Abstract
The recent development of light-activated optogenetic probes allows for the identification and manipulation of specific neural populations and their connections in awake animals with unprecedented spatial and temporal precision. This review describes the use of optogenetic tools to investigate neurons and neural circuits in vivo. We describe the current panel of optogenetic probes, methods of targeting these probes to specific cell types in the nervous system, and strategies of photostimulating cells in awake, behaving animals. Finally, we survey the application of optogenetic tools to studying functional neuroanatomy, behavior, and the etiology and treatment of various neurological disorders.
Optogenetics
The mammalian brain is composed of billions of neurons interconnected into circuits by trillions of synapses [1]. Some neural systems have been difficult to describe anatomically, such as the exquisitely complicated wiring of the cerebral cortex, while other neural systems are relatively well-characterized, such as the circuits that mediate vision, motor movements, breathing/respiration, and sleep/wake architecture. However, even these systems require functional dissection so that the relative contributions of individual cell types and their synaptic connections can be discerned. Perturbing one element in a neural circuit is especially difficult in vivo, where the complex environment of the brain in an awake, behaving animal imposes obstacles to the stimulation, inhibition, or manipulation of biochemical signaling events in specific cell types.
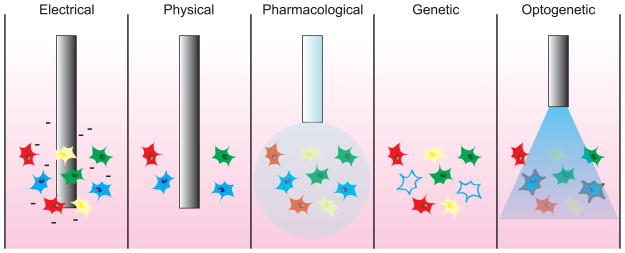
Traditionally, cells and synapses have been manipulated using electrical, physical, pharmacological, and genetic methods (Figure 1) [2]. Although much progress has been made using these classical techniques, considerable drawbacks prevent their use in the study of neural circuits with fine spatial and temporal precision in vivo. Electrical and physical techniques are not spatially precise and can cause stimulation, inhibition, or inactivation of surrounding cells and processes. Pharmacological and genetic methods exhibit improved spatial selectivity but lack temporal resolution at the scale of single action potentials.
Figure 1.
Strategies to manipulate neural activity in vivo. Cells in the nervous system reside in heterogeneous populations composed of different subtypes (here depicted as different colors). Electrical microstimulation techniques have high temporal precision but affect all cells and fibers surrounding the electrode. Physical probes irreversibly ablate or reversibly cool cells to reduce neural activity, but also affect all cells and fibers surrounding the electrode. Pharmacological injection of a drug might be able to target a particular cell-type based on cell-specific protein expression, but the drug could remain in the system for minutes or hours after injection. Genetic inactivation of specific cells (such as the cells depicted in blue) also lacks temporal precision and is often irreversible. Optogenetic stimulation or inhibition of cells using light allows for cell-type specific targeting of optogenetic probes (such as the cells depicted in blue) with millisecond-timescale precision of activation.
To overcome these limitations, a new set of tools collectively referred to as “optogenetics” [3] has been developed to precisely stimulate [4–10], inhibit [11–16], or alter biochemical activity [17, 18] in specific cells or their processes with high temporal precision and rapid reversibility. These probes are activated by light (“opto-“) and are genetically-encoded (“-genetics”), allowing for the direct control of specific populations of cells in vitro and in vivo (Figure 1) [19–23]. The unprecedented spatial and temporal precision of these tools has allowed substantial progress in elucidating the structure and function of previously intractable neural circuits.
This review highlights the use of optogenetic probes to investigate neural circuits in vivo. We survey the diversity of optogenetic tools in behavioral neuroscience, discussing common strategies of cell-type specific targeting and in vivo light delivery. We also describe common and theoretical methods for investigating neural circuits in awake, behaving animals and review some of the optogenetic studies that represent important progress in our understanding of neural circuits in normal behavior and disease. For detail on the discovery and biophysical characterization of optogenetic probes, please see other excellent reviews [24–26].
Optogenetic probes
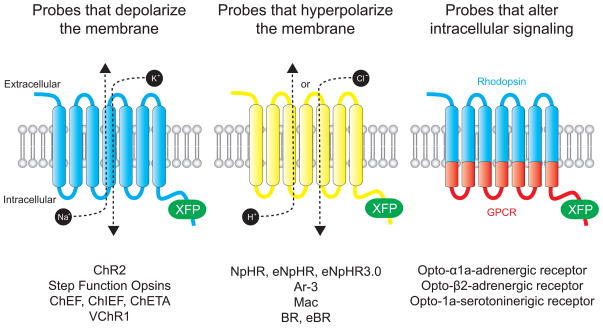
Although there are notable exceptions (Box 1), the most commonly-used optogenetic probes are engineered versions of natural opsins, light-sensitive membrane-bound proteins that translocate ions across the plasma membrane in response to stimulation by specific wavelengths of light [25]. These probes can be classified as either causing a membrane depolarization, membrane hyperpolarization, or a change in intracellular signaling (Figure 2).
Box 1. Nonrhodopsin-based optogenetic modulation of neurons.
Although the majority of optogenetic probes are engineered mutants of natural microbial-opsin proteins, an increasing number of nonrhodopsin-based probes are also enhancing our understanding of neural circuits [1, 25]. For example, SPARK (“Synthetic Photoswitchable Azobenzene-Regulated K+”) channels are modified Shaker potassium channels, genetically engineered to be coupled to a photoswitchable ligand [83]. Illumination with UV light promotes the flux of K+ ions through the channel, which hyperpolarizes the neuron and inhibits action potentials. Visible light blocks SPARK channels, allowing action potential firing. These channels represent the first use of a light-activated ion channel to regulate neural firing, although so far these channels have only been used in vitro.
Photoswitchable ligands have also been genetically combined with glutamate receptors to create LiGluR, a light-gated iontropic glutamate receptor [84]. Stimulation of LiGluR with 500 nm light activates the glutamate receptor, whereas 380 nm illumination turns it off. An alternative channel exhibits reversed activation spectra, with 380 nm light turning the receptor on and 500 nm light turning the receptor off [85]. This channel has been usedin vitroand in vivo[86]. For example, LiGluR was recently used in zebrafish to demonstrate that photostimulation of one neuron, the Kolmer-Agduhr cell, is sufficient to induce a symmetrical tail beating sequence that mimics spontaneous slow forward swimming [87]. Thus, LiGluR-mediated optogenetic modulation demonstrated that a single cell could act as a central pattern generator that generates periodic motor commands for rhythmic movements.
Figure 2.
Optogenetic probes. All optogenetic probes are based on opsins, seven transmembrane domain proteins that interact with a chromophore (retinal or vitamin A) to become light sensitive. Probes that depolarize the membrane act as nonspecific cation channels that open in response to pulses of light. Probes that hyperpolarize the membrane actively pump either Cl− ions (in the case of NpHRs) or protons in response to pulses of light. Probes that alter intracellular activity are chimeric proteins composed of rhodopsin in the extracellular and transmembrane domains and a GPCR in the intracellular domain.
Probes that depolarize the membrane
Channelrhodopsin-2 (ChR2) is a nonspecific cation channel naturally expressed by the green algae Chlamydomonas reinhardtii [10]. ChR2 absorbs blue light with an absorption peak of 480 nm, causing a conformational change from the all-trans-retinal chromophore complex to 13-cis-retinal [27]. This switch causes a subsequent conformational change in the transmembrane protein, opening the pore over 6Å and allowing H+, Na+, K+, and Ca2+ ions to passively diffuse across the membrane [10, 27]. Importantly, 13-cis-retinal relaxes back to the all-trans form within milliseconds, closing the pore and stopping the flow of ions into or out of the cell.
ChR2 has several features that make it particularly attractive as a neuroscience probe to depolarize neurons. First, the channel can be activated very rapidly and closes quickly upon light offset. Therefore, single action potentials can be generated with a brief pulse of blue light, with no long-lasting residual effects of stimulation. Second, standard cell culture media has sufficient retinal to allow for the ChR2 apoprotein to become a light-sensitive haloprotein, and retinal is already present in most vertebrate cells in the form of Vitamin A. Therefore, the addition of exogenous retinal is unnecessary when probing vertebrate neural systems. However, exogenous application is necessary for invertebrate systems. Finally, ChR2 is a genetically-encoded protein, allowing cell-specific targeting with characterized promoter and enhancer elements. Therefore, ChR2 does not need to be loaded into neurons via glass pipettes or iontophoresis like other photosensitive probes. Taken together, these properties have made ChR2 an extremely attractive probe for stimulating neurons with millisecond-precise temporal resolution [4, 9].
After the development and characterization of ChR2 as a neurobiological probe, two strategies were employed to expand the diversity of stimulatory optogenetic tools. One strategy was to search the genomes of other microbial organisms for light-regulated proteins. This approach led to the discovery of Volvox Channelrhodopsin-1 (VChR1) from the multicellular green algae Volvox carteri [5]. Instead of absorbing blue light, VChR1 absorbs light with a peak wavelength of 535 nm, allowing photostimulation of neurons with yellow light. Therefore, VChR1 penetrates deeper into neural tissue, exhibits lower spectral overlap with the Ca2+ indicator dyes used for neural imaging, and can be used in combination with ChR2 to differentially stimulate two neural populations in the same animal.
Another strategy used is genetic modification. For example, in some experiments it is desirable to stimulate neurons over long time periods (seconds to minutes) without constant blue light illumination that can cause undesirable heating in the brain. A series of three step-function opsins (SFOs) were created by mutating Cys 128 in ChR2 to Thr, Ala, or Ser [6]. These mutations effectively mutated ChR2 so that a single brief pulse of blue light causes a stable step in membrane potential for up to 30–60 seconds before the channel eventually closes. A single pulse of yellow light immediately closes the channel. Therefore, SFOs in neurons can be used to precisely turn on (and off) membrane depolarizations, avoid undesirable heating effects from long-term blue light stimulation, and even stimulate neurons at subthreshold levels to allow endogenous synaptic inputs to drive neural activity.
Other mutations to ChR2 seek to improve upon the channel’s inherent temporal limitations, including the occurrence of extra spikes in response to a single light pulse, the inability to fire above ~40 Hz, and the possibility of plateau potentials of 10 mV or more following rapid spike trains. Chimeric combinations of ChR2 and ChR1, another channel from C. reinhardtii [28], has led to the development of the ChEF channel, which undergoes only 33% inactivation during persistent blue light stimulation as compared with 77% for ChR2 [7]. The ChIEF channel is a further mutation of ChEF that accelerates the rate of channel closure while retaining reduced activation, leading to more consistent responses when stimulated above 25 Hz [7]. Finally, the ChETA channel is a mutation of ChR2 in which Glu 123 is replaced with Thr, profoundly reducing extra spikes in response to blue light, eliminating plateau potentials, and allowing for sustained spike trains up to at least 200 Hz [8].
At present, only ChR2 has been used in vivo, but future studies will utilize the larger diversity of stimulatory optogenetic probes. Furthermore, the discovery of additional light-activated cation channels and continued genetic engineering of variants of known channels with altered kinetic, spectral, and trafficking properties will expand the toolkit of optogenetic probes that can precisely affect membrane depolarization in response to various frequencies, wavelengths, and intensities of light.
Probes that hyperpolarize the membrane
The first microbial opsin to gain prominence as a tool for inhibiting neural activity [11, 12] was Halorhodopsin (NpHR), a light-driven Cl− pump naturally expressed by the halobacterium Natronomonas pharaonis [29–31]. NpHR actively pumps Cl− ions into cells in response to yellow light with peak absorption at 570 nm. Similar to ChR2, NpHR utilizes all-trans retinal as its chromophore and therefore can also be used in vertebrate systems without exogenous cofactors. Unlike ChR2, substantial mutagenesis was required in order to achieve high levels of expression in neurons. A second generation of NpHR, Enhanced Halorhodopsin (eNpHR), improved trafficking to the membrane by improving the signal peptide sequence and adding an endoplasmic reticulum export signal [13, 14]. Third generation constructs (eNpHR3.0) provided a 3-fold increase in photocurrent and 2-fold increase in membrane hyperpolarization over eNpHR due to the addition of membrane trafficking sequences [15].
Recently, additional proteins from bacteriorhodopsins have been discovered and characterized that can be used to inhibit neural activity in response to light. Instead of actively pumping Cl− ions into a cell, these proteins function as light-driven proton pumps. Archaerhodopsin-3 (aR-3, also called “Arch”) proteins are derived from Halorubrum sodomense and allow near-100% silencing of neurons in vivo in response to yellow light with efficiency comparable to eNpHR3.0 [16]. Two other bacterial rhodopsins, Mac proteins from Leptosphaeria maculans and Bacteriorhodopsin (BR) from Halobacterium salinarum (and its enhanced, second-generation derivative, eBR), allow silencing of neurons in response to blue-green light [15, 16]. Therefore, hyperpolarizing optogenetic tools now exist in the blue-green and yellow spectra of light, allowing for combinatorial dissection of two neural subtypes in the same preparation. High-throughput genomic screens will hopefully reveal additional channels that can increase the diversity of inhibitory optogenetic tools for future use.
Probes that manipulate intracellular signaling
In addition to electrical signals from ion channels, neurons are also modulated by intracellular signaling events initiated by cell surface receptors that culminate in not only a change in neuronal electrical activity, but also changes in gene expression, synaptic plasticity, and downstream protein cascades. Because rhodopsins belong to the G-protein coupled receptor (GPCR) family of seven-transmembrane domain receptors, it is theoretically possible to design synthetic rhodopsin/GPCR chimeras that combine the light responsive elements of rhodopsin with the biochemical signaling functionality of specific GPCRs [25, 32–34]. The first chimeras introduced light-sensitivity to the Gq-coupled α1a-adrenergic receptor and the Gs-coupled-β2-adrenergic receptor [17]. Termed Opto-XRs, these chimeric receptors demonstrated the potential to regulate appropriate signaling cascades in response to blue light [17]. Recently, this strategy has also been applied to develop an Opto-XR that precisely controls serotonin signaling through the 5-HT(1A) receptor [18]. Future chimeric proteins will hopefully allow for precise control of other GPCR signaling systems including neurotransmitter and neuropeptide receptors.
Taken together, these probes (Figure 2) collectively form an unprecedented toolbox for the genetic dissection of neural circuits. The utility of these probes is dependent on the specificity in which they are genetically targeted to specific neurons of interest. Therefore, we next describe optogenetic gene delivery strategies for in vivo targeting to specific neuronal populations.
Optogenetic gene delivery strategies
The most common method of delivering optogenetic transgenes into the nervous system is to infect cells with a replication deficient virus, typically a lentivirus or adeno-associated virus (AAV), that contains the transgene of interest driven by a short promoter or enhancer element [1]. Cell-type specific promoters that have been used to drive optogenetic transgenes include Ef1α (strong, ubiquitous expression) [35–37], SynapsinI (expression limited to neurons) [11], CamKII (expression limited to excitatory neurons) [4, 38], and GFAP (expression limited to astrocytes) [38]. There are a small number of short (<3kb) promoters that ensure targeting to specific cell-types in the brain, such as the ppHcrt promoter that targets hypocretin (Hcrt)-expressing neurons in the lateral hypothalamus [39], or the synthetic PRSx8 promoter that targets noradrenergic and adrenergic neurons that express dopamine beta hydroxylase [40, 41]. Unfortunately, the short promoters that fit in viral vectors (<3 kb) often do not retain cell-type specificity [1].
Optogenetic constructs with much longer promoter sequences can be utilized in mice generated with transgenic technologies. For example, the Thy-1 promoter is used to drive high expression levels of a transgene in a random subset of neurons [1]. Founders of these Thy-1 lines are selected for expression patterns based on the needs of the investigator. Several transgenic mouse lines [42, 43] and one transgenic rat line [44] carry ChR2 under the Thy-1 promoter. Other transgenic mouse lines use Thy-1 to drive expression of NpHR [14].
In utero electroporation can be used to introduce optogenetic transgenes at specific developmental timepoints. For example, transgenes can be delivered to specific cortical layers of the brain by electroporating mice at embryonic day E12.5 (layers V and VI), E13.5 (layer IV), or E15.5 (layers II and III) [22]. Several studies have used this approach to deliver ChR2 to specific cortical layers for subsequent photostimulation when the mice reach adulthood [20, 45, 46]. In utero electroporation can also be used to deliver optogenetic transgenes to inhibitory neurons of the striatum or hippocampus [1].
Viral, transgenic, and in utero electroporation strategies can be used in combination to overcome the weak transcriptional activity of most endogenous promoters [22]. For example, conditional AAV vectors that carry Cre-dependent transgene cassettes under the control of strong, ubiquitous promoters such as Ef1α have been developed that exploit numerous transgenic mouse lines from individual labs, GENSAT [47], and the Allen Brain Institute [48] that express Cre recombinase in specific cell types. The tissue specificity of optogenetic transgene expression is prescribed by the promoter elements driving Cre, whereas the efficiency of transgene expression is prescribed by the Ef1α promoter [49]. Multiple optogenetic studies have capitalized on this excellent system [35–37].
Finally, it is possible to specify expression of optogenetic transgenes using anatomical-based cell targeting [15]. Proteins such as wheat germ agglutinin or tetanus toxin fragment C exhibit anterograde- and retrograde-transport properties and can be fused to Cre recombinase to be transneuronally delivered to up- or down-stream neurons in other brain regions [1, 15, 22]. Thus, it is possible to restrict expression of optogenetic transgenes to specific cell types even if those cells do not express unique genetic regulatory elements.
The combination of viral, transgenic, physical, and anatomical gene targeting technologies provides scientists with multiple strategies for expressing optogenetic probes in specific neural populations. These technologies also allow for the expression of probes in non-traditional genetic model organisms, such as rats and primates. Often, the first step in optogenetic studies is to validate the correct expression of an optogenetic probe (tagged with a reporter protein) using histological techniques. The next step is to target the brain for in vivo light delivery, which we describe below.
Optogenetic light delivery strategies
In addition to targeting specific cell populations with optogenetic transgenes, it is necessary to deliver light. In vitro light delivery to cultured neurons or brain slices can be achieved using conventional light sources such as halogen/xenon arc lamps, light-emitting diodes (LEDs), and lasers, all of which can be directly coupled to a microscope’s light path [22]. In vivo light delivery is more challenging because surgical implants must be stereotaxically placed adjacent to target regions and therefore must penetrate the skull. Light must be delivered very close to target regions because mammalian brain tissue scatters light exponentially, with only ~10% of light intensity remaining at a distance ~500 μm from the light source [39, 50]. Furthermore, light delivery systems must not weigh more than a freely moving mouse can carry and should cause minimal disruption to natural behaviors.
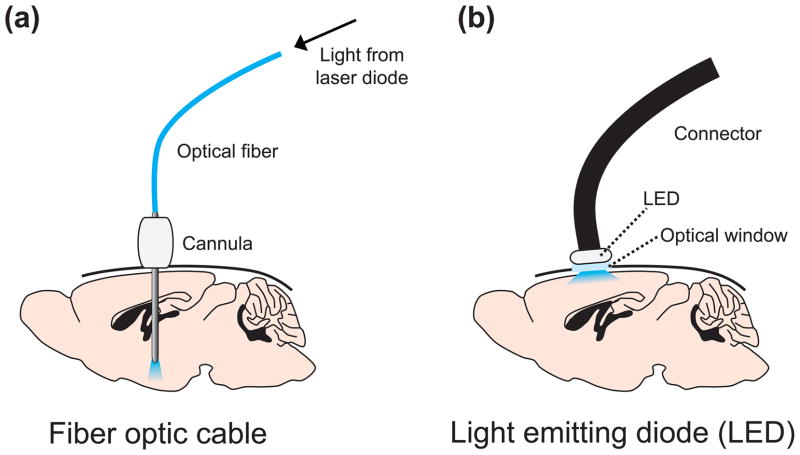
At present, the most common method for delivering light in vivo is to implant a guide cannula for placement of a lightweight fiber optic cable (Figure 3a) [50]. This strategy allows targeting of relatively deep brain structures. Mice can tolerate up to 300 μm diameter fibers and rats can tolerate up to 400 μm fibers. Optical fibers are typically connected to a laser diode, although it is also possible to connect to a LED. The challenge of using optical fibers within guide cannulas is that fiber breakage can result from repeated insertion through the cannula guide, resulting in a clogged cannula and loss of an animal subject. Therefore, many investigators now permanently implant a short fiber segment in the target region [22]. During experiments, this short fiber can be coupled to a longer fiber connected to a laser by a custom fiber-to-fiber connector. This strategy avoids fiber breakage and reduces the likelihood of infection due to environmental exposure through the cannula.
Figure 3.
Strategies to deliver light to transduced neurons in vivo. (a) A guide cannula is stereotaxically implanted above a target region for subsequent placement of an optic fiber. The fiber is attached to a laser diode, which might be attached to a computer or pulse generator for automatic stimulation protocols. (b) An optical window is stereotaxically implanted above a target region. A light-emitting diode (LED) is placed above the window to illuminate surface neurons and is connected to a computer for automatic stimulation protocols.
For optical modulation of superficial cortical neurons, small LEDs can be mounted above the brain over a cranial glass window (Figure 3b) [20, 51]. These LEDs are powerful enough to deliver light to all cortical layers and thin enough to not add significant weight to a mouse.
Additional detail about optogenetic probes, as well as technical details about gene and light delivery in vivo, can be found in other excellent reviews and protocols [19–23]. Detailed information about obtaining and using optogenetic transgenes can be found at the “Optogenetics Resource Center” website (http://www.stanford.edu/group/dlab/optogenetics/) maintained by the lab of Karl Deisseroth.
Strategies for studying neural circuits using optogenetics
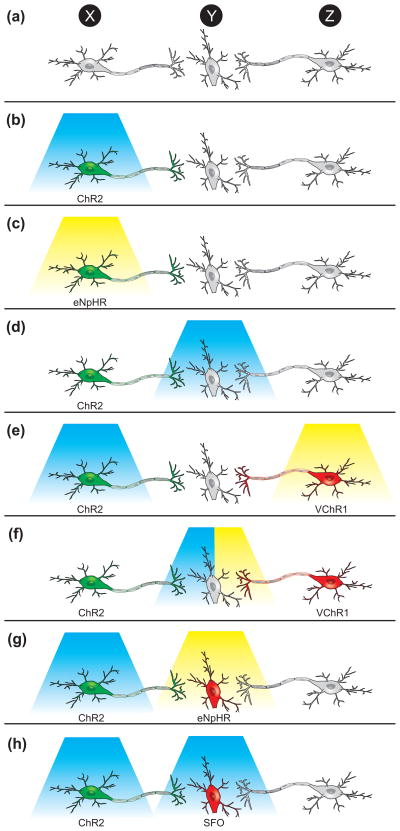
The simplest form of optogenetic experiments (although simple does not imply easy) involves transducing a population of neurons with an optogenetic transgene and investigating the effect of directly stimulating/inhibiting these cells on animal behavior or neural activity in a downstream brain region (Figure 4b,c). For example, photostimulation of Hypocretin-expressing neurons is sufficient to cause sleep-to-wake transitions [39], and microstimulation in the mouse barrel cortex can drive a sensory-induced learning task [51].
Figure 4.
Optogenetic strategies to investigate neural circuits in vivo. (a) A schematic of a neural circuit in which a population of neurons, Population X, projects monosynaptically to Population Y, which also receives projections from Population Z. (b) To determine the effect of stimulating X on behavior, Population X neurons could be transduced with ChR2 (or a variant) and stimulated with blue light. The effect of stimulating X on neural activity in Y could be determined by simultaneously placing an electrode in Y. (c) To determine the effect of inhibiting X on behavior, Population X could be transduced with eNpHR (or another inhibitory probe) and stimulated with yellow light. The effect of inhibiting X on neural activity in Y could be determined by simultaneously placing an electrode in Y. (d) To determine the effect of stimulating the projections of X onto Y on behavior or neural activity in Y, Population X could be transduced with ChR2 and light could be delivered to Y. The blue light will only activate ChR2 on the distal projections and thus the role of the specific synaptic connections can be probed. (e) To determine the relative contributions of activity in X or Z on behavior or activity in Y, Population X could be transduced with ChR2, Population Z could be transduced with VChR1, and X could be stimulated with blue light and Z with yellow light (f) To more specifically determine the relative contributions of activity in X or Z on activity in Y, Population X could be transduced with ChR2, Population Z could be transduced with VChR1, and the projections onto Y could be stimulated with either blue or yellow light while recording with an optrode. (g) To determine the necessity of Y on the ability of X to influence behavior, Population X could be transduced with ChR2, Population Y could be transduced with eNpHR, and X could be stimulated with blue light while Y is inhibited with yellow light. (h) To determine the effect of turning up the gain of activity in Y while stimulating X on behavior, Population X could be transduced with ChR2, and Population Y could be transduced with a step-function opsin (SFO) that converts a single light pulse into a graded increase in membrane potential. Then X could be stimulated while Y is stimulated to subthreshold potential with a single pulse of blue light.
Instead of stimulating the soma of cells transduced with optogenetic transgenes, it is often also possible to stimulate their distal projections (Figure 4d) to measure functional post-synaptic activity in downstream neurons. For example, the relative contributions of thalamocortical versus corticothalamic projections in feedforward excitation/inhibition circuits were studied by transducing thalamic neurons with ChR2 and stimulating their projections in the cortex, or transducing cortical neurons with ChR2 and stimulating their projections in the thalamus [52].
Stimulating fiber projections rather than somas can also be helpful in determining whether behavioral effects of stimulating a specific cell population are mediated by specific synaptic connections in a downstream population. For example, a recent study demonstrated that stimulation of either layer V motor neurons in the motor cortex or their afferent projections to the subthalamic nucleus is sufficient to relieve the symptoms of Parkinson’s disease [38].
If two neural populations project to the same area, it is theoretically possible to dissect their functional role in behavior and anatomy by using combinatorial optogenetics and transducing each with ChR2 or VChR1, as both optogenetic probes are activated by different wavelengths of light (Figure 4e,f).
Finally, it is also possible to investigate whether the gain of activity in one neural population affects the role of another population in mediating behavior or neural activity in downstream regions (Figure 4g,h). For example, if a population of neurons causes a behavior and projects to a downstream region, it might be possible to transduce the neurons with ChR2 and the downstream region with eNpHR. Thus, optogenetics could be used to stimulate one region while simultaneously inhibiting the other. Alternatively, the downstream region could be targeted with a SFO to increase the membrane potential to determine whether the gain of activity in this region affects ChR2-mediated effects of the upstream population.
Many of these strategies have been used to study functional neuroanatomy, behavior, and neurological ailments. We now survey many of these excellent studies to highlight the optogenetic modulation of neural circuits in vivo.
Optogenetic investigation of functional neuroanatomy
The first use of ChR2 as a neurobiological probe was to map anatomical connections in the brain by stimulating presynaptic neurons expressing ChR2 and measuring activity in postsynaptic neurons. This method was first demonstrated in three seminal studies that mapped connections between the olfactory bulb and piriform cortex [42], connections between different layers of cerebral cortex [43], and long-range callosal cortical projections from one hemisphere to the other [53].
These mapping techniques have revealed previously intractable information about functional neuroanatomy. For example, a class of polymodal nociceptive neurons necessary for transducing mechanical but not thermal pain was shown to connect with a broad range of second-order spinal cord neurons rather than preferentially targeting dedicated labeled-line neurons [54]. The hypothesis that synaptically-released glutamate could release GABA from GABAergic interneurons through direct action on GABAergic terminals was disproved, and the functional connectivity between glutamate presynaptic terminals and GABAergic somatodendritic integration was mapped with high spatial precision [45]. In a study of the functional role of adult neurogenesis, it was shown that newborn adult granule cells targeted with ChR2 form functional synapses with interneurons, mossy cells, and CA3 cells of the hippocampus [55]. Activity in horizontal projections from cortical neurons in layer 2/3 were shown to selectively suppress activity in superficial layers while activating deeper layers [46]. The organization of excitatory inputs to pyramidal neurons in layers 3, 5A, and 5B in somatosensory cortex was mapped with exquisite precision and sensitivity, revealing high specificity of functional connectivity from thalamus, motor cortex, and local excitatory interneurons [56].
These studies represent just a few of many examples of high-quality investigations of functional connectivity between different populations of neurons. Furthermore, the application of optogenetics to study connections in the thalamus, peripheral nervous system, spinal cord, cortex, and hippocampus demonstrates the utility of optogenetic experiments in virtually any location in the nervous system.
Optogenetic investigation of neural circuits in health and disease
Many studies have used optogenetic technology to investigate the neural circuitry that underlies mammalian behavior and the etiology of various neurological disorders. Optogenetic probes have also proved useful in studying nonmammalian systems (Box 2). Below, we highlight recent studies that demonstrate the utility of using optogenetics to answering questions about health and disease.
Box 2. Optogenetics in nonmammalian species.
In addition to mice, rats, and primates, optogenetic probes have proven highly useful in studying nonmammalian species as well, including Caenorhabditis elegans[88], Drosophila[89], and zebrafish [90, 91].
In Drosophila, ChR2 was ecpressed in neurons that express a transcription factor called “abnormal chemosensory jump 6” (acj6) [92]. Activity in these neurons was shown to be necessary for a wild-type startle response whereas photostimulation was sufficient to generate an escape response. Optogenetic probes have also been used to study behavior inDrosophilalarvae to show the neuronal basis of odor-aversion behavior [93] and the detection of nociceptive stimuli [94].
ChR2 has also been useful for dissecting the neural basis of behavior in zebrafish, including escape behaviors [95] and generation of saccades [96]. Recently, NpHR was used to localize swim command circuitry to a small hindbrain region [97]. The kinetics of hindbrain-generated control of swimming behavior was studied by the combination of loss-of-function experiments using NpHR and gain-of-function studies using ChR2, demonstrating the ability to interrogate neural circuitry at high spatial and temporal resolution, even in an aquatic species.
Retinal degeneration
Retinal degeneration is characterized by the progressive loss of rod and cone photoreceptors. Previous attempts to restore vision using prosthetic devices that electrically stimulate retinal cells have not yet yielded propitious results [44, 57]. Targeting ChR2 to retinal ganglion cells (RGCs) or bipolar cells could theoretically restore light sensitivity to mammalian models of retinal degeneration in which most rods and cones are genetically ablated [44]. Indeed, expression of ChR2 in either RGCs [58] or bipolar cells [59] can restore visual sensitivity to mice with mutations that cause rod and cone degeneration, as well as in rat models of retinal degeneration caused either by genetic mutation [60] or toxic light exposure [44].
Because photons of light naturally cause hyperpolarization of photoreceptors, NpHR can potentially be used in these cells to restore vision. Interestingly, expression of NpHR in the inner retinal layer can restore OFF responses whereas combined expression of NpHR and ChR2 in RGCs can produce ON, OFF, or even ON-OFF responses depending on the wavelength of light used [61]. The expression of eNpHR in light-insensitive cone photoreceptors (which are intermediate stages of retinal degeneration) can substitute for the native phototransduction cascade and restores light sensitivity in two mouse models of retinal degeneration [62]. Importantly, this treatment leads to normal activity in cone photoreceptors and RGCs in response to yellow light stimulation and allows mice to respond to changes in light intensity and the direction of motion.
In parallel, other studies have demonstrated the utility of using viral vectors to deliver optogenetic transgenes into mammalian retinal cells for long-term expression [63–65]. In human ex vivo retinas, eNpHR expression is non-toxic and can reactivate light-insensitive human photoreceptors [62]. These results demonstrate the promise of using optogenetics to treat various forms of blindness in humans, and various prosthetic devices, such as specialized glasses that increase light intensity, have been proposed to enhance environmental visual stimuli specifically for ChR2- or eNpHR-transduced neurons in the retina [57].
Parkinson’s disease
Parkinson’s disease is a neurodegenerative disorder caused by the selective loss of dopamine neurons in the substantia nigra pars compacta, resulting in muscle rigidity and a loss of physical movement. Two recent studies used optogenetic techniques to dissect the neural circuitry underlying parkinsonian neural circuitry in the basal ganglia [38, 66]. In one study, optogenetic probes were used to systematically drive or inhibit an array of distinct circuit elements in neurons, glia, and fiber projections in the subthalamic nucleus (STN) of freely moving rodent models of Parkinson’s disease [38]. Parkinsonian-like symptoms were relieved only by stimulating afferent axons projecting to the STN, demonstrating a mechanism by which deep brain stimulation might relieve symptoms in humans. In another study, the relative contributions of the dopamine D1-receptor and D2-receptor expressing neurons in the striatum were studied by selectively targeting each with ChR2 [66]. Stimulation of D1-expressing neurons in the striatum reduced parkinsonian-like symptoms in a mouse model of the disease whereas stimulation of D2-expressing neurons caused symptoms in wild-type mice. Taken together, these results enhance our understanding of the functional connections within the basal ganglia and present therapeutic strategies for ameliorating parkinsonian motor deficits, opening the possibility of using optogenetics in human patients [67].
Breathing/Respiration
Spinal cord or brainstem injury can lead to paralysis or, in serious cases, the inability to breath. However, the neural circuitry involved in mediating or repairing respiration is not fully understood. Following cervical dislocation in adult animals, ChR2-mediated photostimulation of phrenic motor neurons was sufficient to recover respiratory diaphragmatic motor activity [68]. In the brainstem, stimulation of the retrotrapezoid nucleus produced long-lasting activation of breathing [40] whereas stimulation of the ventrolateral medulla increased sympathetic nerve activity and blood pressure [41]. Interestingly, it was recently shown that astrocytes in the ventrolateral medulla directly sense changes in pH and regulate breathing by releasing ATP onto adjacent neurons [69]. These studies reveal the contributions of neural and non-neural cell types in the brainstem and spinal cord that regulate a central autonomic process.
Sleep/wake circuitry
The neural basis of the sleep/wake cycle involves a balance between activity in populations of neurons that promote arousal and those that promote sleep [70]. The first use of optogenetics to study these circuits used lentiviral gene delivery to target ChR2 to Hcrt-expressing neurons in the lateral hypothalamus [39]. Dysregulation of the Hcrt system, either the peptides or their receptors, causes the sleep disorder narcolepsy. Photostimulation of ChR2-transduced Hcrt neurons increased the probability of sleep-to-wake transitions [39] and increased neuronal activity in downstream wake-promoting nuclei [71].
Recently, it was also shown that optogenetic stimulation of the locus coeruleus (LC) produces immediate sleep-to-wake transitions while optogenetic inhibition causes a decrease in wakefulness [72]. Furthermore, sustained stimulation of the LC results in a behavioral state resembling cataplexy, a reversible period of muscle atonia that is a hallmark of narcolepsy. These results suggest that the LC maintains wakefulness but that overstimulation can cause behavioral arrests similar to those seen in neuropsychiatric disorders.
Reward
The neural circuits that mediate reward and addiction have been well-characterized, but the relative contributions of distinct tonic versus phasic activity patterns in participating brain structures, as well as the relative contributions of various neurotransmitter systems, have remained unknown [73]. Using optogenetics, selective phasic photostimulation of dopaminergic neurons in the ventral tegmental area was sufficient to drive behavioral conditioning whereas tonic activity was not [35]. Stimulation of dopaminergic neurons in the VTA co-released glutamate into the nucleus accumbens in addition to dopamine, demonstrating that mesolimbic reward signaling might involve glutamatergic transmission [74, 75]. Optical stimulation of α1-adrenergic receptors in the nucleus accumbens but not optical stimulation of β2-adrenergic receptors or ChR2-transduced accumbens neurons showed a robust increase in place preference during behavioral conditioning [17]. Future studies will probe this circuitry in paradigms of self-stimulation and addiction.
Gamma rhythms and cortical circuit performance
Cortical gamma oscillations (20–80 Hz) predict increases in focused attention, but the neural basis of these rhythms, as well as their role in mediating cortical circuit performance, remain unknown. ChR2-mediated photostimulation of Pv interneurons amplified gamma oscillations whereas eNpHR-mediated photoinhibition suppressed gamma oscillations [36, 37]. Furthermore, gamma-frequency modulation of excitatory input enhanced signal transduction in cortical regions, reducing circuit noise and amplifying circuit signals. These studies provide the first causal evidence that distinct network activity states can be induced in vivo by cell-type-specific activation of Pv neurons and suggest a potential mechanism for the altered gamma-frequency synchronization and cognition in schizophrenia and autism [36, 37].
In addition to the neurological studies described above, optogenetic probes have been used to investigate many other aspects of health and disease, including associative fear memory [76–78], epilepsy [79], the blood-oxygen level dependent (BOLD) effect during fRMI [80], and more. The application of optogenetics to primates [81] opens the possibility of using these tools to investigate higher-order cognitive tasks such as perception and decision-making.
Concluding remarks
Although optogenetic technology has only existed for 5 years, dozens of studies have utilized these tools to answer important questions about anatomy, behavior, and disease. These methods permit direct investigation and dissection of complicated neural circuits that would otherwise be intractable for in vivo study.
However, there is still substantial room for growth. For example, to allow for “combinatorial optogenetics” and the ability to stimulate multiple cell types in the same tissue preparation, it will be necessary to engineer a panel of channels that each respond to different wavelengths of light. This will be similar to the mutations made to green fluorescent protein (GFP) that allow variants of this protein to exhibit excitation and emission wavelengths across the visible spectrum of light [82]. To limit the potential side effects of heat, it will also be desirable to increase the conductance of various channels so that less light stimulation is necessary. To better mimic endogenous neural activity, it will be interesting to record natural patterns of neural firing and then use these recordings to “play-back” neural stimulation in patterns of light pulses. Finally, to use optogenetics therapeutically, it will be necessary to develop gene delivery strategies and light delivery implants that are amenable to human patients.
Even though there is room for growth, the contemporary optogenetic toolkit can be applied to many previously intractable questions ranging from what motivates us to eat and drink to the neural basis of complex decision making. The use of light to study the brain has proved to be a remarkably useful strategy, and indeed the future of optogenetics seems very bright.
Acknowledgments
M.E.C. received financial support from an NSF Graduate Research Fellowship and the NIH National Research Service Award (F31MH83439). L.d.L. is supported by the NIMH (MH083702), NIDA (DA021880), and NARSAD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luo L, et al. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter M, Shieh JC. Guide to research techniques in neuroscience. Elsevier/Academic Press; 2010. [Google Scholar]
- 3.Deisseroth K, et al. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyden ES, et al. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 5.Zhang F, et al. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat Neurosci. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berndt A, et al. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 7.Lin JY, et al. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunaydin LA, et al. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 9.Li X, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci U S A. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 12.Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS One. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gradinaru V, et al. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao S, et al. Improved expression of halorhodopsin for light-induced silencing of neuronal activity. Brain Cell Biol. 2008;36:141–154. doi: 10.1007/s11068-008-9034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Airan RD, et al. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 18.Oh E, et al. Substitution of 5-HT1A receptor signaling by a light-activated G protein-coupled receptor. J Biol Chem. 2010 doi: 10.1074/jbc.M110.147298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang F, et al. Channelrhodopsin-2 and optical control of excitable cells. Nat Methods. 2006;3:785–792. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- 20.Gradinaru V, et al. Targeting and readout strategies for fast optical neural control in vitro and in vivo. J Neurosci. 2007;27:14231–14238. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F, et al. Circuit-breakers: optical technologies for probing neural signals and systems. Nat Rev Neurosci. 2007;8:577–581. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- 22.Zhang F, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc. 2010;5:439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardin JA, et al. Targeted optogenetic stimulation and recording of neurons in vivo using cell-type-specific expression of Channelrhodopsin-2. Nat Protoc. 2010;5:247–254. doi: 10.1038/nprot.2009.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bamann C, et al. Microbial rhodopsins in the spotlight. Curr Opin Neurobiol. 2010;20:610–616. doi: 10.1016/j.conb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Kramer RH, et al. New photochemical tools for controlling neuronal activity. Curr Opin Neurobiol. 2009;19:544–552. doi: 10.1016/j.conb.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JY. A User’s Guide to Channelrhodopsin Variants: Features, Limitations and Future Developments. Exp Physiol. 2010 doi: 10.1113/expphysiol.2009.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bamann C, et al. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J Mol Biol. 2008;375:686–694. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 28.Nagel G, et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 29.Schobert B, Lanyi JK. Halorhodopsin is a light-driven chloride pump. J Biol Chem. 1982;257:10306–10313. [PubMed] [Google Scholar]
- 30.Lanyi JK. Halorhodopsin: a light-driven chloride ion pump. Annu Rev Biophys Biophys Chem. 1986;15:11–28. doi: 10.1146/annurev.bb.15.060186.000303. [DOI] [PubMed] [Google Scholar]
- 31.Bamberg E, et al. Light-driven proton or chloride pumping by halorhodopsin. Proc Natl Acad Sci U S A. 1993;90:639–643. doi: 10.1073/pnas.90.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karnik SS, et al. Activation of G-protein-coupled receptors: a common molecular mechanism. Trends Endocrinol Metab. 2003;14:431–437. doi: 10.1016/j.tem.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Pierce KL, et al. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 34.Kim JM, et al. Light-driven activation of beta 2-adrenergic receptor signaling by a chimeric rhodopsin containing the beta 2-adrenergic receptor cytoplasmic loops. Biochemistry. 2005;44:2284–2292. doi: 10.1021/bi048328i. [DOI] [PubMed] [Google Scholar]
- 35.Tsai HC, et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sohal VS, et al. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cardin JA, et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gradinaru V, et al. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324:354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adamantidis AR, et al. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abbott SB, et al. Photostimulation of retrotrapezoid nucleus phox2b-expressing neurons in vivo produces long-lasting activation of breathing in rats. J Neurosci. 2009;29:5806–5819. doi: 10.1523/JNEUROSCI.1106-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbott SB, et al. Photostimulation of channelrhodopsin-2 expressing ventrolateral medullary neurons increases sympathetic nerve activity and blood pressure in rats. J Physiol. 2009;587:5613–5631. doi: 10.1113/jphysiol.2009.177535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arenkiel BR, et al. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, et al. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proc Natl Acad Sci U S A. 2007;104:8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomita H, et al. Visual properties of transgenic rats harboring the channelrhodopsin-2 gene regulated by the thy-1.2 promoter. PLoS One. 2009;4:e7679. doi: 10.1371/journal.pone.0007679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hull C, et al. Neocortical disynaptic inhibition requires somatodendritic integration in interneurons. J Neurosci. 2009;29:8991–8995. doi: 10.1523/JNEUROSCI.5717-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adesnik H, Scanziani M. Lateral competition for cortical space by layer-specific horizontal circuits. Nature. 2010;464:1155–1160. doi: 10.1038/nature08935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gong S, et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atasoy D, et al. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aravanis AM, et al. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4:S143–156. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- 51.Huber D, et al. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cruikshank SJ, et al. Pathway-specific feedforward circuits between thalamus and neocortex revealed by selective optical stimulation of axons. Neuron. 2010;65:230–245. doi: 10.1016/j.neuron.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petreanu L, et al. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 54.Wang H, Zylka MJ. Mrgprd-expressing polymodal nociceptive neurons innervate most known classes of substantia gelatinosa neurons. J Neurosci. 2009;29:13202–13209. doi: 10.1523/JNEUROSCI.3248-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toni N, et al. Neurons born in the adult dentate gyrus form functional synapses with target cells. Nat Neurosci. 2008;11:901–907. doi: 10.1038/nn.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petreanu L, et al. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cepko C. Neuroscience. Seeing the light of day. Science. 2010;329:403–404. doi: 10.1126/science.1194086. [DOI] [PubMed] [Google Scholar]
- 58.Bi A, et al. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron. 2006;50:23–33. doi: 10.1016/j.neuron.2006.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lagali PS, et al. Light-activated channels targeted to ON bipolar cells restore visual function in retinal degeneration. Nat Neurosci. 2008;11:667–675. doi: 10.1038/nn.2117. [DOI] [PubMed] [Google Scholar]
- 60.Tomita H, et al. Channelrhodopsin-2 gene transduced into retinal ganglion cells restores functional vision in genetically blind rats. Exp Eye Res. 2010;90:429–436. doi: 10.1016/j.exer.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, et al. Ectopic expression of multiple microbial rhodopsins restores ON and OFF light responses in retinas with photoreceptor degeneration. J Neurosci. 2009;29:9186–9196. doi: 10.1523/JNEUROSCI.0184-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Busskamp V, et al. Genetic reactivation of cone photoreceptors restores visual responses in retinitis pigmentosa. Science. 2010;329:413–417. doi: 10.1126/science.1190897. [DOI] [PubMed] [Google Scholar]
- 63.Ivanova E, Pan ZH. Evaluation of the adeno-associated virus mediated long-term expression of channelrhodopsin-2 in the mouse retina. Mol Vis. 2009;15:1680–1689. [PMC free article] [PubMed] [Google Scholar]
- 64.Ivanova E, et al. Evaluation of AAV-Mediated Expression of Chop2-GFP in the Marmoset Retina. Invest Ophthalmol Vis Sci. 2010 doi: 10.1167/iovs.10-5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ivanova E, et al. Retinal channelrhodopsin-2-mediated activity in vivo evaluated with manganese-enhanced magnetic resonance imaging. Mol Vis. 2010;16:1059–1067. [PMC free article] [PubMed] [Google Scholar]
- 66.Kravitz AV, et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernstein JG, et al. Prosthetic systems for therapeutic optical activation and silencing of genetically-targeted neurons. Proc Soc Photo Opt Instrum Eng. 2008;6854:68540H. doi: 10.1117/12.768798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alilain WJ, et al. Light-induced rescue of breathing after spinal cord injury. J Neurosci. 2008;28:11862–11870. doi: 10.1523/JNEUROSCI.3378-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gourine AV, et al. Astrocytes control breathing through pH-dependent release of ATP. Science. 2010;329:571–575. doi: 10.1126/science.1190721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saper CB, et al. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 71.Carter ME, et al. Sleep homeostasis modulates hypocretin-mediated sleep-to-wake transitions. J Neurosci. 2009;29:10939–10949. doi: 10.1523/JNEUROSCI.1205-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carter ME, et al. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat Neurosci. 2010 doi: 10.1038/nn.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stuber GD. Dissecting the neural circuitry of addiction and psychiatric disease with optogenetics. Neuropsychopharmacology. 2010;35:341–342. doi: 10.1038/npp.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tecuapetla F, et al. Glutamatergic signaling by mesolimbic dopamine neurons in the nucleus accumbens. J Neurosci. 2010;30:7105–7110. doi: 10.1523/JNEUROSCI.0265-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stuber GD, et al. Dopaminergic terminals in the nucleus accumbens but not the dorsal striatum corelease glutamate. J Neurosci. 2010;30:8229–8233. doi: 10.1523/JNEUROSCI.1754-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ciocchi S, et al. Encoding of conditioned fear in central amygdala inhibitory circuits. Nature. 2010;468:277–282. doi: 10.1038/nature09559. [DOI] [PubMed] [Google Scholar]
- 77.Johansen JP, et al. Optical activation of lateral amygdala pyramidal cells instructs associative fear learning. Proc Natl Acad Sci U S A. 2010;107:12692–12697. doi: 10.1073/pnas.1002418107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Haubensak W, et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tonnesen J, et al. Optogenetic control of epileptiform activity. Proc Natl Acad Sci U S A. 2009;106:12162–12167. doi: 10.1073/pnas.0901915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee JH, et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 2010;465:788–792. doi: 10.1038/nature09108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han X, et al. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62:191–198. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 83.Banghart M, et al. Light-activated ion channels for remote control of neuronal firing. Nat Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Volgraf M, et al. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Biol. 2006;2:47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Numano R, et al. Nanosculpting reversed wavelength sensitivity into a photoswitchable iGluR. Proc Natl Acad Sci U S A. 2009;106:6814–6819. doi: 10.1073/pnas.0811899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Szobota S, et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–545. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 87.Wyart C, et al. Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature. 2009;461:407–410. doi: 10.1038/nature08323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo ZV, et al. Optical interrogation of neural circuits in Caenorhabditis elegans. Nat Methods. 2009;6:891–896. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang W, et al. A toolbox for light control of Drosophila behaviors through Channelrhodopsin 2-mediated photoactivation of targeted neurons. Eur J Neurosci. 2007;26:2405–2416. doi: 10.1111/j.1460-9568.2007.05862.x. [DOI] [PubMed] [Google Scholar]
- 90.Zhu P, et al. Optogenetic Dissection of Neuronal Circuits in Zebrafish using Viral Gene Transfer and the Tet System. Front Neural Circuits. 2009;3:21. doi: 10.3389/neuro.04.021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baier H, Scott EK. Genetic and optical targeting of neural circuits and behavior--zebrafish in the spotlight. Curr Opin Neurobiol. 2009;19:553–560. doi: 10.1016/j.conb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zimmermann G, et al. Manipulation of an innate escape response in Drosophila: photoexcitation of acj6 neurons induces the escape response. PLoS One. 2009;4:e5100. doi: 10.1371/journal.pone.0005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bellmann D, et al. Optogenetically Induced Olfactory Stimulation in Drosophila Larvae Reveals the Neuronal Basis of Odor-Aversion behavior. Front Behav Neurosci. 2010;4:27. doi: 10.3389/fnbeh.2010.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hwang RY, et al. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Douglass AD, et al. Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons. Curr Biol. 2008;18:1133–1137. doi: 10.1016/j.cub.2008.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schoonheim PJ, et al. Optogenetic localization and genetic perturbation of saccade-generating neurons in zebrafish. J Neurosci. 2010;30:7111–7120. doi: 10.1523/JNEUROSCI.5193-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arrenberg AB, et al. Optical control of zebrafish behavior with halorhodopsin. Proc Natl Acad Sci U S A. 2009;106:17968–17973. doi: 10.1073/pnas.0906252106. [DOI] [PMC free article] [PubMed] [Google Scholar]