Abstract
Spinal muscular atrophy (SMA) is a common neuromuscular disorder in humans. In fact, it is the most frequently inherited cause of infant mortality, being the result of mutations in the survival of motor neuron 1 (SMN1) gene that reduce levels of SMN protein. Restoring levels of SMN protein in individuals with SMA is perceived to be a viable therapeutic option, but the efficacy of such a strategy once symptoms are apparent has not been determined. We have generated mice harboring an inducible Smn rescue allele and used them in a model of SMA to investigate the effects of turning on SMN expression at different time points during the course of the disease. Restoring SMN protein even after disease onset was sufficient to reverse neuromuscular pathology and effect robust rescue of the SMA phenotype. Importantly, our findings also indicated that there was a therapeutic window of opportunity from P4 through P8 defined by the extent of neuromuscular synapse pathology and the ability of motor neurons to respond to SMN induction, following which restoration of the protein to the organism failed to produce therapeutic benefit. Nevertheless, our results suggest that even in severe SMA, timely reinstatement of the SMN protein may halt the progression of the disease and serve as an effective postsymptomatic treatment.
Introduction
Spinal muscular atrophy (SMA) is a common (~1:6400 newborns), often fatal, pediatric neuromuscular disorder caused by mutations in the survival of the motor neuron 1 (SMN1) gene (1) and, consequently, a paucity of its translated product, the survival of motor neuron (SMN) protein (2, 3). There is currently no cure for the disease. Available treatments for affected individuals are palliative at best. SMA patients inevitably harbor one or more copies of SMN2 (4), a virtually identical SMN1 homolog that nevertheless fails to fully compensate for the absence of SMN1 due to a translationally silent nucleotide difference in exon 7 (5, 6). The C→T base transition in SMN2 disrupts an exonic splicing enhancer and/or creates an exonic splicing silencer (7, 8) that induces exon 7 skipping and results in reduced levels of the full-length (FL) SMN mRNA transcript and protein. It is not yet clear why spinal motor neurons selectively degenerate in response to reduced SMN protein.
The SMA patient population is commonly stratified based on disease severity and age at onset into severe (type 1), intermediate (type 2), and mild (type 3) forms (9). In general, there is an inverse correlation between SMN2 copy number and SMA severity, suggesting that increasing the copy gene number can eventually result in sufficiently high levels of FL-SMN to rescue the SMA phenotype (10). Indeed, in model mice lacking murine Smn, an SMN1 equivalent gene, but transgenic for 8 copies of the uniquely human SMN2 gene, no disease phenotype is observed, providing direct proof of principle that restoring SMN protein through SMN2 or otherwise is a viable therapeutic option (11).
Given the presence of the SMN2 gene in all SMA patients and the appeal of modulating its expression as a therapeutic strategy, a number of attempts have been made using SMA cell lines as well as model organisms to identify agents that stimulate production of increased functional SMN protein (reviewed in ref. 12). These attempts have been complemented by studies involving the direct restoration of functional SMN using viral vectors (13–15). Indeed, adeno-associated virus–mediated (AAV-mediated) SMN augmentation appears to hold considerable promise as a means of treating SMA. Yet, given that any strategy will first be tested in symptomatic patients, success depends critically upon the stage of the disease at which SMN is still able to confer functional benefit. Importantly, it is currently unclear whether there is a precise window of opportunity during the course of the disease for the initiation of a treatment. Determining if such a window exists and identifying whether it is limited to presymptomatic stages or whether it extends to a point in time when cellular pathology and an overt phenotype are apparent is of particular relevance. This is doubly important since the disease will almost inevitably have progressed variably in different patients when a diagnosis is made.
To address this important, clinically relevant question in a model organism, we have generated what we believe is a novel line of transgenic mice that harbor an inducible Smn allele. We have used the allele to restore SMN expression to affected animals at various time points over the course of the disease. We show for the first time, to our knowledge, that SMN restoration, even after the appearance of cellular pathology and overt muscular weakness, results in dramatic phenotypic rescue. Motor neuron loss in treated mice is arrested, neuromuscular synapse pathology, a hallmark of the disease, mitigated, snRNP assembly, an important SMN function augmented, muscle strength restored, and most importantly, survival considerably enhanced. In a severe SMA mouse model, restoring SMN early during the course of the disease is more effective than augmenting the protein during late stages of the disease. Yet even during an advanced stage of the disease, increasing SMN expression provides marked therapeutic benefit. We demonstrate that the efficacy of overt phenotypic rescue correlates with the ability of increased SMN to mitigate the distal pathology of the motor unit and that there is a limited time frame during the course of the disease within which the protein must be restored for optimal effect. Nevertheless, our study raises the distinct possibility that an SMN-mediated SMA treatment can be effective, even if initiated at relatively advanced stages of the disease.
Results
Generation and characterization of an inducible Smn rescue allele.
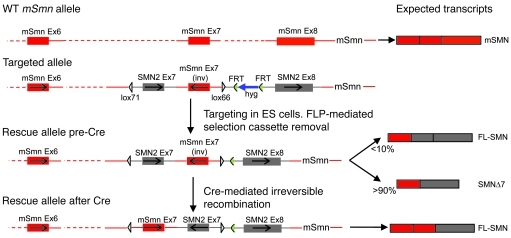
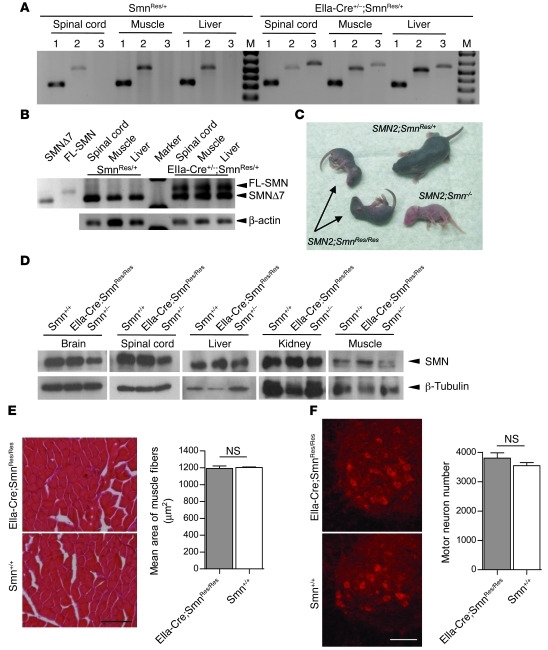
The SMN2 gene is unique to humans (16) and always present in at least one copy in SMA patients (10). Rodents possess a single Smn gene, which is the equivalent of human SMN1 (17). To genetically mimic human SMA in rodents, we created a mouse in which the endogenous Smn gene was engineered to behave like human SMN2 by replacing Smn exons 7 and 8 with a cassette consisting of SMN2 exons 7 and 8. In addition, an inverted, and thus silent, copy of Smn exon 7 was placed in the intron between SMN2 exons 7 and 8. The resulting hybrid allele was also engineered to contain mutant loxP sites (18, 19) placed in a head-to-head orientation and flanking the 2 exons 7 so that it could be conditionally and irreversibly activated by cassette inversion to express WT amounts of the functional FL-SMN protein and thus model the effects of a theoretical therapeutic in patients expressing only the SMN2 gene (Figure 1). Most of the transcripts from the human SMN2 gene lack exon 7 (SMNΔ7), and only a small proportion are full length (20). To characterize the transcriptional profile of the hybrid inducible allele, henceforth referred to as the “rescue allele” (Res), before as well as after activation, tissue from mice expressing it either alone (SmnRes/+) or together with an embryonically active EIIa-Cre allele (EIIa-Cre;SmnRes/+) was subjected to genomic PCR and then RT-PCR. We found that the rescue allele undergoes appropriate recombination in the presence of Cre recombinase and that there is no evidence of any spontaneous inversion of the allele (Figure 2A). Unexpectedly, though, RT-PCR demonstrated that, prior to recombination, the allele expressed no detectable full-length (FL-SMN) transcript in somatic tissue (Figure 2B). This is indicative of a functional null which, as expected, failed to produce any live mice homozygous (SmnRes/Res) for it. However, coexpressing an SMN2 transgene with the allele rescued the lethality, resulting in a phenotype similar to one we have previously described (ref. 11; see Figure 2C) and, furthermore, indicating that the unrecombined rescue allele does not intrinsically alter the disease state.
Figure 1. Construction of the inducible Smn rescue allele.
The structure of the Smn inducible rescue allele prior to and following Cre-mediated recombination, depicting the expected transcripts from the 2 different conformations. The WT murine Smn allele and its single FL-SMN transcript are included for reference. Drawing is not to scale.
Figure 2. In vivo characterization of the inducible Smn rescue allele.
(A) Recombination of the rescue allele is observed in the presence but not in the absence of Cre recombinase in P18 mice. WT murine Smn-specific primers were used to amplify samples in lanes numbered 1. Pre- and postrecombination-specific primers were used to amplify DNA in lanes numbered 2 and 3, respectively. (B) In the absence of Cre recombinase, the rescue allele fails to express FL-SMN transcript in mouse tissue. Perdurance of the SMNΔ7 transcript in EIIa-Cre+/–;SmnRes/+ mice indicates incomplete recombination, perhaps due to mosaic expression of Cre recombinase. The first 2 lanes are amplified products from bacterial clones into which were cloned the SMNΔ7 and FL-SMN fragments, respectively. (C) Human SMN2 rescues SmnRes/Res embryonic lethality but does not prevent appearance of SMA symptoms in neonates (arrows). (D) Cre-mediated recombination of the rescue allele in the germline results in near normal levels of SMN protein by Western blotting and prevents (E) muscle pathology and (F) loss of spinal motor neurons in EIIaCre;SmnRes/Res adult (P60-70) mice. n ≥ 2 for each genotype, P > 0.05, Student’s t test. Scale bars: 170 μm (E); 117 μm (F). Data are represented as mean ± SEM.
To assess whether the inverted, recombined conformation of the rescue allele was truly capable of restoring normal SMN levels and rescuing embryonic lethality, mice coexpressing it and EIIa-Cre (EIIa-Cre;SmnRes/+) were bred to rescue allele carriers (SmnRes/+). EIIa-Cre;SmnRes/Res progeny mice were found to express WT SMN protein levels in all tissues examined (Figure 2D). In addition, animals homozygous for the inverted form of the rescue allele were found to be phenotypically normal (Supplemental Figure 1, A and B; supplemental material available online with this article; doi: 10.1172/JCI57291DS1). Consistent with these findings, there was no evidence of muscle pathology in the EIIa-Cre;SmnRes/Res, mice and spinal motor neuron numbers were similar to those of WT animals (Figure 2, E and F). These results clearly indicated that the rescue allele fully compensates for lack of WT Smn when embryonically activated, thereby allowing us to begin testing the effects of turning on protein expression in a temporally regulated manner.
Restoring SMN postsymptomatically rescues the neuromuscular disease phenotype of severely affected model mice.
To control the activation of the rescue allele in a disease context, we introduced it onto the genetic background of previously described severe SMA (Δ7) model mice (21) in combination with a tamoxifen-inducible (TM-inducible), ubiquitously expressed Cre transgene (Cre-ER) (22). Such mice when homozygous for the rescue allele (SMN2;Δ7;Cre-ER;SmnRes/Res) and prior to TM treatment appeared very similar to severely affected Δ7 SMA mice and, like them, expressed greatly reduced SMN protein in P4 spinal cord tissue (Figure 3A). A phenotypic assessment of the mice at this age indicated obvious disease symptoms (Supplemental Figure 2, A–D) that were reflected in neuromuscular junction (NMJ) defects characterized by abnormal neurofilament (NF) infiltrates in nerve terminals (Figure 3B). To ensure that the behavioral and NMJ defects in the mutants were not merely a result of retarded growth, we examined the junctions and assessed the righting ability of runts that nevertheless bore at least one WT Smn allele. Neither parameter differed significantly from those of larger, control littermates (righting reflex score: controls, 22 ± 1.95; runts, 21 ± 1.41; nerve terminal thickness: controls, 0.87 ± 0.03 μm; runts, 0.90 ± 0.04 μm; n = 4 and P < 0.05, t test in all cases), suggesting that the abnormalities observed in our mutant mice were a direct consequence of reduced SMN.
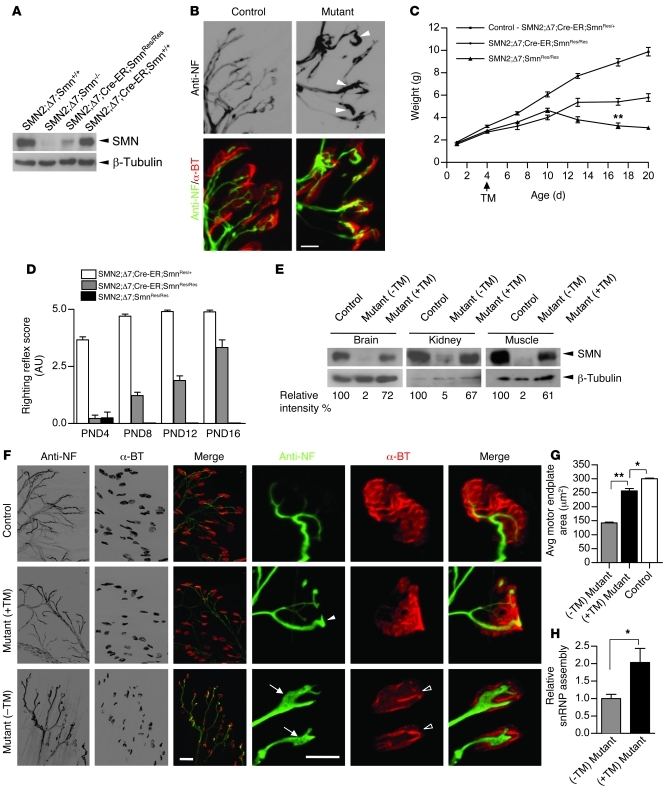
Figure 3. Restoration of the SMN protein in symptomatic SMA model mice rescues the disease phenotype.
(A) Western blotting reveals disease state levels of spinal cord SMN protein in nontreated P4 animals homozygous for the hybrid allele. (B) NMJ pathology in untreated P4 mutants manifests as poor terminal arborization and NF engorged nerve terminals (arrowheads). (C) Postsymptomatic TM treatment of Cre-ER–positive but not Cre-ER–negative mutant mice overcomes disease-related downturn in growth at P13 and accounts for a significant increase in weight by P17 (n ≥ 16, **P < 0.01, Student’s t test). (D) Improved motor performance in a righting reflex assay (n ≥ 4) following TM treatment at P4. (E) Mutant SMN2;Δ7;Cre-ER;SmnRes/Res mice treated at P4 with TM and sacrificed at P16–P17 express increased SMN protein in nervous as well as nonnervous tissue. Controls were WT (Smn+/+) at the murine Smn locus. (F) NMJs of gastrocnemius muscle of TM-treated mutants exhibit a reduction but not complete absence of pathology at P17 (solid arrowhead), whereas those of untreated mutants become progressively worse, displaying NF swollen nerve terminals (arrows) and immature, plaque-like motor endplates (open arrowheads). (G) Significant increase in the size of the motor endplate of TM-treated mutants compared with those of untreated mutants (100 endplates assessed in each of 3 mice; *P < 0.05; **P < 0.01, Student’s t test). (H) An increase in SMN-mediated snRNP assembly is observed in TM-treated mutants at 4 days after treatment. n = 4, *P < 0.05, Student’s t test. Scale bars: 10 μm (B); 40 μm and 10 μm (enlarged images of NMJs). (F). Data are represented as mean ± SEM.
Given the readily apparent disease phenotype at P4, and assuming that the symptoms mirror those of many SMA patients following an initial clinical diagnosis, we sought to determine whether restoring SMN at this stage would be beneficial to mutant mice. Accordingly, mutants with or without the Cre-ER transgene were administered a single dose of 75 mg/kg TM and the phenotype monitored over the following 2 weeks. Mutants without Cre-ER followed a typical disease course, peaking in weight at P10, but becoming progressively weaker until death at approximately P17, indicating that TM treatment itself has no effect on the phenotype. In contrast, Cre-ER–positive mutants gradually gained strength, performed better in a righting reflex assay, and were significantly larger by P17 (Figure 3, C and D, and Supplemental Video 1). As expected, untreated Cre-ER–positive mutants were phenotypically identical to Cre-ER–negative mutants treated with TM and did not differ from the latter with respect to survival (Supplemental Figure 3), since neither genotype is able to effect recombination of the rescue allele. This allowed us to use either genotype as a relevant disease control. To explore the molecular and cellular basis of the rescue, we assessed SMN protein levels and NMJ cytoarchitecture in treated mice and controls at P16–P17. Western blot analysis showed that whereas untreated mice continued to express low SMN, TM-treated animals expressed a relative abundance (60%–70% of WT) of the protein (Figure 3E). Consistent with this, there was considerable improvement in NMJ cytoarchitecture characterized by decreased preterminal NF accumulation, greater terminal arborization, and larger, relatively sophisticated AChR clusters compared with those observed in vehicle-treated animals (Figure 3, F and G). Reduced SMN protein in mouse tissue results in a severely compromised ability to assemble spliceosomal snRNP particles, and the extent of this defect correlates with phenotypic severity (23). Compared with spinal cord extracts from vehicle-treated mutants, extracts from TM-treated mice prepared 4 days after treatment exhibited a significant increase in their ability to assemble snRNPs in an in vitro assay (Figure 3H), indicative of an increase in functional SMN protein. Collectively, these results suggest that restoring SMN following disease onset is sufficient to mitigate the SMA phenotype by the third week of life, arrest the development of NMJ pathology, and enhance the most widely recognized molecular function of the SMN protein.
Restoring SMN postsymptomatically enhances survival of severely affected SMA model mice.
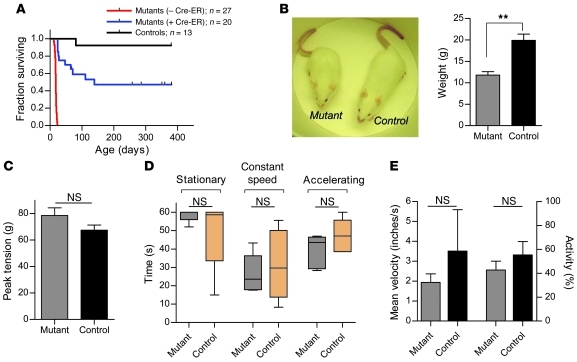
Type 1 (severe) SMA often proves fatal in humans before the age of 2 (24). Δ7 SMA mice, which model severe SMA, succumb to the disease in the second or third postnatal week of life. To determine whether postsymptomatic SMN restoration not only mitigated cellular pathology but also enhanced lifespan in this model, a cohort of mutants treated with TM at P4, with or without the Cre-ER transgene were allowed to age normally. Whereas no Cre-ER–negative mutant survived beyond P19, approximately 75% of Cre-ER–positive mutants survived to P28 and approximately 50% survived to 300 days or more (Figure 4A).
Figure 4. Postsymptomatic restoration of SMN enhances survival of SMA model mice.
(A) Restoration of SMN at P4 results in a 23-fold increase in survival of approximately 50% of mutant mice. Control animals were also treated with TM at P4 and are WT for murine Smn on at least 1 allele. χ2 (treated versus untreated mutants) = 48.65, P < 0.0001, log-rank test. (B) Adult TM-treated mutants have approximately 60% the weight of WT controls (**P < 0.01) but are phenotypically indistinguishable when tested in (C) grip strength (corrected for body weight), (D) rotarod, and (E) open-field assays. n ≥ 5, P > 0.05 in each case for C–E, Student’s t test. Data are represented as mean ± SEM.
A phenotypic assessment of the rescued mice at P50–P70 showed that, except for being smaller and having a lower body weight, the animals could not be distinguished from WT littermate controls (Figure 4B and Supplemental Video 2). Moreover, they did not develop any vascular necrosis of the extremities, a symptom that others have reported in gene therapy–based rescue experiments (13–15). An analysis of grip strength, rotarod, and open field assays demonstrated that the rescued animals performed equivalently to WT controls (Figure 4, C–E), attesting to the degree of phenotypic rescue. The near-normal phenotypic characteristics of the mutants treated with TM at P4 were reflected in the absence of any discernible spinal motor neuron loss and WT muscle morphology in either proximal or distal muscle groups (Supplemental Figure 4, A and B). While mean muscle fiber area of rescued mice was approximately 20% smaller than that of WT controls, the reduction could be accounted for by the smaller size of the former. Similarly, despite a general reduction in the size of the rescued NMJs, they appeared to be fully developed, pretzel-shaped structures with terminal axons devoid of pathologic NF infiltrates (Supplemental Figure 4C). These results not only reinforce our behavioral assessment of the rescued mice at P17, but also suggest that early (~P4) pathogenic defects of the NMJs in the form of preterminal NF aggregates may in fact be reversed following the timely restoration of SMN protein during the course of the disease.
Despite the rescue of the neuromuscular disease phenotype following restoration of SMN at P4, the smaller size of the mutants suggested a fetal or perinatal requirement of the protein. To test this hypothesis, a Sox2-Cre transgene, which is expressed in the epiblast (25), was used to induce SMN expression at E6 from the rescue allele in SMA embryos. Our analysis of adult SMN2;Δ7;Sox2-Cre;SmnRes/– mice indicated that restoring SMN during early gestation did indeed rescue the size/weight phenotype (mutants, 20.39 ± 1.47 g; controls, 24.63 ± 1.25 g, P > 0.09, n = 3, t test) as well as any evidence of neuromuscular pathology (motor endplate area: mutants, 788.5 ± 19.66 μm2; controls, 806 ± 13.18 μm2; muscle fiber size: mutants, 1328 ± 65 μm2; controls, 1273 ± 27 μm2, approximately 50 NMJs and muscle fibers analyzed in 2 mice of each genotype; P > 0.5 in each case, t test). Thus, although postsymptomatic restoration of SMN can rescue those elements of the Δ7 SMA phenotype that most closely resemble the human disease, earlier intervention is required to reverse the mouse-specific deficit in overall growth.
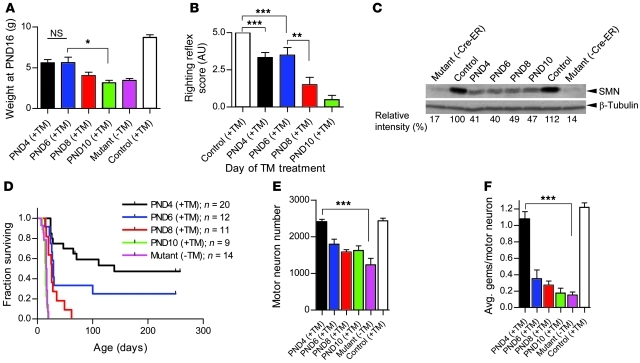
A limited therapeutic window of opportunity in severely affected SMA model mice is defined by the extent of neuromuscular synapse pathology.
The SMA phenotype in the Δ7 model mice used in this study becomes inexorably worse with age until mutants succumb to the disease at approximately P17. Restoration of SMN at P4 resulted in substantial rescue. To further investigate the temporal requirements for the SMN protein during disease progression, we chose 3 time points subsequent to P4 at which to restore the protein. The first cohort of mutant (SMN2;Δ7;Cre-ER;SmnRes/Res) mice was administered 75 mg/kg TM at P6, a second group was treated at P8, and a third at P10. By P16, mice treated at P6 tended to be larger than age-matched mutants treated at P8, and the difference in weight became significant when a comparison was made to P10 treated mutants (Figure 5A). However, the P6 treated mutants appeared similar in body weight at P16 to age-matched mutants treated at P4. These differences in body size were reflected in muscle strength. Whereas mutants treated at P4 and P6 performed equally well when tested in the righting reflex assay at 16 days of age, P8 and P10 treated mutants performed equivalently to each other but significantly less well than the first 2 groups (Figure 5B). None of the animals performed as proficiently as WT littermate controls, indicative of a lag in time before the effect of SMN restoration is observed.
Figure 5. A limited therapeutic window in SMA model mice harboring the rescue allele.
SMN restoration at earlier time points generally correlated with (A) greater increases in body weight (n ≥ 4 for each genotype except P10 mice = 3; *P < 0.05, Student’s t test) and (B) enhanced motor performance assessed in righting ability tests (n ≥ 4 for each genotype, **P < 0.01; ***P < 0.001; 1-way ANOVA). (C) Equivalent increases in spinal cord SMN protein expression are observed in mutants bearing the rescue allele treated between P4 and P10. (D) Earlier restoration of SMN protein translates into greater phenotypic rescue in Kaplan-Meier survival curves. (E) Loss of spinal motor neurons and (F) gems is significantly mitigated following treatment only at the earliest (P4) time point (n = 3 for each cohort, ***P < 0.001; 1-way ANOVA). Data are represented as mean ± SEM.
To determine whether the phenotypic observations reflected altered SMN protein levels, spinal cord tissue was harvested at P13–P16 and subjected to Western blot analysis. Interestingly, although there was a noticeable increase in protein in spinal cord tissue from treated animals compared with untreated mutants, there appeared to be no significant difference in protein levels between the various treated mice, all of which expressed approximately 40% of WT SMN protein levels (Figure 5C). This suggests that TM treatment was equally effective in raising spinal cord SMN levels at the time points considered for SMN restoration. Consistent with this result, we found equivalent levels of FL-SMN in tissue harvested from the different cohorts of mice at 48 hours after TM administration (Supplemental Figure 5A). In mutants treated at P4, elevated protein was observed within 48 hours of drug administration (Supplemental Figure 5B). Intriguingly, considering the equivalent levels of spinal cord SMN protein in the different cohorts of mice, survival appeared highly dependent upon the timing of SMN restoration (Figure 5D). In particular, SMN restoration at P4 (median survival, 139 days) was the most effective, whereas by P10 (median survival, 15 days), TM treatment had ceased to bring about measurable rescue and, in fact, appeared to exacerbate mortality somewhat, perhaps due to the fragility of the mutants when dosed at this late stage of the disease. Increasing the amount of TM administered to 150 mg/kg at P8, the latest time point at which SMN restoration enhanced survival, failed to provide additional therapeutic benefit to that observed at the same age with half as much drug and, once again, tended to enhance mortality (Supplemental Figure 6). This precludes the possibility that failure of therapeutic benefit at later stages of the disease is due to an insufficient drug dose.
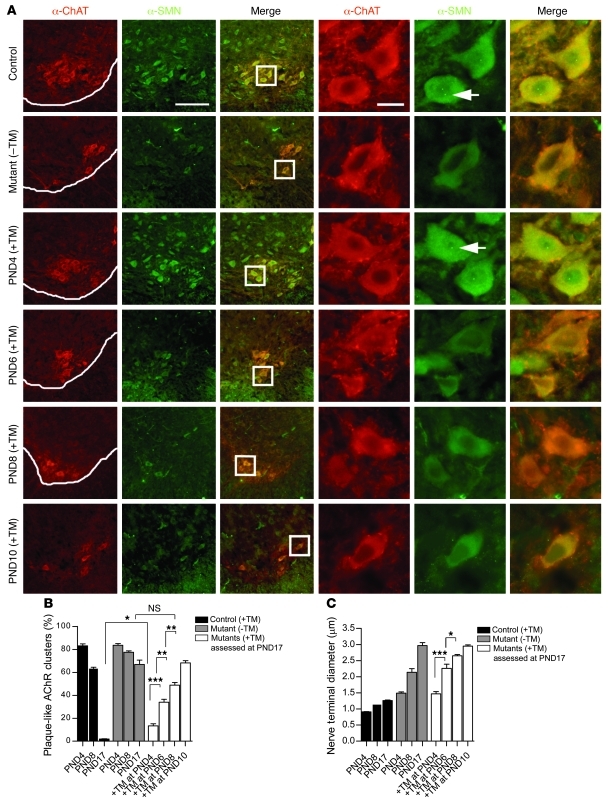
To investigate the cellular basis of the observed survival differences in the different mouse cohorts, we quantified motor neuron numbers and assessed NMJ defects at 17 days. Although all 3 groups of TM-treated animals had greater numbers of motor neurons than vehicle-treated mutants, the numbers reached statistical significance for only P4 treated mutants (Figure 5E). Consistent with these findings, only P4 treated mutants had normal numbers of motor neuronal gems (Figure 5F), nuclear bodies to which the SMN protein localizes (26), and appeared to express normal amounts of cytoplasmic SMN as assessed immunohistochemically (Figure 6A). Considering an increase and apparent equivalence in overall spinal cord SMN in mice treated at the different time points, this result suggests that the ventral horn motor neurons of mutants selectively fail to respond to TM administration if left untreated beyond P6, a likely consequence of their unique vulnerability to the disease. Equivalent levels of SMN staining in the dorsal horn cells of 2-week-old mice treated at P4 and P8, respectively, lends credence to this notion (Supplemental Figure 7). The incomplete correlation between levels of SMN in the motor neuron cell body and enhanced survival of P6 and P8 treated mutants could be explained by a small and undetectable increase that is discernible only in the form of reduced defects seen peripherally at the distal end of the motor unit. We therefore examined the NMJs of these mice. We have previously shown that defects of the neuromuscular synapse are a hallmark of SMA and that morphological changes in these structures precede motor neuronal cell body loss (27). Accordingly, we examined muscle from cohorts of P4, P6, P8, and P10 TM-treated mice as well as vehicle-treated controls and WT littermates. Interestingly, analysis of the complexity of the postsynaptic AChR clusters indicated that, while none of the treatment regimens restored the sophistication of these postsynaptic structures to that observed in WT control littermates, in every instance, TM treatment diminished the immature, plaque-like state of the NMJs compared with those of vehicle-treated controls (Figure 6B). Moreover, there appeared to be a graded effect on AChR complexity that was dependent upon the time point at which SMN was restored. Thus, the earlier the protein was restored, the lower the proportion of plaque-like AChR clusters we observed and the more developed they appeared. This result was reflected in the extent of presynaptic NF accumulation, as assessed by the amount of protein that had infiltrated the nerve terminals to cause swelling and affect terminal axon diameter (Figure 6C). Collectively, these findings indicate that under conditions of reduced SMN, the motor unit does indeed exhibit a temporal sensitivity, gradually accumulating defects at the distal end that reach a threshold at approximately P8 beyond which restoration of the protein fails to mitigate or reverse the SMA phenotype.
Figure 6. Level of phenotypic rescue is inversely correlated with the extent of neuromuscular synapse pathology.
(A) Immunohistochemistry of cervical spinal cord sections from mice administered TM at various time points during the course of the disease revealed gems (arrows) and normal cytoplasmic staining only in P4-treated mutants. Scale bars: 125 μm and 5 μm (insets). The timing of SMN restoration correlates closely with (B) AChR cluster development as well as (C) the ability to clear presynaptic NF aggregates by P17. Early treatment resulted in greater sophistication of the postsynapse and reduced NF in nerve terminals. *P < 0.05; **P < 0.01; ***P < 0.001; 1-way ANOVA. Note: more than 500 AChR clusters (for postsynaptic specialization) and more than 50 endplates (for NF accumulation) were examined in at least 3 mice per time point. Data are represented as mean ± SEM.
Discussion
SMA is caused by a deficiency of the SMN protein. Abolition of the protein, however, is incompatible with life (28). Accordingly, the existence of the disease in humans is owed to the presence in most normal individuals and all SMA patients of SMN2, a copy gene that is unable except in multiple copies to compensate for loss of SMN1. Despite its inability to offset SMN1 loss, the presence of the SMN2 gene in all patients and the observation that the SMA phenotype generally correlates with copy gene number raise the expectation that SMN2 expression may be modulated as a means to a treatment. Direct SMN restoration is a second therapeutic option. Indeed, these 2 treatment approaches have become the focus of scientists at academic institutions as well as industry. Yet the efficacy of either strategy relies critically on a sound assessment of the tissue and temporal requirements for the SMN protein. In this study, we have used severely affected model mice harboring an inducible Smn rescue allele, which we believe to be novel, to address the latter issue. Our findings confirm first that it is possible to rescue SMA postnatally. More critically, from a therapeutic standpoint, they indicate that such rescue can be brought about postsymptomatically. Mice homozygous for the inactive form of the rescue allele develop the characteristic neuromuscular disease phenotype observed in SMA. However, induction of the allele and timely restoration of the SMN protein even after the disease phenotype and cellular pathology ensue is sufficient to bring about near-complete rescue. Second, our results indicate that despite the ability to rescue the disease phenotype in symptomatic animals, there is a limited therapeutic window of opportunity. The extent of therapeutic benefit appears to wane as the disease progresses. Still, we show that even in advanced stages of the disease, treated animals derive significant benefit from SMN expression. Finally, we have demonstrated that the progression of the neuromuscular disease is reflected in the extent of damage at the neuromuscular synapses and that this damage dictates the ability and the degree to which SMN restoration can bring about therapeutic benefit. Collectively, our results provide what we believe to be the most compelling data yet showing that augmenting SMN following the appearance of an overt phenotype can nevertheless mitigate disease conditions and may, in certain cases, serve as a potent treatment in symptomatic SMA patients.
The concept of modulating SMN2 to restore sufficient functional SMN protein to the cells of affected individuals is not a new one. To this end, much effort has been devoted to identifying agents that enhance SMN2 expression in cell-free assays (29), cultured cells (30–33), model organisms (29, 34–36), and even in human patients (37, 38). These studies have established the feasibility not just of SMN2 modulation but also of direct SMN augmentation. Examples that illustrate the first strategy involved the use of antisense oligonucleotides (ASOs) (35, 39) and small molecules (29) to correct the splicing defect of the SMN2 gene. While none of the studies relying on this strategy reported extending survival in mouse models, in every case a significant increase in SMN was observed. Direct SMN augmentation using AAV-based vectors did on the other hand demonstrate enhanced survival in SMA mice and provided evidence that presymptomatic restoration of the SMN protein is effective in treating the disease. In the single study that attempted to explore the effects of later-stage treatment, the authors admitted that failure to achieve benefit was linked to the inability to efficiently target motor neurons due to the altered tropism of the virus to these cells in older animals (13). The study therefore did not resolve questions about the temporal requirements for SMN.
The approach described in this report allowed an investigation of the effect of restoring SMN protein not only early in the disease but also in older animals as the phenotype worsened. We previously showed that muscle weakness in Δ7 SMA mice becomes apparent at P2 and that neuromuscular synapse pathology initially discerned at this age progresses rapidly (27). Not surprisingly, therefore, P4 mutants homozygous for the rescue allele on the Δ7 SMA background displayed an obvious disease phenotype and marked NMJ pathology in the form of abnormal presynaptic NF aggregates. Considering the symptoms observed, we were struck that restoring protein at this stage achieved such robust rescue in a subset of the animals studied. Some of these mice are more than 500 days of age at the time of article submission and, except for reduced body size, no different from controls. This level of postsymptomatic rescue is significantly greater than those reported in earlier studies in which small molecules enhanced the survival of neonatal mutant mice by up to 30% (36, 40, 41). While we are uncertain precisely why a certain proportion of our treated mice perished at earlier ages, the most likely explanation derives from variable phenotypes, including those arising from heart abnormalities (42–44), even among age-matched affected animals, rendering more severely affected mutants less responsive to restoration of SMN protein. Indeed, an examination of the body weights of mutants that responded only partially to treatment revealed significantly lower values at P10, P20, and P30 compared with those of mice that eventually lived beyond 6 months of age (Supplemental Figure 8). Nevertheless, we emphasize that all animals treated at P4 derived benefit from enhanced protein levels, since none of the “nonrescued” group of mice survived to the extent that their shortest surviving “rescued” counterparts did.
An encouraging outcome, to the extent that it can be extrapolated to the clinical context, was the degree of rescue achieved following SMN augmentation at later stages of the disease. A fourth of all mutants treated at P6 survived beyond 250 days, and approximately a third of mice treated at P8 survived to 1 month of age. Although we observed variability in the degree of rescue within cohorts, there was a gradation in therapeutic benefit as the disease progressed. It is unlikely that the decrement in phenotypic rescue is due to a gradual loss in ability to target motor neurons for 2 reasons. First, TM is known to efficiently activate reporter genes in mice coexpressing the Cre-ER transgene in all CNS cells, including neurons of adult animals (22). Second, our own experiments demonstrated similar levels of recombination of the rescue allele 48 hours after treatment in the different cohorts of animals and similar protein levels in 2-week-old spinal cords from the mice. A catastrophic loss of motor neurons, especially between P6 and P10, is also an unlikely explanation for the large difference in therapeutic benefit between these time points, since there was no significant difference in the numbers of these cells at 2 weeks of age. On the other hand, we did observe a correlation between the degree of pathology at the distal (neuromuscular synapse) end of the motor unit at P17 following treatment at the various time points and the extent of phenotypic rescue, suggesting that accumulating distal defects eventually preclude a therapeutic effect. Despite the correlation between distal motor neuronal defects and the ability to achieve phenotypic rescue, it is unlikely that restoration of SMN protein exclusively to motor neurons is responsible for the eventual benefit seen to the animals. In the model mice used in the study, SMN was restored to all tissues, and one possibility, as has been previously suggested (45), is that the expression of the protein in nonneuronal cell types that contribute to the overall SMA phenotype plays a role in the rescue. Ascertaining this possibility will require a follow-up investigation of the tissue-specific requirements for the SMN protein. The Smn rescue allele we report here may be useful for these sorts of studies.
The disparity between the survival of mice in our study and those treated with AAV vectors (13) likely originates in the different treatment times (P4 versus P1, respectively) of the 2 groups of animals and affirms our conclusion that the earlier the protein is restored to affected individuals, the more effective the therapeutic outcome. Moreover, the fact that in every study reported to date, treated mice continue to remain smaller than WT littermates argues for a fetal/perinatal requirement of the SMN protein. Although it is uncertain precisely how early SMN is needed, the requirement is satisfied by E6 when the expression of a Sox2-Cre transgene ensures restoration of the SMN protein from the rescue allele.
In conclusion, this study has revealed that even when SMN is severely depleted, as typically observed in type I SMA, motor neurons, and possibly other cell types that contribute to the phenotype, are not irreversibly damaged until later stages of neonatal life. This conclusion is consistent with electrophysiological studies demonstrating normal motor unit numbers in a handful of presymptomatically diagnosed SMA patients (46) and appears to constitute part of an emerging appreciation of the ability of neurons subject to genetic insults to demonstrate remarkable resilience that allows for their postnatal rescue (47). In the case of SMA, it suggests that the SMN protein serves more as a motor neuron maintenance factor and/or an element required for the elaboration of the adult neuromuscular synapse rather than being a critical component of the neurodevelopmental process. Importantly, our data have also shown that neuronal and phenotypic rescue in SMA may be brought about even after the appearance of cellular pathology and disease symptoms. These results raise the expectation that the disease may be treated postsymptomatically and thus provide the rationale and impetus to develop effective ways of restoring SMN by modulating the SMN2 gene and/or delivering the SMN1 gene or protein to young infants. Whether an effective therapeutic will have to be administered chronically or acutely remains to be determined. It is possible that WT levels of SMN protein are required only during a specific period of neuromuscular development, following which cells become refractory to reduced protein. If so, a future therapeutic, especially a small molecule-type drug that upregulates the SMN2 gene, may be administered acutely. However, ascertaining the duration of a treatment will require further experiments that involve temporally depleting the SMN protein.
Methods
Inducible rescue allele construct
BAC clones 939i7 (containing human SMN2) and 148d4 (containing murine Smn) were isolated from the RPCI-11d and RPCI-23 libraries, respectively (48, 49), and served as the starting material for the vector used to target the endogenous Smn allele. The BAC-based targeting vector (50) was constructed using homologous recombination and involved replacing Smn exons 7 and 8 with a hybrid donor cassette (Supplemental Figure 9). The cassette, assembled by standard PCR and ligation steps, consisted of, in order, a short upstream region (homology upstream [HU]) homologous to the mouse BAC, a lox71 site, a 380-bp fragment containing SMN2 exon 7 and flanking intronic sequences, a 387-bp fragment containing murine Smn exon 7 and flanking intronic sequences in an inverted orientation, a lox66 site inverted with respect to the lox71 site, a hygromycin resistance gene with prokaryotic and eukaryotic promoters flanked by FRT sites, a 928-bp fragment containing SMN2 exon 8 including the 3′ UTR and polyA signal. and a short downstream region (homology downstream [HD]) homologous to the mouse BAC. The modified BAC vector, now consisting of murine Smn exons 1–6 fused to the hybrid sequence, was linearized and electroporated into VGF1 ES cells; targeted hygromycin-resistant ES colonies were then identified by the LONA assay (48). The hygromycin resistance gene was removed by transient FLPe (51) expression in ES cells, and the cells were then microinjected into donor 8-cell embryos (52) to generate mice. Mice heterozygous for the rescue allele (SmnRes/+) were deposited at the Jackson Laboratory (stock no. 007249) and are referred to there by the strain name B6;129-Smn1tm3(SMN2/Smn1)Mrph/J. Allele designations for the hybrid rescue allele (a) containing the hygromycin resistance gene, (b) after FLP-mediated removal of the resistance gene, and (c) after inversion “rescue” of the SMN2 exon 7/Smn exon 7 segment are VG5065, VG5075, and VG5080, respectively.
Mice
SmnRes/+ mice were crossed to animals harboring EIIa-Cre (Jax stock no. 003314; Jackson Laboratory) to generate progeny carrying both alleles (EIIa-Cre;SmnRes/+). Since EIIa-Cre is mosaic in its expression, these progeny were outcrossed to nontransgenic mice to select for inverted alleles in the germline. Animals harboring the fully inverted rescue allele in the absence of EIIa-Cre were then intercrossed to generate SmnRes/Res mice for phenotypic assessment. To generate severely affected “rescuable” SMA model mice, SmnRes/+ animals were first bred to Δ7 SMA carriers (Jax stock no. 005025; Jackson Laboratory). SMN2;Δ7;SmnRes/+ progeny (Jax stock no. 007951; Jackson Laboratory) were then crossed to animals harboring a TM-inducible (Cre-ER) allele (Jax stock no. 004682; Jackson Laboratory) and SMN2;Δ7;Cre-ER;SmnRes/+ mice intercrossed to generate experimental cohorts homozygous for SMN2, Δ7, and SmnRes with or without Cre-ER as required. The same breeding strategy was employed to generate Δ7 SMA mice carrying the rescue allele in combination with Sox2-Cre (Jax stock no. 004783; Jackson Laboratory), except that Smn+/– heterozygotes were used instead of SmnRes/+ mice. All experiments described in this study included both male and female mice. To preclude strain background effects, littermate controls were used in all experiments. Additionally, all experiments were conducted in accordance with the protocols described in the NIH Guide for the Care and Use of Animals and were approved by the Jackson Laboratory and Columbia University’s respective Institutional Laboratory Animal Care and Use Committees.
TM treatment
A stock solution of 20 mg/ml TM (Sigma-Aldrich) was prepared in corn oil. A 75 mg/kg dose of this solution was administered to litters by oral gavage on P4, P6, P8, or P10. Where appropriate, controls were administered corn oil alone.
Genotyping
To detect the native (noninverted) rescue allele, either of the following 2 sets of primers was used: 5′-GGCAGTTTTAGACTCATCATGTATCTG-3′ and 5′-ACTTATGGAGATCCCTCGAGATAAC-3′ or RESC2F 5′-TATCTGAAGATGACTTTGAACTC-3′ and RESC1R 5′-CAGAAGAGGACATTGGATCCCG-3′. The primers amplified 103-bp and 503-bp fragments, respectively. The inverted form of the allele was detected in a similar fashion using either of the following 2 sets of primers: 5′-GTTTTAGACTCATCATGTATCTG-3′ and 5′-GTGTGAGTGAACAATTCAAGCC-3′ or 5080F2, 5′-CAACCAGTTAAGTATGAGAATTC-3′ and 5080R2 5′-CATTGTAACCATAAAGAGAAAGGCAGGATG-3′, which amplified 190-bp and 564-bp fragments, respectively. The SMN2, Δ7, WT Smn, and Cre alleles were amplified and detected as previously described (21, 22, 39).
PCR, Western blot analysis, and snRNP assembly assays
To assess the types of RNA transcripts generated from the rescue allele before and after Cre-mediated recombination, tissue from SmnRes/+ or EIIa-Cre;SmnRes/+ was dissected and RNA extracted with Trizol (Invitrogen) according to the manufacturer’s instructions. Following cDNA synthesis using oligo(dT) primers and RevertAid M-MuLV reverse transcriptase (Fermentas), transcripts from the rescue allele were amplified with the following primers: SmnEx6F 5′-GTCTGGATGACACTGATGCCC-3′ and SMN2Ex8R 5′-CAATGAACAGCCATGTCCAC-3′. β-actin amplified with the primers 5′-TTGTGATGGACTCCGGAGAC-3′, reverse: 5′-CACCAGACAGCACTGTGTTG-3′ served as an internal control. The rescue allele transcripts were subsequently confirmed by sequencing. Note that FL-SMN was detectable in ES cells but not somatic tissue of mice with the rescue allele.
Quantification of FL-SMN expression from the rescue allele in mice treated with TM at different time points was carried out using a TaqMan qPCR assay. Primers exon67fwdm101, 5′-GCTACTATATGGGTTTCAGACAAAATAAAA-3′, exon8hrev233, 5′-GCTTCACATTCCAGATCTGTCT-3′, and exon8hprobe173, 5′-FAM-CATAGAGCAGCTCTAAATGACACCACTAAAGAA-TAMRA-3′ were used to amplify and detect the PCR product. The PCR was set up as follows: 12.5 μl of 2x TaqMan Universal PCR Master Mix (Applied Biosystems), 0.25 μl of each primer (40 μm), 0.75 μl of each probe (5 μm), and 10 μl of diluted template cDNA. RT-PCR for first-strand cDNA synthesis was carried out using Invitrogen Superscript (Invitrogen) first strand cDNA kit with approximately 4 μg of total RNA and oligo(dT) in a final volume of 20 μl reaction. cDNA from this reaction was then used as a template to amplify products under the following conditions: 50°C, 2 minutes; 95°C, 10 minutes; followed by 40 cycles of 95°C, 15 seconds and 60°C, 1 minute on an ABI Prism 7000 sequence detection system (Applied Biosystems). Duplicate reactions were done for each sample.
SMN protein levels in mutant and control mice were determined by Western blot analysis using standard procedures described previously (11). SMN (1:10,000; BD Biosciences) and β-tubulin (1:5000; Sigma-Aldrich) monoclonal antibodies were visualized using the ECL Detection Kit (RPN 2109; GE Healthcare). Quantification of band intensities was performed using ImageJ software (NIH).
In vitro snRNP assembly experiments with radioactive U1 snRNA and mouse spinal cord extracts followed by immunoprecipitation with anti-Sm antibodies were carried out as previously described (21, 53). Quantification of immunoprecipitated U1 snRNA was performed using a STORM 860 Phosphorimager (Molecular Dynamics) and ImageQuant version 4.2 software. P values were calculated using unpaired Student's t test.
Spinal motor neuron and muscle histology
Muscle pathology was investigated in the proximal (biceps) as well as distal (gastrocnemius) muscles. Animals were euthanized and whole muscle dissected, oriented in Tissue-Tek OCT embedding medium (Fisher Scientific) for cross-sectional analysis, and flash-frozen in liquid nitrogen–cooled (N2) isopentane. 8-μm–thick transverse sections from the belly of the muscle were cut on a cryostat (Leica) and the sections stained with H&E (Sigma-Aldrich) for examination of fiber morphology as previously described (54). Fiber size was determined using SPOT v4.5 software (Diagnostic Instruments) following image acquisition of the stained sections.
To carry out spinal motor neuron and gem counts and to assess motor neuron morphology, spinal cord tissue was extracted from mice following transcardial perfusion with 1× PBS and then 4% PFA. Following postfixation in 4% PFA (4°C for 4 hours), tissue was cryoprotected (30% sucrose for 3 days at 4°C), embedded in Tissue-Tek OCT, and then frozen in liquid N2-cooled isopentane for sectioning. 20-μm transverse sections from the cervical spinal cord (C6–C8) were cut, the sections permeabilized and blocked (0.2% Triton-X 100, 1% BSA, 5% donkey serum for 30 minutes at room temperature), and then dual labeled with antibodies against Choline Acetyltransferase (ChAT) (1:100, Millipore) and SMN (7F3, 1:100) (55) for 48 hours at 4°C. After washing, the sections were incubated in appropriate fluorescently conjugated secondary antibodies (24 hours at 4°C), overlaid with a mixture of DAPI and mounting medium (Vectashield), and imaged on an E80i Nikon microscope equipped with a Spot Flex digital camera (Diagnostic Instruments). Motor neuron numbers were assessed in 8 sections of each of 3 (C6–C8) cervical segments and the raw number extrapolated to the entire region based on the length of the region and the thickness of each section. Gem numbers were determined by counting intranuclear SMN-positive dot-like structures within ChAT-positive motor neurons. SMN staining intensity in the dorsal horns of mutant mice was assessed in a 60-μm2 area and expressed as a percentage of the fluorescence in control animals.
Quantification of vGlut1 puncta in the ventral horns of mutant mice and control littermates was carried out using protocols described previously (45).
Cytoarchitecture of NMJs
The analysis of NMJ cytoarchitecture was carried out on whole muscle, which was fixed and permeabilized (100% methanol for 5 minutes), then incubated in 3% BSA to preclude nonspecific binding of antibodies. The tissue was incubated in an antibody against NF (NF-M, 1:1000; Millipore) for 48 hours at 4°C, washed with PBS-Tween, and incubated for 24 hours at 4°C with rhodamine-α-bungarotoxin (α-BT) (1:1000; Invitrogen) and Alexa Fluor 488 anti-rabbit IgG (1:1000; Invitrogen). After further washing in PBS-Tween, muscles were teased to separate individual fibers before mounting in Vectashield (Vector Laboratories) for visualization. Immature, plaque-like motor endplates were defined as those lacking perforations. Motor endplate size was assessed using a procedure similar to that applied to muscle fibers stained with H&E. The extent of NF accumulation was quantified based on the diameter of the thickest portion of a nerve terminal at the presynapse. All images were acquired on a Nikon E80i fluorescent microscope.
Phenotypic assessment of mice
Weight/righting reflex/survival.
Mice were weighed daily from birth and weekly thereafter. Righting reflex was used to evaluate muscle strength as previously described (56). Briefly, pups were turned over onto their backs, and latency to stably place all 4 paws on the bench top was recorded. The following arbitrary scores were used to quantify righting ability: 5 for 0–2 seconds, 4 for 3–5 seconds, 3 for 6–10 seconds, 2 for 11–30 seconds, 1 for 31–60 seconds, and 0 for more than 60 seconds. The procedure was repeated three times for each individual and the sum of scores designated the righting ability score. Survival of mice was determined by noting the day on which a mouse succumbed to the disease or had to be euthanized due to the disease condition. Data were plotted as Kaplan-Meier curves.
Hind limb suspension.
To carry out hind limb suspension or “tube test,” P4 mice were suspended by their hind limbs from the mouth of a 50-ml tube and posture adopted was scored as previously described (57).
Grip strength.
Grip strength was assessed in mutant and control littermates at P50 using the Chatillon-Ametek Digital Force Gauge, DFIS-2 (Columbus Instruments). Briefly, the mouse was raised toward a triangular metal force transducer and instinctively grasped it with its forelimbs. The animal was then pulled downward and peak force (in kg) at the point of release recorded over 3 consecutive trials. An average of the 3 trials served as the final score.
Rotarod analysis.
Motor performance was also evaluated by rotarod analysis using the Economex Accelerating Rotarod (Columbus Instruments). Following a period of acclimatization, P50 “rescued” animals and control littermates were placed on the rotarod in 3 modes: (a) in the stationary phase, (b) at a constant speed of 4 rpm, and (c) while a 0.1 rpm/s acceleration was applied over a 5-minute period. Latency to fall was recorded over 3 consecutive trials and an average calculated for each mouse.
Open field assay.
Exploratory behavior and mean velocity were determined using equipment for open field assays (Ugo Basile). Briefly, following a period of acclimatization, mice were placed in a 24” × 24” arena equipped with infrared beam detectors and a Sony digital video camera. Activity was recorded over a 5-minute period and calculated using BiObserve software provided with the equipment.
Statistics
Kaplan-Meier survival curves were compared and assessed for differences using the log-rank test equivalent to the Mantel-Haenszel test. The unpaired, 2-tailed Student’s t test or 1-way ANOVA followed by Tukey’s post hoc comparison, where indicated, were used to compare means for statistical differences. Data are represented as mean ± SEM unless otherwise indicated. P < 0.05 was considered significant. Statistical analyses were performed with GraphPad Prism v4.0 (GraphPad Software) or JMP (SAS Statistical Software).
Supplementary Material
Acknowledgments
We thank D.C. De Vivo for invaluable advice and G-H Park for expert technical assistance. T.M. Jessell and L. Eng provided important insight during the conception of this strategy. This study was supported by SMA Foundation grants to C.M. Lutz, L. Pellizzoni, C.E. Henderson, U.R. Monani, and Regeneron Pharmaceuticals. Additionally, U.R. Monani was supported by the Muscular Dystrophy Association, SMA-Europe, AFM-France and NIH grants R01 NS057482 and P01 NS055923. L. Pellizzoni was supported by NIH grant R01 NS069601.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(8):3029–3041. doi:10.1172/JCI57291.
See the related Commentary beginning on page 2978.
References
- 1.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 2.Coovert DD, et al. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6(8):1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre S, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16(3):265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 4.Monani UR. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48(6):885–896. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monani UR, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8(7):1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 7.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30(4):377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 8.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34(4):460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 9.Pearn J. Classification of spinal muscular atrophies. Lancet. 1980;1(8174):919–922. doi: 10.1016/s0140-6736(80)90847-8. [DOI] [PubMed] [Google Scholar]
- 10.Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70(2):358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monani UR, et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(–/–) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet. 2000;9(3):333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 12.Lorson CL, Rindt H, Shababi M. Spinal muscular atrophy: mechanisms and therapeutic strategies. Hum Mol Genet. 2010;19(R1):R111–R118. doi: 10.1093/hmg/ddq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foust KD, et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28(3):271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Valori CF, et al. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci Transl Med. 2010;2(35):35ra42. doi: 10.1126/scitranslmed.3000830. [DOI] [PubMed] [Google Scholar]
- 15.Passini MA, et al. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J Clin Invest. 2010;120(4):1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rochette CF, Gilbert N, Simard LR. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum Genet. 2001;108(3):255–266. doi: 10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- 17.DiDonato CJ, Chen XN, Noya D, Korenberg JR, Nadeau JH, Simard LR. Cloning, characterization, and copy number of the murine survival motor neuron gene: homolog of the spinal muscular atrophy-determining gene. Genome Res. 1997;7(4):339–352. doi: 10.1101/gr.7.4.339. [DOI] [PubMed] [Google Scholar]
- 18.Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7(4):649–659. doi: 10.1046/j.1365-313X.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- 19.Oberdoerffer P, Otipoby KL, Maruyama M, Rajewsky K. Unidirectional Cre-mediated genetic inversion in mice using the mutant loxP pair lox66/lox71. Nucleic Acids Res. 2003;31(22):e140. doi: 10.1093/nar/gng140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gennarelli M, et al. Survival motor neuron gene transcript analysis in muscles from spinal muscular atrophy patients. Biochem Biophys Res Commun. 1995;213(1):342–348. doi: 10.1006/bbrc.1995.2135. [DOI] [PubMed] [Google Scholar]
- 21.Le TT, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14(6):845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244(2):305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 23.Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2007;2(9):e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zerres K, Rudnik-Schoneborn S. Natural history in proximal spinal muscular atrophy. Clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch Neurol. 1995;52(5):518–523. doi: 10.1001/archneur.1995.00540290108025. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev. 2002;119 suppl 1:S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15(14):3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 27.Kariya S, Park GH, Maeno-Hikichi Y, Leykekhman O, Lutz C, Arkovitz MS, Landmesser LT, Monani UR. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17(16):2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrank B, et al. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci U S A. 1997;94(18):9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hastings ML, et al. Tetracyclines that promote SMN2 exon 7 splicing as therapeutics for spinal muscular atrophy. Sci Transl Med. 2009;1(5):5ra12. doi: 10.1126/scitranslmed.3000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreassi C, et al. Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet. 2001;10(24):2841–2849. doi: 10.1093/hmg/10.24.2841. [DOI] [PubMed] [Google Scholar]
- 31.Lunn MR, et al. Indoprofen upregulates the survival motor neuron protein through a cyclooxygenase-independent mechanism. Chem Biol. 2004;11(11):1489–1493. doi: 10.1016/j.chembiol.2004.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grzeschik SM, Ganta M, Prior TW, Heavlin WD, Wang CH. Hydroxyurea enhances SMN2 gene expression in spinal muscular atrophy cells. Ann Neurol. 2005;58(2):194–202. doi: 10.1002/ana.20548. [DOI] [PubMed] [Google Scholar]
- 33.Jarecki J, et al. Diverse small-molecule modulators of SMN expression found by high-throughput compound screening: early leads towards a therapeutic for spinal muscular atrophy. Hum Mol Genet. 2005;14(14):2003–2018. doi: 10.1093/hmg/ddi205. [DOI] [PubMed] [Google Scholar]
- 34.Coady TH, Lorson CL. Trans-splicing-mediated improvement in a severe mouse model of spinal muscular atrophy. J Neurosci. 2010;30(1):126–130. doi: 10.1523/JNEUROSCI.4489-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams JH, Schray RC, Patterson CA, Ayitey SO, Tallent MK, Lutz GJ. Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J Neurosci. 2009;29(24):7633–7638. doi: 10.1523/JNEUROSCI.0950-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avila AM, et al. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J Clin Invest. 2007;117(3):659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swoboda KJ, et al. Phase II open label study of valproic acid in spinal muscular atrophy. PLoS One. 2009;4(5):e5268. doi: 10.1371/journal.pone.0005268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brichta L, Holker I, Haug K, Klockgether T, Wirth B. In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate. Ann Neurol. 2006;59(6):970–975. doi: 10.1002/ana.20836. [DOI] [PubMed] [Google Scholar]
- 39.Hua Y, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24(15):1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butchbach ME, et al. Effects of 2,4-diaminoquinazoline derivatives on SMN expression and phenotype in a mouse model for spinal muscular atrophy. Hum Mol Genet. 2010;19(3):454–467. doi: 10.1093/hmg/ddp510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riessland M, et al. SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum Mol Genet. 2010;19(8):1492–1506. doi: 10.1093/hmg/ddq023. [DOI] [PubMed] [Google Scholar]
- 42.Bevan AK, et al. Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum Mol Genet. 2010;19(20):3895–3905. doi: 10.1093/hmg/ddq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shababi M, Habibi J, Yang HT, Vale SM, Sewell WA, Lorson CL. Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum Mol Genet. 2010;19(20):4059–4071. doi: 10.1093/hmg/ddq329. [DOI] [PubMed] [Google Scholar]
- 44.Heier CR, Satta R, Lutz C, DiDonato CJ. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum Mol Genet. 2010;19(20):3906–3918. doi: 10.1093/hmg/ddq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park GH, Maeno-Hikichi Y, Awano T, Landmesser LT, Monani UR. Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J Neurosci. 2010;30(36):12005–12019. doi: 10.1523/JNEUROSCI.2208-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swoboda KJ, et al. Natural history of denervation in SMA: relation to age, SMN2 copy number, and function. Ann Neurol. 2005;57(5):704–712. doi: 10.1002/ana.20473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315(5815):1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osoegawa K, et al. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10(1):116–128. [PMC free article] [PubMed] [Google Scholar]
- 49.Osoegawa K, et al. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11(3):483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valenzuela DM, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21(6):652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 51.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16(7):657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 52.Poueymirou WT, et al. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat Biotechnol. 2007;25(1):91–99. doi: 10.1038/nbt1263. [DOI] [PubMed] [Google Scholar]
- 53.Gabanella F, Carissimi C, Usiello A, Pellizzoni L. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum Mol Genet. 2005;14(23):3629–3642. doi: 10.1093/hmg/ddi390. [DOI] [PubMed] [Google Scholar]
- 54.Monani UR, et al. A transgene carrying an A2G missense mutation in the SMN gene modulates phenotypic severity in mice with severe (type I) spinal muscular atrophy. J Cell Biol. 2003;160(1):41–52. doi: 10.1083/jcb.200208079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carissimi C, et al. Unrip is a component of SMN complexes active in snRNP assembly. FEBS Lett. 2005;579(11):2348–2354. doi: 10.1016/j.febslet.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 56.Butchbach ME, Edwards JD, Burghes AH. Abnormal motor phenotype in the SMNDelta7 mouse model of spinal muscular atrophy. Neurobiol Dis. 2007;27(2):207–219. doi: 10.1016/j.nbd.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.El-Khodor BF, et al. Identification of a battery of tests for drug candidate evaluation in the SMNDelta7 neonate model of spinal muscular atrophy. Exp Neurol. 2008;212(1):29–43. doi: 10.1016/j.expneurol.2008.02.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.