Abstract
Collagen cross-linking mechanically strengthens tissues during development and aging, but there is limited data describing how force transmitted across cross-links affects molecular conformation. We used Steered Molecular Dynamics (SMD) to model perpendicular force through a side chain. Results predicted collagen peptides have negligible bending resistance and that mechanical force causes helix disruption below covalent bond failure strength, suggesting alternative molecular conformations precede cross-link rupture and macroscopic damage during mechanical loading.
Keywords: Collagen, Steered Molecular Dynamics, Enzyme Mechanokinetics, Tumor microenvironment
1. Introduction
Collagen is the major contributor to the mechanical properties of many tissues, and must resist in vivo mechanical forces and deformations. Individual collagen molecules are enzymatically cross-linked to each other to form micro fibrils, which aggregate and link to form sub fibrils, then larger fibrils and in some tissues continuing to assemble into larger structures, such as fascicles (Hansen et al., 2002; Haut, 1986). During aging, these collagen networks undergo spontaneous non-enzymatic chemical cross-linking through processes such as glycation, which results in covalent cross-links bridging reactive lysine and arginines (Bailey et al., 1998; Sell and Monnier, 2004; Verzijl et al., 2002).
Collagen contributes to the mechanical properties of many tissues throughout the body and must resist mechanical forces and deformations such as the expansion and contraction of blood vessels, tension across tendons and ligaments, and compression of cartilage in vivo. Velocities for these tissue deformations vary widely, on the order of 0.06 m/s in the aorta (Vitarelli et al., 2006), 1.0 m/s in the musculoskeletal system (In ‘t Veld and Stevens, 2008), and may exceed 20 m/s during limited traumatic or catastrophic events such as motor vehicle accidents or blast trauma (Wightman and Gladish, 2001).
Intramolecular hydrogen bonds and structural water surrounding collagen help to stabilize the three α-chains into the characteristic triple helical structure (Bella et al., 1995; Kramer et al., 1999). Intermolecular cross-links are formed between collagen molecules and orientated away from the long axis of the molecule to form bridges between amino acid side chains (Fig 1A). Reported X-ray diffraction data suggests that molecular gliding is a major part of fibril failure, and that these intermolecular cross-links are engaged when collagen fibers are in tension (Puxkandl et al., 2002). This implies that tension results in forces directed through the amino acid functional groups in directions that are non-parallel to the long axis of the collagen molecule.
Figure 1.
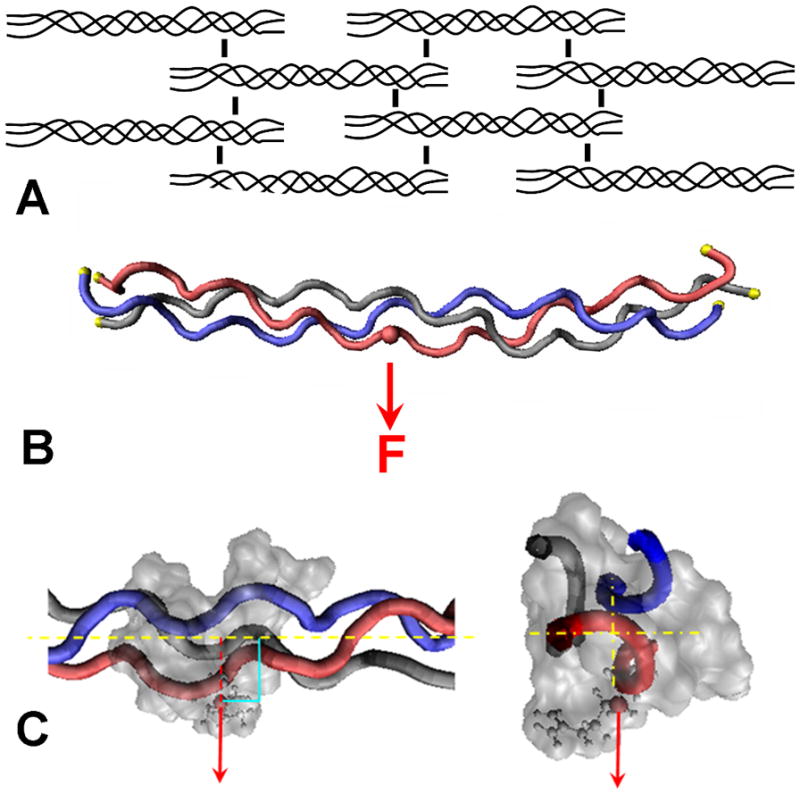
Schematic of the collagen network and location of applied force to a single chain.
(A) Simplified illustration of the collagen fibril network and the intermolecular cross-links between adjacent collagen molecules. (B) Steered Molecular Dynamics (SMD) model of the load applied to a single side chain. The C-α of each residue at the ends of the molecule were fixed in space, indicated with yellow spheres. The force, F, was applied to the C-β of an arginine residue (pseudo cross-link) through a theoretical spring (not shown). The force moved at a constant velocity in the direction indicated by the arrow. (C) Magnified view of the force applied to the C-β of the side chain, perpendicular and away from the long axis of the collagen molecule.
Experimental studies have measured mechanical properties such as the elastic modulus and stiffness of individual collagen molecules using optical beads (Luo et al., 1997) and atomic force microscopy (AFM) (Bozec and Horton, 2005). However, mechanical loading is currently incompatible with x-ray crystallography or nuclear magnetic resonance imaging techniques, resulting in limited details about the molecule’s conformation during mechanical loading. Recent in silico work by Uzel and Buehler described the role of intermolecular enzymatic cross-links between two lysines on the mechanical response of collagen during tensile loading (elongation). In their model the helical domain of one collagen molecule was connected to the telopeptide region of a second collagen molecule. During tensile elongation the telopeptide region was the first to fail (Uzel and Buehler, 2011). However, the effect of mechanical force in directions that are not parallel with the long axis of the molecule (tension) remains unclear.
The purpose of our study was to elucidate the effect of mechanical force acting through a side chain, directed perpendicular and away from the long axis. We hypothesized that mechanical force applied through a simulated cross-link would locally disrupt the native conformation and result in localized microunfolding of the triple helix. We tested our hypothesis by conducting SMD simulations to model the conformation of the collagen peptide when subjected to an external force applied through the C-β atom of an arginine. In this study, the force vector was directed perpendicular to the long axis, normal and away from the molecule’s surface as illustrated in Figure 1. This models transmission of mechanical force in a cross-linked structure through side chains and uses the arginine functional group because of the potential for these residues to be cross-linked during aging in vivo.
2. Results
2.1 Collagen Microunfolding
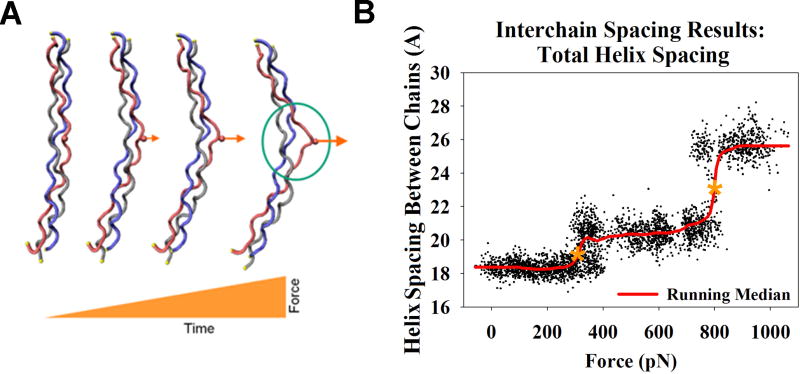
When the SMD results were visualized, a representative video is available as Animation 1 online, distinct conformations were observed. The initial structure was linear (straight) at t=0 and then obtained an overall bent configuration with no distinguishable change in helical structure as the force slowly increased. Further increases in force qualitatively correlated with intramolecular hydrogen bonds breaking and led to visual separation of the pulled chain from the other two chains of the triple helix with the pulled chain forming a loop (microunfolding) while the other two chains appeared to reverse their displacement and began to return to their approximate original linear positions.
The perpendicular force resulted in a distinct loop, with increased spacing between the chains which was readily observable by visual analysis of the trajectory. Quantitative data analysis indicated three specific force regimes; at very low forces the collagen peptide was linear and helical, followed by a regime where the peptide bent without unfolding, and then a regime where collagen bent and microunfolding occurred within the helix (Fig. 2A). To better resolve the microunfolding event we used the Interchain Spacing (ICS) analysis to measure helix geometry across repeated simulations (n=5) and plots were generated of helical spacing vs. applied force for each simulation (representative plot, Fig. 2B). Due to short timescale (<1 ps) fluctuations in the data, a smoothing function was applied using a running median.
Figure 2.
Results from a representative loading simulation.
(A) Representative images of the helical conformation at t=0 (left, unloaded) and as the force increases (right, illustrated with larger arrows). Region exhibiting microunfolding is indicated within the circle.
(B) Plot of Interchain Spacing (ICS) measurements as the force increases to bend and unfold the triple helix, indicated within the left and right rectangles respectively. ICS measured the distance between one C-α from each chain of the peptide backbone in a plane approximately perpendicular to the long axis of the molecule and near the loaded pseudo cross-link. A running median was fit to the data, and the magnitude of force was measured at the mid-point of the minor and major transitions (marked with an * in figure).
Based on the ICS analysis, we identified several regions that represented distinct conformational changes occurring within the triple helix as the force increased. In the first region (<350 pN) the collagen peptide bent until a distinct transition region (minor transition) was reached, concomitant with small changes in the triple helix spacing. Between approximately 350 and 900 pN a second region occurred in which the helix spacing increased as the collagen peptide continued to bend but the chains did not separate. At forces >900 pN a second major transition region occurred (microunfolding) and loss of the characteristic collagen triple helix conformation. The magnitude of force was measured at the mid-point of the minor and major transitions and averaged for the six different simulations, yielding transitions at 355 ± 43 pN and 904 ± 132 pN, respectively (mean ± standard deviation; range 305 pN to 430 pN and 778 pN to 1126 respectively).
2.2 Bending Stiffness
Based on the peptide length (L = 9 nm), applied bending force (F) and resulting bending displacement (d) of the collagen peptide, we calculated the peptide’s persistence length (ξ) from the bending stiffness (EI) as described by Buehler in a previous study (Buehler, 2006). In that model the persistent length ξ was calculated from the bending stiffness (EI) of the force-displacement measurements using the structural mechanics equation for simple three-point bending (simply supported beam with the ends pinned but free to rotate) given by
| (1) |
Since we fixed all three chains at both ends of the collagen peptide, we used the equation for bending of a beam rigidly fixed at both ends (ends not allowed to rotate) given by
| (2) |
where EIfree / EIfixed = 4.
Very early in the simulations before a significant amount of load is applied (F<10 pN) to cause peptide bending (d<0.5 A), in what appears to be an almost free solution regime, EIfixed = 5.49 × 10−29 N·m2 and the persistence length was calculated to be ξ = 13.3 nm. At bending displacements larger than d~0.75 A, the force-displacement response (F-d) of the peptide increased in a linear manner, EIfixed = 15.19 × 10−29 N·m2 and the bent or curved persistence length was calculated to be ξ = 36.7 nm. The solution-like value for ξ from the low bending displacement was in good agreement with the experimentally and computationally determined persistence length of collagen of ξ = 14.5 ± 7.3 nm (Sun et al., 2002) and ξ ≈ 16 nm (Buehler and Wong, 2007) respectively.
The persistence length of ξ = 36.7 nm at the larger bending displacement agreed less with the experimental value. However, this was not completely unexpected as this divergence likely reflected atomic interactions, conformational changes and local inhomogeneity within the collagen structure which would not considered in the structural mechanics solutions for beam bending. In addition our persistence length was also different from that calculated by Buehler (ξ = 23.4 nm) (Buehler, 2006) but this was probably due in part to the different methods used to calculate ξ (equations 2 and 1, respectively) and from the use of different boundary and loading conditions and sequence variations between the collagen peptides.
Of interest is that when we used the force and bending displacement values at the minor transition around 350 pN, the persistence length was calculated to be ξ ~ 800 nm. This likely reflects in part the inability of the structural mechanics equations for beam bending to adequately describe the molecular mechanics of these loading simulations, and also may reflect the mechanical role the collagen peptide plays as a structural element in physiologic structures.
3. Discussion
3.1 SMD discussion
Several recent computational studies have utilized SMD to model the effect of tensile mechanical forces on collagen conformation (Buehler, 2006; Gautieri et al., 2009; In ‘t Veld and Stevens, 2008; Lorenzo and Caffarena, 2005; Uzel and Buehler, 2011). Many of these studies have focused on the tensile properties of the single molecule utilizing proline rich collagen models (Buehler, 2006; Buehler and Wong, 2007; Lorenzo and Caffarena, 2005). However, due to the rigid nature of the proline and hydroxyproline residues’ structures, the generalizability of data predicted from these peptide models to the overall collagen structure is unclear. Lorenzo and Caffarena applied a linear force along the long axis of a collagen model to measure the Young’s modulus (Lorenzo and Caffarena, 2005). The structure used in that study, 1CAG: (Pro-Hyp-Gly)4-Pro-Pro-Ala-(Pro-Hyp-Gly)5, models the highly ordered proline rich triple helical regions of fibrillar collagen. Of interest, Lorenzo and Caffarena reported that the length of the unloaded 1CAG collagen peptide fluctuated in solution by 1.5–2% strain (Lorenzo and Caffarena, 2005). At the time, the authors did not specify the mechanism for these changes in molecular strain. We observed similar strain changes (2%–2.5%) with our equilibration (unloaded) structure which we attributed to bending of the structure in solution.
SMD studies were also performed using a second proline rich collagen peptide, 1QSU: (Pro-Hyp-Gly)4-Glu-Lys-Gly-(Pro-Hyp-Gly)5 (Buehler, 2006). Several types of molecular loading were simulated; including tension, compression, “three-point bending”, intermolecular shear between two molecules, and intramolecular shear between chains within one molecule. In the three-point bending simulations no internal unwinding was reported, in direct contrast to our microunfolding results. This difference may be related to differences in boundary (loading) conditions used, but more likely is due to the high proline content of the 1QSU collagen model. These results suggest sequence specificity or sequence dependence of these local microunfolding events.
In contrast to the two collagen models described above, the 1BKV structure we used contains a flexible region of collagen type 3 (Kramer et al., 1999). This structure represents flexible low proline regions of collagen and models the enzyme cleavage site (Kramer et al., 1999). Recent work by In ‘t Veld and Stevens used that structure to estimate the Young’s modulus of collagen in tension, and described strand separation due to a force perpendicular to the axis at the terminus (In ‘t Veld and Stevens, 2008). Other recent SMD studies used the same model to study tensile loading and reported that the tensile unwinding pathways exhibit a velocity dependence (Gautieri et al., 2009). Our work reported here provides additional details about local unwinding and the conformational response to mechanical forces applied in non-tensile directions.
3.2 Nanomechanics of crosslinks and structural failure
Due to the structural network in collagenous tissues, especially cross-links, tensile forces at the macro and micro levels appear to be transmitted via cross-links at the molecular level and likely result in mechanical loads being applied in a variety of different directions. Recent computational models by Buehler and colleagues have described microfibril tensile mechanics (Gautieri et al., 2011), the role of cross-linking in collagen fibrils (Buehler, 2008) and collagen-collagen interactions (Uzel and Buehler, 2011). These studies help provide new insights into the role of cross-links on stress transfer between molecules in tension. The fibril models emphasized the role of cross-links in resisting shearing between molecules (Buehler, 2008), while atomistic data further supported these findings and indicated that the telopeptide unwinding was a major feature in the collagen – collagen tensile properties (Uzel and Buehler, 2011).
We know that mechanical overload at tissue scales are capable of causing tissue rupture, most likely breaking the collagen structural network via an intermolecular shear or slippage mechanism (Puxkandl et al., 2002; Tang et al., 2010). Although not measured experimentally, predicted failure forces for collagen indicate globular telopeptide disruption at approximately 4,000 pN (Uzel and Buehler, 2011), failure within an α-chain requires approximately 7,000 pN and α-chain pullout requires approximately 11,500 pN (Buehler, 2006). Further, covalent bonds have been measured experimentally to fail at approximately 4,000 pN (Grandbois et al., 1999). These data support a model in which collagenous networks fail as the covalent cross-links break and collagen molecules then begin to slip (Buehler, 2006; Tang et al., 2010). It is therefore of great interest to note that the predicted minor and major microunfolding events identified in our study occur at forces well below these failure forces. Such conformational transitions may serve as an additional mechanism to store or dissipate mechanical energy in the helical regions of collagen. Further, these results suggest the possibility of mechanically induced conformational changes, or what we termed mechano-conformations, during routine loading and may provide new insights for published experimental observations.
3.3 Potential role of “mechano-conformations” in experimental biology
Several studies, including our own work, described a protective effect of tensile forces on degradation of collagen substrates by collagenases (Huang and Yannas, 1977; Nabeshima et al., 1996; Ruberti and Hallab, 2005; Wyatt et al., 2009). We termed the slowing of collagenase enzyme kinetics resulting from mechanical forces an enzyme mechanokinetic (EMK) affect, though the exact mechanism for this effect is still unknown. The computational data reported here further support a proposed EMK mechanism whereby mechanical forces induce a direct conformational change at the molecular level of the collagen substrate to effect enzyme-substrate interactions (Nabeshima et al., 1996; Ruberti and Hallab, 2005; Wyatt et al., 2009).
Collagen matrix stiffness was recently identified as the critical factor in converting an in vitro mammary epithelial cell cancer model to a malignant phenotype by an unknown mechanism (Baker et al., 2010). In addition, previous work has shown that stem cell differentiation can be affected by bulk collagen arrangement (gel vs. sponge) (Chen et al., 2003) or matrix stiffness (Hui et al., 2008). Our work suggests a proposed mechanism for these observations might be the presence of mechano-conformations providing differential cell-matrix attachment sites as collagen matrix mechanical properties change. If experimentally validated, this model may help provide new mechanistic insights into the role of matrix mechanics and tumor microenvironments on cellular mechanobiology.
4. Experimental procedures
4.1. Equilibration and SMD procedures
We downloaded the 1BKV pdb structure file from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (www.pdb.org), and then visualized and manipulated the file using VMD 1.8.6 (Humphrey et al., 1996). CHARMM chemical parameters (MacKerell et al., 1998) were revised to include treatment for hydroxyproline (In ‘t Veld and Stevens, 2008) and hydrogen atoms added to the structure using VMD.
The peptide was surrounded with a minimum of 1 nm of water on all sides, and Na+ and Cl− ions added to bring the solution to 15 mM. The total system of water, protein, and ions contained 18,267 atoms. Using NAMD 2.6 molecular dynamics software package (Phillips et al., 2005), the system was minimized using the included conjugate gradient minimization algorithm for 5,000 steps (Phillips et al., 2005). Hydrogen atoms were constrained using rigid bonds and a time step of 2 femtoseconds was employed to minimize computational time (Gautieri et al., 2009).
The protein was equilibrated at 310 Kelvin in several stages in an adiabatic (NVE) ensemble. First, the C-α atom of the first residue of chain A and last residue of chain C were fixed for 250 picoseconds (ps), released for the next 500 ps, and the peptide further equilibrated while free floating in solution. Due to the linear and flexible nature of the collagen peptide, the system underwent slow fluctuations of bending while maintaining its characteristic helical structure.
The structure at 750 ps was then used as a restart point for five separate 250 ps duration simulations restarts, representing a total duration of 1,000 ps of equilibration. This approach was used to obtain an equilibrated starting structure of collagen in an extended and unloaded conformation, thus avoiding the use of an external steering force to straighten the molecule. The stable equilibration trajectory was determined from the RMSD and coordinates of the resulting structure was used as the initial relaxed structure for the SMD simulations (Fig. 3).
Figure 3.
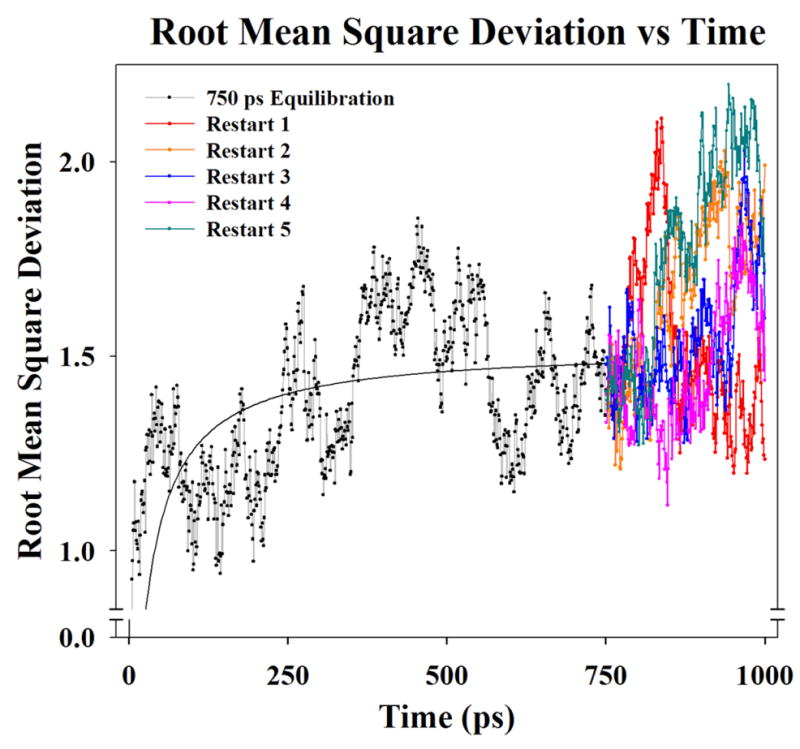
A plot of Root Mean Square Deviation showing peptide backbone movement during equilibration of collagen 1bkv peptide in solution. A two parameter hyperbola was fit to the data (solid black line from 0 – 750 ps) to help visualize the equilibration. The structure at 750 ps was then used as a restart point for five separate 250 ps simulations restarts (total duration of 1,000 ps of equilibration). Restarts were used to obtain an equilibrated starting structure of the unloaded collagen triple in a linear extended conformation without the use of an external steering force to straighten the molecule.
Equilibration simulations showed the collagen peptide’s general flexibility in the water bath. Visualizing individual trajectories at picosecond intervals illustrated that the overall peptide molecule slowly bent when placed unrestrained in a water box during equilibration. When correlated with these equilibration trajectories, the RMSD measurements of the peptide backbone also showed deviations from the average stable structure corresponding with the peptide bending in solution (Fig. 2).
In silico loading SMD simulations were performed at constant velocity and conducted in an isovolumetric and isothermal (NVT) ensemble as described previously in literature (Buehler, 2006). As a simplified model of the long range interactions on the peptide, the C-α atom of each of the N and C termini residues of all three chains were fixed in space to limit overall displacement of the entire structure (Fig. 1B). These residues would normally be connected to the neighbors within the triple helix in the overall linear structure, other structural cross-links, and external forces that would likely resist translation of the overall collagen molecule. The mechanical force, F, was applied to the C-β atom of an arginine side chain near the middle of the peptide, and was selected due to arginine’s involvement in cross-linking in vivo. The force was applied to this atom, designated a pseudo cross-link, through a theoretical spring (spring constant k = 1 kcal/mol/Å2) and was oriented perpendicular to and away from the long axis of the molecule (Fig. 1).
4.2. Interchain Spacing (ICS) Analysis
The complex conformational changes within the collagen peptide structure in space required an alternative analytical tool to describe local changes within the triple helix. We therefore used a geometric description of the atomic positions to analyze conformational changes, and have termed this approach an Interchain Spacing (ICS) analysis. In this analytic technique, we measured the spatial distances between the C-α’s of the peptide backbone of each of the three chains within a plane approximately perpendicular to the long axis of the molecule near the pseudo cross-link. When these distances are plotted as a function of time or force, distinct regions of the plot can be associated with local conformational changes within the structure and allowed us to achieve higher sensitivity to triple helix microunfolding than other measures such as Root Mean Square Deviation (RMSD) or force-displacement response of the peptide.
A potential limitation of the ICS analysis is the relatively low number of points, 1 per α-chain, used in this specific study. We opted to use fewer points to avoid potential confounding from various possible changes the structure might exhibit during loading. For example, tracking a volume (e.g., 2 or 3 points per α-chain) can capture elongation of the structure that would be difficult to separate from true microunfolding. In a best case scenario, higher spatial resolution might improve detection of the microunfolding events if separable from other structural events. Assuming separation of the events is possible, the threshold force values identified with higher resolution are expected to be similar to or slightly lower forces than those identified in this study but would not alter the conclusions drawn from the analysis used herein.
Supplementary Material
Force was applied to the C-β atom of an arginine side chain near the middle of the peptide, indicated by the red sphere. The C-α atom of each of the N and C termini residues of all three chains were fixed in space, indicated by the yellow spheres.
Acknowledgments
Support for this investigation was provided by the Weill Cornell Graduate School of Medical Sciences and Weill Medical College’s Clinical and Translational Science Center NIH-NCRR TL1RR024998 (J.W.B.) and NIH NIAMS R21AR051636 & R01AR45748 (P.A.T.). NAMD and VMD was developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06-RR12538-01 from the NIH-NCRR. We thank Sheela Damle for assistance in editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey AJ, Paul RG, Knott L. Mechanisms of maturation and ageing of collagen. Mech Ageing Dev. 1998;106:1–56. doi: 10.1016/s0047-6374(98)00119-5. [DOI] [PubMed] [Google Scholar]
- Baker EL, Lu J, Yu D, Bonnecaze RT, Zaman MH. Cancer cell stiffness: integrated roles of three-dimensional matrix stiffness and transforming potential. Biophys J. 2010;99:2048–2057. doi: 10.1016/j.bpj.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella J, Brodsky B, Berman HM. Hydration structure of a collagen peptide. Structure. 1995;3:893–906. doi: 10.1016/S0969-2126(01)00224-6. [DOI] [PubMed] [Google Scholar]
- Bozec L, Horton M. Topography and mechanical properties of single molecules of type I collagen using atomic force microscopy. Biophys J. 2005;88:4223–4231. doi: 10.1529/biophysj.104.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehler MJ. Atomistic and continuum modeling of mechanical properties of collagen: Elasticity, fracture, and self-assembly. J Mater Res. 2006;21:1947–1961. [Google Scholar]
- Buehler MJ. Nanomechanics of collagen fibrils under varying cross-link densities: atomistic and continuum studies. J Mech Behav Biomed Mater. 2008;1:59–67. doi: 10.1016/j.jmbbm.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Buehler MJ, Wong SY. Entropic elasticity controls nanomechanics of single tropocollagen molecules. Biophys J. 2007;93:37–43. doi: 10.1529/biophysj.106.102616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Revoltella RP, Papini S, Michelini M, Fitzgerald W, Zimmerberg J, Margolis L. Multilineage differentiation of rhesus monkey embryonic stem cells in three-dimensional culture systems. Stem Cells. 2003;21:281–295. doi: 10.1634/stemcells.21-3-281. [DOI] [PubMed] [Google Scholar]
- Gautieri A, Buehler MJ, Redaelli A. Deformation rate controls elasticity and unfolding pathway of single tropocollagen molecules. Journal of the Mechanical Behavior of Biomedical Materials. 2009;2:130–137. doi: 10.1016/j.jmbbm.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Gautieri A, Vesentini S, Redaelli A, Buehler MJ. Hierarchical structure and nanomechanics of collagen microfibrils from the atomistic scale up. Nano Lett. 2011;11:757–766. doi: 10.1021/nl103943u. [DOI] [PubMed] [Google Scholar]
- Grandbois M, Beyer M, Rief M, Clausen-Schaumann H, Gaub HE. How strong is a covalent bond? Science. 1999;283:1727–1730. doi: 10.1126/science.283.5408.1727. [DOI] [PubMed] [Google Scholar]
- Hansen KA, Weiss JA, Barton JK. Recruitment of tendon crimp with applied tensile strain. J Biomech Eng. 2002;124:72–77. doi: 10.1115/1.1427698. [DOI] [PubMed] [Google Scholar]
- Haut RC. The influence of specimen length on the tensile failure properties of tendon collagen. J Biomech. 1986;19:951–955. doi: 10.1016/0021-9290(86)90190-9. [DOI] [PubMed] [Google Scholar]
- Huang C, Yannas IV. Mechanochemical studies of enzymatic degradation of insoluble collagen fibers. J Biomed Mater Res. 1977;11:137–154. doi: 10.1002/jbm.820110113. [DOI] [PubMed] [Google Scholar]
- Hui TY, Cheung KM, Cheung WL, Chan D, Chan BP. In vitro chondrogenic differentiation of human mesenchymal stem cells in collagen microspheres: influence of cell seeding density and collagen concentration. Biomaterials. 2008;29:3201–3212. doi: 10.1016/j.biomaterials.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- In ‘t Veld P, Stevens MJ. Simulation of the Mechanical Strength of a Single Collagen Molecule. Biophys J. 2008;95:33–39. doi: 10.1529/biophysj.107.120659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer RZ, Bella J, Mayville P, Brodsky B, Berman HM. Sequence dependent conformational variations of collagen triple-helical structure. Nat Struct Biol. 1999;6:454–457. doi: 10.1038/8259. [DOI] [PubMed] [Google Scholar]
- Lorenzo AC, Caffarena ER. Elastic properties, Young’s modulus determination and structural stability of the tropocollagen molecule: a computational study by steered molecular dynamics. J Biomech. 2005;38:1527–1533. doi: 10.1016/j.jbiomech.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Luo ZP, Bolander ME, An KN. A method for determination of stiffness of collagen molecules. Biochem Biophys Res Commun. 1997;232:251–254. doi: 10.1006/bbrc.1997.6268. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins†. The Journal of Physical Chemistry B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y, Grood ES, Sakurai A, Herman JH. Uniaxial tension inhibits tendon collagen degradation by collagenase in vitro. J Orthop Res. 1996;14:123–130. doi: 10.1002/jor.1100140120. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puxkandl R, Zizak I, Paris O, Keckes J, Tesch W, Bernstorff S, Purslow P, Fratzl P. Viscoelastic properties of collagen: synchrotron radiation investigations and structural model. Philos Trans R Soc Lond B Biol Sci. 2002;357:191–197. doi: 10.1098/rstb.2001.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti JW, Hallab NJ. Strain-controlled enzymatic cleavage of collagen in loaded matrix. Biochem Biophys Res Commun. 2005;336:483–489. doi: 10.1016/j.bbrc.2005.08.128. [DOI] [PubMed] [Google Scholar]
- Sell DR, Monnier VM. Conversion of arginine into ornithine by advanced glycation in senescent human collagen and lens crystallins. J Biol Chem. 2004;279:54173–54184. doi: 10.1074/jbc.M408946200. [DOI] [PubMed] [Google Scholar]
- Sun YL, Luo ZP, Fertala A, An KN. Direct quantification of the flexibility of type I collagen monomer. Biochem Biophys Res Commun. 2002;295:382–386. doi: 10.1016/s0006-291x(02)00685-x. [DOI] [PubMed] [Google Scholar]
- Tang Y, Ballarini R, Buehler MJ, Eppell SJ. Deformation micromechanisms of collagen fibrils under uniaxial tension. Journal of The Royal Society Interface. 2010;7:839–850. doi: 10.1098/rsif.2009.0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzel SG, Buehler MJ. Molecular structure, mechanical behavior and failure mechanism of the C-terminal cross-link domain in type I collagen. J Mech Behav Biomed Mater. 2011;4:153–161. doi: 10.1016/j.jmbbm.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Verzijl N, DeGroot J, Ben ZC, Brau-Benjamin O, Maroudas A, Bank RA, Mizrahi J, Schalkwijk CG, Thorpe SR, Baynes JW, Bijlsma JW, Lafeber FP, TeKoppele JM. Crosslinking by advanced glycation end products increases the stiffness of the collagen network in human articular cartilage: a possible mechanism through which age is a risk factor for osteoarthritis. Arthritis Rheum. 2002;46:114–123. doi: 10.1002/1529-0131(200201)46:1<114::AID-ART10025>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Vitarelli A, Conde Y, Cimino E, D’Angeli I, D’Orazio S, Stellato S, Padella V, Caranci F. Aortic wall mechanics in the Marfan syndrome assessed by transesophageal tissue Doppler echocardiography. Am J Cardiol. 2006;97:571–577. doi: 10.1016/j.amjcard.2005.09.089. [DOI] [PubMed] [Google Scholar]
- Wightman JM, Gladish SL. Explosions and blast injuries. Ann Emerg Med. 2001;37:664–678. doi: 10.1067/mem.2001.114906. [DOI] [PubMed] [Google Scholar]
- Wyatt KE, Bourne JW, Torzilli PA. Deformation-Dependent Enzyme Mechanokinetic Cleavage of Type I Collagen. J Biomech Eng. 2009;131:051004. doi: 10.1115/1.3078177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Force was applied to the C-β atom of an arginine side chain near the middle of the peptide, indicated by the red sphere. The C-α atom of each of the N and C termini residues of all three chains were fixed in space, indicated by the yellow spheres.