Abstract
The catalytic subunit α isoform of protein phosphatase 2A (PP2Acα) activity, protein, and mRNA have been found increased in systemic lupus erythematosus (SLE) T cells and to contribute to decreased IL-2 production. The PP2Acα promoter activity is controlled epigenetically through the methylation of a CpG within a cAMP response element (CRE) motif defined by its promoter. We considered that hypomethylation may account for the increased expression of PP2Acα in patients with SLE. Using bisulfite sequencing, we found that SLE T cells displayed decreased DNA methylation in the promoter region compared with normal T cells. More importantly, we found that the CRE-defined CpG, which binds p-CREB, is significantly less methylated in SLE compared with normal T cells, and the levels of methylation correlated with decreased amounts of DNA methyltransferase 1 transcripts. Methylation intensity correlated inversely with levels of PP2Acα mRNA and SLE disease activity. Chromatin immunoprecipitation assays revealed more binding of p-CREB to the CRE site in SLE T cells, resulting in increased expression of PP2Acα. We propose that PP2Acα represents a new methylation-sensitive gene that, like the previously reported CD70 and CD11a, contributes to the pathogenesis of SLE.
Systemic lupus erythematosus (SLE) is an autoimmune disease primarily affecting women and is characterized by autoantibody formation against a host of nuclear Ags and immune complex deposition in multiple organ systems such as the kidney and blood vessels (1, 2). Although multiple genetic loci have been reported to be involved in determining SLE susceptibility, incomplete concordance in monozygotic twins who carry the same SLE-susceptibility genes suggests that environmental factors are also important for its pathogenesis (3–5). Established examples of exogenous agents affecting lupus include drugs like procainamide and hydralazine that cause lupus-like symptoms (6, 7) and exposure to UV light, which may initiate disease flares (8, 9). Several studies have shown that these agents may induce DNA demethylation, which plays an important role in transcriptional regulation by altering the accessibility of several transcription factors to the targeted gene-encoding promoters, genomic imprinting, and X-chromosome inactivation (10–13). Therefore, study of epigenetic mechanisms may provide important clues on how environmental factors may contribute to the expression of autoimmunity related pathology.
Indeed, several studies have suggested that impairment of DNA methylation may account for several T cell abnormalities in patients with SLE and to be involved in the pathogenesis of the disease (14). Treatment of normal T cells with DNA methylation inhibitor 5-azacytidine (5-azaC) induces overexpression of several methylation-sensitive genes, such as LFA-1 (CD11a/CD18) (15, 16), CD70 (17), which is known as a member of TNF superfamily member 7 as well as a ligand for B cell CD27, and CD40L (18), all of which are hypomethylated and overexpressed in T cells from SLE patients (19). Abnormal enhancement of costimulatory signaling pathways initiated or modulated by these molecules may contribute to autoimmunity. Some other methylation-sensitive genes, like perforin 1 (20), or cytokines (IL-4, IL-6, and IFN-g) have been implicated in the expression of autoimmune disease like SLE (21–25). In addition, defective signaling through ERK-1/ERK-2 in lupus T cells has been claimed to contribute to DNA hypomethylation (25–27) because of the reduction of DNA methyltransferase subsequently. Decreased expression of DNA methyltransferase 1 (DNMT1), which is responsible for the methylation of newly replicated daughter DNA strands during mitosis, has been also linked to hypomethylation and SLE expression (8, 27, 28).
The catalytic subunit of protein phosphatase 2A (PP2Ac) is overexpressed in SLE T cells (29). It is a highly abundant and ubiquitously expressed serine-threonine protein phosphatase in eukaryotic cells with various important roles including cell cycle progression and signal transduction (30–32). p-CREB, which is an important transcription factor in the regulation of the expression of IL-2, is a well-known PP2Ac substrate (33). We have shown that increased PP2Ac expression suppresses IL-2 production in SLE T cells by decreasing binding of p-CREB to IL-2 promoter (29). Recently, we identified a core promoter region of the α isoform of PP2Ac (PP2Acα), which contains a cAMP response element (CRE) motif flanked by three GC boxes. The fact that p-CREB can only bind to demethylated CRE motif in the PP2Acα promoter revealed that transcriptional regulation is tightly coordinated in an epigenetic manner (34).
In this study, we investigated DNA methylation patterns in the PP2Acα promoter region in SLE T cells and compared it to that in normal T cells. We show evidence that the PP2Acα promoter is hypomethylated in SLE T cells due to reduced DNMT1 expression and allows enhanced binding of p-CREB, which results in the overexpression of this molecule. Thus, PP2Acα represents a new methylation-sensitive gene, which, like the previously reported CD70, CD40L and CD11a, and probably others, contributes to the pathogenesis of SLE.
Materials and Methods
Patients and T lymphocyte purification
A total of 24 SLE patients (23 female and 1 male) who fulfilled at least 4 of the 11 revised criteria of the American College of Rheumatology for the classification of SLE (35) were enrolled in this study.
The demographics of the patients are shown in Table I. Nine patients were studied at different time points, and their demographics are shown in Table II. The patients shown in Table II were studied in multiple occasions. Thus, total sample number from lupus patients studied in this work is 34. SLE disease activity was assessed by the SLE disease activity index (SLEDAI) (36). Age ranged between 19 and 49 y (average: 35), and SLEDAI ranged between 0 and 36 (average: 7). We classified samples from patients into two groups: the high disease activity group was defined when the SLEDAI score was >6(n = 19), and the low disease activity group was defined when the SLEDAI score was ≤6(n = 15). Appropriate age-, ethnicity-, and sex-matched 16 healthy volunteers were also used as controls. Studies were approved by the Human Use Committee of our institution.
Table I.
Patient demographics and treatment
| Medications |
|||||
|---|---|---|---|---|---|
| Age (y) | Race | Sex | SLEDAI | Predonisolone (mg) | Others |
| 35 | Asian | F | 36 | 1200 (i.v.) | None |
| 38 | African American and Hispanic | F | 14 | 50 | H400 + M2 |
| 27 | White | F | 12 | 1.25 | M3 |
| 36 | African American | F | 8 | 0 | None |
| 27 | African American | F | 8 | 0 | H400 + M3 |
| 52 | White | F | 6 | 10 | None |
| 34 | African American | F | 6 | 30 | M3 |
| 29 | African American and Hispanic | F | 4 | 10 | H400 + M2.5 |
| 29 | African American | F | 3 | 2 | H400 + A125 |
| 44 | African American | F | 1 | 0 | H400 |
| 34 | White | F | 0 | 0 | None |
| 35 | White | F | 0 | 0 | MTX15 |
| 43 | White | F | 0 | 0 | H400 |
| 49 | White | F | 0 | 0 | None |
| 49 | African American | F | 0 | 17.5 | M3 |
A, azathioprine (mg); C, cyclosporine (mg); Cyc, cyclophosphamide (i.v.); F, female; H, hydroxychloroquine (mg); m, mycophenolate mofetil (g); MTX, methotrexate (mg).
Table II.
Time course of SLEDAI and treatment from nine different patients
| Medications |
|||||||
|---|---|---|---|---|---|---|---|
| Patient No. | Age (y) | Race | Sex | Date | SLEDAI | Predonisolone (mg) | Others |
| 1 | 34 | African American and Native American | Female | 09/15/2008 | 14 | 40 | M1 |
| 35 | 02/11/2009 | 10a | 5 | M1.5 | |||
| 35 | 04/16/2009 | 0 | 5 | M1 | |||
| 2 | 22 | African American | Female | 01/22/2009 | 35 | 50 | None |
| 06/26/2009 | 0 | 20 | H200 + Cyc | ||||
| 3 | 23 | Indian | Female | 10/15/2008 | 0 | 0 | H200 + A100 |
| 06/16/2009 | 4 | 0 | H200 + A100 | ||||
| 4 | 26 | African American | Female | 09/29/2009 | 10 | 40 | H400 + A100 |
| 27 | 03/30/2010 | 26 | 40 | M3 | |||
| 5 | 39 | African American | Female | 10/15/2008 | 8 | 10 | H400 + A100 |
| 03/18/2009 | 0 | 5 | H400 | ||||
| 6 | 24 | African American | Male | 02/17/2009 | 2 | 20 | H400 + A150 |
| 25 | 02/11/2010 | 12 | 5 | H400 + A150 | |||
| 7 | 52 | White | Female | 03/03/2009 | 14 | 0 | H400 |
| 04/01/2010 | 0 | 0 | H400 | ||||
| 8 | 19 | White | Female | 07/14/2009 | 12 | 40 | H400 + Cyc |
| 03/22/2010 | 4 | 0 | H400 + M2.5 + C200 | ||||
| 9 | 37 | White and Hispanic | Female | 03/25/2008 | 0 | 0 | H400 |
| 01/23/2009 | 10 | 40 | H400 | ||||
This sample was excluded in prospective analysis in Fig. 6.
A, azathioprine (mg); C, cyclosporine (mg); Cyc, cyclophosphamide (i.v.); H, hydroxychloroquine (mg); M, mycophenolate mofetil (g); MTX, methotrexate (mg).
CD3+ T lymphocytes were purified using a rosette T cell purification kit (Stem Cell Technologies) as described before (34). Subsequently, both RNA and DNA were extracted from T cells (3 × 106) using the AllPrep RNA/DNA/Protein mini kit (Qiagen) according to the manufacturer's protocol.
DNMTs plasmid transfection
After the purification of CD3+ T lymphocytes from normal individuals' peripheral blood, a plasmid encoding DNMT1 (Invivogen) and DNMT3a (Invivogen) or corresponding empty vector (pORF9) were transfected into primary T cells. The total amount of plasmid used in each sample was 6 μg per 6 × 106 T cells. Following transfection, 6 × 106 T cells were resuspended into 1 ml RPMI 1640 medium (Mediatech) supplemented with 10% heat-inactivated FBS (Quality Biological) and 2 mM L-glutamine, placed onto 12-well plates, and harvested after 24 h incubation for several experiments.
Bisulfite sequencing
Genomic DNA isolated from T cells of three lupus patients and three matched control subjects was bisulfite converted using the EpiTect kit (Qiagen) following the manufacturer's protocol. The fragments of 547 bp PP2Acα promoter were amplified by nested PCR and cloned into a pCR 2.1-TOPO vector using the TOPO TA Cloning kit (Invitrogen) after the PCR amplicons were gel purified. Ten clones from each sample were selected by blue/white screening, and plasmid DNA was extracted using the Qiagen Miniprep kit (Qiagen). Subsequently, sequence analysis was conducted to ascertain the methylation patterns of each locus. CpG within the sequences were mapped and labeled as methylated if cytosine remained unconverted and unmethylated if a thymidine was in a cytosine position. The percentage of methylation is calculated as the number of methylated cytosines divided by the total number of cytosines in all of the amplicons analyzed.
The following primers were used: round I, forward primer (−651 to −630): 5′-GGATTGTTAGGTTTTAGGTGAGG-3′ and reverse primer (+8 to +37): 5′-ATCCACTAATCCAACTCCTTAATAAACACCTTC-3′; and round II, forward primer (−517 to −495): 5′-TGGAATAGTGCTCAGGGATTAGT-3′ and reverse primer (+6 to +29): 5′-CAACTCCTTAATAAACACCTTCTC-3−.
Methylation-specific PCR
Genomic DNA (1 μg) from primary T cells was purified and treated with the methylation-sensitive enzyme AatII (New England Biolabs). After DNA repurification, 50 ng DNA was used as a template for PCR. To distinguish between methylated and nonmethylated status within the CRE motif in the PP2Acα promoter, two sets of primers were used. Forward primer F (−468) was the oligonucletotide: 5′-CCGCTAGCATGCTCCAGCTCCATCCTTC-3′. Reverse primer for methylation-sensitive product, R (−83), was the oligonucleotide 5′-CCAAGCTTGCCGGTTCCTCGTGTACTTCT-3′. Reverse primer for the control product, R (′286), was the oligonucleotide 5′-CGAAGCTTATGCCACCCGCCCCAG-3′. The AatII recognition site was defined within the fragment generated by F and R (−83), but not in those by F and R (−286). PCR products were electrophoresed on 1% agarose gels, visualized by ethidium bromide staining, and semiquantified with QuantityOne software.
Reverse transcription and real-time PCR
Total RNA (300 ng) was transcribed in cDNA in a conventional thermocycler using AMV reverse transcriptase and oligo(dT) primer (RT-PCR kit; Promega). Real-time RT-PCR was performed in duplicate for every sample with a LightCycler 480 System by adding SYBR green (Roche) to the reaction mixture. Primers used were: DNMT1 forward 5′-GTGGGGGACTGTGTCTCTGT-3′ and reverse 5′-TGAAAGCTGCATGTCCTCAC-3′; PP2Acα forward 5′-TCCGAGTCCCAGGTCAAGAG-3′ and reverse 5′-GCTACAAGCAGTGTAACTGTTTCA-3′; and GAPDH forward 5′-CAACTACATGGT TTACATGTTCC-3′ and reverse 5′-GGACTGTGGTCATGAGTCCT-3′. The averaged cycle threshold values of each reaction derived from the target gene, determined with LightCycler 480 System software (Roche), were normalized to GAPDH levels. Cycle threshold values were used to calculate relative mRNA expression by the ΔΔCt relative quantification method.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) analysis was done using ChIP Assay kit (Upstate Biotechnology) according to the manufacturer's instructions. T cells (3 × 106) were used per immunoprecipitating Ab. The cells were fixed with 1% formaldehyde for 10 min, lysed, and sonicated. The DNA–protein complexes were immunoprecipitated with anti–p-CREB Ab (Upstate Biotechnology) or its control Ab (rabbit normal IgG; Santa Cruz Biotechnology), and, after a series of washing steps, the cross-linking was reversed, and the protein was digested with proteinase K (Qiagen). The DNA was extracted by the QIAquick PCR Purification kit (Qiagen). Immunoprecipitated and purified DNA was analyzed by PCR using the primers F (−468) and R (−83) described as above, which are specific for the human PP2Acα promoter and were used to amplify a promoter fragment containing a CRE motif. The corresponding nonimmunoprecipitated DNA (input DNA) was also analyzed. PCR products were semiquantified using the same method as above in the methylation-specific PCR.
Statistics
Data are presented as mean value ± SEM. The paired two-tailed Student t test and the Pearson product moment correlation coefficient were used for statistical analysis. Statistical significance was defined as p < 0.05.
Results
Cotransfection of DNMT1 and DNMT3a into normal T cells reduced mRNA expression of PP2Acα by blocking p-CREB binding to methylated promoter region
We previously showed that 5-azaC, a DNA methylation inhibitor, influenced the methylation pattern in PP2Acα promoter region and induced increased activity (34). In this study, we examined whether DNA methyltransferase DNMT1 (maintenance methyltransferase) and DNMT3a (de novo methyltransferases) regulate PP2Acα expression by modifying the methylation pattern on the PP2Acα core promoter region. First, we confirmed that the DNA methylation levels at a specific site that is essential for the binding of transcription factor p-CREB were significantly elevated when the DNMTs were transfected into normal T cells (Fig. 1A). We compared the methylation effect in T cells with DNMT1 or DNMT3a alone or in combination. Cells transfected with both DNMTs produced the strongest methylation effect, and these were used in all subsequent experiments.
FIGURE 1.

Forced expression of DNMTs in human T cells causes DNA methylation and suppresses the expression of PP2Acα mRNA by blocking p-CREB binding to the promoter. A, Plasmid of DNMT1 (3 μg) and DNMT3a (3 μg) or corresponding empty vector (EV; 6 μg) were transfected into normal T cells and harvested after 24 h incubation. Extracted DNA was treated with methylation-sensitive restriction enzyme AatII, which recognizes only unmethylated CRE motifs, then PCR was conducted using two sets of primers to distinguish unmethylated or methylated status at −238 dC (sequence information shown in Fig. 2A). The methylated band (upper band, M) can be detected only when deoxymethylcytosine remains in CRE motif because it cannot be digested by AatII. The control bands (lower band, C) were generated by another set of primers defining an area of the PP2Acα promoter, which did not contain any AatII sensitive motifs. Each OD value of the bands was measured by densitometry. The ratio of the methylation-sensitive product to the control was calculated for the semiquantification of DNA methylation status within CRE of the promoter. Compared to control subjects (set as 1.0, shown as a white bar), the level of DNA methylation was significantly increased following overexpression of DNMTs (shown as a black bar). The results represent the mean ± SEM of five independent experiments. *p = 0.001. B, DNA binding of p-CREB to the PP2Acα promoter was examined using ChIP assays. T cells transfected with DNMT1 and DNMT3a or empty vector were fixed with formalin and sonicated. The anti–p-CREB Ab and its control Ab precipitates were subjected to PCR using PP2Acα promoter-specific primers and visualized on a 1% agarose gel. The corresponding nonimmunoprecipitated DNA (Input) was also analyzed using the same method. Five independent experiments were performed, and the results represent the mean ± SEM normalized to the input DNA. Compared to control (set as 1.0, shown as a white bar), p-CREB binding to the promoter was significantly decreased in T cells transfected with DNMTs (shown as a black bar). *p = 0.001. C, PP2Acα transcripts were measured and normalized to GAPDH by real-time RT-PCR. RNA was purified from T cells transfected DNMT1 and DNMT3a or empty vector. The expression levels of the transcripts were significantly decreased in T cells transfected with DNMTs (shown as a black bar) rather than in those from control subjects (set as 1.0, shown as a white bar). The results represent the mean ± SEM from six independent experiments. *p = 0.0027. D, DNMT1 transcripts in T cells from SLE patients (n = 34) or corresponding control subjects (n = 16) were measured and normalized to GAPDH by real-time RT-PCR. The samples from patients were divided into two groups according to the SLEDAI score. High disease activity was defined as a SLEDAI score of>6 (n = 15), and low disease activity was defined as a SLEDAI score of≤6 (n = 19). The expression levels of the transcripts were significantly reduced in T cells from active lupus patients (shown as a black bar) compared with those from normal subjects (shown as a white bar) or patients with inactive disease (shown as a gray bar). The results represent the mean ± SEM.
Next, we determined the levels of p-CREB binding to the CRE motif in the PP2Acα core promoter region in T cells transfected with DNMTs. As shown in Fig. 1B, using ChIP assay, we found that overexpression of DNMTs inhibited p-CREB binding to the region. We also quantified PP2Acα mRNA expression levels and found them decreased in cells transfected with DNMTs (Fig. 1C). These results showed that DNMTs have an important role in promoting hypermethylation in the promoter region and silencing the methylation-sensitive gene PP2Acα.
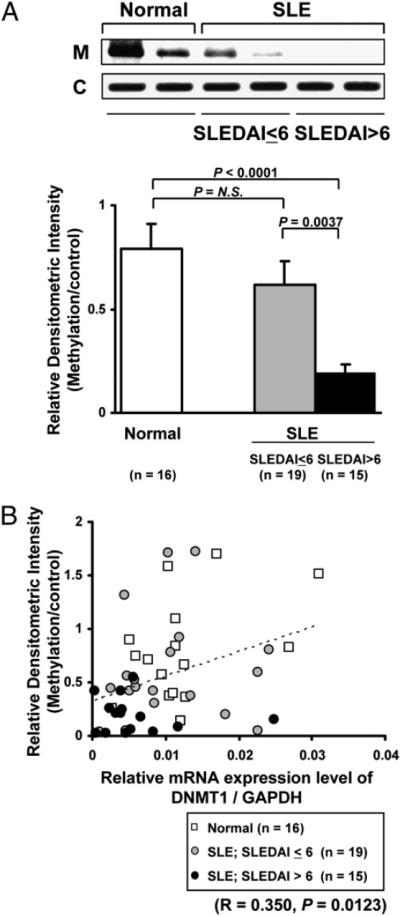
Several studies have demonstrated that DNMT1 is reduced in SLE patients (25–27), but we confirmed this observation using T cells from SLE patients and corresponding control subjects. The demographics of the patients used in the experiments are all shown in Tables I and II. We divided the samples from patients into two groups: the high disease activity group included patients with an SLEDAI score of >6, and the low disease activity group included patients with an SLEDAI score ≤6. Fig. 1D showed that T cells from patients with active disease have a reduced amount of DNMT1 compared with inactive patients or normal subjects. There was a negative correlation between SLEDAI and DNMT1 expression (r = −0.398, p = 0.0192; n = 34).
These data suggested that decreased levels of DNMT1 in SLE T cells may account for the global DNA hypomethylation and contribute to regulation and expression of the methylation-sensitive gene.
SLE T cells display a highly hypomethylated pattern around the CRE motif defined by the PP2Acα promoter
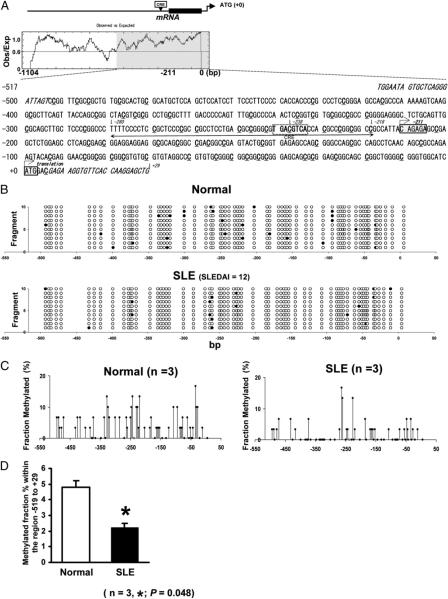
To investigate PP2Acα transcriptional mechanisms through epigenetic mechanisms, we focused on the essential regulatory region of this gene. As shown in Fig. 2A, the region that started at the −517 position from the translation initiation start site (ATG) was characterized by a high GC content and revealed the existence of several potential stable protein 1 transcription factor sites and a complete CRE motif located around the −238 position. In this region, there are 63 CpG. Among them, one of the CpGs is located at the center of the CRE motif (−238 position).
FIGURE 2.
Global hypomethylation within the essential PP2Acα promoter region in T cells from SLE patients. A, Nucleotide sequence within bp −517/+29 of the 5′-flanking region of the human PP2Acα gene. The translation initiation codon, ATG, is indicated as +0. The major transcription factor is located at −211 position. Each primer sequence for round II in nested PCR is shown in italics. The essential promoter region for gene regulation, which we previously determined is located from −280 to −218, is shown with arrows. At the center, there is a complete CRE motif surrounded by dashed box where the transcription factor CREB/p-CREB can bind. Whole region qualifies as a CpG island using CpG finder at the European Bioinformatics Institute (http://www.ebi.ac.uk/). All dC in CpG included in this region are highlighted with underlining. Within this region, there are 63 dCs in CpG. Among them, a dC in CpG included CRE motif exists at −238 position. B, One example of PP2Acα promoter methylation pattern in normal (upper panel) or SLE T cells (lower panel). T cells were isolated from a lupus patient and a matched control individual, and DNA was treated by sodium bisulfite. The region shown in Fig. 1A was amplified by PCR, cloned, and 10 colonies were picked up and sequenced from each subject. The fragment number is shown on the y-axis and the location of each CpG pair on the x-axis. Open circles, unmethylated cytosines; closed circles, methylated cytosines. C, Summary of the promoter DNA methylation pattern in T cells isolated from three different normal individuals (left panel) or three SLE patients (right panel). The value of y-axis showed the mean fraction methylated of 30 cloned and sequenced fragments from three different individuals. Open circles showed a dC at −238 position, which located in a CpG within a CRE motif of the core promoter region. Both patterns showed hypomethylated status, but lupus T cells showed higher demethylation of DNA, especially around core promoter region. D, Overall methylation percentage within the promoter region. The results represent the mean 6 SEM of three independent experiments. *p = 0.048.
We first searched for differences in the DNA methylation pattern between T cells from three SLE patients and three matched control subjects. By bisulfite sequence analysis, both groups showed a global hypomethylated pattern; however, SLE T cells displayed a relatively lower methylation pattern than normal T cells, especially around the CRE motif shown in Fig. 2B and 2C. The total percentage of DNA methylated fragments in this region (−519 to +29) was significantly decreased in SLE T cells compared with normal T cells shown in Fig. 2D.
This method, although very useful in determining global gene methylation patterns, is limited when quantification is required. Therefore, we used a methylation-sensitive PCR to quantify the levels of DNA methylation at the CpG in the CRE motif (−238 position). Briefly, we used the methylation-sensitive restriction enzyme AatII, which recognizes only unmethylated CRE motifs, and applied PCR using two sets of primers to distinguish unmethylated and methylated status at this deoxycytosine (dC) in the CRE motif. A band can be detected only when deoxymethylcytosine exists in the CRE motif because it cannot be digested by AatII. A control band was generated by another set of primers defining an area of the PP2Acα promoter, which did not contain any AatII-sensitive motifs. We measured the densitometric intensity for each band, and the ratio of the methylation-sensitive product to the control was calculated to semiquantify DNA methylation status within the CRE of the promoter. As shown in Fig. 3A, T cells from SLE patients had significantly lower levels of DNA methylation at this nucleotide than those from healthy controls. This was especially true among patients with high (SLEDAI >6) disease activity who had significantly lower methylation status compared with patients with low SLEDAI activity (≤6) and healthy controls. We could also find a moderate negative correlation between SLEDAI and DNA methylation level (r = −0.352, p = 0.040; n = 34).
FIGURE 3.
Quantification of DNA methylation levels at the CpG within a CRE motif of PP2Acα promoter in SLE and normal T cells. A, Each DNA sample isolated from SLE or normal T cells were conducted with methylation-sensitive PCR by the same method in Fig. 1A. We analyzed DNA methylation level at the specific base pair by dividing OD value of the methylated band (upper band, M) into control bands (lower band, C). We purified DNA sample from T cells from the exact same individuals who were used for mRNA quantification in Fig. 1D. We divided lupus samples by the same method in Fig. 1D. Compared to normal subjects (shown as a white bar) or inactive patients (shown as a gray bar), the level of DNA methylation was significantly reduced in active lupus patients (shown as a black bar). The results represent the mean ± SEM. B, Correlation between the DNMT1 transcript levels and DNA methylation levels at a CpG in CRE motif within the promoter. The x-axis represents the relative expression level of DNMT1 transcript shown in Fig. 1D, and y-axis represents the DNA methylation levels shown in Fig. 3A. There was a positive correlation between these two parameters with significant difference. p = 0.0123; r = 0.350; n = 50.
In addition, we determined whether the expression levels of DNMT1 reflected DNA methylation levels at the CpG within the PP2Acα promoter (Fig. 3B). The data show that increases in SLE disease activity result in reduction of DNMT1 expression and hypomethylation of the PP2Acα promoter, especially at the site that defines the CRE.
p-CREB binding to the CRE motif in the PP2Acα promoter was stronger in lupus T cells than in normal T cells
Next, we investigated predicted differences of p-CREB binding to the PP2Acα promoter in SLE and normal T cells. ChIP assays revealed that p-CREB bound to PP2Acα promoter more intensely in SLE T cells compared with normal T cells (Fig. 4). Previously, we had noted increased p-CREB binding to the PP2Acα promoter (area defining the CRE motif) in T cells treated with the DNA methylation inhibitor 5-azaC (33), suggesting that disease flares initiate a process that leads to inhibition of DNA methylation. In this study, we confirmed the same phenomena between SLE T cells and T cells treated with DNA methylation inhibitor.
FIGURE 4.

Enhanced p-CREB binding to CRE motif in the promoter in T cells from SLE patients. DNA binding of p-CREB to the PP2Acα promoter was examined using ChIP assays. The method detail is as in Fig. 1B. Each band showed the binding ability to PP2Ac promoter. Four independent experiments were performed, and the results represent the mean ± SEM normalized to the input DNA. Compared to normal T cells, p-CREB binding to the promoter was significantly increased in SLE T cells. *p = 0.016.
The levels of the PP2Acα transcript are increased in T cells from patients with active SLE and reflects DNA methylation of CRE
Previously, we had determined that mRNA and protein levels of PP2Ac were higher in SLE T cells compared with normal T cells, yet we had failed to show a correlation with disease activity (29). In this study, we focused on the major isoform of PP2Ac, PP2Acα, and re-examined the expression levels of its transcripts. As shown in Fig. 5A, the expression levels of PP2Acα mRNA were significantly higher in the patient group with a high SLEDAI score (>6), but not in the group of patients with a low SLEDAI score (≤6) compared with normal controls. There was a positive correlation between SLEDAI and PP2Acα expression level (r = 0.369, p = 0.0308; n = 34). In addition, we noted a significant negative correlation between the levels of transcripts and the levels of DNA methylation (Fig. 5B). Our data, taken together, indicate that disease activity affects DNA methylation status within the PP2Acα promoter, which can result in the upregulation of its expression.
FIGURE 5.

DNA methylation levels at a CpG within a CRE motif of PP2Acα promoter affected the expression level of PP2Acα mRNA. A, PP2Acα transcripts were measured and normalized to GAPDH by real-time RT-PCR. These RNA samples are same in the analysis of DNMT1 transcript level in Fig. 1D. The expression levels of the transcripts were significantly increased in T cells from active lupus patients (shown as a black bar) rather than in those from normal subjects (shown as a white bar) or inactive patients (shown as a gray bar). B, Correlation between the transcript levels and DNA methylation levels at a CpG in CRE motif within the promoter. The x-axis represents the DNA methylation levels shown in Fig. 3A, and y-axis represents the relative expression level of transcript shown in Fig. 5A. There was a negative correlation between these two parameters with significant difference. p = 0.0101; r = −0.359; n = 50.
Prospective study of PP2Acα promoter methylation status in patients with SLE
Because our data pointed out that DNA hypomethylation is primarily observed in patients with active disease, we wanted to confirm this observation in the same patients as their disease activity changed over time. The demographics of nine patients used in this experiment are shown in Table II. We defined the time point at which disease activity was low as phase 1 (SLEDAI was ranged between 0 and 10, average; 1.78) and the time point when the disease activity was relatively high compared with phase 1 as phase 2 (SLEDAI ranged between 4 and 35; average: 15). The difference of SLEDAI between phase 1 and phase 2 was at least 4 (shown in Table II). The mean duration between phase 1 and phase 2 was 8.3 mo (ranged between 5 and 13 mo). We compared three independent parameters within same individuals. Two representative patients are shown in Fig. 6A, one experiencing increasing disease activity and the other going into remission. Cumulative data from nine patients (classifying them arbitrarily in phase 1 [low activity] and 2 [high activity]) are shown in Fig. 6B. Clearly, as disease activity increases, DNMT1 expression and methylation levels decrease, whereas the levels of PP2Ac mRNA increase.
FIGURE 6.
Prospective analysis of the levels of DNA methylation in the promoter and transcript levels of DNMT1 and PP2Acα. A DNA methylation and transcripts of DNMT1 and PP2Acα were assessed in nine individuals prospectively. The left panel (patient 7 in Table II) is one example of flare during the course. The right panel (patient 9 in Table II) is also the example of well controlled by treatment. The quantification of DNA methylation in the promoter was conducted in a similar method as shown in Figs. 1 and 3. The ratio of methylation band (upper band, M) to control band (lower band, C) represented by lines (right y-axis) in the graph and the change of each transcript level was shown by bars (right y-axis; PP2Acα, white bar; DNMT1, black bar). B, We defined relatively inactive time point as a phase 1 and active time point as a phase 2. The difference of SLEDAI between phase 1 and phase 2 was at least 4. The demographics of all patients used this experiment are shown in Table II. Each parameter was measured at two time points and set as 1.0 at phase 1. DNMT1 transcripts level (left panel) and DNA methylation level (middle panel) were significantly decreased when disease was active. PP2Acα transcript levels (right panel) displayed the opposite trend. The results represent the mean ± SEM of nine independent experiments. *p < 0.0001 (left), *p < 0.0001 (middle), *p = 0.00012 (right).
Discussion
In this study, we present the first evidence, to our knowledge, that PP2Acα, a newly identified methylation-sensitive gene, is involved in the pathogenesis of SLE. Specifically, T cells from patients with active SLE display had increased binding of the transcriptional enhancer p-CREB to the PP2Acα promoter-defined CRE motif because of its low methylation status, reflected by a decreased amount of DNMT1 transcripts. Importantly, disease activity variation correlated with the methylation pattern and PP2Acα expression.
Epigenetic mechanisms have been implicated in the pathogenesis of various disorders, including cancer, immunodeficiency, and autoimmunity (19, 24, 37). DNA methylation involves the covalent modification of the fifth carbon in cytosine residues of CG dinucleotides. Most CG pairs in mammalian DNA are methylated, with exceptions in areas that are located in or near the promoters of active genes, in which expression requires a chromatin configuration permissive of transcription factor binding (10, 38). Thus, abnormalities in DNA methylation could disturb the normal regulation of gene silencing, X chromosome inactivation, and lineage specification (10–13, 39–41). The relationship between DNA methylation and the immune system has been particularly well studied in SLE because it may account for several T cell abnormalities involved in the pathogenesis of the disease by providing useful clues on the mechanisms whereby environmental factors including UV light and drugs contribute to the expression of autoimmunity and related pathology (19, 26, 42–45).
DNMT1 is a major DNA methyltransferase responsible for DNA methylation following DNA replication during cell division (26, 27, 46), which has been found to be decreased in SLE T cells and claimed to be involved in the immunopathogenesis of the disease. Defective ERK signaling has been demonstrated to account for the decreased DNMT1 expression in SLE T cells (27). In addition, a defect in the phosphorylation of protein kinase Cδ, which is located downstream of ERK, results in DNA hypomethylation and overexpression of methylation-sensitive genes such as CD70 (47). SLE T cells have been reported to express higher amounts of microRNAs 21 and 148a, and these are known to downregulate DNMT1 (48). In addition, DNMT1 polymorphisms, V120L in exon 4, have been associated with the production of anti-La Ab in SLE patients (49).
Several genes involved in the DNA methylation pathway have been linked to the expression of SLE. RFX1, a transcription factor that affects DNA methylation and histone acetylation by recruiting the corepressors DNMT1 and histone deacetylase 1 to the promoters of methylation-sensitive genes, was reported to be decreased in SLE T cells (50). Gene polymorphisms of methionine synthase, which is located in chromosome 1q43, a region confirmed to confer risk for the development of SLE, have been linked to SLE (51). Binding of methyl binding domain proteins or methyl cytosine-binding proteins (MECPs) to demethylated promoter regions is known to represent an indirect gene-silencing mechanism by preventing the binding of transcription factors to the targeted gene promoter (52, 53). MECP2 also recruits histone deacetylase, which increases chromatin density and induces a chromatin structure inaccessible for transcription (52, 53). Some MECP2 single nucleotide polymorphisms were reported to be associated with the development of SLE (54).
CD4+ T cells form active SLE patients as well as T cells treated with DNA methylation inhibitors displayed increased expression of integrin α L (CD11a), which was linked to patchy demethylation within the promoter (47). Hypomethylated, autoreactive CD4+ T cells in SLE patients also spontaneously kill autologous macrophages, causing release of antigenic nucleosomes through mechanisms including demethylation and subsequent overexpression of the perforin 1 gene (20). The costimulatory molecules CD70 (TNF superfamily member 7) and CD40L are also known as methylation-sensitive genes, and their overexpression leads to increased stimulation of autoreactive B cells and increased Ab production in patients with SLE (17, 18, 55). Demethylation of CD40L on the inactive X chromosome may also contribute to the striking female predilection of lupus (18). In addition, some cytokines such as IL-4, IL-6, and IFN-γ are regulated by DNA methylation, and abnormal demethylation may disturb appropriate T cell subset development and normal immune response in SLE (23, 25). Moreover, the abundance of hypomethylated DNA derived from apoptotic cells in lupus patients would possibly contribute to autoimmunity via TLR9 stimulation, which is activated by hypomethylated CpG DNA (56–58).
The catalytic subunit of PP2A has been identified as a player in the pathogenesis of SLE. Increased PP2Ac activity results in reduced binding of p-CREB to the IL-2 core promoter and decreased production of IL-2 (29). Abnormal overexpression of PP2Ac in T cells from patients with SLE also contribute to decreased expression of CD3ζ-chain and increased expression of FcRγ-chain by dephosphorylation of eukaryotic transcription elongation factor, which becomes part of the CD3 complex and contributes to aberrant signaling (59).
PP2A is a major serine/threonine phosphatase with complex composition and is involved in many essential aspects of cell function. The heterodimeric PP2A core enzyme consists of a well-conserved 36-kDa catalytic subunit (C subunit; PP2Ac) and a 65-kDa scaffold subunit (A subunit). To gain full activity toward specific substrates, the PP2A core enzyme associates with a variable regulatory subunit (B subunit) to form a heterotrimeric holoenzyme (30–32). PP2Ac has two isoforms, α and β, but α represents the major isoform because of the evidence that the activity of the PP2Acα promoter and mRNA levels are 7 ± 10-fold higher than those of β isoform (60). The PP2Acα gene is composed of seven exons and six introns encoded on chromosome 5q23–q31. It is characterized by an extremely GC-rich sequence and the lack of TATA and CCAAT boxes that are frequently found in many housekeeping genes (61). The expression of PP2Acα is tightly controlled through autoregulation to ensure the presence of relatively constant levels of PP2A (62). One mechanism of this autoregulation involves p-CREB, which regulates gene expression and is also dephosphorylated by PP2Ac itself (34, 63, 64). However, the autoregulation of PP2Acα appeared to be disturbed in T cells from SLE patients because the expression levels of this molecule are significantly higher in SLE T cells than in normal T cells. The control of the expression of PP2Acα levels in SLE T cells is broken at the methylation level of the CRE motif of its promoter. In addition, because PP2A can dephosphorylate p-ERK and p-JNK, which control DNMT1 expression, it may lead to further positive regulation of itself (positive feedback) (65). We previously reported that total PP2Ac (PP2Acα and PP2Acβ iso-forms) is increased in SLE T cells compared with T cells from healthy individuals; yet, we had failed to find a correlation with disease activity. In the current study, we have focused on the regulation of the promoter activity in the PP2Acα, the major isoform of the enzyme, and we have recorded its levels rather than total levels of PP2Ac. The fact that the levels of PP2Acα correlate with disease activity suggests the importance of this isoform in the expression of the disease and that our previous data (29) were confounded by the indiscriminate determination of both isoforms together. In addition, in the current study, we used mRNA samples to examine the correlation between expression levels of PP2Ac and disease activity because we focused on the regulation of the transcription of the PP2Ac gene rather than on the translation of the gene. The regulation of PP2Ac mRNA translation is still unclear and remains to be investigated fully in future (29). In this work, we demonstrated that abnormal hypomethylation of this promoter in lupus T cells allows the overexpression of this molecule by increasing accessibility of p-CREB to the CRE motif in the promoter.
In conclusion, we presented the evidence that hypomethylation of the CRE site of the PP2Acα promoter enables ample binding of p-CREB, which results in increased expression of PP2Acα in SLE T cells. Our results add PP2Acα to the list of genes that are altered epigenetically in patients with SLE. The fact that PP2Acα promoter methylation paralleled disease activity urges a larger prospective study to explore its potential to serve as a disease bio-marker.
Acknowledgments
This work was supported by Public Health Service National Institutes of Health Grants RO1 AI068787 (to G.C.T.) and K23 AR55672 (to V.C.K.) and a Postdoctoral Fellowship for Research Abroad by the Japan Society for the Promotion of Science.
Footnotes
Abbreviations used in this article: 5-azaC, 5-azacitidine; ChIP, chromatin immunoprecipitation; CRE, cAMP response element; dC, deoxycytosine; DNMT1, DNA methyltransferase 1; MECP, methyl cytosine-binding protein; PP2A, protein phosphatase 2A; PP2Ac, catalytic subunit of protein phosphatase 2A; PP2Acα, α isoform of protein phosphatase 2A catalytic subunit; SLE, systemic lupus erythematosus; SLEDAI, systemic lupus erythematosus disease activity index.
Disclosures The authors have no financial conflicts of interest.
References
- 1.Sawalha AH, Harley JB. Antinuclear autoantibodies in systemic lupus erythematosus. Curr. Opin. Rheumatol. 2004;16:534–540. doi: 10.1097/01.bor.0000135452.62800.8f. [DOI] [PubMed] [Google Scholar]
- 2.Harley JB, Kelly JA, Kaufman KM. Unraveling the genetics of systemic lupus erythematosus. Springer Semin. Immunopathol. 2006;28:119–130. doi: 10.1007/s00281-006-0040-5. [DOI] [PubMed] [Google Scholar]
- 3.Järvinen P, Aho K. Twin studies in rheumatic diseases. Semin. Arthritis Rheum. 1994;24:19–28. doi: 10.1016/0049-0172(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 4.Cooper GS, Parks CG, Treadwell EL, St Clair EW, Gilkeson GS, Dooley MA. Occupational risk factors for the development of systemic lupus erythematosus. J. Rheumatol. 2004;31:1928–1933. [PubMed] [Google Scholar]
- 5.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J. Immunol. 1988;140:2197–2200. [PubMed] [Google Scholar]
- 7.Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, Richardson B. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- 8.Wang GS, Zhang M, Li XP, Zhang H, Chen W, Kan M, Wang YM. Ultraviolet B exposure of peripheral blood mononuclear cells of patients with systemic lupus erythematosus inhibits DNA methylation. Lupus. 2009;18:1037–1044. doi: 10.1177/0961203309106181. [DOI] [PubMed] [Google Scholar]
- 9.Rao T, Richardson B. Environmentally induced autoimmune diseases: potential mechanisms. Environ. Health Perspect. 1999;107(Suppl 5):737–742. doi: 10.1289/ehp.99107s5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singal R, Ginder GD. DNA methylation. Blood. 1999;93:4059–4070. [PubMed] [Google Scholar]
- 11.Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 12.Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annu. Rev. Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 13.Nakao M, Sasaki H. Genomic imprinting: significance in development and diseases and the molecular mechanisms. J. Biochem. 1996;120:467–473. doi: 10.1093/oxfordjournals.jbchem.a021434. [DOI] [PubMed] [Google Scholar]
- 14.Kammer GM, Perl A, Richardson BC, Tsokos GC. Abnormal T cell signal transduction in systemic lupus erythematosus. Arthritis Rheum. 2002;46:1139–1154. doi: 10.1002/art.10192. [DOI] [PubMed] [Google Scholar]
- 15.Richardson BC, Strahler JR, Pivirotto TS, Quddus J, Bayliss GE, Gross LA, O'Rourke KS, Powers D, Hanash SM, Johnson MA. Phenotypic and functional similarities between 5-azacytidine-treated T cells and a T cell subset in patients with active systemic lupus erythematosus. Arthritis Rheum. 1992;35:647–662. doi: 10.1002/art.1780350608. [DOI] [PubMed] [Google Scholar]
- 16.Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D, Richardson B. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 2002;46:1282–1291. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- 17.Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J. Immunol. 2005;174:6212–6219. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 18.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J. Immunol. 2007;179:6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 19.Richardson BC. Epigenetics and autoimmunity. Overview. Autoimmunity. 2008;41:243–244. doi: 10.1080/08916930802024129. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J. Immunol. 2004;172:3652–3661. doi: 10.4049/jimmunol.172.6.3652. [DOI] [PubMed] [Google Scholar]
- 21.Kwon NH, Kim JS, Lee JY, Oh MJ, Choi DC. DNA methylation and the expression of IL-4 and IFN-gamma promoter genes in patients with bronchial asthma. J. Clin. Immunol. 2008;28:139–146. doi: 10.1007/s10875-007-9148-1. [DOI] [PubMed] [Google Scholar]
- 22.Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58:2686–2693. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- 23.Mi XB, Zeng FQ. Hypomethylation of interleukin-4 and -6 promoters in T cells from systemic lupus erythematosus patients. Acta Pharmacol. Sin. 2008;29:105–112. doi: 10.1111/j.1745-7254.2008.00739.x. [DOI] [PubMed] [Google Scholar]
- 24.Janson PC, Marits P, Thörn M, Ohlsson R, Winqvist O. CpG methylation of the IFNG gene as a mechanism to induce immunosuppression [correction of immunosupression] in tumor-infiltrating lymphocytes. J. Immunol. 2008;181:2878–2886. doi: 10.4049/jimmunol.181.4.2878. [DOI] [PubMed] [Google Scholar]
- 25.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, Osban J, Knowlton N, Johnson K, Richardson B. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorelik G, Richardson B. Aberrant T cell ERK pathway signaling and chromatin structure in lupus. Autoimmun. Rev. 2009;8:196–198. doi: 10.1016/j.autrev.2008.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng C, Kaplan MJ, Yang J, Ray D, Zhang Z, McCune WJ, Hanash SM, Richardson BC. Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 2001;44:397–407. doi: 10.1002/1529-0131(200102)44:2<397::AID-ANR59>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 28.Balada E, Ordi-Ros J, Serrano-Acedo S, Martinez-Lostao L, Rosa-Leyva M, Vilardell-Tarrés M. Transcript levels of DNA methyltransferases DNMT1, DNMT3A and DNMT3B in CD4+ T cells from patients with systemic lupus erythematosus. Immunology. 2008;124:339–347. doi: 10.1111/j.1365-2567.2007.02771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katsiari CG, Kyttaris VC, Juang YT, Tsokos GC. Protein phosphatase 2A is a negative regulator of IL-2 production in patients with systemic lupus erythematosus. J. Clin. Invest. 2005;115:3193–3204. doi: 10.1172/JCI24895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virshup DM. Protein phosphatase 2A: a panoply of enzymes. Curr. Opin. Cell Biol. 2000;12:180–185. doi: 10.1016/s0955-0674(99)00074-5. [DOI] [PubMed] [Google Scholar]
- 31.Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lechward K, Awotunde OS, Swiatek W, Muszyńska G. Protein phosphatase 2A: variety of forms and diversity of functions. Acta Biochim. Pol. 2001;48:921–933. [PubMed] [Google Scholar]
- 33.Wadzinski BE, Wheat WH, Jaspers S, Peruski LF, Jr., Lickteig RL, Johnson GL, Klemm DJ. Nuclear protein phosphatase 2A dephosphorylates protein kinase A-phosphorylated CREB and regulates CREB transcriptional stimulation. Mol. Cell. Biol. 1993;13:2822–2834. doi: 10.1128/mcb.13.5.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sunahori K, Juang YT, Tsokos GC. Methylation status of CpG islands flanking a cAMP response element motif on the protein phosphatase 2Ac alpha promoter determines CREB binding and activity. J. Immunol. 2009;182:1500–1508. doi: 10.4049/jimmunol.182.3.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 36.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH, The Committee on Prognosis Studies in SLE Derivation of the SLEDAI. A disease activity index for lupus patients. Arthritis Rheum. 1992;35:630–640. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 37.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 38.Richardson B, Yung R. Role of DNA methylation in the regulation of cell function. J. Lab. Clin. Med. 1999;134:333–340. doi: 10.1016/s0022-2143(99)90147-6. [DOI] [PubMed] [Google Scholar]
- 39.Sawalha AH. Epigenetics and T-cell immunity. Autoimmunity. 2008;41:245–252. doi: 10.1080/08916930802024145. [DOI] [PubMed] [Google Scholar]
- 40.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 41.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim. Biophys. Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Pan Y, Sawalha AH. Epigenetic regulation and the pathogenesis of systemic lupus erythematosus. Transl. Res. 2009;153:4–10. doi: 10.1016/j.trsl.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Januchowski R, Prokop J, Jagodziński PP. Role of epigenetic DNA alterations in the pathogenesis of systemic lupus erythematosus. J. Appl. Genet. 2004;45:237–248. [PubMed] [Google Scholar]
- 44.Sekigawa I, Kawasaki M, Ogasawara H, Kaneda K, Kaneko H, Takasaki Y, Ogawa H. DNA methylation: its contribution to systemic lupus erythematosus. Clin. Exp. Med. 2006;6:99–106. doi: 10.1007/s10238-006-0103-x. [DOI] [PubMed] [Google Scholar]
- 45.Patel DR, Richardson BC. Epigenetic mechanisms in lupus. Curr. Opin. Rheumatol. 2010;22:478–482. doi: 10.1097/BOR.0b013e32833ae915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PubMed] [Google Scholar]
- 47.Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J. Immunol. 2007;179:5553–5563. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 48.Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, Li J, Zhou H, Tang Y, Shen N. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol. 2010;184:6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 49.Park BL, Kim LH, Shin HD, Park YW, Uhm WS, Bae SC. Association analyses of DNA methyltransferase-1 (DNMT1) polymorphisms with systemic lupus erythematosus. J. Hum. Genet. 2004;49:642–646. doi: 10.1007/s10038-004-0192-x. [DOI] [PubMed] [Google Scholar]
- 50.Zhao M, Sun Y, Gao F, Wu X, Tang J, Yin H, Luo Y, Richardson B, Lu Q. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J. Autoimmun. 2010;35:58–69. doi: 10.1016/j.jaut.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 51.Burzynski M, Duriagin S, Mostowska M, Wudarski M, Chwalinska-Sadowska H, Jagodzinski PP. MTR 2756 A > G polymorphism is associated with the risk of systemic lupus erythematosus in the Polish population. Lupus. 2007;16:450–454. doi: 10.1177/0961203307077988. [DOI] [PubMed] [Google Scholar]
- 52.Hite KC, Adams VH, Hansen JC. Recent advances in MeCP2 structure and function. Biochem. Cell Biol. 2009;87:219–227. doi: 10.1139/o08-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matouk CC, Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circ. Res. 2008;102:873–887. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- 54.Sawalha AH, Webb R, Han S, Kelly JA, Kaufman KM, Kimberly RP, Alarcon-Riquelme ME, James JA, Vyse TJ, Gilkeson GS, et al. Common variants within MECP2 confer risk of systemic lupus erythematosus. PloS One. 2008;3:e1727. doi: 10.1371/journal.pone.0001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, Yuan J, Pan Y, Fei Y, Qiu X, Hu N, Luo Y, Lei W, Li Y, Long H, et al. T cell CD40LG gene expression and the production of IgG by autologous B cells in systemic lupus erythematosus. Clin. Immunol. 2009;132:362–370. doi: 10.1016/j.clim.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogasawara H, Okada M, Kaneko H, Hishikawa T, Sekigawa I, Hashimoto H. Possible role of DNA hypomethylation in the induction of SLE: relationship to the transcription of human endogenous retroviruses. Clin. Exp. Rheumatol. 2003;21:733–738. [PubMed] [Google Scholar]
- 57.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 58.Yasuda K, Richez C, Uccellini MB, Richards RJ, Bonegio RG, Akira S, Monestier M, Corley RB, Viglianti GA, Marshak-Rothstein A, Rifkin IR. Requirement for DNA CpG content in TLR9-dependent dendritic cell activation induced by DNA-containing immune complexes. J. Immunol. 2009;183:3109–3117. doi: 10.4049/jimmunol.0900399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Juang YT, Wang Y, Jiang G, Peng HB, Ergin S, Finnell M, Magilavy A, Kyttaris VC, Tsokos GC. PP2A dephosphorylates Elf-1 and determines the expression of CD3zeta and FcRgamma in human systemic lupus erythematosus T cells. J. Immunol. 2008;181:3658–3664. doi: 10.4049/jimmunol.181.5.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khew-Goodall Y, Mayer RE, Maurer F, Stone SR, Hemmings BA. Structure and transcriptional regulation of protein phosphatase 2A catalytic subunit genes. Biochemistry. 1991;30:89–97. doi: 10.1021/bi00215a014. [DOI] [PubMed] [Google Scholar]
- 61.Stone SR, Hofsteenge J, Hemmings BA. Molecular cloning of cDNAs encoding two isoforms of the catalytic subunit of protein phosphatase 2A. Biochemistry. 1987;26:7215–7220. doi: 10.1021/bi00397a003. [DOI] [PubMed] [Google Scholar]
- 62.Baharians Z, Schönthal AH. Autoregulation of protein phosphatase type 2A expression. J. Biol. Chem. 1998;273:19019–19024. doi: 10.1074/jbc.273.30.19019. [DOI] [PubMed] [Google Scholar]
- 63.Schild A, Ittner LM, Götz J. Altered phosphorylation of cytoskeletal proteins in mutant protein phosphatase 2A transgenic mice. Biochem. Biophys. Res. Commun. 2006;343:1171–1178. doi: 10.1016/j.bbrc.2006.03.066. [DOI] [PubMed] [Google Scholar]
- 64.Conkright MD, Guzmán E, Flechner L, Su AI, Hogenesch JB, Montminy M. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell. 2003;11:1101–1108. doi: 10.1016/s1097-2765(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 65.Huang PH, Wang D, Chuang HC, Wei S, Kulp SK, Chen CS. alpha-Tocopheryl succinate and derivatives mediate the transcriptional repression of androgen receptor in prostate cancer cells by targeting the PP2AJNK-Sp1-signaling axis. Carcinogenesis. 2009;30:1125–1131. doi: 10.1093/carcin/bgp112. [DOI] [PMC free article] [PubMed] [Google Scholar]