Abstract
Huntington disease (HD) is a devastating neurodegenerative disorder for which there are no disease-modifying treatments. Previous studies have proposed that activation of the heat shock response (HSR) via the transcription factor heat shock factor 1 (HSF1) may be of therapeutic benefit. However, the effect of disease progression on the HSR and the therapeutic potential of this pathway are currently unknown. Here, we used a brain-penetrating HSP90 inhibitor and physiological, molecular, and behavioral readouts to demonstrate that pharmacological activation of HSF1 improves huntingtin aggregate load, motor performance, and other HD-related phenotypes in the R6/2 mouse model of HD. However, the beneficial effects of this treatment were transient and diminished with disease progression. Molecular analyses to understand the transient nature of these effects revealed altered chromatin architecture, reduced HSF1 binding, and impaired HSR accompanied disease progression in both the R6/2 transgenic and HdhQ150 knockin mouse models of HD. Taken together, our findings reveal that the HSR, a major inducible regulator of protein homeostasis and longevity, is disrupted in HD. Consequently, pharmacological induction of HSF1 as a therapeutic approach to HD is more complex than was previously anticipated.
Introduction
Huntington disease (HD) is an inherited, devastating, late-onset, neurodegenerative disorder that affects approximately 5–10 individuals per 100,000 worldwide (1). The average age of onset is 40 years, with HD patients suffering from movement disorders, personality changes, cognitive impairment, and irreversible weight loss over the course of 15–20 years before death (2). At the molecular level, HD is caused by a CAG trinucleotide repeat expansion in exon 1 of the HTT gene (3). The encoded mutant huntingtin has an inherent propensity for aggregation and forms a spectrum of oligomeric species and insoluble aggregates both in vitro and in vivo (4–7).
To counteract the toxicity of misfolded and aggregated proteins, eukaryotic cells have developed an elegant network of molecular chaperones that can act independently or synergistically to identify and refold any misfolded proteins that arise (8, 9). In addition, eukaryotic cells are able to rapidly upregulate chaperone levels via the heat shock response (HSR), unfolded protein response (UPR), and mitochondrial UPR (10–12) upon exposure to acute environmental stress, thereby maintaining proteome integrity even under challenging conditions.
Studies have shown that the overexpression of chaperones, either alone or in combination, can reduce aggregate load and toxicity in multiple models of polyglutamine disease (8, 13–17). However, despite encouraging results in a mouse model of spinocerebellar ataxia 1 (18), overexpression of individual chaperones has had limited success in the amelioration of disease phenotypes in HD mice (19–21). It has therefore been proposed that synchronized upregulation of multiple chaperones may provide greater therapeutic benefit than overexpression of single chaperones alone.
Pharmacological inhibition of the ATPase activity of HSP90 results in the upregulation of multiple chaperones by activating the HSR via the transcription factor HSF1 (22, 23). HSP90 inhibitors, such as geldanamycin, 17-AAG, or 17-DMAG, have been shown to reduce aggregate load and toxicity in cell, fly, and slice culture models of neurodegeneration via a mechanism that is dependent on HSF1 activation (19, 24–28). Whereas the HSP90 inhibitors 17-AAG and geldanamycin have been shown to have beneficial effects in mouse models of spinal and bulbar muscular atrophy (29, 30), the effects of HSF1 activation on HD progression in the mammalian brain have so far remained unknown. This is primarily due to the fact that existing HSF1 activators display high toxicity (geldanamycin) or an inability to efficiently cross the blood-brain barrier (17-AAG) (31).
To attempt to address these questions, we used the 2-amino-7,8-dihdro-6H-pyrido[4,3-D]pyrimidin-5-one compound NVP-HSP990 (referred to herein as HSP990), a brain-penetrant HSP90 inhibitor (32–34), as a tool to study the HSR in the brains of HD mouse models in vivo. We found that chronic treatment of R6/2 mice with HSP990 transiently reduced aggregate load and improved phenotype, which suggests that induction of the HSR could be beneficial in HD. However, we also found that in both R6/2 exon 1 transgenic (35) and HdhQ150 knockin (HdhQ150/Q150) (36, 37) mouse models, the ability to upregulate major heat shock proteins (HSPs) became impaired with disease progression. Whereas HSF1 expression, dissociation from HSP90, hyperphosphorylation, and nuclear translocation were all comparable between HD and WT mice, ChIP revealed that reduced levels of tetra-acetylated histone H4 (Tetra AcH4), a known regulator of HSF1 DNA binding (38), were associated with the heat shock (HS) promoters of HD mice. As a consequence, reduced HSF1 binding, RNA polymerase 2 engagement, and mRNA production were observed at HS genes upon HSP990 treatment.
Results
Systemic HSP990 administration activates the HSR in mouse brain tissue.
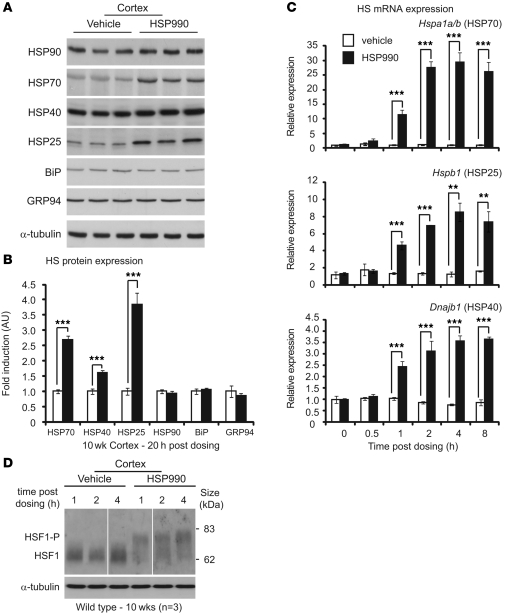
To investigate the effects of HSP990 on the HSR in mouse brain in vivo, the expression levels of major HSPs were assessed in WT mouse cortex 20 hours after HSP990 administration. A single acute oral dose of HSP990 (12 mg/kg) significantly increased levels of HSP70 (2.7-fold), HSP25 (3.8-fold), and HSP40 (1.6-fold) relative to levels in vehicle-treated mice at 10 weeks of age (Figure 1, A and B). In contrast, the expression of HSP90 remained unchanged. In addition, no change in expression was observed in the UPR chaperones BiP and GRP94, which suggests that upregulation of stress proteins after HSP990 treatment is specific to the HSR arm of the chaperone network. A similar pattern of HSP expression was observed in striatum and cerebellum 20 hours after dosing (Supplemental Figure 1, A–D; supplemental material available online with this article; doi: 10.1172/JCI57413DS1).
Figure 1. NVP-HSP990 elicits an HSR in mouse brain after acute oral administration.
(A) Western blots of HSR and UPR proteins in 10-week-old WT mouse cortex 20 hours after treatment with HSP990 (12 mg/kg) or vehicle. (B) Fold upregulation of HSPs in HSP990-treated WT mice (black) was calculated relative to vehicle-treated WT mice (white) by densitometry. Values are mean ± SEM fold induction (n = 6 per treatment group). (C) Mouse cortices (WT) were harvested 0, 0.5, 1, 2, 4, and 8 hours after a single dose of HSP990 (12 mg/kg) or vehicle. Taqman RT-qPCR was used to determine the fold induction of HS genes relative to expression in the vehicle group 0 hours after dosing. Values (mean ± SEM) were calculated by the ΔCt method, normalized to the housekeeping gene Atp5b (n = 4 per treatment group). (D) Western blotting for HSF1 in cortices of 10-week-old WT mice harvested 1, 2, and 4 hours after an acute dose of HSP990 (12 mg/kg) or vehicle (n = 3 per treatment group). HSF1-P, hyperphosphorylated form of HSF1. Lanes were run on the same gel but were noncontiguous (white lines). **P < 0.01, ***P < 0.001, Student’s t test.
To determine the dynamics of HS gene induction, cortex was harvested 0, 0.5, 1, 2, 4, or 8 hours after treatment with HSP990 (12 mg/kg) or vehicle. Significant induction in Hspa1a/b (encoding HSP70; 11.3-fold), Hspb1 (encoding HSP25; 4.6-fold), and Dnajb1 (encoding HSP40; 2.4-fold) mRNA was observed as early as 1 hour after treatment (Figure 1C). Interestingly, although the magnitude of induction was markedly different between HS genes, the dynamics of induction were highly comparable, with all 3 HS genes presenting maximum induction 4 hours after HSP990 treatment.
The definitive mark of a bona fide HSR is activation of the transcription factor HSF1. The activation state of HSF1 was assessed in mouse brain regions 1, 2, and 4 hours after HSP990 treatment. Hyperphosphorylation of HSF1 correlates with its transcriptional activation and can be observed by Western blotting as a retardation of HSF1 migration during SDS-PAGE. Hyperphosphorylated HSF1 was observed in mouse cortex, striatum, and cerebellum 1, 2, and 4 hours after treatment (Figure 1D and Supplemental Figure 2, A and B), which suggests that HSP990 acts through HSF1 to induce an HSR in mouse brain tissue. We conclude that 1 acute oral dose of HSP990 at 12 mg/kg is able to efficiently induce a bona fide HSR in mouse brain in vivo.
Establishment of a well-tolerated HSP990 chronic dosing regimen.
To evaluate whether pharmacological induction of the HSR might prove beneficial in the context of the HD mutation, we established an HSP990 dosing regimen that was well tolerated in mice. Compounds frequently show increased toxicity in WT and R6/2 mice at 4–6 weeks of age compared with older mice (39). Therefore, we assessed the ability of lower doses to induce an HSR and found that HSP70, HSP25, and HSP40 were effectively upregulated by a single 1.2 mg/kg dose in 4-week-old R6/2 mice, although a 7.2 mg/kg dose was required to elicit the same effect in mice of 8 weeks of age (Supplemental Figure 3, A and B). To ascertain the duration of HSP70 induction, 7.2 mg/kg HSP990 was administered to 8-week-old R6/2 mice, which were sacrificed 0, 4, 8, 12, 16, 24, 72, 96, and 120 hours after treatment (Supplemental Figure 3C). The level of HSP70 in the cortex peaked at 12 hours and returned to baseline by 120 hours. Therefore, we devised a dose escalation scheme whereby HSP990 was administered weekly: 1.2 mg/kg at 5 weeks of age, 2.4 mg/kg at 6 weeks, and 7.2 mg/kg thereafter.
Induction of the HSR with HSP990 improves phenotype and reduces aggregate load in R6/2 mice.
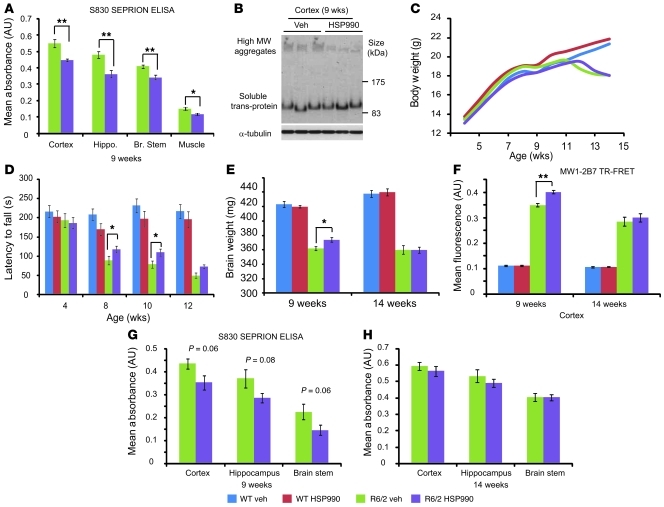
We first assessed whether the induction of molecular chaperone levels via HSP990 treatment decreased the aggregate load in R6/2 mice. WT and R6/2 mice (n = 6) were treated with HSP990 or vehicle from 5 to 9 weeks of age, after which aggregate load was measured by Seprion ligand ELISA (7). Levels of aggregated huntingtin were significantly decreased in cortex, hippocampus, brain stem, and muscle, which was confirmed in the cortex by Western blotting and immunohistochemistry (Figure 2, A and B, and Supplemental Figure 4, A and B).
Figure 2. HSP990 treatment transiently improves rotarod performance and reduces aggregate load in R6/2 mice.
(A) Seprion ligand ELISA was used to quantify aggregate load in tissues of R6/2 mice after treatment for 4 weeks with vehicle (green) or HSP990 (purple). Values were plotted as mean absorbance ± SEM (n = 6 per treatment group). (B) Western blotting and immunodetection with S830 was used to obtain visual representation of results in A. (C and D) Assessment of (C) body weight and (D) rotarod ability with age. (E) Brain weight in 9- and 14-week-old mice after vehicle or HSP990 treatment. Values are presented as mean ± SEM (n = 12 per group). (F–H) R6/2 brain tissues were harvested from 9-week-old satellite or end-of-trial mice (14 weeks). (F) 2B7-MW1 TR-FRET was used to determine levels of soluble exon 1 in mouse cortices after vehicle or HSP990 treatment. Values are presented as mean ± SEM for each group (n ≥ 4 per treatment group). (G and H) S830 Seprion ligand ELISA was used to measure aggregate load after HSP990 treatment at (G) 9 weeks (n = 4 per treatment group) and (H) 14 weeks of age (n ≥ 6 per treatment group). *P < 0.05, **P < 0.01, 1-way ANOVA (C and D) or Student’s t test (A, B, and E–H).
Mutant huntingtin has been proposed to form a spectrum of oligomeric species both in vitro and in vivo (7), with different huntingtin oligomers exhibiting different levels of cytotoxicity (40, 41). Changes in aggregate size and morphology can be observed as alterations in huntingtin antibody reactivity (40, 41) and/or by altered migration of aggregates through an agarose gel (42). To determine whether HSP990 treatment promotes the formation and/or removal of a specific subpopulation of huntingtin aggregates in R6/2 brain tissue, cortical protein lysates were subjected to agarose gel electrophoresis for resolution of aggregates (AGERA) followed by Western blotting and immunodetection with antibodies that have differential affinities for huntingtin aggregates depending on their conformation (40, 41). Immunoblotting with S830, 1C2, and 3B5H10 revealed aggregate smears greater than 500 kDa in size in R6/2, but not WT, cortical lysates (Supplemental Figure 4C). HSP990 treatment did not alter the size distribution of 1C2, 3B5H10, or S830 reactive aggregates in R6/2 mice. However, HSP990 treatment did result in a reduction in aggregate smear intensity that was comparable for all 3 huntingtin antibodies (Supplemental Figure 4C). These data suggest that HSP990 treatment leads to a uniform reduction in aggregate load rather than the reduction of a specific aggregate subpopulation. However, we cannot rule out the possibility that HSP990 treatment leads to subtle conformational changes that are not identifiable by these techniques.
We next used a set of established quantitative tests to determine whether prolonged HSP990 treatment might improve R6/2 behavioral phenotypes (36, 43, 44). Prior to treatment onset, female WT and R6/2 littermates (born over a 3-day period) were assorted into treatment groups matched for weight, grip strength, rotarod performance, and activity level, as previously described (43). WT and R6/2 mice were treated with HSP990 or vehicle (n ≥ 12 per treatment group) according to the dosing regimen devised above, and phenotypic parameters were measured from 5 weeks to 14 weeks of age. R6/2 vehicle- and HSP990-treated groups had well-matched CAG repeats (199 ± 3 and 200 ± 4, respectively; mean ± SD). In parallel, WT and R6/2 satellite groups (n = 4) were treated with HSP990 or vehicle from 5 to 9 weeks of age as in the initial dose-finding study.
Mice were weighed weekly from 4 to 14 weeks of age (Figure 2C). As expected, R6/2 mice weighed less than WT mice (F1,46 = 5.22; P = 0.027) and gained weight at a slower rate (F4,460 = 21.36; P < 0.001). Treatment with HSP990 had no effect on weight (F1,46 = 0.02; P = 0.888) or rate of weight gain (F4,460 = 0.94; P = 0.433) in WT mice, or on weight (F1,46 = 2.54; P = 0.118) or rate of weight loss (F4,460 = 1.18; P = 0.321) in R6/2 mice. Therefore, HSP990 was well tolerated but did not improve the weight loss phenotype in the R6/2 mice.
Rotarod performance, a sensitive indicator of balance and motor coordination, has been reliably shown to decline in R6/2 mice (43). Consistent with previous results, R6/2 rotarod performance was impaired (F1,47 = 58.38; P < 0.001) and deteriorated with age (F2,705 = 31.74; P < 0.001) (Figure 2D). Treatment with HSP990 did not modify the rotarod performance of WT mice (F1,47 = 0.11; P = 0.746) or how this changed with age (F2,705 = 0.24; P = 0.768). However, HSP990 improved the rotarod performance of R6/2 mice at 8 (F1,47 = 6.75; P = 0.012) and 10 (F1,47 = 4.60; P = 0.037) weeks of age, but the effect diminished by 12 weeks (F1,47 = 2.32; P = 0.134). The effect of HSP990 treatment on overall rotarod performance of R6/2 mice was close to statistical significance (F1,47 = 3.63; P = 0.063).
Forelimb grip strength was assessed at 4, 11, 12, and 13 weeks of age. Consistent with previous data, the grip strength of R6/2 mice was impaired (F1,48 = 64.09; P < 0.001) and deteriorated with age (F3,288 = 17.43; P < 0.001). Treatment with HSP990 had no overall effect on the grip strength of WT mice (F1,48 = 0.69; P = 0.410) or over the course of the experiment (F3,288 = 0.22; P = 0.866). HSP990 did not improve R6/2 grip strength (F1,48 = 0.575; P = 0.452) or its rate of decline (F3,288 = 0.12; P = 0.934) (Supplemental Figure 5).
Exploratory activity was assessed fortnightly from 5 to 13 weeks of age, as described previously (44). Mice were assessed for a period of 30 minutes for total activity, mobility, rearing, and unsupported rearing (Supplemental Figure 6 and Supplemental Table 1). The pattern of R6/2 hypoactivity was consistent with previous data (45), but HSP990 treatment had no effect on the activity measures of WT or R6/2 mice (Supplemental Table 1).
R6/2 brain weight decreases with disease progression (4). At 9 weeks of age, HSP990 treatment led to an increase in R6/2 brain weight (P = 0.029) (Figure 2E). In addition, soluble exon 1 huntingtin (46, 47), as measured by time-resolved Forster resonance energy transfer (TR-FRET), was increased in the cortex of treated mice (Figure 2F), and Seprion ligand ELISA showed that there was a trend toward a concomitant decrease in aggregation in the cortex (P = 0.061), hippocampus (P = 0.084), and brain stem (P = 0.062) (Figure 2G), consistent with the results obtained in the initial dose-finding study (Figure 2A). However, these effects were not maintained at 14 weeks of age (Figure 2H). In addition, AGERA revealed that aggregate size distribution and huntingtin antibody immunoreactivity were also unaltered at 14 weeks (Supplemental Figure 4D). Taken together with rotarod performance, which was improved at 8 and 10 but not 14 weeks of age, these data suggest the beneficial effects of chronic HSP990 treatment diminish with disease progression.
HSP990-mediated HSP induction is impaired in HD mouse models.
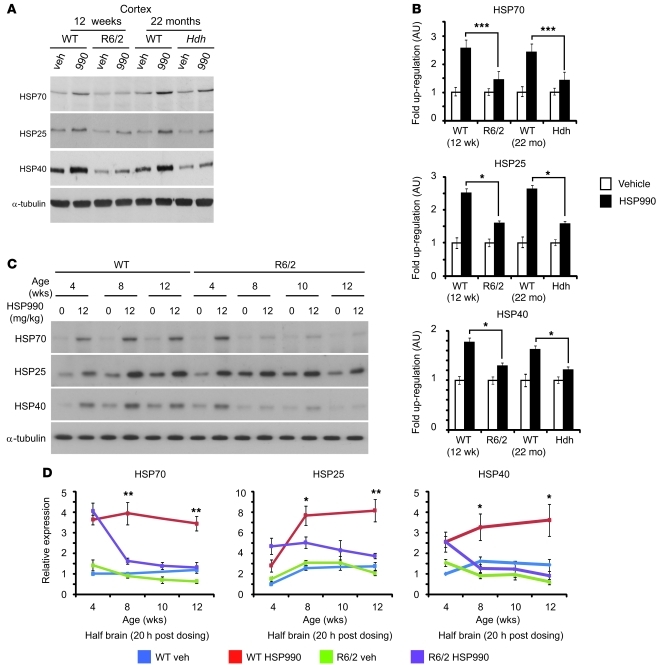
As the beneficial effects of HSP990 treatment were transient in R6/2 mice, we hypothesized that impairment of the HSR might occur with disease progression. Therefore, we investigated whether HSP990 treatment induced an HSR in the brains of R6/2 and/or HdhQ150/Q150 mice in late-stage disease. In our colonies, R6/2 mice and HdhQ150/Q150 homozygotes reached end-stage disease at approximately 15 weeks and 24 months of age, respectively. Treatment with HSP990 induced a 2.5-fold increase in HSP70 in cortical tissues from 12-week-old and 22-month-old WT mice 20 hours after dosing (Figure 3, A and B). In addition, both HSP25 (2.5-fold) and HSP40 (1.5-fold) levels were also significantly elevated. Interestingly, both R6/2 and HdhQ150/Q150 models showed markedly lower fold induction of all 3 HSPs in cortex (Figure 3, A and B), striatum (Supplemental Figure 7, A and C), cerebellum (Supplemental Figure 7, B and D), and muscle (Supplemental Figure 8, A and B). These data suggest that at late-stage disease, both HD mouse models may have an impaired HSR in both brain and periphery.
Figure 3. HSP upregulation is impaired in HD mouse models.
(A) Representative Western blots of HSP70, HSP25, HSP40, and α-tubulin expression in WT, R6/2 or HdhQ150/Q150 (Hdh) cortex 20 hours after treatment with vehicle or HSP990 (12 mg/kg). (B) Immunoblotting and densitometry were used to calculate the expression levels of HSP70, HSP25, and HSP40 in mouse cortex relative to levels of α-tubulin. Fold expression of each chaperone was calculated for HSP990 relative to vehicle. Values are mean fold ± SEM (n ≥ 4 per treatment group). (C) Representative Western blots of HSP70, HSP25, HSP40, and α-tubulin in half brain of 4-, 8-, 10-, and 12-week-old WT or R6/2 mice 20 hours after treatment with vehicle or HSP990 (12 mg/kg). (D) Relative expression of HSP70, HSP25, and HSP40 at 20 hours after treatment with HSP990 (12 mg/kg) or vehicle, calculated relative to α-tubulin by densitometry. Fold expression relative to vehicle-treated 4-week-old WT mice was calculated for each chaperone and age, and values are mean ± SEM (n = 4). *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test.
HSR impairment is progressive and is pronounced in early-stage disease.
In order to determine the relationship between HSR impairment and disease progression, WT and R6/2 mice were sacrificed 20 hours after an acute dose of vehicle or HSP990 (12 mg/kg) at 4, 8, 10, and 12 weeks of age. Given that HSR impairment had been observed in all brain regions studied, subsequent experiments were performed on half brains. As previously reported (19), we found significantly reduced levels of HSP70 and HSP40 in R6/2 compared with WT brain tissue by 12 weeks of age (Figure 3, C and D). This was not observed for HSP25, although interestingly, levels of HSP25 were generally lower in both WT and R6/2 mice at 4 weeks of age. WT mice showed consistent upregulation of HSP70 (≈3-fold), HSP25 (≈3-fold), and HSP40 (≈2-fold) in response to 12 mg/kg HSP990 treatment at all ages tested. At 4 weeks of age, induction of HSP70, HSP25, and HSP40 was extremely comparable between R6/2 and WT mice (Figure 3, C and D). However, upregulation of all 3 HSPs was compromised in R6/2 brain tissue by 8 weeks of age (HSP70, 1.7-fold; HSP25, 1.8-fold; HSP40, 1.3-fold), with impaired HSP upregulation continuing to 12 weeks despite equivalent levels of HSP990 reaching WT and R6/2 brain tissue 0.5, 1, 2, 4, and 24 hours after dosing (Supplemental Figure 9). These data suggest that HD mice are able to elicit an HSR early in disease, but this becomes impaired with disease progression.
HSR impairment occurs at the level of transcription.
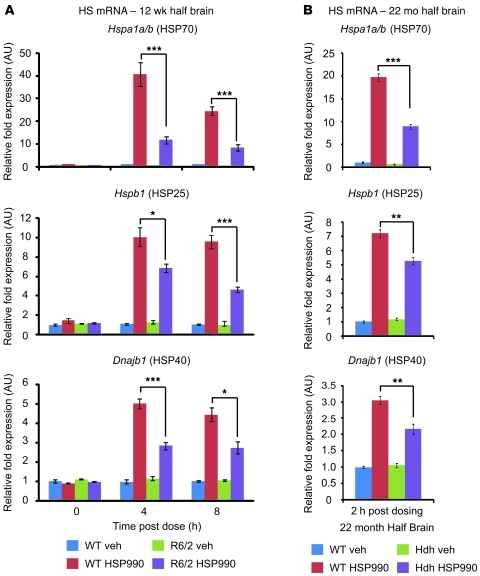
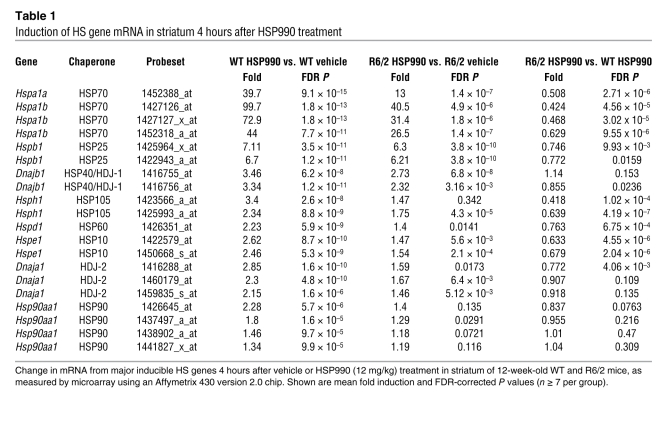
To determine whether impairment of the HSR occurs at the level of transcription, 12-week-old WT and R6/2 mice were sacrificed 4 and 8 hours after treatment with vehicle or 12 mg/kg HSP990, and levels of HS gene mRNA were assessed by real-time quantitative PCR (RT-qPCR). An increase in HS gene mRNA after HSP990 treatment was found in both WT and R6/2 mice. However, R6/2 mice exhibited significantly lower fold induction of Hspa1a/b, Hspb1, and Dnajb1 mRNA compared with WT mice at both 4 and 8 hours after treatment (Figure 4A). This effect was also observed in HdhQ150/Q150 mice 2 hours after dosing (Figure 4B). Consistent with a progressive decline in the HSR in HD mice, impaired Hspa1a/b, Hspb1, and Dnajb1 mRNA production after HSP990 treatment was observed 4 hours after dosing in R6/2 mice at 8 weeks of age, but not at 4 weeks (Supplemental Figure 10, A and B). In contrast, basal levels of Hspa1a/b only decreased progressively in HD mice, whereas basal levels of Hspb1 or Dnajb1 were not dysregulated (Supplemental Figure 11, A and B). To determine the extent of this impaired induction, WT and R6/2 mice were dosed with vehicle or 12 mg/kg HSP990 (n ≥ 7 per treatment group), and striata were removed 4 hours after dosing. Expression profiling using Affymetrix microarrays (see Methods) showed that the fold induction in the expression of all major inducible HSPs was reduced in R6/2 compared with WT striatum (Table 1).
Figure 4. Impaired upregulation of HSPs occurs at the level of transcription.
Taqman RT-qPCR of Hspa1a/b, Hspb1, and Dnajb1 was performed on half brains of 12-week-old WT and R6/2 mice 0, 4, or 8 hours after treatment with vehicle or HSP990 (12 mg/kg) (A), or on half brains of 22-month-old WT and HdhQ150/Q150 mice 2 hours after treatment with vehicle or HSP990 (B). (Due to the reduced availability of mice aged 22 months, HdhQ150/Q150 analysis was performed at 2 hours after dose to allow investigation of mRNA levels and chromatin architecture on the same samples.) Chaperone mRNA expression levels were normalized to the housekeeping gene Atp5b. Fold induction of each HS gene after HSP990 treatment was calculated relative to expression levels of WT vehicle groups 0 hours after treatment, and expressed as mean fold ± SEM (n = 4 per treatment group). *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test.
Table 1 .
Induction of HS gene mRNA in striatum 4 hours after HSP990 treatment
HSF1 dissociation from HSP90, hyperphosphorylation, and nuclear translocation are not impaired in HD mice upon HSP990 treatment.
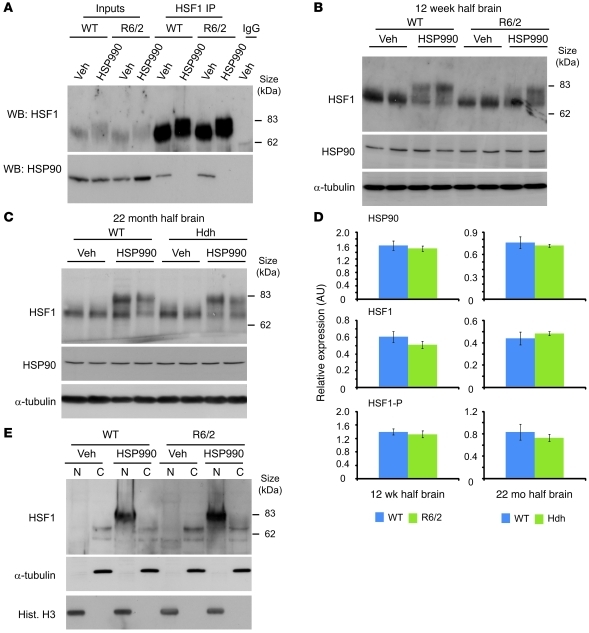
Levels of HSF1 and HSP90 expression were not significantly different between 12-week-old WT and R6/2 or 22-month-old WT and HdhQ150/Q150 brain lysates (Figure 5, A–D). Therefore, the impaired HSR observed in HD mice is not simply caused by differences in the expression levels of HSF1 or HSP90. One possible explanation for the impairment is that upon HSP990 treatment, HSF1 cannot dissociate efficiently from its repressive HSP90 complex. To test whether this was the case, HSF1 was immunoprecipitated from 12-week-old WT and R6/2 brain tissue that had been harvested 2 hours after treatment with HSP990 or vehicle. Western blotting for HSP90 after HSF1 IP revealed that HSP90 coimmunoprecipitated with HSF1 in brain tissue from both WT and R6/2 vehicle-treated mice. This is consistent with previous findings (22), which suggests that the majority of inactive HSF1 is found sequestered in a repressive HSP90 complex. In contrast, 2 hours after treatment with HSP990, no HSP90 was detected after HSF1 IP from either WT or R6/2 brain tissue (Figure 5A). Therefore, HSF1 is able to efficiently dissociate from HSP90 in both WT and R6/2 mice.
Figure 5. HSF1 dissociates from HSP90, becomes hyperphosphorylated, and translocates to the nucleus upon HSP990 treatment.
(A) Western blots of HSF1 and HSP90 after HSF1 IP from 12-week-old WT and R6/2 mouse brains 2 hours after treatment with vehicle or HSP990 (12 mg/kg). (B and C) Representative Western blots of HSF1 and HSP90 in (B) 12-week-old WT and R6/2 or (C) 22-month-old WT and HdhQ150/Q150 mouse half brains 2 hours after treatment with vehicle or HSP990 (12 mg/kg). α-Tubulin was used as a loading control. (D) Expression of HSP90 and HSF1 relative to α-tubulin, or phosphorylated HSF1 (HSF1-P) relative to unphosphorylated HSF1, in 12-week-old WT and R6/2 or 22-month-old WT and HdhQ150/Q150 mice. Densitometry values are mean ± SEM (n = 4 per treatment group). (E) Representative Western blots for HSF1 in nuclear and cytoplasmic fractions derived from 12-week-old WT and R6/2 mouse half brains 2 hours after treatment with vehicle or HSP990 (12 mg/kg). Purity of nuclear (N) and cytoplasmic (C) fractions was demonstrated by immunoblotting for α-tubulin and histone H3.
An alternative explanation for the impaired HSR is that upon HSP990 treatment, HSF1 hyperphosphorylation does not occur, and subsequently, HSF1 is less potent at inducing HS gene expression. Western blotting revealed that 2 hours after treatment with HSP990, HSF1 was hyperphosphorylated in brain tissues of 12-week- and 22-month-old WT, 12-week-old R6/2, and 22-month-old HdhQ150/Q150 mice (Figure 5, B–D). Furthermore, HSF1 hyperphosphorylation was observed as early as 30 minutes after treatment with HSP990 and continued to 8 hours after dosing in both WT and R6/2 brain tissue (Supplemental Figure 12). This suggests that the HSR impairment in HD mice is not simply caused by an alteration in the dynamics of HSF1 activation.
Finally, we investigated whether HSF1 can effectively translocate to the nucleus. Nuclear and cytoplasmic fractions were obtained from 12-week-old WT and R6/2 mouse brains harvested 2 hours after HSP990 treatment. HSF1 was not observed in the nuclear fractions of WT or R6/2 mice treated with vehicle; however, hyperphosphorylated HSF1 resided predominantly in the nuclear fraction of both WT and R6/2 brain tissue (Figure 5E).
Taken together, these findings demonstrate successful target engagement of HSP990 in the mouse brain and also suggest that upon HSP990 treatment, HSF1 is able to dissociate from its HSP90 repressor complex, become hyperphosphorylated, and translocate to the nucleus. Furthermore, the capacity of HSF1 to perform these functions is highly comparable in both WT and HD mouse brain tissue at late-stage disease.
Reduced HSF1 promoter binding and chromatin accessibility in R6/2 versus WT mice 2 hours after HSP990 treatment.
Given the unimpaired nuclear translocation of hyperphosphorylated HSF1 (Figure 5), we reasoned that diminished binding of HSF1 to HS promoters may be the cause of impaired HSR (Figures 3 and 4). To investigate this possibility, SYBR green quantitative PCR (qPCR) using primers specific for HS gene promoters was coupled to HSF1 ChIP in mouse brains harvested 2 hours after vehicle or HSP990 treatment. HSF1 ChIP qPCR revealed a significant increase in HSF1 promoter binding 2 hours after HSP990 treatment in mouse brain tissue compared with tissue from vehicle-treated mice. However, the level of HSF1 binding at all 3 HS promoters after HSP990 treatment was significantly lower in the brains of 12-week-old R6/2 mice (Figure 6A) and 22-month-old HdhQ150/Q150 mice (Figure 6D).
Figure 6. Reduced HSF1 promoter binding and altered nucleosome landscapes are observed at HS loci in R6/2 mice, but do not correlate with chromatin accessibility.
(A–E) Levels of HSF1 (A and D), RNA polymerase 2 (RNApol2), H3AcK9, H3AcK27 (B), and Tetra AcH4 (C and E) bound to HS promoters 2 hours after vehicle or HSP990 treatment (12 mg/kg) was determined by ChIP in 12-week-old WT and R6/2 (A–C) and 22-month-old WT and HdhQ150/Q150 (D and E) mouse half brains. Chromatin was immunoprecipitated, and SYBR green qPCR was performed on the resulting DNA with primers specific for the Hspa1b, Hspb1, and Dnajb1 promoters. Signal was normalized to 10% of the input for each sample. Values are mean ± SEM (n = 5 per treatment group). Black lines in A–E indicate mean signal obtained after pulldown with rabbit IgG alone (n = 2). (F) MNase digestion was performed on chromatin extracted from mouse brain tissue 2 hours after treatment with vehicle or HSP990. SYBR green was then performed using primers spanning the Hspa1b gene. Digested signal was normalized to undigested signal, and values are mean ± SEM (n = 4 per treatment group). *P < 0.05 vs. WT HSP990, #P < 0.05 vs. WT vehicle, Student’s t test.
As equivalent levels of a hyperphosphorylated form of HSF1 were found to reside in the nucleus of both WT and R6/2 mice (Figure 5E), we postulated that reduced HSF1 promoter binding is most likely caused by either the DNA binding competency of HSF1 itself or the chromatin architecture and accessibility of HS promoters. In order to determine how the nucleosome landscape influences HSF1 promoter binding, we conducted further ChIP analysis at the Hspa1b promoter. It has recently been published that the nucleosome landscape can significantly affect inducible binding of HSF1 to target promoters; thus, we focused our analysis on histone marks that had shown the most robust effects on the inducible DNA binding ability of HSF1 (38).
ChIP analysis at the Hspa1b promoter revealed that significant levels of RNA polymerase 2, H3AcK9, H3AcK18, H3AcK27, H3triMeK4, and Tetra AcH4 were all associated with the Hspa1b promoter region in vehicle-treated mice (Figure 6, B and C, and Supplemental Figure 13). Upon treatment with HSP990, the levels of all these factors (except H3AcK18) increased at the Hspa1b promoter in both WT and R6/2 brain tissue (Figure 6, B and C, and Supplemental Figure 13, A and B).
The increase in the level of RNA polymerase 2 was significantly lower in R6/2 than WT brain tissue, consistent with reduced HSF1 binding (Figure 6A) and reduced mRNA production (Figure 4A). No difference in H3 acetylation was observed between WT and R6/2 mice treated with vehicle or HSP990 (Figure 6B and Supplemental Figure 13B). In contrast, significant hypoacetylation of histone H4 was observed at all 3 HS genes between WT and R6/2 mice, whether treated with HSP990 or vehicle (Figure 6C). An equivalent pattern of hypoacetylation was also observed when comparing WT with HdhQ150/Q150 mice, regardless of treatment (Figure 6E). Furthermore, this effect was not observed in 4-week-old mice (Supplemental Figure 14). We therefore conclude that in the brains of HD mice, HS loci become hypoacetylated with disease progression. This phenomenon is specific to histone H4 (a known modifier of HSF1 DNA binding activity) and correlates with the reduced ability of HSF1 to bind target promoters following HSP990 treatment in these mice.
We reasoned that H4 hypoacetylation may reduce chromatin accessibility across HS genes, thereby reducing the ability of HSF1 to bind promoters and subsequently impairing the HSR. To investigate whether this was the case at the Hspa1b promoter, we performed a SYBR green qPCR-coupled micrococcal nuclease (MNase) digestion assay on chromatin isolated from mouse brains 2 hours after treatment with vehicle or HSP990. The Hspa1b promoter region was hypersensitive to MNase treatment in all treatment groups (Figure 6F), which suggests that the promoter is equally accessible in both WT and R6/2 mice and that this accessibility does not change upon HSP990 treatment. To investigate the chromatin accessibility downstream of the promoter region, we performed SYBR green qPCR for regions spanning the transcription start site to the 3′ untranslated region (48). As was observed for the promoter region, the chromatin accessibility of all regions of Hspa1b was highly comparable between WT and R6/2 mice at 12 weeks of age. However, upon treatment with HSP990, WT mice — but not R6/2 mice — showed increased MNase digestion along the length of the gene (Figure 6F), a feature coupled to HSF1 promoter binding (48).
These data strongly suggest that there is reduced HSF1 promoter binding and subsequent nucleosome dissociation from the Hspa1b gene in R6/2 mice compared with WT littermates after treatment with HSP990. However, this does not appear to be due to the accessibility of the chromatin itself around the Hspa1b promoter, despite the presence of hypoacetylated H4 in HD mice (Figure 6, C and E). Tetra AcH4 has been suggested to be a strong modulator of HSF1 binding (38), and it is conceivable that the acetylation state of H4 could dictate the inducible binding of HSF1 to target DNA sequences independently of promoter accessibility.
Discussion
The HSR is a powerful evolutionary mechanism that allows cells to maintain proteome integrity in the face of proteotoxic insults (10). Given that initiation of an HSR has the potential to neutralize the toxicity exerted by misfolded and aggregated proteins, it is not surprising that activation of this pathway is an attractive therapeutic target for multiple protein misfolding diseases. In the present study, we showed that the HSP90 inhibitor HSP990 is a potent inducer of the HSR in brain. We devised a treatment strategy in which weekly dosing resulted in the pulsed induction of the HSR without any apparent toxicity. Treatment of R6/2 mice with HSP990 resulted in improved brain weight, a 20% reduction in aggregate load in brain tissues, and a 30% improvement in rotarod performance, effects comparable to those mediated by other treatment strategies (39, 49–51). However, HSP990 did not restore all aspects of disease progression, as grip strength, body weight and hypoactivity remained unchanged. Consistent with previous data from cells, flies, and slice culture systems treated with HSP90 inhibitors (19, 24, 25, 52), the observed reduction in aggregate load was accompanied by a reciprocal increase in levels of soluble mutant huntingtin in brain tissue upon HSF1 activation. However, the beneficial effects of HSP990 treatment were not maintained throughout disease progression.
Whereas oral administration of HSP990 induced the HSR in WT mice at all ages tested, induction of all major HS genes became impaired with disease progression in R6/2 mice. This disease-related attenuation is most likely responsible, at least in part, for the transient nature of the beneficial effects mediated by HSP990. This vastly diminished HSR occurred at the level of transcription and was also apparent in the HdhQ150/Q150 model of HD. The decrease in HS gene transcription occurred despite the effective dissociation of HSF1 from HSP90 followed by its translocation to the nucleus and hyperphosphorylation, features that infer that HSF1 trimerization and its ability to drive HS gene expression are intact (10). Similarly, the levels of HSP90 and hyperphosphorylated HSF1 were not changed between the brains of WT and R6/2 or WT and HdhQ150/Q150 mice at late-stage disease, which indicates that the impairment did not result from decreased levels of activated HSF1.
ChIP analysis revealed that in HD mice, less HSF1 became inducibly bound to the promoters of HS genes after HSP990 treatment. Consistent with previous studies in Drosophila melanogaster, ChIP analysis also revealed that the nucleosome landscape at the Hspa1b promoter is complex and dynamic with multiple histone marks residing at this locus both before and after treatment with HSP990.
Tetra AcH4 has been proposed to be a strong modulator of the inducible binding of HSF1 to target DNA sequences (38). We found that H4 was hypoacetylated in symptomatic HD mice compared with WT mice, both before and after HSP990 treatment, at all HS promoters tested. Given that levels of Tetra AcH4 strongly influence the inducible binding of HSF1 to target DNA sequences (38), we propose a model whereby disease progression leads to hypoacetylation of histone H4 at HS promoters. As a consequence, HSF1 is unable to efficiently bind to these promoters and release/recruit RNA polymerase 2 to transcribe HS genes, thereby resulting in an impaired HSR (Figure 7). Given that an increase in H4 acetylation may occur via direct recruitment of histone acetyl transferase complexes by HSF1 (53–55), reduced initial binding of HSF1 may exacerbate the amount of HSF1 that can further associate with promoters upon subsequent rounds of binding. In addition, CREB binding protein (CBP) has been shown to associate with nuclear inclusions in HD mouse and human brain tissue, and loss of CBP-mediated transcription is a proposed feature of HD progression. It is therefore possible that loss of CBP acetyl transferase activity contributes to the hypoacetylation of HS promoters observed in HD mice (56, 57).
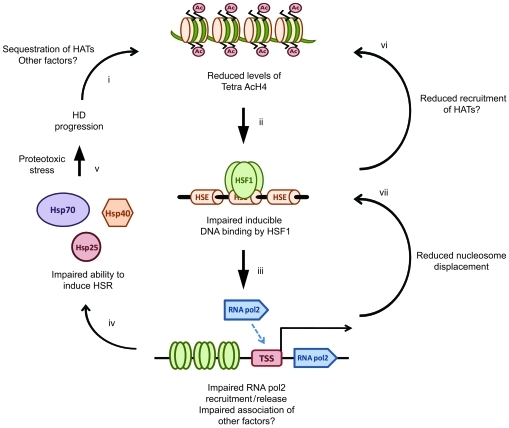
Figure 7. Proposed model for impairment of the HSR in HD.
Disease pathogenesis in HD leads to a progressive reduction in levels of Tetra AcH4 at HS promoters (i), possibly as a consequence of reduced histone acetyl transferase (HAT) activity. Reduced levels of Tetra AcH4 impair the ability of HSF1 to bind its target consensus sequences at HS genes (ii). Reduced HSF1 binding leads to reduced release and recruitment of RNA polymerase 2 (iii), which in turn results in an inability to efficiently upregulate HSPs (iv). Impairment of the HSR may be augmented by reduced H4 acetylation and nucleosome displacement as a consequence of impairment of HSF1 binding, RNA polymerase 2 release and/or recruitment, and association of other factors (e.g., PARP) (vi and vii). As a consequence, cells become vulnerable to acute proteotoxic stress, thereby accelerating disease progression (v).
Strikingly, despite H4 hypoacetylation and transcriptional dysregulation, we were unable to detect any difference in chromatin accessibility across the Hspa1b gene between WT and R6/2 mice. Interestingly, HSP990 treatment did increase chromatin accessibility across the Hspa1b gene in WT mice, but not R6/2 mice. In Drosophila melanogaster, nucleosome displacement across the Hspa1b gene upon HS has been proposed to be caused by recruitment of PARP and release of paused RNA polymerase 2 for transcription (48, 58). Thus, the difference in chromatin accessibility between WT and R6/2 mice after HSP990 treatment may reflect differences in the recruitment of RNA polymerase 2 and other factors as a consequence of impaired HSF1 binding in HD mice.
The significance of our data is 2-fold. First, we showed that the compound HSP990 potently elicited an HSR in mouse brains and that treatment with this compound was able to transiently reduce aggregate load and improve phenotype in a mouse model of HD. We consider this to be a major proof of principle that activation of HSF1 in a mammalian model of HD can be of therapeutic benefit. However, given the multitude of processes controlled by HSP90 (59), it is likely that brain penetrant compounds that can activate HSF1 independently of HSP90 inhibition (26) will prove a more promising avenue for translation of compounds for HD into the clinic.
Second, we provide the first evidence to our knowledge that the HSR becomes impaired in a mouse model of neurodegeneration; consequently, the collapse in protein homeostasis associated with HD is not simply limited to constitutive chaperone networks. Given that a collapse in protein homeostasis has been proposed to be a major pathogenic mechanism in HD (60) and that the ability to respond to stress is intimately coupled to longevity (61–63), loss of the HSR may contribute significantly to disease progression in HD by leaving cells vulnerable to environmental insults encountered on a daily basis (e.g., viral infection, inflammation, and production of reactive oxygen species). Unfortunately, it is difficult to determine whether the HSR becomes progressively impaired in HD patients, as available tissues are limited to postmortem brain samples or peripheral cell lines. In our experience, the extent of the HSR is extremely variable between fibroblast lines, regardless of whether they are derived from HD patients or control subjects (our unpublished observations), and it is well documented that the HSR is activated by EBV transformation to generate lymphoblastoid lines (64), making them highly sensitive to HSP90 inhibitors (65).
We conclude that impairment of the HSR with disease progression is an unforeseen complication of devising therapeutic strategies based on improving protein folding capacity via HSF1 activation. In order for HSR induction to have major beneficial consequences, treatment before onset or during early-stage disease would have to maintain protein homeostasis to the extent that the HSR impairment is prevented from becoming established. This could include the development of compounds that allow the induction of a sustained HSR. Although the effects of prolonged HSF1 activation are unknown and could be detrimental (66, 67), the constitutive expression of a dominant-positive HSF1 mutant in peripheral tissues was shown to have beneficial effects in R6/2 mice (68). In the long term, it will be important to determine the exact mechanism by which the HSR is impaired in order to develop compounds that can act synergistically with HSF1 activators (e.g., by increasing histone acetylation), act as novel HSR inducers that bypass the impairment, or restore this pathway by as-yet unidentified mechanisms.
Methods
Mouse maintenance, breeding, genotyping, and CAG repeat sizing.
All experiments were approved by the King’s College London Ethical Review Process Committee and performed under project and personal licenses approved and issued by the UK Home Office (project license no. 70/6545). Hemizygous R6/2 mice (35) were bred by backcrossing R6/2 males to (CBA × C57BL/6) F1 females (B6CBAF1/OlaHsd; Harlan Olac). HdhQ150/Q150 homozygous mice (36, 37) on a (CBA × C57BL/6) F1 background were generated by intercrossing HdhQ150/Q7 heterozygous CBA/Ca and C57BL/6J congenic lines (inbred lines from Harlan Olac). All animals were subjected to a 12-hour light/12-hour dark cycle and had unlimited access to water and breeding chow (Special Diet Services). Housing conditions and environmental enrichment were as previously described (43). R6/2 mice were always housed with WT mice. R6/2 and HdhQ150/Q150 mice were genotyped and CAG repeat was sized as previously described (7). The mean repeat size (± SD) for all mice used in the entire study was 165 ± 10 for HdhQ150/Q150 mice and 204 ± 7 for R6/2 mice. Mouse brains or brain regions were snap-frozen in liquid nitrogen and stored at –80°C.
Behavioral assessment and phenotyping.
Mouse behavioral assessment was performed as previously described, with the specific exceptions that rotarod and activity assessment was conducted over the course of 300 seconds and 30 minutes, respectively. All data were collected and analyzed by repeated-measures general linear model ANOVA with Greenhouse Geisser post-hoc analysis as described previously (44).
HSP990 dosing.
HSP990, a 2-amino-7,8-dihdro-6H-pyrido[4,3-d]pyrimidin-5-one compound (34), was obtained from Novartis and can be purchased from Active Biochem. HSP990 can be formulated using either a propriety formulation solution from Novartis Pharma AG or 2% methyl cellulose diluted with 9 volumes of saline solution (water with 0.9% NaCl). The HSP990 vehicle mixture was briefly sonicated at high frequency in a water bath and mixed thoroughly to form a uniform suspension. Compound or vehicle alone was freshly prepared for each round of treatment and administered to mice by oral gavage, with thorough mixing performed between dosing to ensure the suspension remained even. Mice were weighed and dosed in the afternoon and typically dissected at the end of their light cycle. For all experiments, each treatment group contained age- and sex-matched mice.
Nuclear cytoplasmic fractionations.
Nuclear and cytoplasmic fractions were obtained from mouse half brains as previously published (69). The purity of isolated fractions was confirmed by immunoblotting with antibodies specific for α-tubulin and histone H3.
Antibodies.
See Supplemental Table 2 for primary and secondary antibodies. One of the anti-HSF1 antibodies used was a gift from R. Morimoto (Northwestern University, Evanston, Illinois, USA).
IP.
IP was performed as previously described (47). 2 mg protein isolated from half brains was incubated with 2 μg anti-HSF1 antibody (H311-X; Santa Cruz).
SDS-PAGE and immunoblotting.
Frozen mouse brain tissue was homogenized in ice cold-RIPA buffer (150 mM NaCl; 1% NP40; 0.5% Na deoxycholate; 0.1% SDS; 50 mM Tris-HCl, pH 8.0; 1 mM 2-mercaptoethanol; 1 mM PMSF; complete protease inhibitors [1 tablet per 10 ml; Roche]). Protein concentration was determined by BCA assay (Thermo Scientific), and 30 μg protein was added to 2× Laemmli loading buffer before being subjected to SDS-PAGE and Western blotting as described previously (36). Membranes were incubated with primary antibody overnight at 4°C in PBS with 0.02% Tween (PBST) and 0.5% nonfat milk for all antibodies except anti-HSP40, anti-HSP90, anti-GRP94, and anti-S830, which were incubated for 45 minutes at room temperature, and anti–α-tubulin and anti–histone H3, which were incubated for 20 minutes at room temperature. Blots were washed 3 times for 5 minutes in PBS with 0.2% Tween, incubated with secondary antibodies in PBST for 1 hour at room temperature, washed 3 times for 5 minutes in PBS with 0.2% Tween, and exposed to ECL according to the manufacturer’s recommendations (Amersham). Signal was developed using Amersham hyperfilm and a Xenograph developer.
Densitometry.
Densitometry of Western blots was performed using a BioRad GS-800 densitometer. Developed films were scanned, and the average pixel density (OD) for each band was measured. The OD of an area devoid of bands was subtracted from the values obtained for bands of interest in order to normalize OD against background. Relative expression was determined by dividing the normalized OD of bands of interest by the OD of α-tubulin for each sample.
Primers and probes for RT-qPCR.
See Supplemental Table 3.
Taqman RT-qPCR.
RNA extraction, cDNA synthesis, Taqman, and SYBR green RT-qPCR and ΔCt analysis were performed as previously described (70).
Affymetrix gene expression arrays.
Biotinylated cRNAs were prepared from 200 ng total RNA using the GeneChip 3′ IVT Express Kit (Affymetrix) following the manufacturer’s instructions. cRNA (15 μg) was hybridized to GeneChip Mouse Genome 430 version 2.0 Arrays (Affymetrix) and processed, stained, and scanned according to the manufacturer’s recommendations. The quality of input RNAs and cRNAs was verified with the Bioanalyzer 2100 (Agilent Technologies) before use in each step of the procedure. Microarray quality control was performed using the software package provided on RACE (71). Chips with a median normalized unscaled standard error greater than 1.05 were excluded. Affymetrix annotations (version 3.0) were used for probeset-to-gene assignments. 2-tailed t test was performed to assess the differences in gene expression between treated and untreated groups for each genotype (n ≥ 7 per treatment group). Corrections for multiple testing were performed using the false discovery rate (FDR) according to Benjamini and Hochberg (72), with a significance threshold of FDR P < 0.05. All microarray data were deposited in the National Center for Biotechnology Information (NCBI) gene expression omnibus (accession no. GSE29681).
ChIP.
Frozen half brains were homogenized in 1 ml PBS containing protease inhibitors (1 complete protease inhibitor tablet per 10 ml; Roche) and then spun at 500 g for 5 minutes to remove supernatant. Pellets were washed twice with PBS, resuspended in 1 ml of 1% formaldehyde, and incubated at 37°C for 10 minutes. Samples were pelleted and washed 3× with PBS before being resuspended in 1 ml ChIP lysis buffer (50 mM Tris-HCl, pH 8.0; 1% SDS; 10 mM EDTA). Samples were then sonicated at high frequency for 9 minutes (30 seconds on, 30 seconds off) with a Diogenode Bioruptor sonicator to yield chromatin fragments of 500–1,000 bp in size. After sonication, samples were spun at 10,000 g for 15 minutes at 4°C, and supernatant was retained. 200 μg chromatin was added to 20 μl blocked protein-G dynabeads (Invitrogen) with 2 μg antibody or rabbit IgG in 1 ml dilution buffer (17 mM Tris-HCl, pH 8.0; 167 mM NaCl; 1% Triton-X100; 1.2 mM EDTA). Samples were left to rotate overnight at 4°C. Beads were then washed twice in wash buffer 1 (20 mM Tris-HCl, pH 8.0; 150 mM NaCl; 0.1% SDS; 1% Triton-X100; 2 mM EDTA), wash buffer 2 (20 mM Tris-HCl, pH 8.0; 500 mM NaCl; 0.1% SDS; 1% Triton-X100; 2 mM EDTA), and wash buffer 3 (10 mM Tris-HCl, pH 8.0; 250 mM LiCl; 1% NP-40; 1% Na deoxycholate; 1 mM EDTA) before being resuspended in 120 μl elution buffer (1% SDS and 0.1 M NaHCO3). Beads were placed on a rotating wheel (SB3 rotating wheel; Stuart Instruments) for 20 minutes at room temperature before incubation at 65°C overnight in the presence of 0.2 M NaCl. Proteinase K digestion was then performed (1 μg per 250 μl proteinase K; 10 mM EDTA; 40 mM Tris-HCl, pH 6.8) at 45°C for 1 hour. Chromatin was purified using the Qiaquick min-elute PCR purification kit (QIAGEN) per the manufacturer’s instructions. 10% input chromatin was also de–cross-linked and purified as described above. SYBR green qPCR was then carried out on pulldown and input samples, and the ΔCt method was used to determine relative amounts of DNA.
MNase protection assay.
MNase assay was essentially performed as previously described (48), except that conditions were modified so as to obtain 100- to 500-bp fragments from 4 μg chromatin after MNase digestion. Briefly, chromatin was isolated from frozen mouse half brains as described above for ChIP. After mock or MNase digestion (500U MNase at 37°C for 30 minutes), DNA was purified as for ChIP, and SYBR green qPCR was carried out on digested and undigested DNA with primers specific for regions of the Hspa1b gene.
Seprion ligand ELISA.
Seprion ligand ELISA was performed as previously described (7).
TR-FRET.
Huntingtin TR-FRET was performed as previously described (46).
AGERA.
AGERA was essentially performed as previously described (42). Briefly, 100 μg protein in nonreducing Laemmli buffer was loaded onto a 1.5% agarose, 0.1 SDS gel and run at 250 mA until the dye front had reached the bottom of the gel. Western blotting and immunodetection with anti-huntingtin antibodies was then used to detect aggregate material.
Quantification of HSP990 in mouse brain tissue.
Mouse brain tissue was homogenized in 2 volumes of 2 Mm KH2PO4, pH 7.2, using an acoustic wave homogenizer (Covaris). A 50-μl aliquot of brain homogenate was spiked with 5 μl of a structurally related internal standard (CHIR623562) and then deproteinated by addition of acetonitrile (200 μl). After centrifugation, an aliquot of the supernatant was subjected to high-performance liquid chromatography/mass spectrometry/mass spectrometry (HPLC/MS/MS) analysis. Quantification of HSP990 in brain samples was performed by LC/MS/MS analysis using a CTC autosampler (CTC Analytics), a Rheos Allegro UHPLC pump (Flux/Thermo Fischer Scientific), and an LTQ Velos MS detector (Thermo Fischer Scientific). Chromatographic separation from interfering endogenous and exogenous contaminants was achieved on a Kinetex C18 LC column (2.6 μm, 2.1 × 50 mm; Phenomenex) using a multistep gradient with 1% formic acid and acetonitrile/methanol (1:1 v/v) supplemented with 1% formic acid (AMF) as solvents (0.0–3.5 minutes, 5%–80% AMF; 3.5–3.6 minutes, 80%–98% AMF). The column oven was set to room temperature. The flow from the HPLC system at 400 μl/min was directly introduced into the ion source of a LTQ Velos MS detector (ion trap mass analyzer) and subjected to heated electrospray ionization (HESI; positive ion mode). HSP990 was specifically detected using single reaction monitoring from its protonated quasimolecular ion [M+H]+ at m/z 380.2 Da to its daughter ion of m/z 363.3 Da, applying a collision energy of 41 eV. The chromatographic retention time under these conditions for HSP990 was 3.04 minutes. The internal standard eluted after 3.17 minutes. Quantification of brain levels of the parent compound was based on a 6-level calibration curve in brain homogenate (in triplicate) using a cubic equation after logarithmic transformation of the values. The dynamic range was set from 1.2 ng/g (LLOQ, lower limit of quantification) to 18.7 μg/g. The bias of all calibration standards and quality control samples were within the acceptance criteria of ±20.
Statistics.
P values were calculated by 2-tailed Student’s t test in Microsoft Excel. Statistical analysis of the mouse behavioral work was analyzed by GLM ANOVA in SPSS, and microarray data are described above. P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Angela Bithell and Noel Buckley for help with ChIP experiments; Grit Laue, Claudia Textor, and Sandrine Desrayaud for carrying out HSP990 PK analysis in mouse brain tissue; David Taylor for help with Affymetrix microarrays; and Donna Groom and members of the Neurogenetics lab for mouse breeding and CAG repeat sizing. We thank Dave Borchelt, Ron Kopito, Rick Morimoto, and Erich Wanker for insightful discussion. This work was supported by an MRC studentship to J. Labbadia and by funding from the MRC (801314), the CHDI Foundation, and the Huntington’s Disease Society of America Coalition for the Cure.
Footnotes
Conflict of interest: The R6/2 mice are licensed by King’s College London for commercial use. Andreas Weiss, Saliha Moussaoui, and Stefan Frentzel are employees of Novartis Pharma AG.
Citation for this article: J Clin Invest. 2011;121(8):3306–3319. doi:10.1172/JCI57413.
Paolo Paganetti’s present address is: AC Immune SA, Lausanne, Switzerland.
See the related Commentary beginning on page 2972.
References
- 1. Bates G, Harper P, Jones L, eds.Huntington’s Disease . 3rd ed. New York, New York, USA: Oxford University Press; 2002. [Google Scholar]
- 2.Novak MJ, Tabrizi SJ. Huntington’s disease. BMJ. 2010;340:c3109. doi: 10.1136/bmj.c3109. [DOI] [PubMed] [Google Scholar]
- 3.The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 4.Davies SW, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90(3):537–548. doi: 10.1016/S0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 5.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 6.Scherzinger E, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90(3):549–558. doi: 10.1016/S0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 7.Sathasivam K, et al. Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington’s disease. Hum Mol Genet. 2010;19(1):65–78. doi: 10.1093/hmg/ddp467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6(1):11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 9.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 10.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11(8):545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123(pt 22):3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 12.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 13.Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet. 2000;9(19):2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 14.Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23(4):425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 15.Kazemi-Esfarjani P, Benzer S. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 2000;287(5459):1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 16.Jana NR, Tanaka M, Wang G, Nukina N. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum Mol Genet. 2000;9(13):2009–2018. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- 17.Howarth JL, et al. Hsp40 molecules that target to the ubiquitin-proteasome system decrease inclusion formation in models of polyglutamine disease. Mol Ther. 2007;15(6):1100–1105. doi: 10.1038/sj.mt.6300163. [DOI] [PubMed] [Google Scholar]
- 18.Cummings CJ, et al. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum Mol Genet. 2001;10(14):1511–1518. doi: 10.1093/hmg/10.14.1511. [DOI] [PubMed] [Google Scholar]
- 19.Hay DG, et al. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet. 2004;13(13):1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 20.Zourlidou A, et al. Hsp27 overexpression in the R6/2 mouse model of Huntington’s disease: chronic neurodegeneration does not induce Hsp27 activation. Hum Mol Genet. 2007;16(9):1078–1090. doi: 10.1093/hmg/ddm057. [DOI] [PubMed] [Google Scholar]
- 21.Hansson O, Nylandsted J, Castilho RF, Leist M, Jaattela M, Brundin P. Overexpression of heat shock protein 70 in R6/2 Huntington’s disease mice has only modest effects on disease progression. Brain Res. 2003;970(1–2):47–57. doi: 10.1016/S0006-8993(02)04275-0. [DOI] [PubMed] [Google Scholar]
- 22.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94(4):471–480. doi: 10.1016/S0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 23.Kim HR, Kang HS, Kim HD. Geldanamycin induces heat shock protein expression through activation of HSF1 in K562 erythroleukemic cells. IUBMB Life. 1999;48(4):429–433. doi: 10.1080/713803536. [DOI] [PubMed] [Google Scholar]
- 24.Sittler A, et al. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington’s disease. Hum Mol Genet. 2001;10(12):1307–1315. doi: 10.1093/hmg/10.12.1307. [DOI] [PubMed] [Google Scholar]
- 25.Fujikake N, Nagai Y, Popiel HA, Okamoto Y, Yamaguchi M, Toda T. Heat shock transcription factor 1-activating compounds suppress polyglutamine-induced neurodegeneration through induction of multiple molecular chaperones. J Biol Chem. 2008;283(38):26188–26197. doi: 10.1074/jbc.M710521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 2010;8(1):e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auluck PK, Bonini NM. Pharmacological prevention of Parkinson disease in Drosophila. Nat Med. 2002;8(11):1185–1186. doi: 10.1038/nm1102-1185. [DOI] [PubMed] [Google Scholar]
- 28.Auluck PK, Meulener MC, Bonini NM. Mechanisms of suppression of {alpha}-synuclein neurotoxicity by geldanamycin in Drosophila. J Biol Chem. 2005;280(4):2873–2878. doi: 10.1074/jbc.M412106200. [DOI] [PubMed] [Google Scholar]
- 29.Waza M, et al. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat Med. 2005;11(10):1088–1095. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- 30.Katsuno M, et al. Pharmacological induction of heat-shock proteins alleviates polyglutamine-mediated motor neuron disease. Proc Natl Acad Sci U S A. 2005;102(46):16801–16806. doi: 10.1073/pnas.0506249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egorin MJ, Zuhowski EG, Rosen DM, Sentz DL, Covey JM, Eiseman JL. Plasma pharmacokinetics and tissue distribution of 17-(allylamino)-17-demethoxygeldanamycin (NSC 330507) in CD2F1 mice1. Cancer Chemother Pharmacol. 2001;47(4):291–302. doi: 10.1007/s002800000242. [DOI] [PubMed] [Google Scholar]
- 32.Vaughan CK, Neckers L, Piper PW. Understanding of the Hsp90 molecular chaperone reaches new heights. Nat Struct Mol Biol. 2010;17(12):1400–1404. doi: 10.1038/nsmb1210-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10(8):537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Machajewski T, Shafer C, McBride C, inventors; Novartis Pharma AG, assignee. 2–amino–7,8–dihydro–6H–pyrido[4,3–D]pyrimidin–5–one compounds. US patent 20070123546. May 31, 2007. [Google Scholar]
- 35.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. doi: 10.1016/S0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 36.Woodman B, et al. The Hdh(Q150/Q150) knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Res Bull. 2007;72(2–3):83–97. doi: 10.1016/j.brainresbull.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Lin CH, et al. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum Mol Genet. 2001;10(2):137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 38.Guertin MJ, Lis JT. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 2010;6(9):e1001114. doi: 10.1371/journal.pgen.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hockly E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci U S A. 2003;100(4):2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nekooki-Machida Y, Kurosawa M, Nukina N, Ito K, Oda T, Tanaka M. Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proc Natl Acad Sci U S A. 2009;106(24):9679–9684. doi: 10.1073/pnas.0812083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol. 2004;11(12):1215–1222. doi: 10.1038/nsmb860. [DOI] [PubMed] [Google Scholar]
- 42.Weiss A, et al. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington’s disease. J Neurochem. 2008;104(3):846–858. doi: 10.1111/j.1471-4159.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- 43.Hockly E, Woodman B, Mahal A, Lewis CM, Bates G. Standardization and statistical approaches to therapeutic trials in the R6/2 mouse. Brain Res Bull. 2003;61(5):469–479. doi: 10.1016/S0361-9230(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 44.Hockly E, et al. Evaluation of the benzothiazole aggregation inhibitors riluzole and PGL-135 as therapeutics for Huntington’s disease. Neurobiol Dis. 2006;21(1):228–236. doi: 10.1016/j.nbd.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Benn CL, et al. Genetic knock-down of HDAC7 does not ameliorate disease pathogenesis in the R6/2 mouse model of Huntington’s disease. PLoS One. 2009;4(6):e5747. doi: 10.1371/journal.pone.0005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss A, et al. Single-step detection of mutant huntingtin in animal and human tissues: a bioassay for Huntington’s disease. Anal Biochem. 2009;395(1):8–15. doi: 10.1016/j.ab.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Landles C, et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J Biol Chem. 2010;285(12):8808–8823. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134(1):74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong HK, et al. Blocking acid-sensing ion channel 1 alleviates Huntington’s disease pathology via an ubiquitin-proteasome system-dependent mechanism. Hum Mol Genet. 2008;17(20):3223–3235. doi: 10.1093/hmg/ddn218. [DOI] [PubMed] [Google Scholar]
- 50.Li M, Huang Y, Ma AA, Lin E, Diamond MI. Y-27632 improves rotarod performance and reduces huntingtin levels in R6/2 mice. Neurobiol Dis. 2009;36(3):413–420. doi: 10.1016/j.nbd.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 51.Bauer PO, et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28(3):256–263. doi: 10.1038/nbt.1608. [DOI] [PubMed] [Google Scholar]
- 52.Herbst M, Wanker EE. Small molecule inducers of heat-shock response reduce polyQ-mediated huntingtin aggregation. A possible therapeutic strategy. Neurodegener Dis. 2007;4(2–3):254–260. doi: 10.1159/000101849. [DOI] [PubMed] [Google Scholar]
- 53.Fritah S, et al. Heat-shock factor 1 controls genome-wide acetylation in heat-shocked cells. Mol Biol Cell. 2009;20(23):4976–4984. doi: 10.1091/mbc.E09-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomson S, Hollis A, Hazzalin CA, Mahadevan LC. Distinct stimulus-specific histone modifications at hsp70 chromatin targeted by the transcription factor heat shock factor-1. Mol Cell. 2004;15(4):585–594. doi: 10.1016/j.molcel.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Smith ST, et al. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6(2):162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- 56.Nucifora FC, Jr, et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291(5512):2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 57.Steffan JS, et al. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A. 2000;97(12):6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kulaeva OI, Hsieh FK, Studitsky VM. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc Natl Acad Sci U S A. 2010;107(25):11325–11330. doi: 10.1073/pnas.1001148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11(7):515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 60.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311(5766):1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 61.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106(35):14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300(5622):1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 63.Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37(8):894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young P, Anderton E, Paschos K, White R, Allday MJ. Epstein-Barr virus nuclear antigen (EBNA) 3A induces the expression of and interacts with a subset of chaperones and co-chaperones. J Gen Virol. 2008;89(pt 4):866–877. doi: 10.1099/vir.0.83414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun X, et al. Hsp90 inhibitors block outgrowth of EBV-infected malignant cells in vitro and in vivo through an EBNA1-dependent mechanism. Proc Natl Acad Sci U S A. 2010;107(7):3146–3151. doi: 10.1073/pnas.0910717107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130(6):1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feder JH, Rossi JM, Solomon J, Solomon N, Lindquist S. The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes Dev. 1992;6(8):1402–1413. doi: 10.1101/gad.6.8.1402. [DOI] [PubMed] [Google Scholar]
- 68.Fujimoto M, et al. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J Biol Chem. 2005;280(41):34908–34916. doi: 10.1074/jbc.M506288200. [DOI] [PubMed] [Google Scholar]
- 69.Davies SW, et al. Detection of polyglutamine aggregation in mouse models. Methods Enzymol. 1999;309:687–701. doi: 10.1016/s0076-6879(99)09045-x. [DOI] [PubMed] [Google Scholar]
- 70.Benn CL, Fox H, Bates GP. Optimisation of region-specific reference gene selection and relative gene expression analysis methods for pre-clinical trials of Huntington’s disease. Mol Neurodegener. 2008;3:17. doi: 10.1186/1750-1326-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Psarros M, Heber S, Sick M, Thoppae G, Harshman K, Sick B. RACE: Remote Analysis Computation for gene Expression data. Nucleic Acids Res. 2005;33(Web Server issue):W638–W643. doi: 10.1093/nar/gki490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.