Abstract
MicroRNAs modulate cellular phenotypes by inhibiting expression of mRNA targets. In this study, we have shown that the muscle-specific microRNAs miR-133a-1 and miR-133a-2 are essential for multiple facets of skeletal muscle function and homeostasis in mice. Mice with genetic deletions of miR-133a-1 and miR-133a-2 developed adult-onset centronuclear myopathy in type II (fast-twitch) myofibers, accompanied by impaired mitochondrial function, fast-to-slow myofiber conversion, and disarray of muscle triads (sites of excitation-contraction coupling). These abnormalities mimicked human centronuclear myopathies and could be ascribed, at least in part, to dysregulation of the miR-133a target mRNA that encodes dynamin 2, a GTPase implicated in human centronuclear myopathy. Our findings reveal an essential role for miR-133a in the maintenance of adult skeletal muscle structure, function, bioenergetics, and myofiber identity; they also identify a potential modulator of centronuclear myopathies.
Introduction
Human centronuclear myopathies (CNMs) are a group of congenital myopathies characterized by muscle weakness and abnormal centralization of nuclei in muscle myofibers (1, 2). CNMs can be classified into 3 main forms: the recessive X-linked myotubular myopathy (XLMTM), with a severe neonatal phenotype, caused by mutations in the myotubularin gene (MTM1); the classical autosomal-dominant form, with mild, moderate, or severe phenotypes, caused by mutations in the dynamin 2 gene (DNM2); and an autosomal-recessive form presenting severe and moderate phenotypes, caused by mutations in the amphiphysin 2 gene (BIN1) (1, 2). Despite their heterogeneous clinical phenotypes, all 3 forms of CNMs present the following common pathological characteristics: (a) type I myofiber predominance and small fiber sizes; (b) abnormal NADH–tetrazolium reductase (NADH-TR) staining patterns, indicative of mitochondrial abnormalities; and (c) absence of necrosis, myofiber death, or regeneration (2).
XLMTM, the most severe and most common form of CNM, has been extensively studied in mice and zebrafish (3–6). Mice with homozygous mutations of the Mtm1 gene develop a progressive CNM that recapitulates the pathological characteristics of XLMTM in humans (5). Mtm1-deficient mice also display disorganized triads and defective excitation-contraction coupling, which may be responsible for the impaired muscle function in XLMTM (3).
The autosomal-dominant form of CNM is associated with a wide clinical spectrum of slowly progressive CNMs, from those beginning in childhood or adolescence to more severe sporadic forms with neonatal onset (7–9). Multiple missense mutations in the DNM2 locus have been identified in recent years, hence, the autosomal-dominant CNM is also called DNM2-associated CNM. Dynamin 2 is a ubiquitously expressed large GTPase involved in many cellular functions, including endocytosis and membrane trafficking (10, 11). However, the precise mechanism whereby multiple missense mutations in the DNM2 gene cause CNM remains unknown. Currently, there is no mouse model for DNM2-related CNM, and a knockin mouse model expressing the most frequent CNM-related DNM2 mutation, R465W Dnm2, failed to reproduce the autosomal-dominant form of human CNM (9). Homozygous mice carrying the R465W Dnm2 mutation die within 24 hours after birth, whereas heterozygous mice develop a myopathy followed by atrophy and impaired muscle function without centralized nuclei (9).
microRNAs (miRNAs) are highly conserved small noncoding RNAs that regulate a range of biological processes by inhibiting the expression of target mRNAs with complementary sequences in their 3′ untranslated regions (3′ UTRs) (12). Watson-Crick base pairing of nucleotides 2–8 of a miRNA with the mRNA target results in mRNA degradation and/or translational repression. Recent studies have revealed key roles for miRNAs in the regulation of skeletal muscle differentiation, and changes in miRNA expression are associated with various skeletal muscle disorders (13–15). However, the potential involvement of miRNAs in skeletal myopathies has not been demonstrated.
The miR-1-1/miR-133a-2 and miR-1-2/miR-133a-1 miRNA clusters are expressed in cardiac and skeletal muscle, whereas the miR-206/miR-133b cluster is only expressed in skeletal muscle (16). miR-206 is required for efficient regeneration of neuromuscular synapses after acute nerve injury, and loss of miR-206 accelerates disease progression of amyotrophic lateral sclerosis in mice (17). miR-1 and miR-133a play important roles in heart development and function (18, 19) and have also been shown to regulate myoblast proliferation and differentiation in vitro (20). However, the potential functions of these miRNAs in skeletal muscle development or function in vivo have not been studied.
We showed previously that miR-133a-1 and miR-133a-2 are important for cardiac development and function (18). Mice lacking either miR-133a-1 or miR-133a-2 are normal, whereas approximately 50% of double knockout (dKO) mice lacking both miRNAs die as embryos or neonates from ventricular-septal defects (18). To explore the functions of miR-133a in skeletal muscle, we studied the surviving miR-133a dKO mice for skeletal muscle defects. Here, we show that adult mice lacking miR-133a developed progressive CNM, accompanied by mitochondrial dysfunction and fast-to-slow myofiber conversion. These muscle abnormalities can be attributed, at least in part, to upregulation of dynamin 2, a target for repression by miR-133a-1 and miR-133a-2. Our findings revealed an essential role for miR-133a in the maintenance of adult skeletal muscle structure and function and as a modulator of CNMs.
Results
Expression of miR-133 in skeletal muscle.
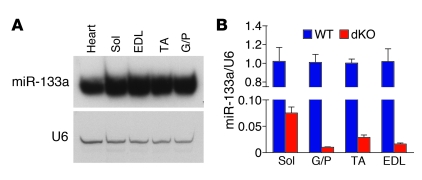
The miR-133 family contains 3 highly homologous miRNAs: miR-133a-1, miR-133a-2, and miR-133b. miR-133a-1 and miR-133a-2 are identical and differ from miR-133b by 2 nucleotides at the 3′ terminus (18). We have previously shown that miR-133a-1 and miR-133a-2 are expressed in cardiac and skeletal muscle, whereas miR-133b is skeletal muscle specific (18). We determined the expression of miR-133 by Northern blot analysis in several skeletal muscles of different myofiber contents. Oxidative, type I (slow-twitch) myofibers are enriched in soleus muscle, and glycolytic type II (fast-twitch) myofibers are enriched in other muscle groups, such as gastrocnemius and plantaris (G/P), tibialis anterior (TA), and extensor digitorum longis (EDL) muscles. miR-133a was expressed at equivalent levels in all of these muscle groups (Figure 1A), indicative of its comparable levels in type I and type II myofibers. miR-133b was cotranscribed with miR-206 and was enriched in soleus muscle, which contains predominantly type I fibers (17).
Figure 1. Expression of miR-133 in skeletal muscle.
(A) Northern blot analysis of miR-133a in adult WT mouse tissues. The blot was stripped and reprobed with 32P-labeled U6 probe as a loading control. Sol, soleus. (B) Expression of miR-133 in skeletal muscle, detected by real-time RT-PCR and expressed relative to U6.
We generated miR-133a–/– (i.e., dKO) mice by interbreeding miR-133a-1+/–;miR-133a-2+/– mice, as described previously (18), and confirmed the loss of miR-133a expression in dKO skeletal muscle by quantitative real-time RT-PCR (Figure 1B). The low level of miR-133 expression detected in dKO skeletal muscle represented the presence of miR-133b, which is detected by miR-133a probes. Based on results from real-time RT-PCR, we estimate that the relative abundance of miR-133a versus miR-133b in WT mice is about 15:1 in soleus and about 50:1 in G/P, EDL, and TA muscle, which confirms that miR-133b is less abundant than miR-133a in skeletal muscle and is enriched in soleus muscle.
Accumulation of centronuclear myofibers in dKO skeletal muscle.
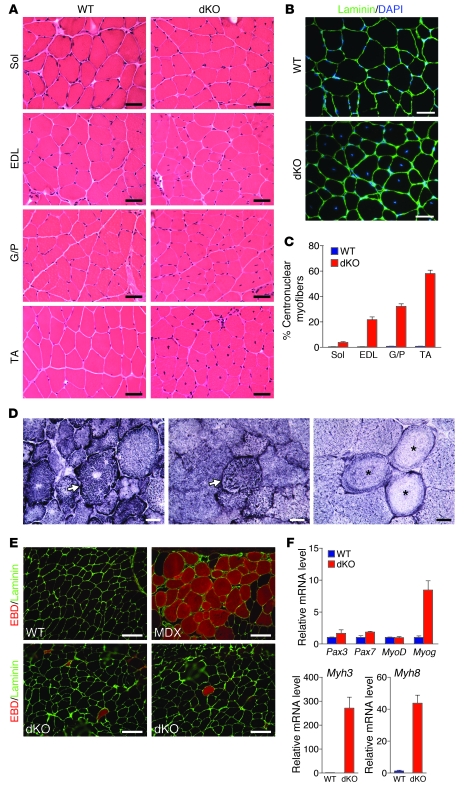
dKO mice did not show apparent abnormalities in mobility. At 4 weeks of age, dKO muscles appeared normal by histological analysis and immunostaining for laminin and DAPI, and myofibers were comparable in size to those of WT muscle (Supplemental Figure 1, A–C; supplemental material available online with this article; doi: 10.1172/JCI46267DS1). However, by 6 weeks of age, myofibers with centralized nuclei began to appear in dKO mice, and the percentage of myofibers with central nuclei in EDL, G/P and TA muscle increased progressively with age (Supplemental Figure 2A). By 12 weeks of age, nearly 60% of myofibers in TA muscle of dKO mice contained centralized nuclei (Figure 2, A–C). In contrast, dKO soleus muscle had relatively few centralized nuclei (Figure 2, A and C). These findings suggest that the phenotype of centrally located nuclei in dKO mice is specific to type II myofibers. In addition, at 12 weeks of age, dKO mice were significantly smaller in both body mass and mass of various muscle groups when normalized to tibia length (Supplemental Figure 2B). TA myofibers of dKO mice also had smaller diameters than normal at this age (Supplemental Figure 2C).
Figure 2. Centronuclear myofibers in dKO skeletal muscle.
(A) H&E staining of soleus, EDL, G/P, and TA muscles of WT and dKO mice at 12 weeks of age. Scale bars: 40 μm. (B) Immunostaining of TA muscle against laminin. Nuclei are stained with DAPI. dKO TA muscle showed central nuclei. Scale bars: 40 μm. (C) Percentage of centronuclear myofibers in 4 WT mice and 10 dKO mice at 12 weeks of age. For each mouse, more than 500 myofibers were counted for TA and G/P muscles and more than 300 myofibers were counted for soleus and EDL muscles. (D) NADH-TR staining of dKO TA muscle revealed abnormal distribution, radiating intermyofibrillary network (arrows), and ring-like fibers (asterisks). Scale bars: 20 μm. (E) EBD uptake of TA muscles of WT, dKO, and mdx mice. Immunostaining with laminin (green) is shown; EBD is detected as a red signal under fluorescence microscopy. Scale bars: 100 μm. (F) Expression of myogenic genes and of embryonic MHC (Myh3) and perinatal MHC (Myh8) in WT and dKO TA muscle, determined by real-time RT-PCR. n = 3 (WT and dKO).
As a further assessment of muscle abnormalities, we analyzed the distribution of mitochondria and sarcoplasmic reticulum (SR) by NADH-TR staining in dKO muscle fibers at 12 weeks. dKO fibers showed more oxidative enzyme activity in G/P, EDL, and TA muscles than did WT myofibers (Supplemental Figure 3A), which may reflect a shift from glycolytic to oxidative myofibers in these muscles. The oxidative enzyme activity within individual fibers was also unevenly distributed, and some myofibers showed radiating intermyofibrillary networks (Figure 2D). Ring-like fibers were also occasionally observed upon NADH-TR staining (Figure 2D). There was no significant difference in NADH-TR staining in soleus muscle between dKO and WT littermates (Supplemental Figure 3A). Interestingly, normal NADH-TR staining patterns were observed in 4-week-old dKO muscle when no centrally located nuclei were present (Supplemental Figure 3B).
Accumulation of centralized nuclei is usually indicative of muscle regeneration in response to disease or injury (21–23). We therefore searched for signs of muscle damage and degeneration in dKO myofibers at 12 weeks of age. Monitoring sarcolemmal integrity by the uptake of Evans blue dye (EBD), which accumulates in damaged cells, showed very few dye-positive fibers (less than 4 per transverse section) (Figure 2E). We examined muscle from mdx mice, which develop muscular dystrophy, for comparison; these mice showed extensive EBD uptake (Figure 2E). We also measured serum levels of creatine kinase (CK) activity, indicative of sarcolemmal leakage, and observed only slightly elevated (2-fold) CK levels in dKO mice at 3 months (data not shown). In addition, dKO myofibers showed no signs of inflammation, fibrosis, or apoptosis (data not shown), which are characteristic of dystrophic muscle fibers. At 12 months of age, we did not observe worsening in myofiber morphologies or signs of inflammation, fibrosis, or cell death in dKO myofibers (Supplemental Figure 3C).
To assay for muscle regeneration, we analyzed expression of mRNAs encoding several myogenic markers of regeneration. Expression of Myog (which encodes myogenin) was upregulated 7-fold in dKO TA muscle, but there was no change in the expression levels of other myogenic markers, such as Pax3, Pax7, and MyoD (Figure 2F). Although there was a strong increase in both embryonic (Myh3) and perinatal MHC (Mhy8) mRNA levels in TA muscle by real-time RT-PCR (Figure 2F), embryonic MHC protein was rarely detected in dKO muscle fibers by immunohistochemistry (data not shown). These data indicate that there is only rare muscle regeneration in dKO mice, which is insufficient to account for the extensive centronuclear fibers observed in these mice. Thus, centronuclear myofibers in dKO mice without apparent necrosis, myofiber death, or significant regeneration are pathological characteristics reminiscent of human CNMs (1, 2).
T-tubule disorganization in muscle fibers of dKO mice.
In skeletal muscle, excitation-contraction coupling occurs at triads, which are composed of a transverse tubule (T-tubule) and a pair of terminal cisternae of the SR (24). In Mtm1-deficient mice, muscle fibers have a decreased number of triads and abnormal organization of T-tubules (3). T-tubule disorganization has also been reported in human CNM patients (6, 25).
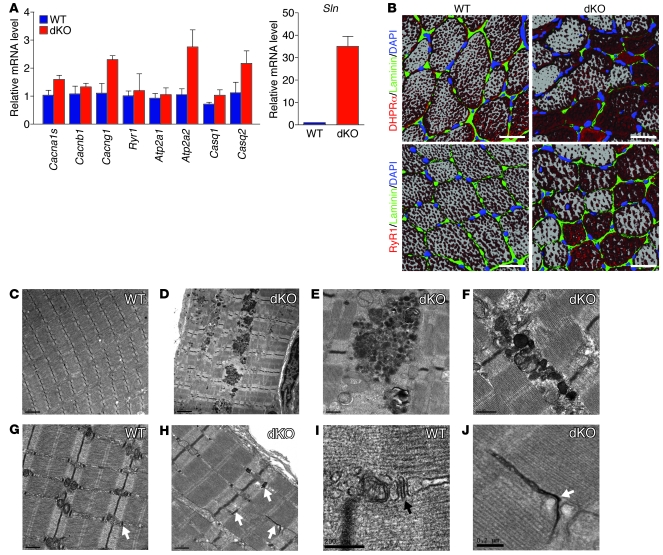
To assess whether T-tubule organization is affected in dKO muscle, we examined the expression of genes encoding components of T-tubules and SR that are important for excitation-contraction coupling, including the α1, β1, and γ1 subunits of the dihydropyridine receptor (DHPR) (encoded by Cacna1s, Cacnb1, and Cacng1, respectively), ryanodine receptor 1 (Ryr1), type 1 and 2 SERCA pumps (Atp2a1 and Atp2a2), and calsequestrin 1 and 2 (Casq1 and Casq2). At the mRNA level, expression of most of the genes was unchanged, except for a 2.5-fold increase in Cancng1 (Figure 3A). We also examined expression of RyR1, DHPRα, calsequestrin, and SERCA2 at the protein level and observed minimal changes (Supplemental Figure 4). In contrast, we observed a 35-fold increase in mRNA levels of Sln, accompanied by a comparable increase in sarcolipin protein (Figure 3A and Supplemental Figure 4). Sarcolipin upregulation is a common feature in skeletal muscle myopathies (ref. 26 and M. Periasamy, unpublished observations), but the significance of this upregulation is unknown. Expression of phospholamban was slightly upregulated in dKO muscle, but the phosphorylated phospholamban was slightly decreased at the protein level (Supplemental Figure 4).
Figure 3. Disorganization of triads in TA muscle fibers in dKO mice.
(A) Expression of mRNA transcripts encoding components of T-tubules and SR was determined by real-time RT-PCR in TA muscles of 12-week-old mice. n = 3 (WT and dKO). (B) Immunostaining of T-tubules and SR in transverse sections of TA muscle from WT and dKO mice at 12 weeks of age. T-tubules were detected by anti-DHPRα, and terminal cisternae of the SR were detected by anti-RyR1. Nuclei were detected by DAPI, and the myofiber perimeter was stained by anti-laminin. Images of multiple levels of the sections were taken and reconstructed to create the 3D effect. Scale bars: 30 μm. (C–J) Electron micrographs of WT and dKO muscle. dKO TA muscle showed accumulation of electron-dense structures (D–F) that were absent in WT TA muscle (C). dKO muscle (H and J) displayed T-tubules (arrows) in abnormal orientations compared with WT muscle (G and I). Scale bars: 2 μm (C and D); 0.5 μm (E–H); 0.2 μm (I and J).
We also analyzed the organization of triads by immunohistochemistry against DHPRα, a marker for T-tubules, and RyR1, a marker for terminal cisternae of SR. In transverse sections of WT myofibers, both T-tubules and terminal cisternae of SR displayed dot-like staining patterns distributed evenly along the myofibers (Figure 3B), which reflected the transverse orientations of triads relative to sarcomeres. In dKO myofibers, however, both T-tubules and SR showed aggregated staining, absence of staining in some regions, and irregular distribution within individual fibers (Figure 3B). In addition, in WT muscle, adjacent myofibers showed the same staining patterns. However, in dKO muscle, the adjacent myofibers often displayed different staining patterns (Figure 3B), suggestive of different orientations of triads in adjacent fibers. At 4 weeks of age, when dKO mice had not yet developed CNM, T-tubule structures were normal, as demonstrated by DHPRα staining (Supplemental Figure 3D).
We further analyzed the morphology of triads at the ultrastructural level by electron microscopy (Figure 3, C–J). In adult dKO TA muscle fibers, some T-tubules (stained dark by potassium ferricyanide) showed abnormal morphologies and longitudinal orientations aligned with the direction of myofibrils; these were rarely observed in WT muscle fibers (Figure 3, G–J). We also observed accumulation of electron-dense membranous structures along the myofibers and at triads in dKO myofibers (Figure 3, D–F). Overall, these findings indicate that miR-133a is important for the organization of T-tubules and triads and that its absence results in T-tubule disorganization.
Mitochondrial dysfunction in dKO skeletal muscle.
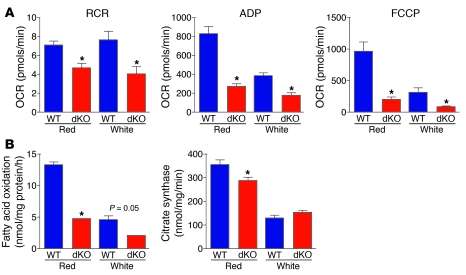
To determine whether lack of miR-133a alters mitochondrial function in skeletal muscle, mitochondria were isolated from red and white portions of the gastrocnemius muscle from dKO and WT mice. Immediately after isolation, mitochondrial respiration and fatty acid oxidation were assessed. Assessments of mitochondrial function include: (a) respiratory control ratio (RCR), the coupling between oxidative phosphorylation and ATP synthesis; (b) ADP-stimulated state 3 respiration, the respiratory rate during which the mitochondria are producing ATP; and (c) carbonylcyanide-p-trifluoromethoxyphenylhydrazone–stimulated (FCCP-stimulated) respiration, the maximal respiratory rate when oxidative phosphorylation is uncoupled from ATP synthesis. A reduction in any of these measures suggests mitochondrial dysfunction, which could be due to altered substrate handling, ATP synthase activity, or a dysfunction in respiratory chain components. Since we observed a reduction in all 3 parameters, dysfunction appears to be due to altered substrate handling or dysfunction in oxidative phosphorylation. The absence of miR-133a resulted in significant declines in RCR, ADP-stimulated state 3 respiration, and FCCP-stimulated maximal respiration in both red and white muscle, although the effects on FCCP-stimulated maximal respiration appeared to be more pronounced in red muscles (Figure 4A). In addition, total fatty acid oxidation was also significantly lower in mitochondria isolated from both red and white portions of gastrocnemius muscle from dKO animals (Figure 4B). There was also a reduction in citrate synthase in red quadricep muscle, but not in white quadricep muscle (Figure 4B). Collectively, these results demonstrate that the absence of miR-133a results in lower intrinsic mitochondrial function and fatty acid oxidation in both red and white skeletal muscle.
Figure 4. Mitochondrial dysfunction in dKO muscle.
(A) Mitochondria were isolated from red and white gastrocnemius muscle, and oxygen consumption rate (OCR) was measured for RCR, ADP-stimulated state 3 respiration (ADP), and FCCP-stimulated respiration (FCCP). n = 6 (WT and dKO). *P < 0.05 vs. WT. (B) Fatty acid oxidation was measured in isolated mitochondria from red and white gastrocnemius muscle. Citrate synthase enzyme activity was measured in isolated mitochondria from red and white quadriceps muscle. n = 6 (WT and dKO). *P < 0.05 vs. WT.
miR-133a targets dynamin 2, a regulator of CNM.
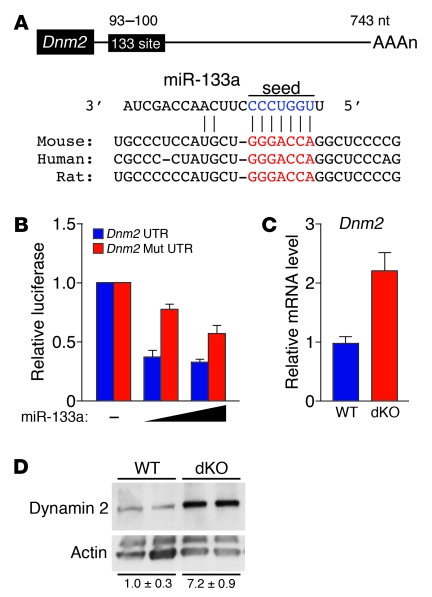
To begin to explore the mechanistic basis of skeletal muscle abnormalities in dKO mice, we searched for targets of miR-133a with potential roles in CNM. Among the strongly predicted targets of miR-133a is Dnm2 mRNA, encoding a large GTPase implicated in endocytosis, membrane trafficking, and regulation of the actin and microtubule cytoskeletons (11). Point mutations in the human DNM2 gene, thought to act in a dominant-negative manner, cause the autosomal-dominant form of CNM (7, 8, 27, 28). The 3′ UTR of Dnm2 mRNA contains an evolutionarily conserved miR-133a binding site (Figure 5A). miR-133a repressed a luciferase reporter gene linked to the 3′ UTR of Dnm2 mRNA, whereas a mutation in the predicted miR-133a binding site in the 3′ UTR prevented repression (Figure 5B), confirming Dnm2 mRNA as a target for miR-133a. Moreover, we observed a 2-fold increase in Dnm2 mRNA by quantitative real-time RT-PCR and an approximate 7-fold increase in dynamin 2 protein in TA muscle of dKO compared with WT mice by Western blot analysis (Figure 5, C and D). These results indicate that miR-133 represses dynamin 2 expression at both mRNA and protein levels.
Figure 5. miR-133a regulates Dnm2 expression in skeletal muscle.
(A) Position of miR-133a target site in Dnm2 3′ UTR and sequence alignment of miR-133a and the Dnm2 3′ UTR from various species are shown. Conserved miR-133a binding sites in Dnm2 3′ UTR are shown in red. Mutations in Dnm2 3′ UTR were introduced to disrupt base-pairing with miR-133a seed sequences (blue). (B) Luciferase reporter constructs containing WT and mutant Dnm2 3′ UTR sequences were cotransfected into COS-1 cells with a plasmid expressing miR-133a. 48 hours after transfection, luciferase activity was measured and normalized to β-galactosidase activity. (C) Real-time RT-PCR showing expression of Dnm2 mRNA in WT and dKO TA muscle. n = 3 (WT and dKO). (D) Western blot showing expression of dynamin 2 protein in TA muscle of WT and dKO mice. n = 2 (WT and dKO). The blot was stripped and reprobed with an antibody against α-actin as a loading control. Quantification of dynamin 2 protein, determined by densitometry and normalized to α-actin, is also shown.
Overexpression of dynamin 2 in skeletal muscle causes CNM in type II myofibers.
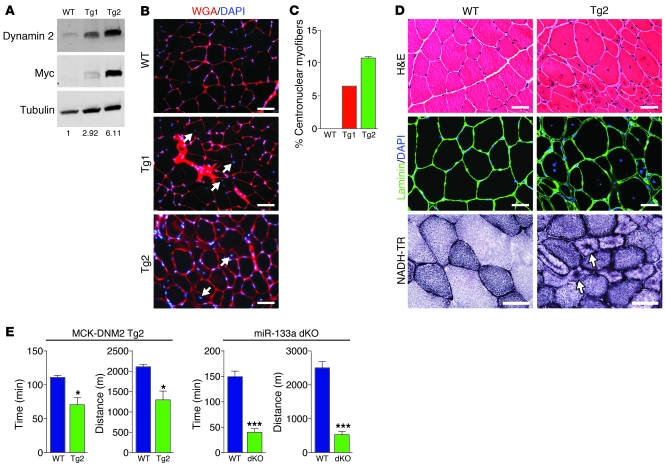
To examine whether elevated expression of dynamin 2, as observed in dKO myofibers, is sufficient to cause CNM, we generated transgenic mice in which dynamin 2 protein (with a myc-tag on the C terminus) was expressed under control of the muscle CK (MCK) promoter (referred to herein as MCK-DNM2 mice) (29, 30). Overexpression of dynamin 2 protein in skeletal muscle of transgenic mice was confirmed by Western blotting using antibodies against dynamin 2 as well as the myc epitope tag (Figure 6A). We obtained 2 MCK-DNM2 transgenic mouse lines, Tg1 and Tg2, which showed 3- and 6-fold overexpression of dynamin 2, respectively, compared with WT levels. At 7 weeks of age, both transgenic lines displayed accumulation of centronuclear myofibers (Figure 6B). Interestingly, Tg2 mice, which overexpressed dynamin 2 at a level similar to that of dKO mice, displayed age-dependent centronuclear myofibers in TA muscle comparable to those of dKO mice (Figure 6C).
Figure 6. Overexpression of Dnm2 in skeletal muscle causes CNM.
(A) Western blot analysis of TA muscle from WT and MCK-DNM2 transgenic mouse lines Tg1 and Tg2 using anti–dynamin 2 and anti-myc to show overexpression of transgene. Anti-tubulin was used as a loading control. Protein quantification, determined by densitometry, is also shown. (B) Transverse sections of TA muscles of WT, Tg1, and Tg2 mice at 6 weeks of age were stained with wheat germ agglutinin (WGA) and DAPI to show central nuclei (arrows) in transgenic mice. Scale bars: 100 μm. (C) Percentage of centronuclear myofibers in TA muscle of transgenic mice at 7 weeks of age. (D) Histological analysis of TA and soleus muscles of WT and Tg2 mice at 11 weeks of age. TA muscle sections were stained with H&E, with anti-laminin and DAPI to show central nuclei, and with NADH-TR to reveal abnormal distribution and radiating intermyofibrillary network (arrows). Scale bars: 40 μm. (E) 10-week-old WT and Tg2 mice (n = 3 per group), as well as 3 month-old WT and dKO mice (n = 5 per group), were subjected to forced downhill running on a treadmill. Muscle performance was measured as time to exhaustion. Total running distance is also shown. *P < 0.05; ***P < 0.001.
At 11 weeks of age, Tg2 mice displayed signs of muscle atrophy, with decreased muscle mass in both TA and G/P muscle (Supplemental Figure 5A). There was no difference in body mass between Tg2 and WT littermates (Supplemental Figure 5A). Histological analysis of TA muscle showed heterogeneous fiber sizes and the presence of centronuclear fibers in Tg2 mice (Figure 6D). The percentage of centronuclear myofibers in TA muscle of Tg2 mice was approximately 23% at this age (data not shown). NADH-TR staining revealed abnormal aggregation of oxidative enzymatic activity and radiating intermyofibrillary networks (Figure 6D). Abnormal organization of T-tubules was also observed in Tg2 TA muscle, as detected by immunohistochemistry against DHPRα (Supplemental Figure 5B).
Dynamin 2 protein was not significantly overexpressed in soleus muscle or heart of Tg2 mice (Supplemental Figure 5C), consistent with the preferential expression of the MCK promoter in type II myofibers (29, 30). Not surprisingly, therefore, we did not observe any abnormalities in soleus muscle or heart function in Tg2 mice (Supplemental Figure 5C and data not shown).
To assess muscle performance, we subjected mice to downhill treadmill running and analyzed running time and distance to exhaustion. At 10 weeks of age, Tg2 mice ran for a significantly shorter time than did WT mice (Figure 6E), indicative of muscle weakness. dKO mice showed a more dramatic decrease in running capacity (Figure 6E). However, the compromised cardiac function in dKO mice may also be a contributing factor to the reduction in exercise capacity.
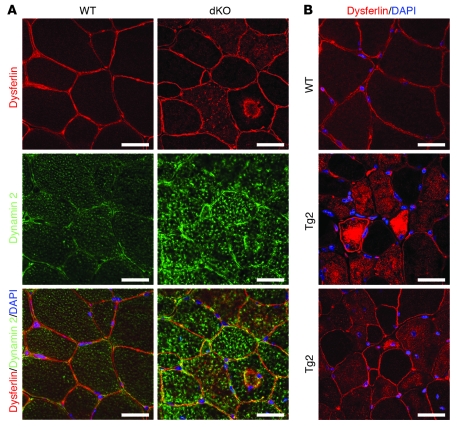
Intracellular accumulation of dysferlin has been recently reported in human DNM2-associated CNM patients, as well as in heterozygous mice carrying the R456W Dnm2 mutation (9). We also analyzed localization of dysferlin in dKO muscle and Tg2 muscle. Interestingly, substantial accumulation of dysferlin inside the myofibers was observed in both dKO and Tg2 muscle fibers (Figure 7, A and B). Furthermore, at least some of the intracellular dysferlin was colocalized with dynamin 2 in dKO muscle fibers (Figure 7A).
Figure 7. Intracellular accumulation of dysferlin in dKO and MCK-DNM2 transgenic mouse myofibers.
(A) Immunostaining of TA muscle from WT and dKO mice to detect dynamin 2 and dysferlin. Intracellular accumulation of dysferlin was observed in dKO myofibers. Overlay images indicate localization of dynamin 2 and dysferlin in the intracellular aggregates in dKO muscle. Scale bars: 30 μm. (B) Immunostaining of TA muscle from WT and Tg2 mice to detect dysferlin. Intracellular accumulation of dysferlin was observed in Tg2 myofibers. Scale bars: 30 μm.
These results demonstrate that elevated expression of Dnm2 in skeletal muscle causes CNM, predominantly in type II fibers, mimicking the dKO phenotype. We conclude that the CNM in dKO muscle can be explained, at least in part, by dysregulation of Dnm2.
dKO mice show increased type I myofibers in soleus muscle.
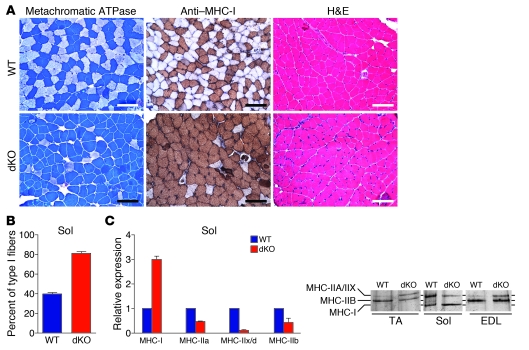
In addition to CNM, dKO mice displayed increased numbers of type I fibers in soleus muscle, which does not show CNM. We analyzed fiber type composition of soleus muscle from adult dKO mice by metachromatic ATPase staining and by immunohistochemistry against type I myosin heavy chain (MHC), shown by dark brown staining. Soleus muscle of WT mice was composed of about 43% type I fibers (Figure 8, A and B). Soleus muscle of dKO mice showed a 2-fold increase in the number of type I fibers (Figure 8, A and B).
Figure 8. Control of skeletal muscle fiber type by miR-133a.
(A) Metachromatic ATPase staining and anti–MHC-1 immunostaining of soleus muscle from WT and dKO mice at 12 weeks of age showed an increase in type I myofibers in dKO soleus muscle. H&E staining of the soleus muscles is also shown. Scale bars: 100 μm. (B) Percentage of type I myofibers in soleus muscles, determined by metachromatic ATPase staining. n = 6 (WT and dKO). (C) Expression of transcripts of MHC isoforms in soleus muscle, determined by real-time RT-PCR. n = 3 (WT and dKO). Expression of MHC isoforms from protein extracts of soleus, EDL, and TA muscles from WT and dKO mice was also determined by glycerol gel electrophoresis followed by silver staining.
Quantitative real-time RT-PCR analysis of the expression of transcripts encoding individual MHC isoforms revealed an increase in type I MHC (MHC-I) and decreases in type II MHCs (MHC-IIa, MHC-IIx/d, and MHC-IIb) in soleus muscle of dKO compared with WT mice (Figure 8C). We examined the protein composition of MHC isoforms in soleus, EDL, and TA muscle by silver staining of glycerol gels: 3 bands were present in protein extracts of soleus muscle isolated from WT mice, corresponding to MHC-IIa/IIx, MHC-IIb, and MHC-I proteins; 2 bands were present in protein extracts of TA and EDL muscles from WT mice, representing MHC-IIb and MHC-IIa/IIx (Figure 8C). Consistent with results from quantitative real-time RT-PCR, soleus muscle of dKO mice displayed an increase in MHC-I protein and a decrease in MHC-IIa/IIx proteins. MHC-IIb protein was not observed in dKO soleus muscle. Interestingly, there was an increase in the oxidative MHC-IIa/IIx protein and a decrease in the glycolytic MHC-IIb protein in TA and EDL muscles of dKO mice compared with WT mice, which indicates that these muscle groups also display a fiber type shift toward more oxidative (type IIa) fibers.
To determine whether loss of miR-133a affects the formation of type I fibers during fetal development, we examined MHC-I expression by immunohistochemistry at P1. There was no obvious difference in the number of MHC-I–positive myofibers in soleus or EDL muscles of dKO mice at P1 (Supplemental Figure 6A), which indicates that miR-133a does not influence embryonic development of type I myofibers. To determine when the fiber type switch takes place in dKO mice, we analyzed fiber type composition in both 2- and 4-week old mice by metachromatic ATPase staining. At both ages, the percentage of type I fibers in soleus was increased by almost 2-fold in dKO mice (Supplemental Figure 6B). We conclude that miR-133a does not influence specification of type I myofibers during embryonic development. Rather, miR-133a represses type I myofibers postnatally, such that the absence of miR-133a results in an increase in type I myofibers of adult mice.
Discussion
The results of this study reveal important roles of miR-133a in maintaining normal structure and function of adult skeletal muscle. The absence of miR-133a resulted in CNM, mitochondrial dysfunction, disarray of muscle triads, and a shift from glycolytic to oxidative myofibers. The skeletal muscle abnormalities in dKO mice were remarkably similar to those of human CNMs, suggestive of an important role of this miRNA in modulation of this disorder.
miR-133a and CNM.
The histological features of dKO muscle, including the presence of centronuclear fibers and absence of necrosis or myofiber death, demonstrated similarities to human CNMs. NADH-TR staining patterns in dKO fibers mimicked the typical NADH-TR staining pattern of DNM-associated CNM, which shows radial distribution of sarcoplasmic strands (2). However, in contrast to human CNMs, centronuclear fibers were only observed in type II fibers, but not in type I fibers in dKO mice. The lack of CNM phenotype in soleus muscle of dKO mice could be due to the expression of miR-133b, which was enriched in soleus muscle. Alternatively, the differences in myofiber distribution of centralized nuclei between mice and humans may reflect species differences in muscle function. Interestingly, we previously reported type II fiber–specific CNM in mice lacking the Srpk3 gene, which encodes a muscle-specific serine, arginine protein kinase (SRPK) regulated by MEF2 (31). Given the histological similarities between skeletal muscle of Srpk3-null mice and the dKO mice of the present study, it is possible that miR-133a and Srpk3 act through common mechanisms to influence muscle structure and function.
Multiple missense mutations within the DNM2 gene have been linked to autosomal-dominant CNMs (7, 8, 27, 28). Interestingly, these mutations are heterozygous missense mutations or small deletions that do not affect DNM2 transcript levels, protein expression, or localization (8, 28). However, the mechanisms whereby CNM-associated mutations affect DNM2 cellular function are unknown.
Our results demonstrated that miR-133a directly regulates Dnm2 mRNA and dynamin 2 protein expression. Moreover, elevated expression of Dnm2 in skeletal muscle, at levels comparable to those in dKO mice, caused CNM, which indicates that skeletal muscle function depends on a precise level of dynamin 2 expression. Although the exact mechanism is unknown, it is possible that increased dynamin 2 protein may cause abnormally strong dynamin assembly and disrupt the energetic balance of efficient assembly and disassembly that is required for proper DNM2 function in skeletal muscle. In this regard, CNM-related DNM2 mutations in humans have been reported to act in a dominant-negative manner to impair membrane trafficking, cytoskeleton-related processes, and centrosomal function (8, 28). It is unclear how Dnm2 gain of function in dKO mice and MCK-DNM2 transgenic mice also cause CNM. However, a recent study showed that specific CNM-related DNM2 mutations cause increased GTPase activity and promote dynamin oligomerization without altering lipid binding (32). Another study also showed that CNM-related DNM2 mutants enhance the stability of dynamin polymers without impairing their ability to bind and/or hydrolyze GTP (33).
Recently, Durieux et al. showed that heterozygous mice expressing the most frequent Dnm2 mutation, R456W, developed a myopathy with muscle atrophy and weakness, but not CNM (9). They hypothesized that the effect of dynamin 2 on contractile properties and nuclear positioning are independent. Intriguingly, our results showed that overexpression of Dnm2 in skeletal muscle affected both muscle function and nuclear position. The difference in these phenotypes could be explained by different model systems used (i.e., overexpression vs. knockin). Nonetheless, our results demonstrated that skeletal muscle is sensitive to dynamin 2 protein level and that elevated dynamin 2 expression results in CNM in mice.
It is also intriguing that loss of miR-133a in mouse skeletal muscle caused CNM only in type II fibers, in contrast to the type I fiber predominance in human DNM2-associated CNM patients. It is likely that, in mice, the soleus muscle is protected from muscle damage. However, we can not rule out the possibility that the soleus-enriched miR-133b, which is highly homologous to miR-133a, protects soleus muscle from CNM.
miR-133a is also predicted to target other genes, such as those encoding profilin 2, calmodulin 1, FGFR1, and mastermind-like 1. We performed luciferase reporter assays with the 3′ UTRs of these mRNAs and confirmed that they were targeted by miR-133a in vitro; however, their regulation by miR-133a in vivo was less prominent in skeletal muscle (data not shown). Therefore, we believe that although miR-133a targets multiple genes in skeletal muscle, the primary effect comes from its regulation of DNM2.
Modulation of myofiber phenotypes and mitochondrial function by miR-133a.
Skeletal muscle is composed of heterogeneous myofibers with distinctive contractile and metabolic properties (34). Adult myofibers are highly plastic and can switch between type I and type II phenotypes in response to work load, hormonal stimuli, and disease. The phenotype of dKO mice indicates that miR-133a suppresses the type I myofiber gene program. Type I myofibers are believed to be more resistant to disease or damage than type II fibers (35). In many muscle diseases, such as Duchenne muscular dystrophy, there is a switch in fiber type toward type I, which may serve as a protective mechanism (36, 37). We cannot rule out the possibility that changes in fiber types in dKO muscle are secondary to the CNM phenotype.
Mitochondrial dysfunction has been implicated in a number of myopathies, including Duchenne muscular dystrophy and metabolic and neurological disorders (38–40), as well as in the aging process (41, 42). Our results are consistent with the previous finding that mitochondrial abnormalities are associated with DNM2-related CNM (43). However, this result seems incompatible with the fast-to-slow myofiber conversion in dKO mice, since type I fibers are believed to have more oxidative enzyme activity. Although the exact mechanism underlying this discrepancy is unclear, there are several possibilities. The switch to type I fiber could be the result of changes in myosin composition that do not affect mitochondria content. In addition, a fast-to-slow myofiber conversion is associated with increases in capillary and mitochondrial density. However, this does not take into account the functional capacity of the individual mitochondria. Finally, impairments in mitochondrial function result in reduced ATP availability to the muscle. Thus, it is possible that the fiber type switch in dKO muscle is a protective mechanism against mitochondrial dysfunction and reduced ATP availability (35).
Role of miR-133a in skeletal muscle and heart.
Our results demonstrated that miR-133a, which is expressed in both heart and skeletal muscle, plays different roles in these tissues. In the heart, miR-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene program during heart development (18). Interestingly, miR-133a was dispensable for skeletal muscle development, as dKO mice did not display any skeletal muscle abnormalities until after 4 weeks of age. The skeletal muscle of dKO mice developed CNM after 4 weeks of age, whereas the heart develops dilated cardiomyopathy at a later age, which leads to heart failure and sudden death in a subset of mice (18). Interestingly, the hearts of dKO mice showed pronounced sarcomere disorganization and disrupted Z-discs, as well as severe mitochondrial abnormalities at 4 months of age (18). On the other hand, sarcomeric structures and mitochondrial morphologies were largely unaffected in dKO skeletal muscle (Figure 3). Rather, miR-133a specifically affects triads in skeletal myofibers. It is unclear why the heart and skeletal muscle show different abnormalities in response to the loss of miR-133a. We speculate that this may reflect the regulation of different target genes by miR-133a in skeletal muscle (such as dynamin 2) and heart (such as cyclin D2 and SRF). Another reason could be the fact that the highly homologous miR-133b is expressed in dKO skeletal muscle, albeit at a lower level, but not in dKO heart. Although it is conceivable that the cardiomyopathy in dKO mice could contribute, CNM is not associated with other mouse models of cardiomyopathy. Therefore, we believe that the skeletal muscle abnormalities in dKO mice are mainly caused by cell-autonomous functions of miR-133a in skeletal muscle.
Therapeutic implications.
The similarities in skeletal muscle abnormalities in dKO mice and human CNM patients raise the possibility that miR-133a plays a modulatory role in human myopathies. In this regard, it may be possible to modulate miR-133a mRNA targets, such as DNM2, through oligonucleotide modulation of miR-133a. The realization that miRNAs such as miR-133a influence muscle structure and function and interface with human disease mechanisms underscores the potential insights that may be gleaned from the analysis of miRNA targets involved in disease. In the future, it will be of interest to explore the potential involvement of other miRNAs in human muscle disease.
Methods
Generation of MCK-DNM2 transgenic mice.
A MCK-DNM2 transgene was generated by placing a C-terminal myc-tagged rat Dnm2 cDNA (gift from J. Albanesi, University of Texas Southwestern Medical Center, Dallas, Texas, USA) downstream of the 4.8-kb MCK promoter. The construct contained a downstream human growth hormone poly(A) signal. Transgenic mice were generated as previously described (44, 45). We analyzed 2 F1 lines, termed Tg1 and Tg2.
Northern blot analysis.
Total RNA was isolated from mouse skeletal muscle tissues using the miRNeasy mini kit (QIAGEN). Northern blots to detect miR-133a and U6 were performed as described previously (18). 32P-labled Star-Fire oligonucleotide probes (IDT) against mature miR-133a and U6 probes were used in the hybridization.
RT-PCR and real-time analysis.
RNA was treated with Turbo RNase-free DNase (Ambion Inc.) prior to the reverse transcription step. RT-PCR was performed using random hexamer primers (Invitrogen). Quantitative real-time RT-PCR was performed using TaqMan probes (ABI) or Sybr Green probes (see Supplemental Methods for Sybr Green primer sequences). Quantitative real-time RT-PCR on miRNA was performed using the TaqMan miRNA assay kits (ABI) according to manufacturer’s protocol.
Histological analysis of skeletal muscle.
Various muscle groups were harvested, flash frozen in embedding medium containing a 3:1 mixture of Tissue Freezing Medium (Triangle Biomedical Sciences) and gum tragacanth (Sigma-Aldrich) or fixed in 4% paraformaldehyde, and processed for routine paraffin histology. Frozen sections were cut on a cryotome and stained with H&E as previously described (45). NADH-TR staining on frozen sections was performed according to standard protocol. Metachromatic ATPase staining on frozen sections was performed as described previously (44, 45).
To determine the number of myofibers with centralized nuclei, more than 500 myofibers were counted for TA and G/P muscles and more than 300 were counted for soleus and EDL muscles of each mouse. Myofiber cross-sectional area was determined using ImageJ, and more than 200 fibers per muscle section were examined.
Electron microscopy.
Mice were anesthetized, then transcardially perfused with 0.1M phosphate buffer (pH 7.3) followed by 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1M sodium cacodylate buffer. TA muscles were dissected and processed for selective staining of T-tubules as described previously (3).
EBD uptake.
EBD uptake was performed as described previously (46). Briefly, EBD (10 mg/ml in PBS) was administered to mice intraperitoneally (0.1 ml per 10 g body mass). Mice were subjected to exercise using a running wheel overnight (all mice underwent wheel running), and muscles were harvested approximately 18 hours later. Gastrocnemius and TA muscles were flash frozen in embedding medium. Frozen sections were immunostained with primary antibody rabbit anti-laminin (Sigma-Aldrich, 1:200), followed by secondary antibody Alexa Fluor 488–conjugated goat anti-rabbit IgG (Invitrogen, 1:400). EBD was detected as red autofluorescence using fluorescence microscopy.
Immunohistochemistry.
Frozen sections were fixed in freshly prepared 4% paraformaldehyde for 20 minutes on ice and were then treated with 0.3% Triton X-100 in PBS at room temperature for 20 minutes. Sections were incubated with mouse IgG blocking solution from the M.O.M. kit (Vector Lab) diluted in 0.01% Triton X-100 in PBS at room temperature for 1 hour. Sections were then incubated with 5% goat serum (Sigma-Aldrich) in M.O.M. protein diluent for 30 minutes. Sections were incubated with primary antibodies diluted in M.O.M. protein diluent at 4°C overnight. The next morning, slides were washed with PBS and incubated with secondary antibodies diluted in M.O.M. protein diluent at room temperature for 45 minutes. Sections were then washed and mounted with VectoShield Mounting Medium with DAPI. Pictures were taken with a Zeiss confocal microscope. Primary and secondary antibodies were as follows: DHPRα (Thermo Scientific, 1:100), RyR1 (clone34C, Sigma-Aldrich, 1:100), Laminin (Sigma-Aldrich, 1:200), MHC-I (clone NOQ7.5.4D, Sigma-Aldrich, 1:5,000), Dysferlin (Hamlet, Novocastra, 1:40), dynamin 2 (Abcam, 1:400), Alexa Fluor 594–conjugated goat anti-mouse IgG1 (Invitrogen, 1:400), Alexa Fluor 488–conjugated goat anti-rabbit IgG (Invitrogen, 1:400). Wheat germ agglutinin staining was performed as previously described (46). MHC-I (clone NOQ7.5.4D, Sigma-Aldrich, 1:5,000) was used for primary detection of type I myosin, and HRP-conjugated secondary antibody (A8924, Sigma-Aldrich) followed by DAB chromagen reaction (DAKO) were used for detection. Samples were then counterstained with hematoxylin.
Western blot analysis.
Total cell lysates were extracted from skeletal muscle tissues and resolved on SDS-PAGE. Western blotting was performed by standard protocol. Antibodies against dynamin 2 (Santa Cruz Biotechnology, 1:100), c-Myc (Santa Cruz Biotechnology, 1:1,000), DHPRα (Thermo Scientific, 1:100), RyR1 (clone 34C, Sigma-Aldrich, 1:100), SERCA2 (BD Biosciences, 1:1,000), sarcolipin (gift from M. Periasamy, Ohio State University, Columbus, Ohio, USA, 1:1,000), phospholamban (Upstate, 1:1,000), phospho-phospholamban (Millipore, 1:1,000), calsequestrin 2 (Santa Cruz, 1:1,000), tubulin (Sigma-Aldrich, 1:5,000) and α-actin (Sigma-Aldrich, 1:2,000) were used. Quantification of Western blots was performed by densitometry using a PhosphoImager.
Cell culture, transfection, and luciferase assays.
1-kb fragments of the Dnm2 3′ UTR containing the miR-133a binding sites were cloned into pMIR-REPORT vector (Ambion). Mutagenesis of the miR-133a binding site, cell culture, and luciferase assay were performed as previously described (18).
Treadmill test.
The treadmill test was performed using the Exer-6M (Columbus Instruments) at 15° downhill. Mice were trained on the treadmill at 5 m/min for 5 minutes for 2 consecutive days. The following day, mice ran on the treadmill at 5 m/min for 2 minutes, 7 m/min for 2 minutes, 8 m/min for 2 minutes, and 10 m/min for 5 minutes. Subsequently, speed was increased by 1 m/min to a final speed of 20 m/min. Exhaustion was defined as the inability of the animal to remain on the treadmill despite electrical prodding.
Electrophoresis of MHC isoforms.
Myosin was isolated from skeletal muscle and was separated by electrophoresis on glycerol-SDS-PAGE gels as previously described (47). Gels were stained with a silver nitrate staining kit (Bio-Rad).
Mitochondrial isolation from gastrocnemius muscle.
Mitochondria were isolated from red and white skeletal muscle dissected from gastrocnemius muscle as previously described (48), with modifications. Tissue samples were collected in buffer containing 67 mM sucrose, 50 mM Tris/HCl, 50 mM KCl, 10 mM EDTA/Tris, and 10% bovine serum albumin. Samples were minced and digested in 0.05% trypsin for 30 minutes. Samples were then homogenized, and mitochondria were isolated by differential centrifugation.
Respiration in isolated mitochondria.
Respirometry of isolated mitochondria was performed using an XF24 extracellular flux analyzer (Seahorse Bioscience). Immediately after isolation and protein quantification, mitochondria were plated on Seahorse cell culture plates at 5 μg/well in the presence of 10 mM pyruvate and 5 mM malate. Experiments consisted of 25-second mixing and 4- to 7-minute measurement cycles. Oxygen consumption was measured under basal conditions, ADP-stimulated (5 mM) state 3 respiration, oligomycin-induced (2 μM) state 4 respiration, and uncoupled respiration in the presence of FCCP (0.3 μM) to assess maximal oxidative capacity. The RCR was calculated as the ratio of state 3/state 4 respiration. All experiments were performed at 37°C.
Fatty acid metabolism.
Fatty acid oxidation was assessed in isolated mitochondria by measuring and summing 14CO2 production and 14C-labeled acid-soluble metabolites from the oxidation of [1-14C]-palmitic acid as previously described (49, 50). Citrate synthase activity was determined as previously described (51).
Statistics.
Data are presented as mean ± SEM. Differences between groups were tested for statistical significance using the unpaired 2-tailed Student’s t test. P values less than 0.05 were considered significant.
Study approval.
All animal experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committees of University of Texas Southwestern Medical Center.
Supplementary Material
Acknowledgments
We thank Beverly Rothermel, Dennis Burns, Joseph Albanesi, Lei Wang, and Andrew Williams for discussion. We are grateful to Joseph Albanesi for the Dnm2 cDNA clone and Muthu Periasamy for the sarcolipin antibody. We thank John McAnally for generation of transgenic mice. We thank Cheryl Nolen, Mi-Sung Kim, Gaile Vitug, and Evelyn Tennison for technical help. We are grateful to Christopher Gilpin (University of Texas Southwestern Electron Microscopy Core Facility) for EM images and Abhijit Bugde (Live Cell Imaging Core Facility) for 3D reconstruction of confocal microscope images. We thank José Cabrera for graphics and Jennifer Brown for editorial assistance. Work in the laboratory of E.N. Olson was supported by grants from the NIH, the Fondation Leducq’s Transatlantic Network of Excellence in Cardiovascular Research Program, and the Robert A. Welch Foundation (grant no. I-0025). N. Liu was supported by a Scientist Development Grant from American Heart Association.
Footnotes
Conflict of interest: Eric N. Olson is cofounder of miRagen Therapeutics, a company focused on developing miRNA-based therapies for cardiovascular disease.
Citation for this article: J Clin Invest. 2011;121(8):3258–3268. doi:10.1172/JCI46267.
References
- 1.Jungbluth H, Wallgren-Pettersson C, Laporte J. Centronuclear (myotubular) myopathy. Orphanet J Rare Dis. 2008;3:26. doi: 10.1186/1750-1172-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romero NB. Centronuclear myopathies: a widening concept. Neuromuscul Disord. 2010;20(4):223–228. doi: 10.1016/j.nmd.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Al-Qusairi L, et al. T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc Natl Acad Sci U S A. 2009;106(44):18763–18768. doi: 10.1073/pnas.0900705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buj-Bello A, et al. AAV-mediated intramuscular delivery of myotubularin corrects the myotubular myopathy phenotype in targeted murine muscle and suggests a function in plasma membrane homeostasis. Hum Mol Genet. 2008;17(14):2132–2143. doi: 10.1093/hmg/ddn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buj-Bello A, et al. The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc Natl Acad Sci U S A. 2002;99(23):15060–15065. doi: 10.1073/pnas.212498399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowling JJ, et al. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009;5(2):e1000372. doi: 10.1371/journal.pgen.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bitoun M, et al. Dynamin 2 mutations cause sporadic centronuclear myopathy with neonatal onset. Ann Neurol. 2007;62(6):666–670. doi: 10.1002/ana.21235. [DOI] [PubMed] [Google Scholar]
- 8.Bitoun M, et al. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat Genet. 2005;37(11):1207–1209. doi: 10.1038/ng1657. [DOI] [PubMed] [Google Scholar]
- 9.Durieux AC, et al. A centronuclear myopathy-dynamin 2 mutation impairs skeletal muscle structure and function in mice. Hum Mol Genet. 2010;19(24):4820–4836. doi: 10.1093/hmg/ddq413. [DOI] [PubMed] [Google Scholar]
- 10.Heymann JA, Hinshaw JE. Dynamins at a glance. J Cell Sci. 2009;122(Pt 19):3427–3431. doi: 10.1242/jcs.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durieux AC, Prudhon B, Guicheney P, Bitoun M. Dynamin 2 and human diseases. J Mol Med. 2010;88(4):339–350. doi: 10.1007/s00109-009-0587-4. [DOI] [PubMed] [Google Scholar]
- 12.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 13.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21(3):461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenberg I, Alexander MS, Kunkel LM. miRNAS in normal and diseased skeletal muscle. J Cell Mol Med. 2009;13(1):2–11. doi: 10.1111/j.1582-4934.2008.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenberg I, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci U S A. 2007;104(43):17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18(4):510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams AH, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326(5959):1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22(23):3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 20.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi X, Garry DJ. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006;20(13):1692–1708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- 22.McNally EM, Pytel P. Muscle diseases: the muscular dystrophies. Annu Rev Pathol. 2007;2:87–109. doi: 10.1146/annurev.pathol.2.010506.091936. [DOI] [PubMed] [Google Scholar]
- 23.Tedesco FS, Dellavalle A, Diaz-Manera J, Messina G, Cossu G. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J Clin Invest. 2010;120(1):11–19. doi: 10.1172/JCI40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franzini-Armstrong C, Jorgensen AO. Structure and development of E-C coupling units in skeletal muscle. Annu Rev Physiol. 1994;56:509–534. doi: 10.1146/annurev.ph.56.030194.002453. [DOI] [PubMed] [Google Scholar]
- 25.Toussaint A, et al. Defects in amphiphysin 2 (BIN1) and triads in several forms of centronuclear myopathies. Acta Neuropathol. 2011;121(2):253–266. doi: 10.1007/s00401-010-0754-2. [DOI] [PubMed] [Google Scholar]
- 26.Babu GJ, et al. Ablation of sarcolipin enhances sarcoplasmic reticulum calcium transport and atrial contractility. Proc Natl Acad Sci U S A. 2007;104(45):17867–17872. doi: 10.1073/pnas.0707722104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bitoun M, et al. A new centronuclear myopathy phenotype due to a novel dynamin 2 mutation. Neurology. 2009;72(1):93–95. doi: 10.1212/01.wnl.0000338624.25852.12. [DOI] [PubMed] [Google Scholar]
- 28.Bitoun M, et al. Dynamin 2 mutations associated with human diseases impair clathrin-mediated receptor endocytosis. Hum Mutat. 2009;30(10):1419–1427. doi: 10.1002/humu.21086. [DOI] [PubMed] [Google Scholar]
- 29.Donoviel DB, Shield MA, Buskin JN, Haugen HS, Clegg CH, Hauschka SD. Analysis of muscle creatine kinase gene regulatory elements in skeletal and cardiac muscles of transgenic mice. Mol Cell Biol. 1996;16(4):1649–1658. doi: 10.1128/mcb.16.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sternberg EA, Spizz G, Perry WM, Vizard D, Weil T, Olson EN. Identification of upstream and intragenic regulatory elements that confer cell-type-restricted and differentiation-specific expression on the muscle creatine kinase gene. Mol Cell Biol. 1988;8(7):2896–2909. doi: 10.1128/mcb.8.7.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagawa O, et al. Centronuclear myopathy in mice lacking a novel muscle-specific protein kinase transcriptionally regulated by MEF2. Genes Dev. 2005;19(17):2066–2077. doi: 10.1101/gad.1338705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenniston JA, Lemmon MA. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010;29(18):3054–3067. doi: 10.1038/emboj.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Barylko B, Byers C, Ross JA, Jameson DM, Albanesi JP. Dynamin 2 mutants linked to centronuclear myopathies form abnormally stable polymers. J Biol Chem. 2010;285(30):22753–22757. doi: 10.1074/jbc.C110.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 35.Webster C, Silberstein L, Hays AP, Blau HM. Fast muscle fibers are preferentially affected in Duchenne muscular dystrophy. Cell. 1988;52(4):503–513. doi: 10.1016/0092-8674(88)90463-1. [DOI] [PubMed] [Google Scholar]
- 36.Wang JF, Forst J, Schroder S, Schroder JM. Correlation of muscle fiber type measurements with clinical and molecular genetic data in Duchenne muscular dystrophy. Neuromuscul Disord. 1999;9(3):150–158. doi: 10.1016/S0960-8966(98)00114-X. [DOI] [PubMed] [Google Scholar]
- 37.Cros D, Harnden P, Pellissier JF, Serratrice G. Muscle hypertrophy in Duchenne muscular dystrophy. A pathological and morphometric study. J Neurol. 1989;236(1):43–47. doi: 10.1007/BF00314217. [DOI] [PubMed] [Google Scholar]
- 38.Naumann M, et al. Mitochondrial dysfunction in adult-onset myopathies with structural abnormalities. Acta Neuropathol. 1995;89(2):152–157. doi: 10.1007/BF00296359. [DOI] [PubMed] [Google Scholar]
- 39.Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ Res. 2008;102(4):401–414. doi: 10.1161/CIRCRESAHA.107.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jongpiputvanich S, Sueblinvong T, Norapucsunton T. Mitochondrial respiratory chain dysfunction in various neuromuscular diseases. J Clin Neurosci. 2005;12(4):426–428. doi: 10.1016/j.jocn.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 41.Hebert SL, Lanza IR, Nair KS. Mitochondrial DNA alterations and reduced mitochondrial function in aging. Mech Ageing Dev. 2010;131(7–8):451–462. doi: 10.1016/j.mad.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanza IR, Nair KS. Mitochondrial function as a determinant of life span. Pflugers Arch. 2010;459(2):277–289. doi: 10.1007/s00424-009-0724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zanoteli E, Vergani N, Campos Y, Vainzof M, Oliveira AS, d’Azzo A. Mitochondrial alterations in dynamin 2-related centronuclear myopathy. Arq Neuropsiquiatr. 2009;67(1):102–104. doi: 10.1590/S0004-282X2009000100023. [DOI] [PubMed] [Google Scholar]
- 44.Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J Biol Chem. 2000;275(7):4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 45.Wu H, et al. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296(5566):349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 46.Millay DP, et al. Genetic and pharmacologic inhibition of mitochondrial-dependent necrosis attenuates muscular dystrophy. Nat Med. 2008;14(4):442–447. doi: 10.1038/nm1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim MS, et al. Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol Cell Biol. 2008;28(11):3600–3609. doi: 10.1128/MCB.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2(2):287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 49.Frisard MI, et al. Toll-like receptor 4 modulates skeletal muscle substrate metabolism. Am J Physiol Endocrinol Metab. 2010;298(5):E988–E998. doi: 10.1152/ajpendo.00307.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hulver MW, et al. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab. 2005;2(4):251–261. doi: 10.1016/j.cmet.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heilbronn LK, Civitarese AE, Bogacka I, Smith SR, Hulver M, Ravussin E. Glucose tolerance and skeletal muscle gene expression in response to alternate day fasting. Obes Res. 2005;13(3):574–581. doi: 10.1038/oby.2005.61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.