Abstract
Metastasis is a predominant cause of death in patients with cancer. It is a complex multistep process that needs to be better understood if we are to develop new approaches to managing tumor metastasis. Tumor cell invasion of the local stroma is suppressed by collapsin response mediator protein-1 (CRMP-1). Recently, we identified a long isoform of CRMP-1 (LCRMP-1), expression of which correlates with cancer cell invasiveness and poor clinical outcome in patients with non-small-cell lung cancer (NSCLC). Here, we report that LCRMP-1 overexpression in noninvasive human cell lines enhanced filopodia formation, cancer cell migration, and invasion via stabilization of actin. This effect required a highly conserved N-terminal region of LCRMP-1 as well as the WASP family verprolin-homologous protein-1/actin nucleation pathway (WAVE-1/actin nucleation pathway). Furthermore, LCRMP-1 appeared to act downstream of Cdc42, a Rho family protein known to be involved in actin rearrangement. In addition, LCRMP-1 associated with CRMP-1, which downregulated cancer cell metastasis by interrupting the association of LCRMP-1 and WAVE-1. Finally, we found that high-level expression of LCRMP-1 and low-level expression of CRMP-1 were associated with lymph node metastasis and poor survival in patients with NSCLC. In sum, we show that LCRMP-1 and CRMP-1 have opposing functions in regulating cancer cell invasion and metastasis and propose that this pathway may serve as a potential anticancer target.
Introduction
Cancer metastasis, which is the major cause of treatment failure in cancer patients, is a complex process that involves basement membrane degradation, cell migration, stromal (local) invasion, angiogenesis, intravasation into the circulatory system, adhesion, extravasation into the parenchyma of distant tissues, and colonization (1–3). These processes are regulated by numerous metastasis-promoting and -suppressing genes (4). Thus, identifying novel metastatic genes and their action mechanisms may provide new insights into the pathogenesis and management of cancer metastasis.
We previously identified collapsin response mediator protein-1 (CRMP-1) as a novel invasion suppressor and showed that CRMP-1 expression is negatively associated with cell invasiveness and positively associated with better clinical outcomes in patients with non-small-cell lung cancer (NSCLC) (5). Recent studies (6, 7) have shown that CRMP-1 is functionally involved in connective tissue growth factor–mediated inhibition of invasion and metastasis in human lung adenocarcinoma.
The CRMPs comprise a family of 5 cytosolic phosphoproteins that inhibit extension of the axonal growth cone during neuronal development (8–11). The members of the CRMP family are closely related 60- to 66-kDa proteins that share 50%–70% amino acid sequence homology and are capable of forming heterotetramers (8, 11–14). These proteins are distributed mainly in the lamellipodia and filopodia of a neuron’s axonal growth cone (14, 15), in which they mediate the signaling pathways that control axonal growth cone collapse (8, 14) and promote growth cone collapse by depolymerizing F-actin (15, 16). Recent studies (8, 12) have shown that the effect of CRMP-2 on growth cone collapse in dorsal root ganglion cells is mediated through signal transduction cascades that involve either heterotrimeric G proteins or a Rho-associated protein kinase.
F-actin reorganization plays a major role in cell movement. Cdc42, Rac, and Rho, which are 3 small GTPases of the Rho family, are key regulators of actin assembly that control the formation of filopodia, lamellipodia, and stress fibers, respectively (17–20). These small GTPases transmit extracellular chemotactic signals to downstream effectors, such as members of the Wiskott-Aldrich syndrome protein (WASP) family, which are key regulators of actin polymerization (20–22). Activated WASPs induce the formation of protrusive membrane structures that are involved in cell migration and degradation of the extracellular matrix. To date, 5 mammalian WASP family proteins have been identified: WASP, neural WASP (N-WASP), and the 3 WASP family verprolin-homologous proteins (WAVEs), WAVE-1, WAVE-2, and WAVE-3. These proteins link Cdc42- and Rac-dependent signaling to the formation of filopodia and lamellipodia, respectively, by activating the Arp2/3 complex that mediates de novo actin polymerization (21, 23–25).
We recently identified a novel human isoform of the CRMP family proteins, called long-form CRMP-1 (LCRMP-1) and showed that LCRMP-1 expressions are associated with poor clinical outcome and lymph node metastasis in patients with NSCLC (26). Here, we characterized the functions of LCRMP-1 in cancer cell invasion, migration, and metastasis. We further dissected the molecular mechanism through which LCRMP-1 promotes cancer cell invasion; our results suggest that the invasion and migration abilities of cancer cells are controlled by the expressions and interactions of CRMP-1 and LCRMP-1.
Results
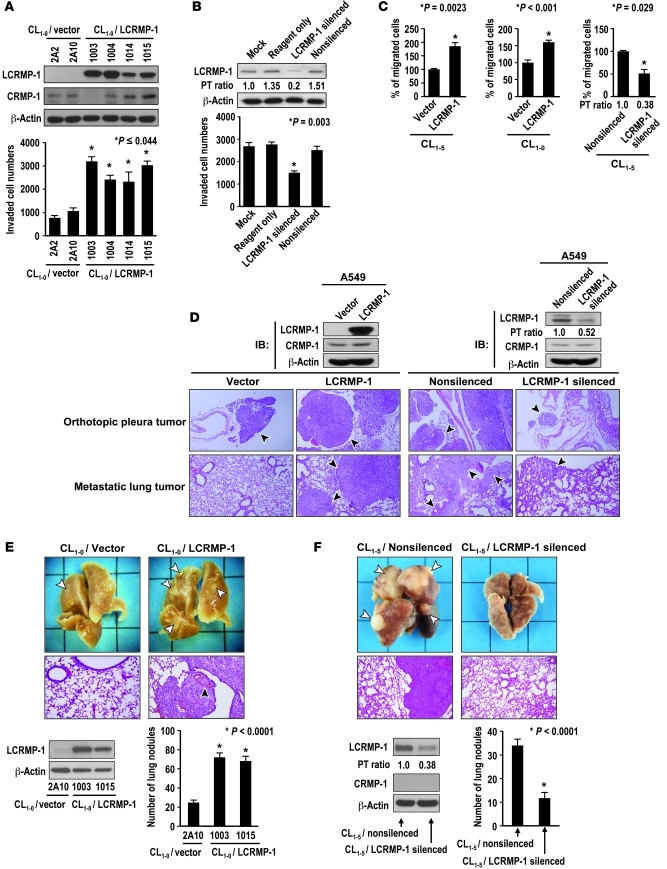
LCRMP-1 expression promotes cancer cell invasion and migration in vitro.
We reported that the expression of LCRMP-1 was correlated with lymph node metastasis in patients with NSCLC (26). To extend this finding, we asked whether ectopic LCRMP-1 expression could promote cancer cell migration, invasion, and metastasis. To examine this possibility, we established Flag-tagged LCRMP-1 stably expressing cell lines in low-invasive CL1-0 cells (27), and the protein expression patterns of both LCRMP-1 and CRMP-1 were confirmed by immunoblotting with specific antibodies (details are shown in Supplemental Results and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI42975DS1). The cells were then used to examine the effect of LCRMP-1 on cell proliferation, migration, and invasion. The results showed that a nonsignificant difference in growth was observed in our established cell lines (Supplemental Figure 2); but in a modified Boyden chamber invasion assay, LCRMP-1 overexpression increased cell invasiveness 2.5–3 fold compared with that of the control (P ≤ 0.044; Figure 1A). Conversely, knockdown of endogenous LCRMP-1 expression in highly invasive CL1-5-F4 cells through the use of small interference RNA (siRNA) reduced cell invasiveness by 50% (P = 0.003; Figure 1B). The effects of LCRMP-1 expression on cell invasiveness were confirmed in H23 (lung adenocarcinoma) and SN12C (renal cell carcinoma) cells (Supplemental Figure 3).
Figure 1. Function of LCRMP-1 in cancer invasion, migration, and metastasis.
(A and B) Protein levels of LCRMP-1 and CRMP-1 in CL1-0/LCRMP-1–overexpressing (4 stable clones, 1003, 1004, 1014, 1015; 2 controls, 2A2, 2A10) and CL1-5-F4/LCRMP-1–silenced (untreated [mock], oligofectamine [reagent only], LCRMP-1–targeting siRNA [LCRMP-1 silenced], or scrambled siRNA [nonsilenced]) cells were examined by immunoblotting. β-Actin was used as internal control. PT ratio, LCRMP-1 protein level normalized to that of mock. (A) Number of invading cells from LCRMP-1–expressing clones and (B) LCRMP-1–silenced cells quantified from modified Boyden chamber invasion assay (n = 3 experiments). (C) Effects of LCRMP-1 on cancer cell migration were examined by wound-healing assay in pools of stably transfected CL1-5/vector control, CL1-5/LCRMP-1, CL1-0/vector, CL1-0/LCRMP-1, CL1-5/nonsilenced, and CL1-5/LCRMP-1 silenced cells. Percentages of migrated cells were quantified from pictures taken at 0 and 12 hours after wounding (n = 6 experiments). (D–F) Effects of LCRMP-1 cancer metastasis in vivo examined by orthotopic implantation with stable (D) A549/LCRMP-1–overexpressing (left) or A549/LCRMP-1–silenced cells (right), and tail vein metastasis assays with (E) stable CL1-0/LCRMP-1–overexpressing (lines 1003 and 1015) or (F) CL1-5/LCRMP-1–silenced cells. Numbers of metastatic tumor nodules were calculated from 10 mice per group (n = 2 experiments). Histology was confirmed by H&E staining (original magnification, ×100). (D–F) Arrowheads indicate orthotopic or metastatic lung tumors. (E) The black arrowhead indicates where tumor cells invade blood vessels and form tumor thrombi. Error bars indicate mean ± SEM, and P values were calculated by 2-sided Student’s t test.
In a wound-healing assay, LCRMP-1 expression increased the migration of both less-invasive CL1-0 cells and highly invasive CL1-5 cells, whereas knockdown of LCRMP-1 protein expression in CL1-5 cells decreased cell migration (all P < 0.029; Figure 1C and Supplemental Figure 4). Collectively, these results indicate that LCRMP-1 expression can promote cancer cell invasion and migration in an in vitro model.
LCRMP-1 expression increases metastasis in vivo.
We further assessed whether LCRMP-1 could promote metastasis in vivo. The cancer cells were orthotopically inoculated directly into the pleural cavities of mice and examined for the formation of metastatic nodules. We found that A549/LCRMP-1 cells formed larger orthotopic tumors (tumor volume, 5.5 ± 2.07 mm for A549/LCRMP-1 and 2.22 ± 1.3 mm for A549/vector; P = 0.003) and more metastatic lung tumor nodules (mean number of nodules, 9.13 ± 3.0 for A549/LCRMP-1 and 3.44 ± 2.24 for A549/vector; P < 0.001) than the vector control cells. Conversely, fewer metastatic lung tumor nodules were formed by A549/LCRMP-1–silenced cells compared with nonsilenced control cells (mean number of nodules, 5.25 ± 3.65 for A549/LCRMP-1–silenced and 10.14 ± 2.61 for A549/nonsilenced cells; P = 0.01; Figure 1D and Table 1).
Table 1 .
LCRMP-1 induces regulation of lung metastasis in an orthotopic inoculation mouse modelA
To further investigate the effects of LCRMP-1 on the later steps of metastatic progression, we injected LCRMP-1–overexpressing cells directly into the circulation of mice, thereby eschewing the initial steps of local invasion and intravasation. We found that mice injected intravenously with CL1-0/LCRMP-1 cells (lines 1003 and 1015) developed more pulmonary nodules than those injected with CL1-0/vector cells (line 2A10) (mean number of nodules, 72.1 ± 13.67 for line 1003, 68.2 ± 14.97 for line 1015, and 24.6 ± 8.33 for control 2A10; both P < 0.0001; Figure 1E). In contrast, mice that were injected with CL1-5/LCRMP-1–silenced cells developed fewer pulmonary nodules than those injected with nonsilenced control CL1-5 cells (mean number of nodules, 11.7 ± 7.66 for LCRMP-1–silenced and 34.0 ± 7.95 for nonsilenced control cells; P < 0.0001; Figure 1F). Metastatic lung tumor nodules formed by CL1-0/LCRMP-1 and CL1-5/nonsilenced control cells had morphologies consistent with that of a typical adenocarcinoma (Figure 1, E and F, middle). All relevant statistical data are shown in Table 2.
Table 2 .
LCRMP-1 induces regulation of lung metastasis in a tail vein metastasis mouse modelA
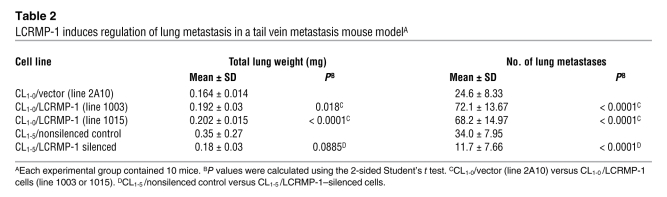
Localization of LCRMP-1 with F-actin and association with filopodia formation.
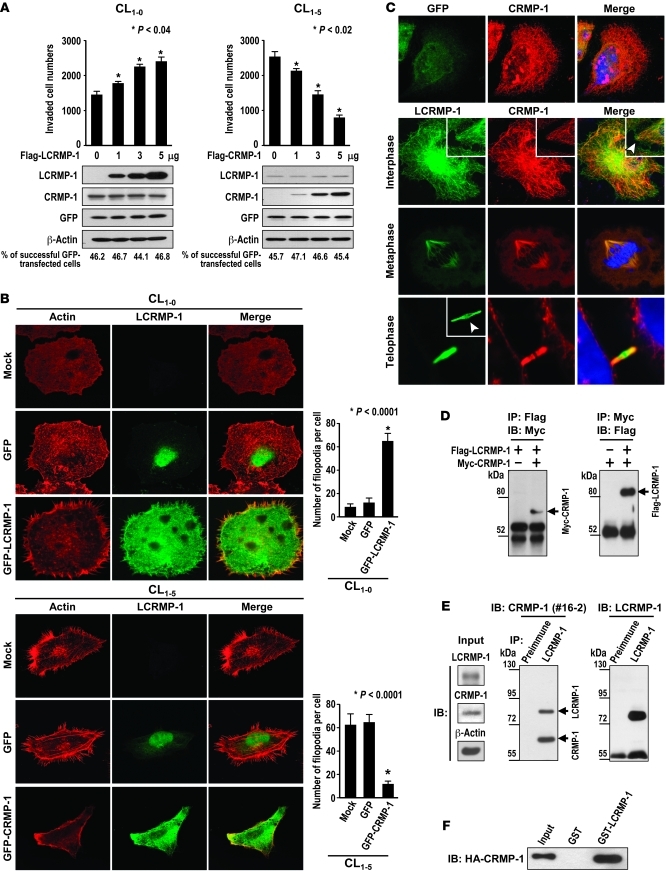
To investigate whether the effect of LCRMP-1 on cancer cell invasion and metastasis is associated with actin filaments, we first examined the localization of LCRMP-1 and actin filaments. Immunofluorescence staining of CL1-5 cells with an anti–LCRMP-1 antibody and rhodamine-conjugated phalloidin (for actin) showed that endogenous LCRMP-1 and actin may share some common compartments in localization, especially in the filopodia (Figure 2A). Similarly, exogenous GFP–LCRMP-1 fusion proteins shared common localization with rhodamine-conjugated phalloidin–stained actin in CL1-0 and CL1-5 cells, especially in the regions of the lamellipodia and filopodia. Morphologically, control CL1-0 cells were rounded and had few filopodia, while GFP–LCRMP-1–expressing CL1-0 cells were elongated and had far more filopodia (P < 0.001; Figure 2B, top right). Similarly, GFP–LCRMP-1–overexpressing CL1-5 cells formed more filopodia than control CL1-5 cells (P < 0.001; Figure 2B, bottom right). Furthermore, cytochalasin D treatment revealed that LCRMP-1–induced filopodia formation was associated with the reorganization of actin filaments (Supplemental Figure 5). Collectively, these findings indicate that LCRMP-1 induces filopodia formation and that this process is associated with the reorganization of actin filaments.
Figure 2. Effects of LCRMP-1 on filopodia formation.
(A and B) Localization of (A) endogenous LCRMP-1 (green), (B) exogenous GFP-LCRMP-1 (green), and actin (red) by immunofluorescent staining. (A) Preimmune serum and (B) CL1-0/CL1-5 mock were used as controls. LCRMP-1 shares some common compartments with actin in its distribution, especially in lamellipodia and filopodia regions. Arrows indicate presence of LCRMP-1 in the filopodia region. (B) In addition, exogenous GFP–LCRMP-1 promotes filopodia formation in both CL1-0 and CL1-5 cells. Numbers of filopodia were counted (n = 20 cells per group; original magnification, ×1,000 [A, top 2 and the bottom rows, and B, top and bottom row]; ×4,000 [A, third row, and B, middle row]). (C) Schematic of GFP-tagged LCRMP-1 N-terminal deletion mutants (LCRMP-1, LCRMP-1–P28AQ30A, LCRMP-1–R29AK31A, LCRMP-1Δ22, LCRMP-1Δ72, LCRMP-1Δ105, LCRMP-1Δ127, and CRMP-1; the double asterisks indicate the 2 mutation sites of each point mutation). (D) CL1-0 cells were transfected with indicated GFP-tagged LCRMP-1 N-terminal deletion constructs and actin stained with rhodamine-conjugated phalloidin (red). Number of filopodia were counted (n = 20 cells per group; original magnification, ×1,000). Data are presented as mean ± SEM, and P values were calculated by 2-sided Student’s t test.
The LCRMP-1 N-terminal conserved region (residues 22–72) controls filopodia formation in vitro.
Our previous sequence analysis showed that the LCRMP-1 and CRMP-1 proteins only differed in their N-terminal regions, including exon 1 (26). Here, we examined whether the 127 N-terminal amino acids of LCRMP-1 could be involved in actin rearrangement. We first performed amino acid sequence alignments of human LCRMP-1 with the other longer CRMP family members (also known as CRMP-A members), like p80 and TUC-4b from mouse (28, 29) and Ch1A ~4A from chicken (Ch1A~4A) (30). The result showed that N-terminal residues 22–72 of LCRMP-1 are highly conserved across species (Supplemental Figure 6). Based on this finding, we constructed various GFP-tagged N-terminal deletion mutants of LCRMP-1 (Figure 2C), overexpressed these constructs in CL1-0 cells, and stained the cells with rhodamine-conjugated phalloidin. Notably, CL1-0 cells expressing LCRMP-1 proteins lacking residues 22–72 failed to form filopodia (P < 0.0002; Figure 2D). We then introduced point mutations into the amino acid regions that showed the highest conservation between LCRMP-1 and the CRMP-A proteins, constructed 4 GFP-tagged LCRMP-1 mutants (P28AQ30A, R29AK31A, V39AE40A, and G34AF36A), and overexpressed these proteins in CL1-0 cells. Overexpression of mutants P28AQ30A and R29AK31A failed to induce filopodia formation, whereas mutants G34AF36A and V39AE40A induced filopodia formation when overexpressed (Figure 2D and Supplemental Figure 7).
Overexpression of LCRMP-1 stabilizes F-actin structures and induces dynamic filopodia formation.
F-actin, which is the polymerized form of G-actin, plays a critical role in filopodia formation and cell migration (17–20). Since LCRMP-1 was found to induce filopodia formation, we next examined the F- and G-actin contents in LCRMP-1–expressing cells to determine whether LCRMP-1 can affect dynamic actin rearrangement. Immunofluorescence staining of G- and F-actin showed that LCRMP-1 overexpression increased the amount of F-actin versus that of G-actin. A ratio-merge analysis was used to identify cells with more F- or G-actin structures. Fluorescence quantification showed that the mean ratio of F-actin to total actin in CL1-0/LCRMP-1 cells was higher than that in CL1-0/vector control cells (54.28% ± 6.23% vs. 31.00% ± 10.01%; P < 0.001; Figure 3A). In contrast, there was no difference in this ratio between LCRMP-1 R29AK31A mutant-expressing cells and controls. Immunoblotting analyses of the G- and F-actin fractions from stable transfectants showed that the relative expression level of F-actin/G-actin was higher in LCRMP-1–overexpressing CL1-0 cells compared with that in vector control CL1-0 cells (P < 0.001; Figure 3B). Although the amount of F-actin was increased in cells expressing R29AK31A mutant LCRMP-1 proteins, the relative expression ratio of F- versus G-actin was lower in these cells compared with that in cells overexpressing wild-type LCRMP-1 (P < 0.001; Figure 3B) These results collectively suggest that LCRMP-1 induces filopodia formation, stabilizes F-actin polymerization, and supports the transition of G-actin to F-actin.
Figure 3. Effects of LCRMP-1 on actin dynamics.
(A) Expressions of G-actin and F-actin in indicated CL1-0 stable cells (vector control, LCRMP-1, and LCRMP-1 R29AK31A mutant), analyzed by immunofluorescence staining with FITC-conjugated DNase I (G-actin, green) and rhodamine-conjugated phalloidin (F-actin, red). For ratio-merge panel, blue and red indicate F-actin and G-actin, respectively. The level of F-actin relative to that of total actin was quantified (n = 100 cells per group; original magnification, ×400). (B) Expressions of G-actin and F-actin in indicated CL1-0 stable cells analyzed by immunoblotting. The protein level of F-actin relative to that of constant G-actin was quantified (n = 3 experiments). (C) F-actin reorganization examined by Dynamic NIWOP tomography in CL1-0/vector (line 2A10) and CL1-0/LCRMP-1 (lines 1003 and 1015) cells. Arrows indicate filopodia. Filopodia were counted every 5 minutes, and the averages over a 25-minute period were calculated from 8 random cells in each group (n = 2 experiments; original magnification, ×1,000). Error bars indicate mean ± SEM, and P values were calculated by 2-sided Student’s t test.
We further observed the dynamics of the lamellipodia and filopodia formed by LCRMP-1–overexpressing and vector-transfected CL1-0 cells using noninterferometric wide-field optical profilometric (NIWOP) super-resolution bright-field optical microscopy (31–33). Our results revealed that LCRMP-1–overexpressing CL1-0 cells produced more filopodia per 5 minutes than the vector controls (mean number of filopodia, 21.9 ± 3.2 for 1003 cells and 7.1 ± 2.1 for control 2A10 cells, P < 0.001; Figure 3C). A 6-minute video of one observation is available in Supplemental Videos 1 and 2. These results suggest that LCRMP-1 induces filopodia formation through a rapid and dynamic reorganization of F-actin filaments.
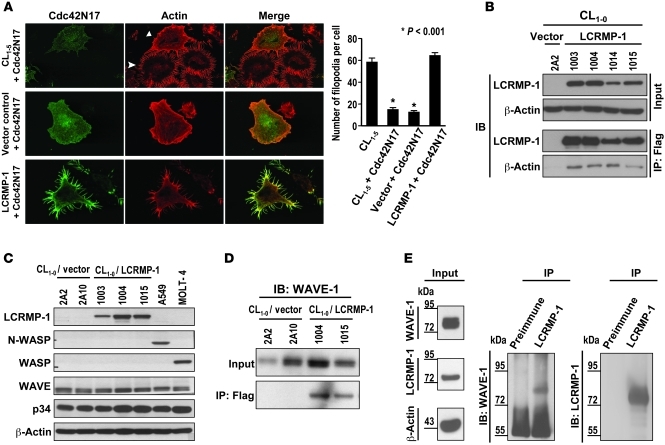
LCRMP-1 acts as a downstream effector of Cdc42.
Since the Rho family proteins Cdc42 and Rac are known to play major roles in actin rearrangement, we evaluated whether there could be a relationship between LCRMP-1 and Cdc42. Transfection with a plasmid expressing Cdc42N17, which is a dominant-negative form of Cdc42, suppressed filopodia formation in CL1-5 cells (P < 0.001; Figure 4A). This effect of Cdc42N17 was blocked by cotransfection of an LCRMP-1–expressing plasmid (Figure 4A) but not the vector control (P < 0.001; Figure 4A), indicating that LCRMP-1 can overcome the suppressive effects of dominant-negative Cdc42 on filopodia formation. When CL1-5 cells were cotransfected with N-terminal–deletion constructs or point mutants of LCRMP-1 plus Cdc42N17, LCRMP-1 mutants lacking residues 22–72 and those harboring point mutations P28AQ30A and R29AK31A were unable to reverse the Cdc42N17-induced regression of filopodia (Supplemental Figure 8). These results suggest that the conserved N-terminal region of LCRMP-1 is the functional domain necessary for filopodia formation.
Figure 4. Molecular mechanism of LCRMP-1–induced filopodia formation.
(A) Effects of LCRMP-1 on Cdc42N17-induced filopodia regression. CL1-5 cells were transfected with pcDNA3-Cdc42N17 (top) or cotransfected with pcDNA3-Cdc42N17 and pCMV-Tag 2A (middle) or pCMV-Tag 2A-LCRMP-1 (bottom). Transfected cells were then stained with anti-Myc antibody (Cdc42N17, green) and rhodamine-conjugated phalloidin (actin, red). Arrow and arrowhead indicate cells with and without Cdc42N17 transfectants, respectively. The numbers of filopodia per cell were calculated from 20 cells in each group (original magnification, ×1,000). (B) Lysates from CL1-0/LCRMP-1 (lines 1003, 1004, 1014, and 1015) and CL1-0/vector (line 2A2) cells were immunoprecipitated with anti-Flag antibody and immunoblotted for LCRMP-1 and β-actin. (C) Actin-nucleation proteins, N-WASP, WASP, WAVE, and p34, were analyzed by immunoblotting in indicated cell lines. β-Actin was used as internal control. (D) Flag-tagged LCRMP-1 binds to WAVE-1. Lysates of CL1-0/LCRMP-1 (lines 1004 and 1015) and CL1-0/vector cells (lines 2A2 and 2A10) were examined for presence of WAVE-1 (input) and WAVE-1 coprecipitated with Flag-tagged LCRMP-1 (IP: Flag) by immunoblotting with anti–WAVE-1 antibody (n = 3 experiments). (E) Endogenous LCRMP-1 interacts with WAVE-1. Total cell lysates (15 mg) from CL1-5 cells were immunoprecipitated with anti–LCRMP-1 (C2) antibody and analyzed by immunoblotting using anti–WAVE-1 and anti–LCRMP-1 (C2) antibodies (n = 3 experiments). Error bars indicate mean ± SEM, and P values were calculated by 2-sided Student’s t test.
The effect of LCRMP-1 on actin nucleation and filopodia formation.
The studies described above suggested that LCRMP-1 may share the same compartment in localization with F-actin filaments and acts as a downstream effector of Cdc42. Next, we further explored the cellular importance of LCRMP-1–induced actin rearrangement. Using monoclonal anti-Flag antibodies to immunoprecipitate lysates from Flag–LCRMP-1–expressing cells, we confirmed that wild-type LCRMP-1 associated with actin, but LCRMP-1 proteins harboring mutations in the conserved N-terminal region did not (Figure 4B and Supplemental Figure 9). These results suggest that the conserved N-terminal region of LCRMP-1 is the functional domain responsible for actin binding.
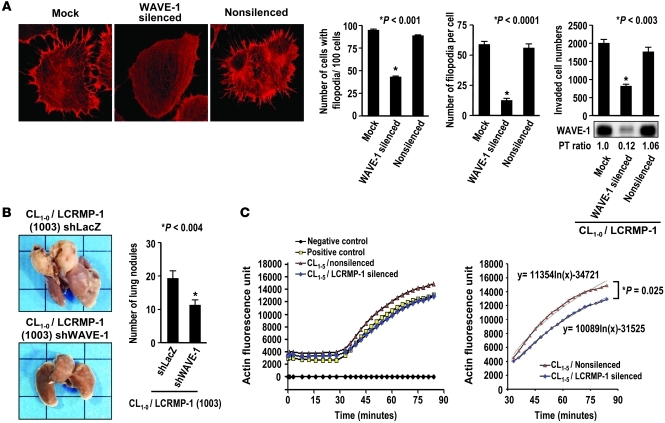
Since LCRMP-1 can associate with actin and promote filopodia formation in vitro, we then asked whether LCRMP-1 is involved in the process of actin nucleation, the initial step in the formation of an actin filament, in which actin monomers combine to form a new filament. We examined the expressions of actin-nucleation proteins in CL1-0/vector and CL1-0/LCRMP-1 stable transfectants. Interestingly, the most common actin-nucleation proteins, N-WASP and WASP, were not expressed in the CL lung cancer cell lines (Figure 4C). Instead, these cell lines expressed another subset of the WASP family proteins: the WAVE proteins. Other actin-nucleation binding proteins, such as p34, which is a component of the Arp complex (34), were present in all tested cells (Figure 4C). These findings suggested that LCRMP-1 may promote filopodia formation through the WAVE/actin-nucleation pathway rather than via the WASP pathway. In support of this, immunoprecipitation studies on both exogenous and endogenous LCRMP-1 confirmed that LCRMP-1 could bind to WAVE-1 (Figure 4, D and E, and Supplemental Figure 10). Further, in experiments to determine the reciprocal interaction domains of LCRMP-1 and WAVE-1 by in vitro transcription/translation and GST pull-down assays and coimmunoprecipitation assays, we found that residues 1–72 of LCRMP-1 were required for WAVE-1 binding and LCRMP-1 may directly bind to the Src homology domain (SHD) and the basic domain of WAVE-1 (Supplemental Figure 11). siRNA-based silencing of endogenous WAVE-1 expression in LCRMP-1–overexpressing CL1-0 cells (line 1003) led to a dramatic regression of filopodia (Figure 5A, left). This not only decreased the number of cells with filopodia (mean number of filopodia, 43.3 ± 1.2 for WAVE-1–silenced cells and 95.0 ± 1.7 for mock-transfected cells; P < 0.001), but the number of filopodia per cell (P < 0.0001) and the degree of cell invasiveness (fold change of invasiveness, WAVE-1 silenced vs. mock = 0.27 and nonsilenced control vs. mock = 0.96; P < 0.003 for both; Figure 5A) were also significantly changed. To confirm this effect in vivo, we established stable WAVE-1–knockdown cells using a shRNA virus infection system in CL1-0/LCRMP-1–overexpressing cells (line 1003). We then injected these cells directly into the circulation of mice and assessed tumor (nodule) formation. Our results showed that mice injected with WAVE-1–knockdown cells developed fewer pulmonary nodules than those mice injected with nonsilenced (shLacZ) control cells (mean number of nodules, 11.3 ± 4.6 for CL1-0/LCRMP-1/shWAVE-1 cells vs. 21.0 ± 11.1 for CL1-0/LCRMP-1/shLacZ cells; P < 0.004; Figure 5B). These findings suggest that LCRMP-1 may associate with actin filaments and regulate filopodia formation through the WAVE-1 nucleation pathway.
Figure 5. WAVE-1 is involved in LCRMP-1–induced cancer metastasis.
(A) In vitro effects of siRNA-mediated silencing of WAVE-1 in CL1-0/LCRMP-1 cells (line 1003). Cells were analyzed for WAVE-1 protein expression by immunoblotting (right bottom) and examined for invasive ability by modified Boyden chamber invasion assay (right top) and filopodia formation by staining with rhodamine-conjugated phalloidin (left; original magnification, ×1,000). Percentage of cells with filopodia and number of filopodia per cell were counted (n = 100 cells per group). (B) In vivo effects of WAVE-1 knockdown in CL1-0/LCRMP-1 cells (line 1003). Mice that were tail vein injected with CL1-0/LCRMP-1/shWAVE-1 cells developed fewer pulmonary nodules than those injected with CL1-0/LCRMP-1/shLacZ cells (10 mice per group; n = 2 experiments). (C) pEGFP–WAVE-1 plasmids were transfected into CL1-5/LCRMP-1–silenced and CL1-5/nonsilenced cells, and then the GFP-WAVE-1 fusion proteins of each group were immunoprecipitated by anti-GFP antibody to examine their ability to promote actin polymerization (Actin Polymerization Biochem Kit; left; n = 2 experiments). Slopes of regression curves were calculated. Error bars indicate mean ± SEM, and P values were calculated by 2-sided Student’s t test.
Silencing LCRMP-1 protein expression decreases the activity of WAVE-1 in actin polymerization.
To determine whether LCRMP-1 is affecting WAVE-1 activity in actin polymerization, the GFP–WAVE-1 protein was overexpressed in CL1-5/nonsilenced and CL1-5/LCRMP-1–silenced cells. Then, the fusion proteins were captured by protein A beads that coupled with anti-GFP antibody and used to examine the effects on initiation of actin polymerization (35). The results showed that the slope of actin polymerization curve significantly decreased when CL1-5 cells lost LCRMP-1 protein expression (P = 0.025; Figure 5C). This implies that LCRMP-1 may regulate the activity of WAVE-1 and then promote actin polymerization and filopodia formation.
The expression levels of LCRMP-1 and CRMP-1 control cancer cell invasion and filopodia formation.
Since we previously showed that CRMP-1 acts as an invasion suppressor (5, 16), and the above-described results illustrated that LCRMP-1 acts as a novel enhancer of cancer metastasis to our knowledge, we next examined whether differences in the expression levels of these 2 proteins could control the invasiveness of cancer cells. To test this, we transfected different amounts of Flag-tagged LCRMP-1 into less-invasive CL1-0 cells (which express endogenous CRMP-1 but not LCRMP-1) and Flag-tagged CRMP-1 into highly invasive CL1-5 cells (which express LCRMP-1 but not CRMP-1) and then assessed the invasiveness of the transfectants. Our results revealed that invasive ability was significantly correlated with the expression levels of LCRMP-1 and CRMP-1 (both P < 0.04; Figure 6A and Supplemental Figure 12). Furthermore, less-invasive CL1-0 cells expressing GFP-tagged LCRMP-1 were observed to form filopodia, whereas filopodia regression was observed among highly invasive CL1-5 cells expressing GFP-tagged CRMP-1 (both P < 0.0001; Figure 6B). The counterregulatory effects of LCRMP-1 and CRMP-1 on cancer cell invasion were further confirmed in H226 (lung squamous cell carcinoma) cells, which do not normally express either protein (Supplemental Figure 13).
Figure 6. Reciprocal regulation of cancer invasion by LCRMP-1 and CRMP-1.
(A) Less-invasive CL1-0 cells, which typically express endogenous CRMP-1 but not LCRMP-1, were transfected with different amounts of pCMV-Tag 2A–LCRMP-1. Protein levels were confirmed by immunoblotting. Equal amounts of pEGFP were cotransfected into the cells as a control of transfection efficiency. Percentage of GFP-transfected cells was quantified from immunofluorescence. Invasiveness was measured by modified Boyden chamber invasion assay (n = 3) (left). The identical experiment, except for plasmid transfected (pCMV-Tag 2A-CRMP-1), performed in highly invasive CL1-5 cells, which endogenously express LCRMP-1 but not CRMP-1 (right). (B) Effects of LCRMP-1 and CRMP-1 on morphology in CL1-0 or CL1-5. Cells were transfected with pEGFP, pEGFP-LCRMP-1, or pEGFP-CRMP-1. Actin was visualized with rhodamine-conjugated phalloidin. Number of filopodia per cell was calculated (n = 20 cells per group). (C) pEGFP-LCRMP-1 or pEGFP were cotransfected with pDsRed-CRMP-1 into CL1-0 cells to detect localization of exogenous LCRMP-1 and CRMP-1 in interphase, metaphase, and telophase (n = 20 cells per group). Arrowheads indicate the place in which only GFP-LCRMP-1 was present. (D and E) Lysates of CL1-0 cells cotransfected with (D) Flag-tagged LCRMP-1 and Myc-tagged CRMP-1 and lysates of (E) H522 cells (15 mg) were immunoprecipitated for (D) Flag-tagged LCRMP-1 and (E) endogenous LCRMP-1. Presence of (D) exogenous or (E) endogenous LCRMP-1 and CRMP-1 was analyzed by immunoblotting. Arrows indicate CRMP-1 and LCRMP-1 proteins (numbers indicate the kDa of each examined protein; “#16-2” refers to the clone number of anti–CRMP-1 antibody). (F) HA-tagged CRMP-1 proteins were produced by in vitro transcription/translation and pulled down with GST–LCRMP-1 proteins. Data are presented as mean ± SEM, and P values were calculated by 2-sided Student’s t test (n = 3 experiments). Original magnification, ×1,000 (B and C); ×4,000 (C, insets).
Localization of LCRMP-1 and CRMP-1.
Since our results suggested that the expression patterns of LCRMP-1 and CRMP-1 could control cancer cell invasiveness, we next explored how these 2 proteins might interact during cancer metastasis. To address this, we first detected the cellular distributions of LCRMP-1 and CRMP-1. After cotransfection of the GFP-tagged LCRMP-1 and DsRed-tagged CRMP-1 fusion proteins, the results suggested that LCRMP-1 and CRMP-1 iso-proteins may share some common compartments in their endogenous distributions in interphase (mainly in the cytoplasm), metaphase, and telophase (Figure 6C). During interphase, GFP-LCRMP-1 was seen in the lamellipodia and filopodia region as compared with the signaling of DsRed-CRMP-1 (Figure 6C, arrow in the merged picture of the second row), whereas the GFP control was not seen (Figure 6C, first). During telophase, only GFP-LCRMP-1 was seen in the middle portion of the midbody (Figure 6C, arrow in the fourth row). These observations were also confirmed by endogenous staining of LCRMP-1 and CRMP-1 proteins in H522 cells (Supplemental Figure 14).
LCRMP-1 forms heterodimers with CRMP-1.
Since the CRMP family proteins are known to form heterotetramers with each other, and our results of proteomic identification also revealed that LCRMP-1 is an associated protein of Flag-tagged CRMP-1 (Supplemental Figure 15), we next asked whether LCRMP-1 can directly associate with CRMP-1. A coimmunoprecipitation assay using Flag-tagged LCRMP-1 and Myc-tagged CRMP-1 revealed that the 2 proteins formed heterodimers in vitro (Figure 6D). We further confirmed this association by showing that LCRMP-1 could immunoprecipitate endogenous CRMP-1 from H522 cell lysates (Figure 6E). Finally, we used in vitro transcription/translation followed by GST pull-down assays to show that GST–LCRMP-1 could directly interact with HA–CRMP-1 in the absence of adaptor proteins (Figure 6F). Collectively, these results suggested that LCRMP-1 may associate and form heterodimers with CRMP-1.
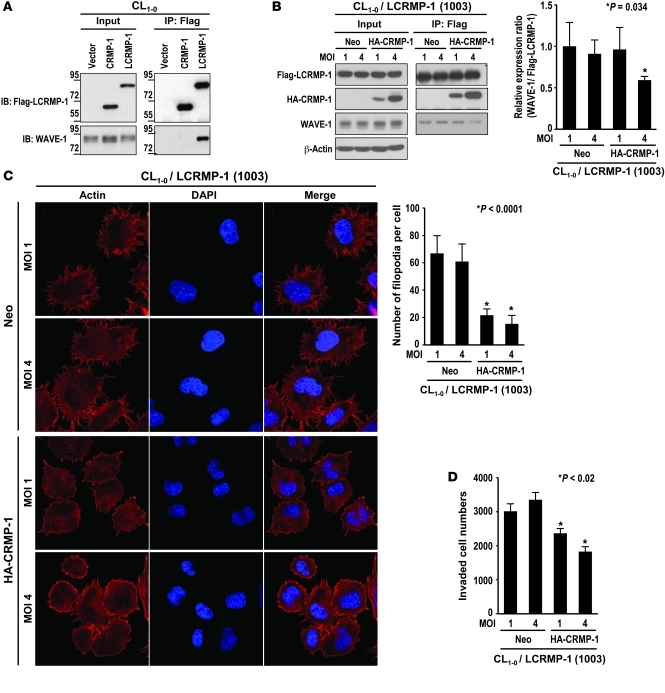
CRMP-1 interrupts the binding between LCRMP-1 and WAVE-1.
Since LCRMP-1 and CRMP-1 can coexist in cells and appear to heterodimerize, we next questioned what mechanism might cause these 2 proteins to have opposing effects on cancer metastasis. Since we showed that LCRMP-1 might promote cancer invasion and filopodia formation through binding to WAVE-1 (Figures 4 and 5), we examined whether CRMP-1 could also bind to WAVE-1 and/or whether it could interfere with the binding between LCRMP-1 and WAVE-1. Coimmunoprecipitation experiments showed that WAVE-1 interacted with LCRMP-1 but not CRMP-1 (Figure 7A). Overexpression of CRMP-1 inhibited the interaction between WAVE-1 and ectopic LCRMP-1 (P = 0.034; Figure 7B) and endogenous LCRMP-1 (Supplemental Figure 16), and this competition phenomena could be partially reversed if cells overexpressed CRMP-1 mutant (CRMP-1 6m) that could not form heterodimer with LCRMP-1 (Supplemental Figure 17). Besides, we also found that overexpressed CRMP-1 protein in CL1-0/LCRMP-1–overexpressing cells may cause regression of filopodia that was induced by LCRMP-1 (P < 0.0001; Figure 7C) and also suppressed the invasive ability of LCRMP-1–overexpressing cells (P < 0.02; Figure 7D). These findings suggest that CRMP-1 and WAVE-1 may compete for binding to LCRMP-1 and that this might be the mechanism through which these proteins affect filopodia formation and cancer cell invasion/migration.
Figure 7. CRMP-1 inhibits the binding between LCRMP-1 and WAVE-1 and further affects the function of LCRMP-1 on filopodia formation and invasion.
(A) Lysates of CL1-0/vector, CL1-0/CRMP-1, and CL1-0/LCRMP-1 cells were examined for the presence of WAVE-1 and Flag-tagged LCRMP-1 or CRMP-1 (left) and WAVE-1 coprecipitated with Flag-tagged LCRMP-1 or CRMP-1 (right) by immunoblotting using anti–WAVE-1 or anti-Flag antibodies (n = 3 experiments). (B) A lentivirus infection-based system was used to express different amounts of HA-tagged CRMP-1 in CL1-0/LCRMP-1 cells (line 1003). Cell lysates were immunoprecipitated with anti-Flag antibody, and binding of WAVE-1 and HA-CRMP-1 was examined by immunoblotting. Amounts of coprecipitated WAVE-1 normalized to the level of precipitated Flag–LCRMP-1 were quantified (n = 3 experiments). (C) Different amounts of HA-tagged CRMP-1 were expressed in CL1-0/LCRMP-1 cells by lentiviral infection, and actin was visualized by staining with rhodamine-conjugated phalloidin (left; original magnification, ×600). Number of filopodia per cell was counted (n = 20 cells per group). (D) In vitro–modified Boyden chamber invasion assay was used to compare the number of invading cells between control (Neo, MOI 1 and 4) and HA-CRMP-1–expressing clones (n = 3 experiments). Data are shown as mean ± SEM, and P value was calculated by 2-sided Student’s t test.
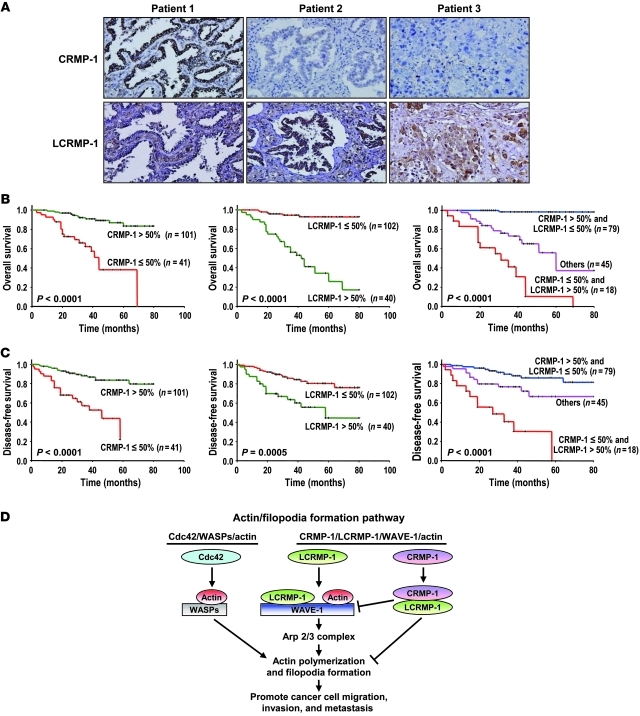
High-level LCRMP-1 and low-level CRMP-1 protein expressions are associated with poor survival in patients with NSCLC.
Although our results consistently suggested that cancer invasion could be counter-regulated by LCRMP-1 and CRMP-1, both in vitro and in vivo, such studies do not fully reflect clinical malignancy. Accordingly, we extended our analysis by examining CRMP-1 and LCRMP-1 expression levels in tumor specimens from 142 patients with NSCLC. The clinical characteristics of these patients are summarized in Supplemental Table 1, and the used positive and negative controls (e.g., the anti–CRMP-1 antibody, Y21, and the anti–LCRMP-1 antibody, C2) are shown in Supplemental Figure 18. Serial sections of each specimen were stained with antibodies against CRMP-1 and LCRMP-1 (Figure 8A). Consistent with our previous reports (5, 16, and 26), patients with low-level CRMP-1 expression had poorer overall (P < 0.0001; Figure 8B, left) and disease-free (P < 0.0001; Figure 8C, left) survival. Similarly, patients with high-level LCRMP-1 expression had poorer overall (P < 0.0001; Figure 8B, middle) and disease-free (P = 0.0005; Figure 8C, middle) survival. Multivariable Cox proportional-hazards regression analyses, with a stepwise selection model, were used to evaluate the associations of various independent prognostic factors with patient survival (Table 3). Our results revealed that the independent prognostic factors were CRMP-1 expression (hazard ratio [HR] = 0.15, 95% CI = 0.06–0.41; P < 0.001), LCRMP-1 expression (HR = 17.56, 95% CI = 5.49–56.17; P < 0.001), histological type (HR for death = 0.07, 95% CI = 0.02–0.25; P < 0.001), age (HR for death = 0.96, 95% CI = 0.92–1.0; P = 0.035), and stage (HR for death = 1.92, 95% CI = 1.23–2.99; P = 0.004). The independent prognostic factors associated with metastasis were CRMP-1 expression (HR = 0.25, 95% CI = 0.12–0.53; P = 0.0003) and LCRMP-1 expression (HR = 2.8, 95% CI = 1.37–5.75; P = 0.0049) (Table 4). Analysis of the combined effect of both proteins on patients’ prognoses revealed that patients with high-level expression of CRMP-1 and low-level expression of LCRMP-1 had better overall (P < 0.0001) and disease-free (P < 0.0001) survival than those with low-level CRMP-1 expression and high-level LCRMP-1 expression (Figure 8, B and C, right). Patients with both high-level LCRMP-1 and CRMP-1 expressions had longer overall survival, as well as disease-free survival (P = 0.002), than those with high-level LCRMP-1 and low-level CRMP-1 expressions (P = 0.012) (Supplemental Figure 19, A and B). In addition, patients with both high-level LCRMP-1 and CRMP-1 expressions had shortened overall survival compared with those with low-level LCRMP-1 and high-level CRMP-1 expressions (P < 0.0001) (Supplemental Figure 19C). A similar result was found in disease-free survival but did not reach the significant level (P = 0.213) (Supplemental Figure 19D). These results confirm that LCRMP-1 and CRMP-1 appear to counter-regulate cancer metastasis in the clinic.
Figure 8. Kaplan-Meier survival plots for patients with NSCLC grouped by LCRMP-1 and CRMP-1 protein expression levels.
(A) Protein expression of CRMP-1 and LCRMP-1 was examined by immunohistochemical staining in serial dissections of primary tumor specimens from 142 patients with NSCLC who underwent surgical resections. Results from 3 patients are shown (original magnification, ×100). (B and C) Patients were designated as having high LCRMP-1 or CRMP-1 expression if more than 50% of the neoplastic cells in their tumor sections were immunoreactive and as having low LCRMP-1 or CRMP-1 expression if fewer than 50% were immunoreactive. The results shown reflect Kaplan–Meier estimates of (B) overall survival and (C) disease-free survival in the 142 patients with NSCLC, according to their expression levels of LCRMP-1, CRMP-1, or both. P values were obtained from 2-sided log-rank tests. (D) Proposed model of the actin/filopodia formation pathway. Cells typically form filopodia through Cdc42/WASPs/actin pathway. Cdc42 activates WASP nucleation proteins, forms Arp2/3 complex, and promotes actin polymerization, filopodia formation, cell migration, and invasion. In some cancer cells, filopodia formation can be regulated through CRMP-1/LCRMP-1/WAVE-1/actin pathway. LCRMP-1 could form a complex with WAVE-1 and actin and potentially act via Arp2/3 complex to promote actin polymerization, filopodia formation, and cell migration, invasion, and metastasis. Furthermore, CRMP-1 could heterodimerize with LCRMP-1 and inhibit the binding of LCRMP-1 to WAVE-1.
Table 3 .
HRs for death (from any cause) among patients with NSCLC, according to multivariable Cox regression analysisA
Table 4 .
HRs for metastasis among patients with NSCLC, according to multivariable Cox regression analysisA
Discussion
Our results demonstrate an interesting example from cell biology, wherein protein isoforms or splicing variants participate in a negative-feedback mechanism in a living cell. We previously identified CRMP-1 as suppressing cancer invasion through F-actin depolymerization and inhibition of filopodia formation (5, 16). Here, we show that the recently reported LCRMP-1, an isoform of CRMP-1 (26), can promote cancer cell migration, invasion, and certain steps in the metastatic cascade. LCRMP-1 forms heterodimers or hetero-oligomers with CRMP-1, functionally antagonizes CRMP-1–mediated suppression of cancer invasion, associates with actin filaments, enhances filopodia formation, and reverses the suppressive effects of dominant-negative Cdc42 on filopodia. Furthermore, LCRMP-1–induced actin polymerization and filopodia formation are mediated via the WAVE-1 pathway, but CRMP-1 overexpression can inhibit the interaction between LCRMP-1 and WAVE-1. Mapping of the putative functional domain showed that N-terminal residues 22–72 of LCRMP-1 are required for filopodia formation. LCRMP-1 overexpression could prompt the less-invasive CL1-0 cells, which are typically rounded and have few filopodia (5, 27), to switch their morphologies and functions to a highly invasive CL1-5 cell-like phenotype of a more elongated cell shape, with an abundance of filopodia. In an in vivo orthotopic implantation and tail vein metastasis assay with LCRMP-1–overexpressing or –silenced cancer cells, LCRMP-1 was found to promote cancer metastasis. Finally, high-level expression of LCRMP-1 and low-level expression of CRMP-1 were found to be associated with worse overall survival and poorer disease-free survival in patients with NSCLC (Figure 8 and Supplemental Figure 19). Collectively, these results suggest that LCRMP-1 and CRMP-1 may counter-regulate each other to mediate cancer cell invasion. The identification of LCRMP-1 as a cancer invasion enhancer and CRMP-1 as a cancer invasion suppressor and the finding that these proteins independently predict the outcomes of patients with NSCLC may have important clinical implications. For example, doctors could potentially use LCRMP-1 and CRMP-1 to identify high-risk patients with NSCLC as candidates for effective adjuvant therapy (36).
The identification of LCRMP-1 as an antagonistic isoform of CRMP-1 is consistent with previous reports that larger CRMP isoforms in other species can functionally oppose their smaller counterparts (28–30, 37). Neurons transfected with TUC-4b (the 75-kDa CRMP “b” isoform in the rat) showed increased branching and elongated neurites, whereas neurons transfected with TUC-4a (the smaller isoform of TUC-4b) showed the opposite effects (29). The CRMP-B isoform in the chicken is an N-terminal variant of the original chicken CRMP subtype. CRMP-2A and CRMP-2B have opposing effects on cell polarity and neurite morphology in developing neurons (30). Here, for the first time to our knowledge, we show that LCRMP-1 and its smaller isoform, CRMP-1, can functionally counter-regulate each other in cancer invasion through actin reorganization.
Sequence analysis previously revealed that the LCRMP-1 mRNA is derived from a unique exon (exon 1A) 4.3-kb upstream of the reported first exon (exon 1B) of CRMP-1 on chromosome 4p16 (26). This unique exon is located upstream of the CRMP-1 promoter region (38), suggesting that LCRMP-1 may be transcribed from an alternative promoter. Therefore, these genes define 2 subtypes of CRMP-1. Here, we compared the amino acid sequences of the CRMP isoform proteins from various species and identified a conserved region (residues 22–72) that differed in LCRMP-1 and the other larger CRMP family members compared with the rest of the family members. To our knowledge, this is the first reported cross-species similarity among CRMP isoforms, which were previously only known to have unique 127-residue N-terminal regions and share a core region with the other CRMPs (28–30). Our functional assessment of various serial deletion constructs of LCRMP-1 revealed that the N-terminal 22–72 amino acid sequence plays an important role in filopodia formation. Fusion of this specific N terminus with the core region of CRMP-2 could also promote filopodia formation in CL1-0 cells (Supplemental Figure 20). This is an important finding on this isoform of the CRMP family. Future studies to clarify the functional motif(s) in this N-terminal region may provide new insights into cancer progression and may offer new potential molecular targets for anticancer therapy.
The complex process of tumor invasion includes cell migration, which depends critically on dynamic reorganization of the actin cytoskeleton. The procession of cell migration and invasion through the extracellular matrix begins with actin polymerization and filopodia formation (39). Gaining a better understanding this process may help scientists design new and more effective means to control cancer invasion and metastasis. Here, we showed that LCRMP-1 can promote cell migration and invasion through actin association and polymerization. In vitro, actin polymerization has various rate-limiting steps, including actin trimer assembly (39, 40). In vivo, however, actin nucleation can be stimulated by regulatory proteins, such as WASP, N-WASP, and the WAVEs, all of which relay signals from Cdc42 and Rac to the actin nucleation machinery of the Arp2/3 complex, thereby mediating actin polymerization (41–44).
Although the N-WASP pathway is well known to mediate actin polymerization and filopodia formation, we were somewhat surprised to discover that LCRMP-1 acts through the WAVE-1 pathway in CL cells. Indeed, our data suggest that WAVE-1 may be an alternative pathway for actin polymerization and filopodia formation in CL lung cancer cells. WAVE was previously shown to auto-inhibit WASP in actin polymerization assays (40, 45), and fibroblasts isolated from N-WASP–deficient mice were found to still be capable of forming filopodia (46, 47), suggesting the presence of an alternative mechanism for actin polymerization. Furthermore, WAVE-2 and WAVE-3 can be recruited to the filopodia through their SCAR homology domains, which are located next to the leucine zipper-like motif; following this recruitment, the WAVE family members can guide actin bundles into the filopodia tips for actin assembly (48). However, the WAVEs do not contain the GTPase binding domain (also known as the Cdc42/Rac-interactive binding region) that is involved in the activation of N-WASP (40). Instead, the WAVEs may be activated by the binding of active Cdc42 and Rac to their SHD domains (44). Here, we showed that LCRMP-1 could reverse the filopodia regression induced by expression of dominant-negative Cdc42. Thus, our results collectively suggest that LCRMP-1 acts downstream of Cdc42 and associates with actin, thereby promoting actin polymerization, filopodia formation, and cancer cell invasion.
On the basis of these findings, we propose a set of action pathways for LCRMP-1–induced filopodia formation, as illustrated schematically in Figure 8D. To summarize, cells typically form filopodia through the Cdc42/WASP/actin pathway. Cdc42 can trigger activation of the WASP nucleation proteins, formation of the Arp2/3 complex, polymerization of actin, and formation of filopodia, thereby promoting cell migration and invasion. In some cancer cells, however, filopodia formation can be regulated through the CRMP-1/LCRMP-1/WAVE-1/actin pathway. In these cells, the LCRMP-1 may form a complex with WAVE-1 and actin, potentially act via the Arp2/3 complex to promote actin polymerization, filopodia formation, and cell migration, invasion, and metastasis. Furthermore, CRMP-1 can heterodimerize with LCRMP-1 and inhibit the binding of LCRMP-1 to WAVE-1, suggesting that cancer cell invasion may be counter-regulated by LCRMP-1 and CRMP-1. We know that LCRMP-1 acts downstream of Cdc42, but whether this small GTPase signaling is necessary for the LCRMP-1/WAVE-1/actin pathway of filopodia formation should be further clarified in the future.
Besides, we also call attention to an interesting phenomena, in that the opposite effects of LCRMP-1 and CRMP-1 on cancer cell invasion not only exist in CL1-0, CL1-5, H23, and SN12C cells, but also in H226 (lung squamous cell carcinoma) cells, which do not endogenously express either LCRMP-1 or CRMP-1 protein (Supplemental Figure 13). This implies that CRMP-1 not only can act via LCRMP-1 to displace WAVE-1 and inhibit invasion, but that it is possible that it may have an LCRMP-1–independent mechanism in cells without LCRMP-1 expression to regulate cancer cell invasion. In addition, the differences in tumor growth were seen in vivo using orthotopic A549/LCRMP-1 injections, while in vitro LCRMP-1 did not appear to have growth advantage, suggesting that LCRMP-1 may involve other mechanisms, such as angiogenesis, that promote tumor progression. In conclusion, we herein show that LCRMP-1 is a cancer metastasis enhancer that counter-regulates the tumor invasion suppressor, CRMP-1. LCRMP-1 promotes actin polymerization, filopodia formation, and cancer invasion through the WAVE-1 pathway, and increased LCRMP-1 expression is associated with poor survival among patients with NSCLC. Collectively, these findings suggest that LCRMP-1 may be a potential therapeutic target for new cancer treatments.
Methods
The plasmids, antibodies, and some detailed protocols are listed in the Supplemental Methods, and the sequence of specific siRNA that target to human LCRMP-1 and WAVE-1 is shown in Supplemental Table 3.
Cell lines and culture conditions.
The human lung adenocarcinoma cell lines (CL1-0 cells) were isolated from a 64-year-old male patient with a poorly differentiated adenocarcinoma and selected in our laboratory by in vitro Transwell invasion to get 5 sublines with progressive invasiveness, with similar genotypic background (designated CL1-1, CL1-2, CL1-3, CL1-4, and CL1-5) as previously described (27). A549 (a human lung adenocarcinoma epithelial cell line), HEK293 (a human embryonic kidney cell line), and HEK293T (a human embryonic kidney cell line that was transformed using sheared HAd5 DNA to make it very sensitive to human adenovirus and permissive to adenovirus DNA) cells were purchased from ATCC. The H522 (human non-small-lung cancer), H23 (lung adenocarcinoma), SN12C (human renal carcinoma), and H226 (human lung squamous carcinoma) cells in the 60 human cancer cell lines used by the National Cancer Institute (NCI-60 cell lines) were purchased from the National Cancer Institute’s Developmental Therapeutics Program (NCI). The cultured conditions of cells are detailed in the Supplemental Methods.
Immunoprecipitation and immunoblotting.
Immunoprecipitation was performed as described previously (49). In brief, transfected CL, A549, or H522 cells were lysed on ice for 5–10 minutes in 20 mM Tris (pH 8.0), 150 mM NaCl, 100 μM Na3VO4, 50 mM NaF, 30 mM sodium pyrophosphate, and 0.5% NP-40 (Sigma-Aldrich), and a 25-fold dilution of a stock solution was treated with 1 Mini Protease Inhibitor Cocktail Tablet (Roche Diagnostics) dissolved in 2 ml of distilled water. The cell lysates were passed several times through a 21-gauge needle and clarified by centrifugation at 8,000 g for 30 minutes at 4°C. The supernatants were taken as the total cell lysates. LCRMP-1 or CRMP-1 was immunoprecipitated using anti-Flag, anti-Myc, anti–LCRMP-1, or anti–CRMP-1 antibodies and protein A-Sepharose beads (Sigma-Aldrich). The immunoprecipitated proteins were separated by SDS-PAGE and transferred to polyvinylidene membranes (Millipore) for immunoblotting with anti-Flag (1:5,000), anti-Myc (1:3,000), anti–CRMP-1 (1:2,000), anti–LCRMP-1 (C2; 1:10,000), anti–N-WASP (1:2,000), anti-WASP (1:2,000), anti-p34 (1:1,000), anti–pan WAVE (1:2,000), anti–WAVE-1 (1:2,000), anti-HA (1:5,000), or anti–β-actin (1:20,000) primary antibodies. Immunoblotting was performed according to the standard procedure, as described previously (5).
Modified Boyden chamber invasion assay.
Modified Boyden chambers with polycarbonate-membrane inserts (pore size 8 μm; Falcon, Becton Dickinson) coated with 30 μg Matrigel (BD) were used for cell invasion assays. Stable transfectants were suspended in RPMI medium containing 10% NuSerum (Invitrogen). 2.5 × 104 cells were placed in the upper chambers, and 1 ml of medium was placed in the lower chambers. After incubation for 24 hours at 37°C, cells were methanol fixed for 10 minutes at room temperature and then stained for 30 minutes at room temperature with a 50 μg/ml solution of propidium iodide (Sigma-Aldrich). The number of cells on each membrane was counted under a microscope at a magnification of ×50, using the Analytical Imaging Station software package (Imaging Research Inc.). Experiments were performed at least twice, and each sample was assayed in triplicate.
Scratch wound-healing assay.
Transfected cells stably expressing LCRMP-1 were seeded to 6-well tissue culture dishes and grown to confluence. Each confluent monolayer was then wounded linearly using a pipette tip and washed 3 times with PBS. Thereafter, cell morphology and migration were observed and photographed at regular intervals for 12 hours. Three independent experiments in triplicate were performed for each cell line.
Experimental metastasis in vivo.
For orthotopic tumor implantation assays, lentivirus-infected A549/vector, A549/LCRMP-1–overexpressing, A549/nonsilenced, and A549/LCRMP-1–silenced cells (105 cells in 20 μl PBS containing 10 ng Matrigel) were injected into the pleural cavity of 6-week-old SCID mice (supplied by the animal center at the College of Medicine, National Taiwan University; n = 10 per group). A preliminary study combined with luciferase image observation (data not shown) had indicated that mice developed many lung metastasis nodules after 4 weeks. Therefore, mice were sacrificed by anesthesia with carbon dioxide 28 days after implantation, all organs were removed and fixed in 10% formalin, and the lung nodules were counted under gross and microscopic examination. The number of mice used for the experiments (n = 10) was based on the goal of having 98% power to detect a 2-fold between-group difference in nodule number at P < 0.05.
For the in vivo tail vein metastasis assay, a single-cell suspension containing 106 cells (CL1-0/vector [line 2A10], CL1-0/LCRMP-1–overexpressing [lines 1003 and 1015], CL1-5/nonsilenced, or CL1-5/LCRMP-1–silenced cells) in 0.1 ml PBS was injected into the lateral tail veins of 6-week-old SCID mice (supplied by the animal center at the College of Medicine, National Taiwan University). Our preliminary study in this animal model (data not shown) had indicated that mice injected with CL1-0 cells developed lung metastasis nodules within 8 weeks, while those injected with CL1-5 cells developed nodules within 5 weeks. Therefore, either 8 weeks after injection (for CL1-0/LCRMP-1–overexpressing groups; n = 10 per group) or 5 weeks after injection (for CL1-5/LCRMP-1–silenced groups; n = 10 per group), the relevant mice were sacrificed by carbon dioxide anesthesia, and their lungs were examined for metastatic nodules (see Supplemental Methods for the details).
All mouse experiments were performed in accordance with the animal guidelines and with the approval of the Department of Animal Care at the Institute of Biomedical Sciences (Academia Sinica).
Immunofluorescence staining for observation of protein localization and filopodia formation.
Transfected or nontransfected CL or H522 cells were fixed for 10 minutes at room temperature in 3.7% cold paraformaldehyde in PBS (pH 7.0), washed 3 times with PBS, and permeabilized for 10 minutes at room temperature in PBS containing 0.1% Triton X-100. The cells were blocked with PBS containing 3% bovine serum albumin and stained overnight at 4°C with polyclonal anti-Myc (Viogene Biotech), anti-Flag (Sigma-Aldrich), anti–LCRMP-1, or monoclonal anti–CRMP-1 antibodies, followed by incubation for 1 hour at 37°C with FITC-, rhodamine-, or Cy5-conjugated secondary antibodies (Molecular Probes), respectively. Negative control sections were stained with preimmune serum, primary antibodies preabsorbed with the immunization antigen, or an unrelated control antibody. The cells were mounted onto microscope slides with 50% glycerol in PBS and then examined and photographed using a Zeiss Axiophot epifluorescence microscope equipped with an MRC-1000 laser scanning confocal imaging system (Bio-Rad Laboratories).
Modified actin polymerization assay.
pEGFP–WAVE-1 plasmids were used to produce N terminus–tagged GFP–WAVE-1 proteins in both LCRMP-1–silenced and nonsilenced control CL1-5 cells. Then, the GFP–WAVE-1 fusion proteins in both cells were immunoprecipitated by protein A beads that were coated with anti-GFP antibody (35), and the GFP–WAVE-1 fusion proteins were then used to determine their activities on actin polymerization as described by the manufacturer (Cytoskeleton). Briefly, 5 μM final concentration of monomeric actin (1:10 pyrene labeled) was incubated on ice for 10 minutes with the immunoprecipitated GFP–WAVE-1 fusion proteins from CL1-5/LCRMP-1–silenced or CL1-5/nonsilenced control cells. Samples were then equilibrated for 10 minutes in a fluorescence spectrophotometer (ISS), after which polymerization was induced by the addition of KCl, MgCl2, and ATP. The increase in fluorescence intensity that occurs when pyrene G-actin forms pyrene F-actin was recorded every 5 seconds at 25°C using the fluorescence spectrophotometer, with the excitation wavelength of 365 nm and emission wavelength of 407 nm.
Patients and tumor specimens.
Lung tumor tissue specimens were obtained from patients (n = 142) with histologically confirmed NSCLC, who had undergone complete surgical resections at the National Taiwan University Hospital (Taipei, Taiwan) between December 28, 1995, and December 26, 2005. This investigation was approved by the Institutional Review Board of the National Taiwan University Hospital. The enrolled patients had not been treated with neoadjuvant chemotherapy or irradiation therapy; patients provided informed consent. All specimens were formalin fixed, sectioned, stained with H&E, and examined by microscopy. Pathological staging was performed by Yih-Leong Chang according to the international staging system for lung cancer (50).
Immunohistochemical analysis of LCRMP-1 and CRMP-1 expression in tumor samples from patients with NSCLC.
Immunohistochemical staining of tumor tissue samples from patients with NSCLC was carried out using a modified avidin-biotin peroxidase complex method. The sections used for immunohistochemical analysis of CRMP-1 or LCRMP-1 protein expression were first autoclaved in Trilogy Solution (Cell Marque Corp.) or Antigen Retrieval Citra Solution (Biogenex) at 121°C for 10 minutes. The samples were then treated with 3% H2O2-methanol and sequentially subjected to the following: incubation with DakoCytomation Dual Endogenous Enzyme Block (DakoCytomation Inc.) for 10 minutes; incubation with Ultra V Block (Lab Vision Corporation) for 10 minutes; incubation with antibody-dilution buffer (Ventana Medical Systems Inc.) for 10 minutes; and incubation with a monoclonal anti–CRMP-1 antibody (Y21) for 6 hours at room temperature or a polyclonal anti–LCRMP-1 antibody (C2; 1:300 dilution) overnight at 4°C. Detection of the immunostaining was carried out using the Super Sensitive Non-Biotin Polymer HRP Detection System (BioGenex), according to the manufacturer’s instructions.
Statistics.
The correlations between various clinical and pathological parameters and the expression levels of CRMP-1 and LCRMP-1 were analyzed by using the Pearson χ2 test. To identify a suitable cut-off point for patient separation, we performed a sensitivity analysis on the immunohistochemical staining results. The cut-off points were checked at 10% intervals, from 20% to 80% for CRMP-1 and LCRMP-1, and Kaplan-Meier analyses and log-rank tests were performed using the different cut-off values. The P values for cut-off values of 40%, 50%, and 60% were smallest for both CRMP-1 and LCRMP-1 (Supplemental Table 2), so we selected 50% expression of CRMP-1 and LCRMP-1 as the cut-off point. High-level LCRMP-1 or CRMP-1 expression in tumors was defined as immunoreactivity in more than 50% of the neoplastic cells. A multivariable Cox proportional-hazards regression model was fitted with the following variables: continuous age, sex, histological type (squamous vs. adenocarcinoma), stage (stage I vs. stage II vs. stage III), CRMP-1 expression (high vs. low), and LCRMP-1 expression (high vs. low). The quantitative in vitro and in vivo data were analyzed using the Student’s t test. All analyses were performed using SPSS software (v10.0; SPSS Inc.) and SAS v9.1 software (SAS Institute Inc.). All statistical tests were 2 sided, and P values < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by grants from the Department of Health, Executive Yuan, Taiwan (DOH-96-TD-G-111-011 and DOH97-TD-G-111-018) and the National Science Council of the Republic of China (NSC97-2314-B-002-146-MY3, NSC97-3112-B-002-033, NSC97-2627-P-001-003, and NSC98-2628-B-002-086-MY3). The shRNA constructs were obtained from the National RNAi Core Facility located at the Institute of Molecular Biology/Genomic Research, Academia Sinica. The authors thank Ruey-Hwa Chen (Institute of Biological Chemistry, Academia Sinica) for providing the dominant-negative mutant Cdc42 plasmids, S.R. Roffler (Institute of Biomedical Sciences, Academia Sinica) for providing the monoclonal anti-Myc antibody, H.K. Sytwu (Graduate Institute of Medical Sciences, National Defense Medical Center) for providing the plasmids for the lentivirus infection system, and Ker-Chau Li (Institute of Statistical Sciences, Academia Sinica) for cooperating with us to purchase the NCI-60 cancer cell lines. In addition, the authors would like to thank the following people for providing technical support: Chung-Wu Lin, Chia-Tung Shun (Department of Pathology and Graduate Institute of Pathology, National Taiwan University), Jin-Yuan Shih, Pei-Ying Lin (Department of Internal Medicine, National Taiwan University Hospital), Win-Ping Deng (Institute of Biomedical Materials and Engineering, Taipei Medical University), Chi-Chung Wang (Graduate e of Basic Medicine, Fu Jen Catholic University), Hsiao-Chun Huang (Institute of Biomedical Sciences, Academia Sinica), Shih-Han Kao, Pui-San Leong, and the National Taiwan University Center of Genomic Medicine and Microarray Core Facility.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(8):3189–3205. doi:10.1172/JCI42975.
References
- 1.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2(8):563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12(8):895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 3.Valastyan S, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137(6):1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3(1):55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 5.Shih JY, et al. Collapsin response mediator protein-1 and the invasion and metastasis of cancer cells. . J Natl Cancer Inst. 2001;93(18):1392–1400. doi: 10.1093/jnci/93.18.1392. [DOI] [PubMed] [Google Scholar]
- 6.Chang CC, et al. Connective tissue growth factor and its role in lung adenocarcinoma invasion and metastasis. J Natl Cancer Inst. 2004;96(5):364–375. doi: 10.1093/jnci/djh059. [DOI] [PubMed] [Google Scholar]
- 7.Chang CC, et al. Effect of connective tissue growth factor on hypoxia-inducible factor 1alpha degradation and tumor angiogenesis. J Natl Cancer Inst. 2006;98(14):984–995. doi: 10.1093/jnci/djj242. [DOI] [PubMed] [Google Scholar]
- 8.Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376(6540):509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- 9.Hamajima N, Matsuda K, Sakata S, Tamaki N, Sasaki M, Nonaka M. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene. 1996;180(1–2):157–163. doi: 10.1016/S0378-1119(96)00445-3. [DOI] [PubMed] [Google Scholar]
- 10.Wang LH, Strittmatter SM. A family of rat CRMP genes is differentially expressed in the nervous system. J Neurosci. 1996;16(19):6197–6207. doi: 10.1523/JNEUROSCI.16-19-06197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L H, Strittmatter SM. Brain CRMP forms heterotetramers similar to liver dihydropyrimidinase. J Neurochem. 1997;69(6):2261–2269. doi: 10.1046/j.1471-4159.1997.69062261.x. [DOI] [PubMed] [Google Scholar]
- 12.Arimura N, et al. Phosphorylation of collapsin response mediator protein-2 by Rho-kinase. Evidence for two separate signaling pathways for growth cone collapse. J Biol Chem. 2000;275(31):23973–23980. doi: 10.1074/jbc.M001032200. [DOI] [PubMed] [Google Scholar]
- 13.Fukada M, et al. Molecular characterization of CRMP5, a novel member of the collapsin response mediator protein family. J Biol Chem. 2000;275(48):37957–37965. doi: 10.1074/jbc.M003277200. [DOI] [PubMed] [Google Scholar]
- 14.Quinn CC, Gray GE, Hockfield S. A family of proteins implicated in axon guidance and outgrowth. J Neurobiol. 1999;41(1):158–164. doi: 10.1002/(SICI)1097-4695(199910)41:1<158::AID-NEU19>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 15.Rosslenbroich V, Dai L, Baader SL, Noegel AA, Gieselmann V, Kappler J. Collapsin response mediator protein-4 regulates F-actin bundling. Exp Cell Res. 2005;310(2):434–444. doi: 10.1016/j.yexcr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Shih JY, Lee YC, Yang SC, Hong TM, Huang CY, Yang PC. Collapsin response mediator protein-1: a novel invasion-suppressor gene. Clin Exp Metastasis. 2003;20(1):69–76. doi: 10.1023/A:1022598604565. [DOI] [PubMed] [Google Scholar]
- 17.Kaibuchi K, Kuroda S, Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu Rev Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- 18.Lee DJ, Cox D, Li J, Greenberg S. Rac1 and Cdc42 are required for phagocytosis, but not NF-kappaB-dependent gene expression, in macrophages challenged with Pseudomonas aeruginosa. J Biol Chem. 2000;275(1):141–146. doi: 10.1074/jbc.275.1.141. [DOI] [PubMed] [Google Scholar]
- 19.Li S, et al. Distinct roles for the small GTPases Cdc42 and Rho in endothelial responses to shear stress. J Clin Invest. 1999;103(8):1141–1150. doi: 10.1172/JCI5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suetsugu S, et al. Regulation of actin cytoskeleton by mDab1 through N-WASP and ubiquitination of mDab1. Biochem J. 2004;384(pt 1):1–8. doi: 10.1042/BJ20041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97(2):221–231. doi: 10.1016/S0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 22.Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J Cell Biol. 2000;150(6):1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15(19):5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 24.Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17(23):6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suetsugu S, Miki H, Takenawa T. Identification of another actin-related protein (Arp) 2/3 complex binding site in neural Wiskott-Aldrich syndrome protein (N-WASP) that complements actin polymerization induced by the Arp2/3 complex activating (VCA) domain of N-WASP. J Biol Chem. 2001;276(35):33175–33180. doi: 10.1074/jbc.M102866200. [DOI] [PubMed] [Google Scholar]
- 26.Pan SH, et al. Long form collapsin response mediator protein-1 (LCRMP-1) expression is associated with clinical outcome and lymph node metastasis in non-small cell lung cancer patients. Lung Cancer. 2010;67(1):93–100. doi: 10.1016/j.lungcan.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 27.Chu YW, et al. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am J Respir Cell Mol Biol. 1997;17(3):353–360. doi: 10.1165/ajrcmb.17.3.2837. [DOI] [PubMed] [Google Scholar]
- 28.Leung T, et al. p80 ROKalpha binding protein is a novel splice variant of CRMP-1 which associates with CRMP-2 and modulates RhoA-induced neuronal morphology. FEBS Lett. 2002;532(3):445–449. doi: 10.1016/S0014-5793(02)03736-5. [DOI] [PubMed] [Google Scholar]
- 29.Quinn CC, et al. TUC-4b, a novel TUC family variant, regulates neurite outgrowth and associates with vesicles in the growth cone. J Neurosci. 2003;23(7):2815–2823. doi: 10.1523/JNEUROSCI.23-07-02815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuasa-Kawada J, Suzuki R, Kano F, Ohkawara T, Murata M, Noda M. Axonal morphogenesis controlled by antagonistic roles of two CRMP subtypes in microtubule organization. Eur J Neurosci. 2003;17(11):2329–2343. doi: 10.1046/j.1460-9568.2003.02664.x. [DOI] [PubMed] [Google Scholar]
- 31.Hsu TH, Liao WY, Yang PC, Wang CC, Xiao JL, Lee CH. Dynamics of cancer cell filopodia characterized by super-resolution bright-field optical microscopy. Opt Express. 2007;15(1):76–82. doi: 10.1364/OE.15.000076. [DOI] [PubMed] [Google Scholar]
- 32.Huang SW, Mong HY, Lee CH. Super–resolution bright–field optical microscopy based on nanometer topographic contrast. Microsc Res Tech. 2004;65(4–5):180–185. doi: 10.1002/jemt.20091. [DOI] [PubMed] [Google Scholar]
- 33.Wang CC, Lin JY, Chen HC, Lee CH. Dynamics of cell membranes and the underlying cytoskeletons observed by noninterferometric widefield optical profilometry and fluorescence microscopy. Opt Lett. 2006;31(19):2873–2875. doi: 10.1364/OL.31.002873. [DOI] [PubMed] [Google Scholar]
- 34.Snapper SB, et al. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat Cell Biol. 2001;3(10):897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- 35.Kang H, Wang J, Longley SJ, Tang JX, Shaw SK. Relative actin nucleation promotion efficiency by WASP and WAVE proteins in endothelial cells. Biochem Biophys Res Commun. 2010;400(4):661–666. doi: 10.1016/j.bbrc.2010.08.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winton T, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. . N Engl J Med. 2005;352(25):2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 37.Li W, Herman RK, Shaw JE. Analysis of the Caenorhabditis elegans axonal guidance and outgrowth gene unc-33. Genetics. 1992;132(3):675–689. doi: 10.1093/genetics/132.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu CC, et al. Modulation of the expression of the invasion-suppressor CRMP-1 by cyclooxygenase-2 inhibition via reciprocal regulation of Sp1 and C/EBPalpha. Mol Cancer Ther. 2008;7(6):1365–75. doi: 10.1158/1535-7163.MCT-08-0091. [DOI] [PubMed] [Google Scholar]
- 39.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/S0092-8674(03)00120-X. [DOI] [PubMed] [Google Scholar]
- 40.Stradal TE, Rottner K, Disanza A, Confalonieri S, Innocenti M, Scita G. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 2004;14(6):303–311. doi: 10.1016/j.tcb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Carlier MF, Ducruix A, Pantaloni D. Signaling to actin: the Cdc42–N–WASP–Arp2/3 connection. Chem Biol. 1999;6(9):R235–R240. doi: 10.1016/S1074-5521(99)80107-0. [DOI] [PubMed] [Google Scholar]
- 42.Egile C, et al. Activation of the CDC42 effector N–WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin–based motility. J Cell Biol. 1999;146(6):1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 44.Takenawa T, Miki H. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J Cell Sci. 2001;114(pt 10):1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- 45.Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8(25):1347–1356. doi: 10.1016/S0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 46.Lommel S, Benesch S, Rottner K, Franz T, Wehland J, Kuhn R. Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO Rep. 2001;2(9):850–857. doi: 10.1093/embo-reports/kve197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snapper SB, et al. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat Cell Biol. 2001;3(10):897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- 48.Nozumi M, Nakagawa H, Miki H, Takenawa T, Miyamoto S. Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J Cell Sci. 2003;116(pt 2):239–246. doi: 10.1242/jcs.00233. [DOI] [PubMed] [Google Scholar]
- 49.Chang YW, et al. CD13 (aminopeptidase N) can associate with tumor-associated antigen L6 and enhance the motility of human lung cancer cells. Int J Cancer. 2005;116(2):243–252. doi: 10.1002/ijc.21089. [DOI] [PubMed] [Google Scholar]
- 50. Sobin L, Wittekind C.TNM Classification of Malignant Tumours . 6th ed. Hoboken, New Jersey, USA: John Wiley & Sons; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.