hepatic steatosis via transcriptional activation of SREBP1 and PPAR-gamma. Gastroenterology. 2007;132:1955–1967. doi: 10.1053/j.gastro.2007.03.039. [DOI] [PubMed] [Google Scholar]
- Kittlesen DJ, Chianese-Bullock KA, Yao ZQ, Braciale TJ, Hahn YS. Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J Clin Invest. 2000;106:1239–1249. doi: 10.1172/JCI10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike K. Pathogenesis of HCV-associated HCC: dual-pass carcinogenesis through activation of oxidative stress and intracellular signaling. Hepatol Res. 2007;37:S115–S120. doi: 10.1111/j.1872-034X.2007.00173.x. [DOI] [PubMed] [Google Scholar]
- Koike K. Steatosis, liver injury, and hepatocarcinogenesis in hepatitis C viral infection. J Gastroenterol. 2009;44(Suppl XIX):82–88. doi: 10.1007/s00535-008-2276-4. [DOI] [PubMed] [Google Scholar]
- Koike K, Moriya K, Iino S, Yotsuyanagi H, Endo Y, Miyamura T, et al. High-level expression of hepatitis B virus HBx gene and hepatocarcinogenesis in transgenic mice. Hepatology. 1994;19:810–819. [PubMed] [Google Scholar]
- Kordes C, Sawitza I, Müller-Marbach A, Ale-Agha N, Keitel V, Klonowski-Stumpe H, et al. CD133+ hepatic stellate cells are progenitor cells. Biochem Biophys Res Commun. 2007;352:410–417. doi: 10.1016/j.bbrc.2006.11.029. [DOI] [PubMed] [Google Scholar]
- Korenaga M, Wang T, Li Y, Showalter LA, Chan T, Sun J, et al. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases ROS production. J Biol Chem. 2005;280:37481–37488. doi: 10.1074/jbc.M506412200. [DOI] [PubMed] [Google Scholar]
- Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene. 2006;25:3823–3833. doi: 10.1038/sj.onc.1209559. [DOI] [PubMed] [Google Scholar]
- Kummee P, Tangkijvanich P, Poovorawan Y, Hirankarn N. Association of HLA-DRB1*13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in Thai population. J Viral Hepatitis. 2007;14:841–848. doi: 10.1111/j.1365-2893.2007.00880.x. [DOI] [PubMed] [Google Scholar]
- Kusano N, Shiraishi K, Kubo K, Oga A, Okita K, Sasaki K. Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology. 1999;29:1858–1862. doi: 10.1002/hep.510290636. [DOI] [PubMed] [Google Scholar]
- Kusano N, Okita K, Shirahashi H, Harada T, Shiraishi K, Oga A, et al. Chromosomal imbalances detected by comparative genomic hybridization are associated with outcome of patients with hepatocellular carcinoma. Cancer. 2002;94:746–751. doi: 10.1002/cncr.10254. [DOI] [PubMed] [Google Scholar]
- Kwun HJ, Jang KL. Dual effects of hepatitis-C virus core protein on the transcription of cyclin-dependent kinase inhibitor p21 gene. J Viral Hepat. 2003;10:249–255. doi: 10.1046/j.1365-2893.2003.00434.x. [DOI] [PubMed] [Google Scholar]
- Kwun HJ, Jung EY, Ahn JY, Lee MN, Jang KL. p53-dependent transcriptional repression of p21 (waf1) by hepatitis C virus NS3. J Gen Virol. 2001;82:2235–2241. doi: 10.1099/0022-1317-82-9-2235. [DOI] [PubMed] [Google Scholar]
- Lan KH, Sheu ML, Hwang SJ, Yen SH, Chen SY, Wu JC, et al. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene. 2002;21:4801–4811. doi: 10.1038/sj.onc.1205589. [DOI] [PubMed] [Google Scholar]
- Lee SG, Rho HM. Transcriptional repression of the human p53 gene by hepatitis B viral X protein. Oncogene. 2000;19:468–471. doi: 10.1038/sj.onc.1203312. [DOI] [PubMed] [Google Scholar]
- Leenders MW, Nijkamp MW, Rinkes IHB. Mouse models in liver cancer research: a review of current literature. World J Gastroenterol. 2008;14:6915–6923. doi: 10.3748/wjg.14.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat H, Honda M, Beard MR, Loesch K, Sun J, Yang Y, et al. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352–365. doi: 10.1053/gast.2002.31001. [DOI] [PubMed] [Google Scholar]
- Lev S, Moreno H, Martinez R, Canoll P, Peles E, Musacchio JM, et al. Protein tyrosine kinase PYK2 involved in Ca(2+)-induced regulation of ion channel and MAP kinase functions. Nature. 1995;376:737–745. doi: 10.1038/376737a0. [DOI] [PubMed] [Google Scholar]
- Li B, Gao B, Ye L, Han X, Wang W, Kong L, et al. Hepatitis B virus X protein (HBx) activates ATF6 and IRE1-XBP1 pathways of unfolded protein response. Virus Res. 2007;124:44–49. doi: 10.1016/j.virusres.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Lin YW, Sheu JC, Liu LY, Chen CH, Lee HS, Huang GT, et al. Chromosomal abnormality in hepatocellular carcinoma by comparative genomic hybridisation in Taiwan. Eur J Cancer. 1999;35:652–658. doi: 10.1016/s0959-8049(98)00430-4. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;930:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Kao JH, Chen DS. Therapeutic implications of hepatitis B virus genotypes. Liver Int. 2005;25:1097–1107. doi: 10.1111/j.1478-3231.2005.01177.x. [DOI] [PubMed] [Google Scholar]
- Liu H, Luan F, Ju Y, Shen H, Gao L, Wang X, et al. in vitro transfection of the hepatitis B virus PreS2 gene into the human hepatocarcinoma cell line HepG2 induces upregulation of human telomerase reverse transcriptase. Biochem Biophys Res Commun. 2007;355:379–384. doi: 10.1016/j.bbrc.2007.01.160. [DOI] [PubMed] [Google Scholar]
- Livingston SE, Simonetti JP, McMahon BJ, Bulkow LR, Hurlburt KJ, Homan CE, et al. Hepatitis B virus genotypes in Alaska native people with hepatocellular carcinoma: preponderance of genotype F. J Infect Dis. 2007;195:5–11. doi: 10.1086/509894. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008a;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Chen Y, Wurmbach E, Roayaie S, Fiel MI, Schwartz M, et al. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology. 2006;131:1758–1767. doi: 10.1053/j.gastro.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008b;24:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- Locarnini S, McMillan J, Bartholomeusz A. The hepatitis B virus and common mutants. Sem Liver Disease. 2003;23:5–20. doi: 10.1055/s-2003-37587. [DOI] [PubMed] [Google Scholar]
- Lonardo A, Adinolfi LE, Loria P, Carulli N, Ruggiero G, Day CP. Steatosis and hepatitis C virus: mechanisms and significance for hepatic and extrahepatic disease. Gastroenterology. 2004;1262:586–597. doi: 10.1053/j.gastro.2003.11.020. [DOI] [PubMed] [Google Scholar]
- Longato L, de la Monte S, Kuzushita N, Horimoto M, Rogers AB, Slagle BL, et al. Overexpression of insulin receptor substrate-1 and hepatitis Bx genes causes premalignant alterations in the liver. Hepatology. 2009;49:1935–1943. doi: 10.1002/hep.22856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo L, Platini F, Scardino A, Alabiso O, Vasapollo G, Tessitore L. Autophagy inhibition enhances anthocyanin-induced apoptosis in hepatocellular carcinoma. Mol Cancer Ther. 2008;7:2476–2485. doi: 10.1158/1535-7163.MCT-08-0361. [DOI] [PubMed] [Google Scholar]
- Luber B, Arnold N, Sturzl M, Höhne M, Schirmacher P, Lauer U, et al. Hepatoma-derived integrated HBV DNA causes multistage transformation in vitro. Oncogene. 1996;12:1597–1608. [PubMed] [Google Scholar]
- Machida K, Cheng KT, Lai CK, Jeng KS, Sung VM, Lai MM. Hepatitis C virus triggers mitochondrial permeability transition with production of reactive oxygen species, leading to DNA damage and STAT3 activation. J Virol. 2006;80:7199–7207. doi: 10.1128/JVI.00321-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood S, Kawanaka M, Kamei A, Izumi A, Nakata K, Niiyama G, et al. Immunohistochemical evaluation of oxidative stress markers in chronic hepatitis C. Antioxid Redox Signal. 2004;6:19–24. doi: 10.1089/152308604771978318. [DOI] [PubMed] [Google Scholar]
- Majumder M, Ghosh AK, Steele R, Ray R, Ray RB. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J Virol. 2001;75:1401–1407. doi: 10.1128/JVI.75.3.1401-1407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder M, Steele R, Ghosh AK, Zhou XY, Thornburg L, Ray R, et al. Expression of hepatitis C virus non-structural 5A protein in the liver of transgenic mice. FEBS Lett. 2003;555:528–532. doi: 10.1016/s0014-5793(03)01337-1. [DOI] [PubMed] [Google Scholar]
- Martindale JL, Holbrook NJ. Cellular response to oxidative stress: signaling for suicide and survival. J Cell Physiol. 2002;192:1–15. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S, Schaeffer C, Robert EI, van Breugel PC, Leupin O, Hantz O, et al. Hepatitis B virus X protein affects S phase progression leading to chromosome segregation defects by binding to damaged DNA binding protein 1. Hepatology. 2008;48:1467–1476. doi: 10.1002/hep.22542. [DOI] [PubMed] [Google Scholar]
- Martín-Vílchez S, Sanz-Cameno P, Rodríguez-Muñoz Y, Majano PL, Molina-Jiménez F, López-Cabrera M, et al. The hepatitis B virus X protein induces paracrine activation of human hepatic stellate cells. Hepatology. 2008;47:1872–1883. doi: 10.1002/hep.22265. [DOI] [PubMed] [Google Scholar]
- Mas VR, Maluf DG, Archer KJ, Yanek K, Kong X, Kulik L, et al. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009;15:85–94. doi: 10.2119/molmed.2008.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K. Modulation of TGF-beta signaling during progression of chronic liver diseases. Front Bioscience. 2009;14:2923–2934. doi: 10.2741/3423. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K, Murata M, Yoshida K, Sekimoto G, Uemura Y, Sakaida N, et al. Chronic inflammation associated with hepatitis C virus infection perturbs hepatic transforming growth factor beta signaling, promoting cirrhosis and hepatocellular carcinoma. Hepatology. 2007;46:48–57. doi: 10.1002/hep.21672. [DOI] [PubMed] [Google Scholar]
- Ming L, Thorgeirsson SS, Gail MH, Lu P, Harris CC, Wang N, et al. Dominant role of hepatitis B virus and cofactor role of aflatoxin in hepatocarcinogenesis in Qidong, China. Hepatology. 2002;36:1214–1220. doi: 10.1053/jhep.2002.36366. [DOI] [PubMed] [Google Scholar]
- Missiha SB, Ostrowski M, Heathcote EJ. Disease progression in chronic hepatitis C: modifiable and nonmodifiable factors. Gastroenterology. 2008;134:1699–1714. doi: 10.1053/j.gastro.2008.02.069. [DOI] [PubMed] [Google Scholar]
- Moinzadeh P, Breuhahn K, Stutzer H, Schirmacher P. Chromosome alterations in human hepatocellular carcinomas correlate with aetiology and histological grade: results of an explorative CGH meta-analysis. Br J Cancer. 2005;92:935–941. doi: 10.1038/sj.bjc.6602448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microb. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Ishibashi K, Matsuura Y, et al. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol. 1997;78:1527–1531. doi: 10.1099/0022-1317-78-7-1527. [DOI] [PubMed] [Google Scholar]
- Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Matsuura Y, et al. Hepatitis C virus core protein induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065–1068. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- Moriya K, Nakagawa K, Santa T, Shintani Y, Fujie H, Miyosh H, et al. Oxidative stress in the absence of inflammation in a mouse model for hepatitis C virus-associated hepatocarcinogenesis. Cancer Res. 2001;61:4365–4370. [PubMed] [Google Scholar]
- Moriyama M, Matsumura H, Fukushima A, Ohkido K, Arakawa Y, Nirei K, et al. Clinical significance of evaluation of serum zinc concentrations in C-viral chronic liver disease. Dig Dis Sci. 2006;51:1967–1977. doi: 10.1007/s10620-005-9051-7. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Saigo K, Takashima H, Minami M, Okanoue T, Brećhot C, et al. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut. 2005;54:1162–1168. doi: 10.1136/gut.2004.054452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Matsuzaki K, Yoshida K, Sekimoto G, Tahashi Y, Mori S, et al. Hepatitis B virus X protein shifts human hepatic transforming growth factor (TGF)-beta signaling from tumor suppression to oncogenesis in early chronic hepatitis B. Hepatology. 2009;49:1203–1217. doi: 10.1002/hep.22765. [DOI] [PubMed] [Google Scholar]
- Myung SJ, Yoon JH, Gwak GY, Kim W, Lee JH, Kim KM, et al. Wnt signaling enhances the activation and survival of human hepatic stellate cells. FEBS Lett. 2007;581:2954–2958. doi: 10.1016/j.febslet.2007.05.050. [DOI] [PubMed] [Google Scholar]
- Na TY, Shin YK, Roh KJ, Kang SA, Hong I, Oh SJ, et al. Liver X receptor mediates hepatitis B virus x protein-induced lipogenesis in hepatitis B virus-associated hepatocellular carcinoma. Hepatology. 2009;49:1122–1131. doi: 10.1002/hep.22740. [DOI] [PubMed] [Google Scholar]
- Naas T, Ghorbani M, Alvarez-Maya I, Lapner M, Kothary R, De Repentigny Y, et al. Characterization of liver histopathology in a transgenic mouse model expressing genotype 1a hepatitis C virus core and envelope proteins 1 and 2. J Gen Virol. 2005;86:2185–2196. doi: 10.1099/vir.0.80969-0. [DOI] [PubMed] [Google Scholar]
- Newell P, Villanueva A, Friedman SL, Koike K, Llovet JM. Experimental models of hepatocellular carcinoma. J Hepatol. 2008;485:858–879. doi: 10.1016/j.jhep.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki T, Pekny M, Hellemans K, Bleser PD, Berg KV, Vaeyens F, et al. Class VI intermediate filament protein Nestin is induced during activation of rat hepatic stellate cells. Hepatology. 1999;29:520–527. doi: 10.1002/hep.510290232. [DOI] [PubMed] [Google Scholar]
- Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908–918. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez O, Fernandez-Martinez A, Majano PL, Apolinario A, Gomez-Gonzalo M, Benedicto I, et al. Increased intrahepatic cyclooxygenase 2, matrix metalloproteinase 2, and matrix metalloproteinase 9 expression is associated with progressive liver disease in chronic hepatitis C virus infection: role of viral core and NS5A proteins. Gut. 2004;53:1665–1672. doi: 10.1136/gut.2003.038364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JC, Jeong DL, Kim IK, Oh SH. Activation of calcium signaling by hepatitis B virus-X protein in liver cells. Exp Mol Med. 2003;35:301–309. doi: 10.1038/emm.2003.41. [DOI] [PubMed] [Google Scholar]
- Ohata K, Hamasaki K, Toriyama K, Matsumoto K, Saeki A, Yanagi K, et al. Hepatic steatosis is a risk factor for hepatocellular carcinoma in patients with chronic hepatitis C virus infection. Cancer. 2003;97:3036–3043. doi: 10.1002/cncr.11427. [DOI] [PubMed] [Google Scholar]
- Okabe H, Ikai I, Matsuo K, Satoh S, Momoi H, Kamikawa T, et al. Comprehensive allelotype study of hepatocellular carcinoma: potential differences in pathways to hepatocellular carcinoma between hepatitis B virus-positive and -negative tumors. Hepatology. 2000;31:1073–1079. doi: 10.1053/he.2000.6409. [DOI] [PubMed] [Google Scholar]
- Okabe H, Satoh S, Kato T, Kitahara O, Yanagawa R, Yamaoka Y, et al. Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 2001;61:2129–2137. [PubMed] [Google Scholar]
- Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, et al. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–375. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- Okuno M, Kojima S, Akita K, Matsushima-Nishiwaki R, Adachi S, Sano T, et al. Retinoids in liver fibrosis and cancer. Front Biosci. 2002;1:d204–d218. doi: 10.2741/A775. [DOI] [PubMed] [Google Scholar]
- Pang R, Lee TK, Poon RT, Fan ST, Wong KB, Kwong YL, et al. Pin1 interacts with a specific serine-proline motif of hepatitis B virus X-protein to enhance hepatocarcinogenesis. Gastroenterology. 2007;132:1088–1103. doi: 10.1053/j.gastro.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Park IY, Sohn BH, Yu E, Suh DJ, Chung YH, et al. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476–1494. doi: 10.1053/j.gastro.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80:827–841. doi: 10.1002/(sici)1097-0215(19990315)80:6<827::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- Parsons CJ, Takashima M, Rippe RA. Molecular mechanisms of hepatic fibrogenesis. J Gastroenterol Hepatology Suppl. 2007;1:S79–S84. doi: 10.1111/j.1440-1746.2006.04659.x. [DOI] [PubMed] [Google Scholar]
- Pekow JR, Bhan AK, Zheng H, Chung RT. Hepatic steatosis is associated with increased frequency of hepatocellular carcinoma in patients with hepatitis C-related cirrhosis. Cancer. 2007;109:2490–2496. doi: 10.1002/cncr.22701. [DOI] [PubMed] [Google Scholar]
- Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Pinzani M, Milani S, Herbst H, DeFranco R, Grappone C, Gentilini A, et al. Expression of platelet derived growth factor and its receptor in normal human liver and during active hepatic fibrogenesis. Am J Pathol. 1996;148:785–800. [PMC free article] [PubMed] [Google Scholar]
- Poon TC, Wong N, Lai PB, Rattray M, Johnson PJ, Sung JJ. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology. 2006;131:1262–1270. doi: 10.1053/j.gastro.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Qadri I, Iwahashi M, Capasso JM, Hopken MW, Flores S, et al. Induced oxidative stress and activated expression of manganese superoxide dismutase during hepatitis C virus replication: role of JNK, p38 MAPK and AP-1. Biochem J. 2004;378:919–928. doi: 10.1042/BJ20031587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadri I, Iwahashi M, Simon F. Hepatitis C virus NS5A protein binds TBP and p53, inhibiting their DNA binding and p53 interactions with TBP and ERCC3. Biochem Biophys Acta. 2002;21:193–204. doi: 10.1016/s0167-4889(02)00315-4. [DOI] [PubMed] [Google Scholar]
- Qi P, Chen YM, Wang H, Fang M, Ji Q, Zhao YP, et al. 509C>T polymorphism in the TGF-beta1 gene promoter, impact on the hepatocellular carcinoma risk in Chinese patients with chronic hepatitis B virus infection. Cancer Immunol Immunother. 2009;58:1433–1440. doi: 10.1007/s00262-009-0660-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RB, Steele R, Meyer K, Ray R. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J Biol Chem. 1997;272:10983–10986. doi: 10.1074/jbc.272.17.10983. [DOI] [PubMed] [Google Scholar]
- Roskams T. Different types of liver progenitor cells and their niches. J Hepatol. 2006;45:1–4. doi: 10.1016/j.jhep.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893–3896. doi: 10.1128/jvi.69.6.3893-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana A, Manns MP, Rudolph KL. Telomeres and telomerase: a dual role in hepatocarcinogenesis. Hepatology. 2004;40:276–283. doi: 10.1002/hep.20308. [DOI] [PubMed] [Google Scholar]
- Schaefer S. Hepatitis B virus: significance of genotypes. J Viral Hepat. 2005;12:111–124. doi: 10.1111/j.1365-2893.2005.00584.x. [DOI] [PubMed] [Google Scholar]
- Schluter V, Meyer M, Hofschneider PH, Koshy R, Caselmann WH. Integrated hepatitis B virus X and 3′ truncated preS/S sequences derived from human hepatomas encode functionally active transactivators. Oncogene. 1994;9:3335–3344. [PubMed] [Google Scholar]
- Severi T, Vander Borght S, Libbrecht L, VanAelst L, Nevens F, Roskams T, et al. HBx or HCV core gene expression in HepG2 human liver cells results in a survival benefit against oxidative stress with possible implications for HCC development. Chem Biol Interact. 2007;168:128–134. doi: 10.1016/j.cbi.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Shafritz DA, Shouval D, Sherman HI, Hadziyannis SJ, Kew MC. Integration of hepatitis B virus DNA into the genome of liver cells in chronic liver disease and hepatocellular carcinoma studies in percutaneous liver biopsies and post-mortem tissue specimens. N Engl J Med. 1981;305:1067–1073. doi: 10.1056/NEJM198110293051807. [DOI] [PubMed] [Google Scholar]
- Shin HD, Park BL, Kim LH, Jung JH, Kim JY, Yoon JH, et al. Interleukin 10 haplotype associated with increased risk of hepato-cellular carcinoma. Hum Mol Genet. 2003;12:901–906. doi: 10.1093/hmg/ddg104. [DOI] [PubMed] [Google Scholar]
- Shin JY, Hur W, Wang JS, Jang JW, Kim CW, Bae SH, et al. HCV core protein promotes liver fibrogenesis via up-regulation of CTGF with TGF-beta1. Exp Mol Med. 2005;37:138–145. doi: 10.1038/emm.2005.19. [DOI] [PubMed] [Google Scholar]
- Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirma H, Weil R, Rosmorduc O, Urban S, Israël A, Kremsdorf D, et al. Cytosol is the prime compartment of hepatitis B virus where it colocalizes with the proteasome. Oncogene. 1998;16:2051–2063. doi: 10.1038/sj.onc.1201737. [DOI] [PubMed] [Google Scholar]
- Sorrell MF, Belongia EA, Costa J, Gareen IF, Grem JL, Inadomi JM, et al. National Institutes of Health consensus development conference statement: management of hepatitis B. Hepatology. 2009;49(5 Suppl):S4–S12. doi: 10.1002/hep.22946. [DOI] [PubMed] [Google Scholar]
- Street A, Macdonald A, McCormick C, Harris M. Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular beta-catenin and stimulation of beta-catenin-responsive transcription. J Virol. 2005;79:5006–5016. doi: 10.1128/JVI.79.8.5006-5016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F, Schneider RJ. HBx protein activates transcription factor NF-kappaB by acting on multiple cytoplasmic inhibitors of rel-related proteins. J Virol. 1996;70:4558–4566. doi: 10.1128/jvi.70.7.4558-4566.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PF, Lee TC, Lin PJ, Lee PH, Jeng YM, Chen CH, et al. Differential DNA methylation associated with hepatitis B virus infection in hepatocellular carcinoma. Int J Cancer. 2007;121:1257–1264. doi: 10.1002/ijc.22849. [DOI] [PubMed] [Google Scholar]
- Sumida Y, Nakashima T, Yoh T, Nakajima Y, Ishikawa H, Mitsuyoshi H, et al. Serum thioredoxin levels as an indicator of oxidative stress in patients with hepatitis C virus infection. J Hepatology. 2000;13:159–163. doi: 10.1034/j.1600-0641.2000.033004616.x. [DOI] [PubMed] [Google Scholar]
- Sy SM, Wong N, Lai PB, To KF, Johnson PJ. Regional over-representations on chromosomes 1q, 3q and 7q in the progression of hepatitis B virus-related hepatocellular carcinoma. Mod Pathol. 2005;18:686–692. doi: 10.1038/modpathol.3800345. [DOI] [PubMed] [Google Scholar]
- Tai AW, Chung RT. Treatment failure in hepatitis C: mechanisms of non-response. J Hepatol. 2009;50:412–420. doi: 10.1016/j.jhep.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai PC, Suk FM, Gerlich WH, Neurath AR, Shih C. Hypermodification and immune escape of an internally deleted middle-envelope (M) protein of frequent and predominant hepatitis B virus variants. Virology. 2002;292:44–58. doi: 10.1006/viro.2001.1239. [DOI] [PubMed] [Google Scholar]
- Takeya R, Sumimoto H. Regulation of novel superoxideproducing NAD(P)H oxidases. Antioxid Redox Signal. 2006;8:1523–1532. doi: 10.1089/ars.2006.8.1523. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Moriya K, Kiyosawa K, Koike K, Aoyama T. Hepatitis C virus core protein induces spontaneous and persistent activation of peroxisome proliferator-activated receptor alpha in transgenic mice: implications for HCV-associated hepatocarcinogenesis. Int J Cancer. 2008a;122:124–131. doi: 10.1002/ijc.23056. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Moriya K, Kiyosawa K, Koike K, Gonzalez FJ, Aoyama T. PPAR-α is essential for severe hepatic steatosis and hepatocellular carcinoma induced by HCV core protein. J Clin Invest. 2008b;118:683–694. doi: 10.1172/JCI33594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- Tang H, Oishi N, Kaneko S, Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977–983. doi: 10.1111/j.1349-7006.2006.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, Kato N, Otsuka M, Goto T, Yoshida H, Shiratori Y, et al. Hepatitis C virus core protein upregulates transforming growth factor-beta 1 transcription. J Med Virol. 2004;72:52–59. doi: 10.1002/jmv.10545. [DOI] [PubMed] [Google Scholar]
- Tardif KD, Mori K, Siddiqui A. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J Virol. 2002;76:7453–7459. doi: 10.1128/JVI.76.15.7453-7459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant B, Toshkov I, Peek SF, Jacob JR, Menne S, Hornbuckle WE, et al. Hepatocellular carcinoma in the Woodchuck model of hepatitis B virus infection. Gastroenterology. 2004;127:S283–S293. doi: 10.1053/j.gastro.2004.09.043. [DOI] [PubMed] [Google Scholar]
- Thakur V, Guptan RC, Kazim SN, Malhotra V, Sarin SK. Profile, spectrum and significance of HBV genotypes in chronic liver disease patients in the Indian subcontinent. J Gastroenterol Hepatol. 2002;17:165–170. doi: 10.1046/j.1440-1746.2002.02605.x. [DOI] [PubMed] [Google Scholar]
- Tien Kuo M, Savaraj N. Roles of reactive oxygen species in hepatocarcinogenesis and drug resistance gene expression in liver cancers. Mol Carcinog. 2006;459:701–709. doi: 10.1002/mc.20240. [DOI] [PubMed] [Google Scholar]
- Tsuchiya H, Akechi Y, Ikeda R, Nishio R, Sakabe T, Terabayashi K, et al. Suppressive effects of retinoids on iron-induced oxidative stress in the liver. Gastroenterology. 2009;136:341–350. doi: 10.1053/j.gastro.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Tsutsumi T, Suzuki T, Shimoike T, Moriya K, Yotsuyanagi H, Matsuura Y, et al. Interaction of hepatitis C virus core protein with retinoid X receptor-α modulates its transcriptional activity. Hepatology. 2002;35:937–946. doi: 10.1053/jhep.2002.32470. [DOI] [PubMed] [Google Scholar]
- Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- Villanueva A, Newell P, Chiang DY. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007;27:55–76. doi: 10.1055/s-2006-960171. [DOI] [PubMed] [Google Scholar]
- Wang HC, Huang W, Lai MD, Shu IJ. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 2006;97:683–688. doi: 10.1111/j.1349-7006.2006.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zindy F, Chenivesse X, Lamas E, Henglein B, Bréchot C, et al. Modification of cyclin A expression by HBV DNA integration in a HCC. Oncogene. 1992;7:1652–1656. [PubMed] [Google Scholar]
- Wang T, Weinman SA. Causes and consequences of mitochondrial reactive oxygen species generation in hepatitis C. J Gastroenterol Hepatol. 2006;21:S34–S37. doi: 10.1111/j.1440-1746.2006.04591.x. [DOI] [PubMed] [Google Scholar]
- Wang XW, Forrester K, Yeh H, Feitelson MA, Gu JR, Harris CC. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity and association with ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CM, Ng IO. Molecular pathogenesis of hepatocellular carcinoma. Liver Int. 2008;28:160–174. doi: 10.1111/j.1478-3231.2007.01637.x. [DOI] [PubMed] [Google Scholar]
- Wu CF, Yu MW, Lin CL, Liu CJ, Shih WL, Tsai KS, et al. Long-term tracking of hepatitis B viral load and the relationship with risk for hepatocellular carcinoma in men. Carcinogenesis. 2008;29:106–112. doi: 10.1093/carcin/bgm252. [DOI] [PubMed] [Google Scholar]
- Wu SC, Chang SC, Wu HY, Liao PJ, Chang MF. Hepatitis C virus NS5A protein down-regulates the expression of spindle gene Aspm through PKR-p38 signaling pathway. J Biol Chem. 2008;283:29396–29404. doi: 10.1074/jbc.M802821200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- Yaginuma K, Kobayashi H, Kobayashi M, Morishima T, Matsuyama K, Koike K. Multiple integration site of hepatitis B virus DNA in hepatocellular carcinoma and chronic active hepatitis tissues from children. J Virol. 1987;61:1808–1813. doi: 10.1128/jvi.61.6.1808-1813.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T, Kodama T, Doi T. Subcellular localization of HCV core protein regulates its ability for p53 activation and p21 suppression. Biochem Biophys Res Commun. 2002;294:528–534. doi: 10.1016/S0006-291X(02)00508-9. [DOI] [PubMed] [Google Scholar]
- Yanagitani A, Yamada S, Yasui S, Shimomura T, Murai R, Murawaki Y, et al. Retinoic acid receptor alpha dominant negative form causes steatohepatitis and liver tumors in transgenic mice. Hepatology. 2004;40:366–375. doi: 10.1002/hep.20335. [DOI] [PubMed] [Google Scholar]
- Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, et al. REVEAL-HBV Study Group Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:1134–1143. doi: 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JC, Teng CF, Wu HC, Tsai HW, Chuang HC, Tsai TF, et al. Enhanced expression of vascular endothelial growth factor-A in ground glass hepatocytes and its implication in hepatitis B virus hepatocarcinogenesis. Hepatology. 2009;49:1962–1971. doi: 10.1002/hep.22889. [DOI] [PubMed] [Google Scholar]
- Yang L, Wang Y, Mao H, Fleig S, Omenetti A, Brown KD, et al. Sonic hedgehog is an autocrine viability factor for myofibroblastic hepatic stellate cells. J Hepatol. 2008;48:98–106. doi: 10.1016/j.jhep.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo YD, Ueda H, Park K, Flanders KC, Lee YI, Jay G, et al. Regulation of transforming growth factor-beta 1 expression by the hepatitis B virus (HBV) X transactivator. Role in HBV pathogenesis. J Clin Invest. 1996;97:388–395. doi: 10.1172/JCI118427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You LR, Chen CM, Yeh TS, Tsai TY, Mai RT, Lin CH, et al. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J Virol. 1999;73:2841–2853. doi: 10.1128/jvi.73.4.2841-2853.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DY, Moon HB, Son JK, Jeong S, Yu SL, Yoon H, et al. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J Hepatol. 1999;31:123–132. doi: 10.1016/s0168-8278(99)80172-x. [DOI] [PubMed] [Google Scholar]
- Yu MW, Horng IS, Hsu KH, Chiang YC, Liaw YF, Chen CJ. Plasma selenium levels and risk of hepatocellular carcinoma among men with chronic hepatitis virus infection. Am J Epidemiol. 1999;150:367–374. doi: 10.1093/oxfordjournals.aje.a010016. [DOI] [PubMed] [Google Scholar]
- Yu MW, Yang YC, Yang SY, Cheng SW, Liaw YF, et al. Hormonal markers and hepatitis B virus-related hepatocellular carcinoma risk: a nested case-control study among men. J Natl Cancer Inst. 2001;93:1644–1651. doi: 10.1093/jnci/93.21.1644. [DOI] [PubMed] [Google Scholar]
- Yu MW, Yang SY, Pan IJ, Lin CL, Liu CJ, Liaw YF, et al. Polymorphisms in XRCC1 and glutathione S-transferase genes and hepatitis B-related hepatocellular carcinoma. J Natl Cancer Inst. 2003;95:1485–1488. doi: 10.1093/jnci/djg051. [DOI] [PubMed] [Google Scholar]
- Yu MW, Yeh SH, Chen PJ, Liaw YF, Lin CL, Liu CJ, et al. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: a prospective study in men. J Natl Cancer Inst. 2005;97:265–272. doi: 10.1093/jnci/dji043. [DOI] [PubMed] [Google Scholar]
- Yuan JM, Gao YT, Ong CN, Ross RK, Yu MC. Prediagnostic level of serum retinol in relation to reduced risk of hepatocellular carcinoma. J Natl Cancer Inst. 2006;98:482–490. doi: 10.1093/jnci/djj104. [DOI] [PubMed] [Google Scholar]
- Yuan JM, Ambinder A, Fan Y, Gao YT, Yu MC, Groopman JD. Prospective evaluation of hepatitis B 1762(T)/1764(A) mutations on hepatocellular carcinoma development in Shanghai, China. Cancer Epidemiol Biomarkers Prev. 2009;18:590–594. doi: 10.1158/1055-9965.EPI-08-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemel R, Gerechet S, Greif H, Bachmatove L, Birk Y, Golan-Goldhirsh A, et al. Cell transformation induced by hepatitis C virus NS3 serine protease. J Viral Hepat. 2001;8:96–102. doi: 10.1046/j.1365-2893.2001.00283.x. [DOI] [PubMed] [Google Scholar]
- Zhao LJ, Wang L, Ren H, Cao J, Li L, Ke JS, et al. Hepatitis C virus E2 protein promotes human hepatoma cell proliferation through the MAPK/ERK signaling pathway via cellular receptors. Exp Cell Res. 2005;305:23–32. doi: 10.1016/j.yexcr.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Zheng DL, Zhang L, Cheng N, Xu X, Deng Q, Teng XM, et al. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J Hepatol. 2009;50:377–387. doi: 10.1016/j.jhep.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Zondervan PE, Wink J, Alers JC, IJzermans JN, Schalm SW, de Man RA, et al. Molecular cytogenetic evaluation of virus-associated and non-viral hepatocellular carcinoma: analysis of 26 carcinomas and 12 concurrent dysplasias. J Pathol. 2000;192:207–215. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH690>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]