Abstract
Background
In years past, methicillin-resistant S. aureus (MRSA) has been frequently detected in pigs in Europe, North America and Asia. Recent, yet sporadic studies have revealed a low occurrence of MRSA in Switzerland. In 2009, a monitoring survey of the prevalence and genetic diversity of methicillin-resistant S. aureus (MRSA) in slaughter pigs in Switzerland was conducted using methods recommended by the EU guidelines, and using a sampling strategy evenly distributed throughout the year and representative of the Swiss slaughter pig population. Monitoring should determine if the overall prevalence of MRSA in the entire country is increasing over the years and if specific multi-resistant MRSA clones are spreading over the country.
Results
In 2009, the nasal cavities of eight out of 405 randomly selected pigs were positive for MRSA, representing a prevalence of 2.0% (95% CI 0.9-3.9). The following year, 23 out of 392 pigs were positive for MRSA [5.9% prevalence (95% CI 3.8-8.7)]. Three multilocus sequence types (ST), four spa types and two types of staphylococcal cassette chromosome mec (SCCmec) elements were detected. The most frequent genotypes were ST398 (MLST)-(spa)t034-V(SCCmec) (n = 18) and ST49-t208-V (n = 7), followed by ST398-t011-V (n = 4), ST398-t1451-V (n = 1), and ST1-t2279-IVc (n = 1). The isolates displayed resistance to ß-lactams [mecA, (31/31); blaZ, (19/31)]; tetracycline [tet(M), (31/31); tet(K), (30/31)] (n = 31); macrolides and lincosamides [erm(C) (4/31) or erm(A) (18/31)] (n = 22); tiamulin [vga(A)v (9/31) or unknown mechanism (18/31)] (n = 27); trimethoprim [dfr(G) (18/31); spectinomycin [ant(9)-Ia (19/31) or unknown mechanism (3/31)] (n = 22); streptomycin [str (19/31)]; sulphamethoxazole (7/31) and ciprofloxacin (n = 1) (mechanisms not determined).
Conclusions
This study is the first to describe the presence of MRSA ST49 in slaughter pigs, and to demonstrate a significant and nearly three-fold increase of MRSA prevalence in pigs within two years. The presence of a specific clonal lineage of MRSA from Switzerland suggests that it has been selected in Swiss pig husbandry. Effective hygiene measures should be enhanced within the entire pig production chain to suppress the spread of these pathogens into the community.
Background
Over the years, methicillin-resistant Staphylococcus aureus (MRSA) has been increasingly reported in animals worldwide [1-4]. A specific clonal lineage, MLST ST398, has been found to be widespread among pigs in Europe and North America, whereas ST9 seemed to be predominant in Asia [4-6]. Several spa types were detected within these lineages with spa types t011, t108, and t034 being the most frequent in MRSA ST398, and t899 in MRSA ST9 [3-6]. Colonisation with MRSA ST398 was also described in other animals like poultry, horses, and calves, and recently, ST398 has been associated with infections in animals and humans [2,4,7]. Humans that come in close contact with farm animals were more likely to be colonised with livestock-associated ST398 and had a higher risk of developing infections with ST398 in case of hospitalisation [8-18]. The pig husbandry environment also represents a large reservoir for MRSA. MRSA ST398 was recovered from dust samples in pig production holdings in Europe with an average prevalence of 26.9% varying between 0% and 51.2% among the EU member states in 2008 [19]. MRSA ST398-t011 was the most dominant type (63.5%), followed by ST398-t108 (9.4%), and ST398-t034 (8.4%). 1.4% of the MRSA from this study were from seven other non-ST398 strains (ST1, ST5, ST8, ST9, ST39, ST97, and ST132). The majority of the non-ST398 strains originated from Germany and Italy [19].
In 2008, dust samples from pig husbandries in Switzerland were analysed and did not contain MRSA [19]. In fact, there was no MRSA detected during previous studies in Swiss slaughter pigs and pig carcasses before 2009 [20,21]. However in 2009, 10 MRSA ST398-t034, which contained the staphylococcal cassette chromosome SCCmecV (ST398-t034-V), were found among 800 nasal swabs from Swiss pigs at two slaughterhouses; these results indicated that MRSA had also emerged in the Swiss pig population [22]. That same year, an official monitoring of MRSA in pigs at the slaughterhouse was launched to determine the overall prevalence in the entire country. A sampling strategy for monitoring antimicrobial resistance was developed in 2008, which considered the geographical distribution and size of the slaughterhouses, and the number of slaughtered pigs in Switzerland [23]. Since 2009, this sampling plan was also used for monitoring MRSA. This study will determine the overall prevalence and dynamics of MRSA colonisation in the slaughtered pig population and will indicate if specific genetic lineages of MRSA are colonising pigs in Switzerland.
Results
Increased prevalence and molecular typing
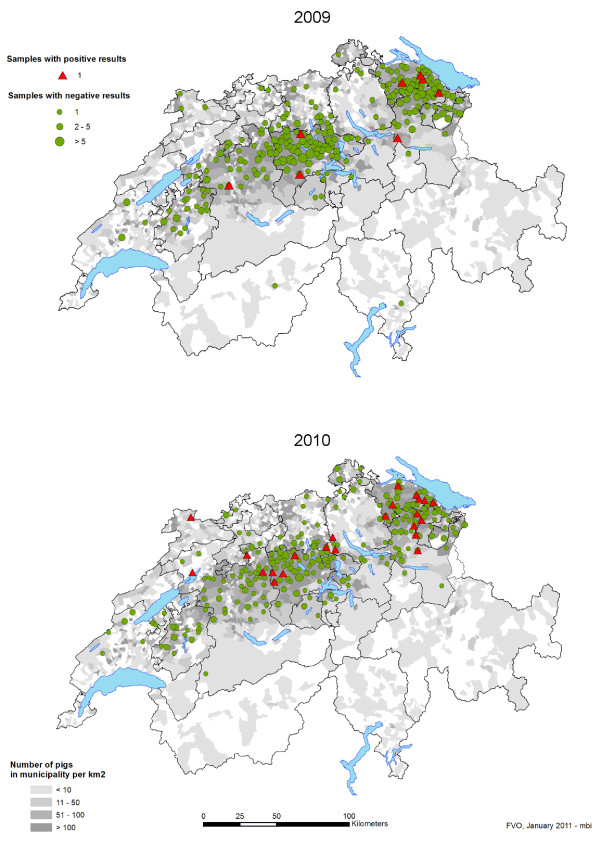
MRSA (n = 31) were detected in samples from 5 of 9 slaughterhouses (A, B, C, E, G) in 2009 and in 7 of 9 slaughterhouses (A-D, G-I) in 2010 (Table 1). MRSA was mostly found in pigs raised in cantons where the pig population is the highest (Figure 1), that is, Thurgau (n = 11), Lucerne (n = 6), St. Gallen (n = 5), Bern (n = 4), and Aargau (n = 2). Single isolates were found in pigs from Appenzell Innen Rhodes, Jura, and Zurich (Table 1 and Table 2). In 2009, the prevalence of MRSA in Swiss slaughter pigs was 2.0% (95% CI 0.9-3.9) with eight out of 405 pig nasal samples being positive and increased significantly (p-value < 0.05) to 5.9% (95% CI 3.8-8.7) in 2010 with 23 out of 392 nasal swabs containing MRSA. MRSA was detected every month in 2010, except in September, with up to five positive samples per month, whereas MRSA was only sporadically detected over the year in 2009.
Table 1.
Distribution of methicillin-resistant Staphylococcus aureus genotypes in slaughterhouses and pig farms in Switzerland.
| 2009 | 2010 | |||||||||
| Abattoir |
Total no. of samples |
No. of MRSA positive samples |
Percentage of MRSA positive samples |
Origina | Genotype (n) |
Total no. of samples |
No. of MRSA positive samples |
Percentage of MRSA positive samples |
Origina | Genotype (n) |
| A | 84 | 2 | 2.4 | TG1, LU2 | ST398-t011-V (2) | 68 | 11 | 16.2 | LU4, LU5, SG3, SG5, TG5, TG8, TG9, BE3, BE4 ZH1, JU1 |
ST398-t034-V (11) |
| B | 90 | 1 | 1.1 | SG1 | ST398-t011-V | 101 | 2 | 2.0 | AG2, SG6 | ST49-t208-V (2) |
| C | 63 | 1 | 1.6 | BE1 | ST398-t034-V | 65 | 4 | 6.2 | TG6 BE2, TG7, LU6 | ST49-t208-V ST398-t034-V (3) |
| D | 62 | 0 | 0 | - | - | 58 | 2 | 3.4 | AG1, TG10 | ST398-t034-V (2) |
| E | 39 | 1 | 2.6 | LU1 | ST398-t1451-V | 37 | 0 | 0 | - | - |
| F | 28 | 0 | 0 | - | - | 22 | 0 | 0 | - | - |
| G | 19 | 3 | 15.8 | TG3, TG2, SG2 | ST1-t2279-IVc ST49-t208-V (2) |
15 | 2 | 13.3 | TG4, SG4 | ST49-t208-V (2) |
| H | 10 | 0 | 0 | - | - | 14 | 1 | 7.1 | LU3 | ST398-t034-V |
| I | 10 | 0 | 0 | - | - | 12 | 1 | 8.3 | AI1 | ST398-t011-V |
| A-I (all) | 405 | 8 | 2.0 | 392 | 23 | 5.9 | ||||
a) Geographical origin of the farms: letters indicate the Cantons (AI, Appenzell Innen Rhodes; AG, Aargau; BE, Bern; JU, Jura; LU, Luzern; SG, St Gallen; TG, Thurgau; ZH, Zurich). and numbers indicate individual farms.
Figure 1.
Pig density and geographical distribution of the samples and results for MRSA isolated from the nasal cavities of slaughter pigs in Switzerland in 2009 and 2010. The pig density appears in grey. Samples with positive results appear as red triangles, samples with negative results appear as green dots.
Table 2.
The clonal lineage and resistance profile of methicillin-resistant Staphylococcus aureus from pig noses in Switzerland.
| Antibiotic resistance genes and resistance breakpoints (mg/L)b | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MLST | spa type | SCCmec type | Number of isolates | Farma | Representative strain | FOX | PEN | TET | ERY | CLI | STR | TIA | SPC | TMP | SMX | CIP |
| (>4) | (>0.125) | (>2) | (>2) | (>0.5) | (>16) | (>2) | (>128) | (>2) | (>128) | (>1) | ||||||
| 2009 | ||||||||||||||||
| ST1 | t2279 | IVc | 1 | TG3 | IMD887-09 | mecA | mecA blaZ | tet(M) | erm(A) | erm(A) | ant(9)-Ia | + | ||||
| ST398 | t011 | V | 1 | SG1 | IMD358-09 | mecA | mecA | tet(M) tet(K) | vga(A)v | |||||||
| ST398 | t011 | V | 1 | TG1 | IMD355-09 | mecA | mecA | tet(M) tet(K) | erm(C) | erm(C) | + | |||||
| ST398 | t011 | V | 1 | LU2 | IMD1753-09 | mecA | mecA | tet(M) tet(K) | + | |||||||
| ST398 | t034 | V | 1 | BE1 | IMD1116-09 | mecA | mecA blaZ | tet(M) tet(K) | erm(A) | erm(A) | str | + | ant(9)-Ia | dfr(G) | ||
| ST398 | t1451 | V | 1 | LU1 | IMD1270-09 | mecA | mecA | tet(M) tet(K) | erm(C) | erm(C) | str | vga(A)v | ||||
| ST49 | t208 | V | 1 | TG2 | IMD1000-09 | mecA | mecA | tet(M) tet(K) | erm(C) | erm(C) | vga(A)v | + | ||||
| ST49 | t208 | V | 1 | SG2 | IMD1771-09 | mecA | mecA | tet(M) tet(K) | vga(A)v | + | ||||||
| 2010 | ||||||||||||||||
| ST398 | t034 | V | 16 | TG5 TG8 TG9 TG10 LU3 LU4 LU5 LU6 BE2 BE3 BE4 AG1 JU1 SG3 SG5 ZH1 | IMD49-10 | mecA | mecA blaZ | tet(M) tet(K) | erm(A) | erm(A) | str | + | ant(9)-Ia | dfr(G) | ||
| ST398 | t034 | V | 1 | TG 7 | IMD704-10 | mecA | mecA blaZ | tet(M) tet(K) | erm(A) | erm(A) | + | ant(9)-Ia | dfr(G) | |||
| ST398 | t011 | V | 1 | AI1 | IMD233-10 | mecA | mecA | tet(M) tet(K) | erm(C) | erm(C) | + | |||||
| ST49 | t208 | V | 3 | SG4 SG6 TG4 | IMD426-10 | mecA | mecA | tet(M) tet(K) | vga(A)v | + | ||||||
| ST49 | t208 | V | 1 | TG6 | IMD603-10 | mecA | mecA | tet(M) tet(K) | erm(A) | erm(A) | vga(A)v | + | ||||
| ST49 | t208 | V | 1 | AG2 | IMD2002-10 | mecA | mecA | tet(M) tet(K) | str | vga(A)v | + | |||||
a) Geographical origin of the farm: letters indicate the Cantons (AI, Appenzell Innen Rhodes; AG, Aargau; BE, Bern; JU, Jura; LU, Luzern; SG, St Gallen; TG, Thurgau; ZH, Zurich). and numbers indicate individual farms.
b) CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; FOX, cefoxitin; PEN, penicillin; SPC, spectinomycin; SMX, sulphamethoxazole; STR, streptomycin; TET, tetracycline; TIA, tiamulin; TMP, trimethoprim. The MIC breakpoints (in micrograms per millilitre) that determine resistance were recommended from EUCAST for S. aureus http://www.eucast.org. Resistance breakpoints for tiamulin, spectinomycin, streptomycin and sulphamethoxazole were tentatively derived from epidemiological MIC cut-off values from EUCAST.
Antibiotic resistance genes and their functions are indicated as follows: ant(9)-Ia, spectinomycin adenylnucleotidyltransferase; blaZ, β-lactamase; dfr(G), dihydrofolate reductase; erm(A) and erm(C), macrolides, lincosamides and streptogramins B rRNA methylase; mecA, penicillin-binding protein PBP2a; str, streptomycin adenyltransferase; tet(K), tetracycline efflux protein; tet(M), tetracycline ribosomal protection protein; vga(A)v, pleuromutilins and streptogramins A ATP binding transporter.
ND, not detected; +, The MIC values were greater than the resistance breakpoint, but the resistance mechanism remained uncharacterised; blank spaces indicate no resistance
With the exception of one isolate (SCCmecIVc), most of the MRSA contained the SCCmecV element (Table 2). The eight MRSA isolated in 2009 belonged to five genotypes: ST398-t011-V (n = 3), ST398-t034-V (n = 1), ST398-t1451-V (n = 1), ST49-t208-V (n = 2), and ST1-t2279-IVc (n = 1); each isolate displayed a different resistance profile (Table 2). In 2010, only three clonal lineages were identified with ST398-t034-V (n = 17) and ST49-t208-V (n = 5) being the most common, whereas ST398-t011-V was only detected once (Table 1 and Table 2).
MRSA isolated from a same slaughterhouse in 2009 and 2010 displayed a different genetic profile and were isolated at different days, except for some MRSA ST398-t034 and ST49-t208 isolates. They were also detected several times from samples from the same slaughterhouses at different days [slaughterhouses A, C, D (ST398-t034)] as well as during the same sampling day in 2010 [slaughterhouses A (ST398-t034) and G (ST49-t208)]. All samples originated from pigs raised in different husbandry (Table 1 and Table 2).
Resistance profiles
Isolates belonging to the most commonly detected genotype, ST398-t034-V, shared the same resistance profile; however, one isolate was susceptible to streptomycin and did not contain the streptomycin adenyltransferase gene str (Table 2). Otherwise, ST398-t034-V isolates showed resistance to ß-lactams specified by mecA and blaZ, tetracycline [tet(K), tet(M)], macrolide-lincosamide-streptogramin B (MLSB) antibiotics [erm(A)], spectinomycin [ant(9)-Ia], streptomycin [str], trimethoprim [dfr(G)], and tiamulin. The tiamulin resistance could not be attributed to either of the five known pleuromutilin-streptogramin A resistance genes vga(A), vga(A)v, vga(B), vga(C), vga(D) [24], or to the cfr rRNAmethylase transferase gene which confers cross resistance to phenicols, lincosamides, oxazolidones, pleuromutilins, and streptgramin A antibiotics [25]. Mutations in the 23 rRNA and rplC gene were not investigated [26].
Different resistance profiles were observed among the ST49-t208-V isolates, which was the second most prevalent lineage. The difference was mainly due to the presence, or absence, of either of the MLSB resistance genes erm(A) or erm(C) (Table 2). In general, ST49 isolates displayed resistance to ß-lactams [mecA], tetracycline [tet(K), tet(M)], tiamulin [vga(A)v], and sulphonamides (these mechanisms were not characterised). Notably, ST49-t208 isolates were susceptible to trimethoprim and resistant to sulphonamides, whereas ST398-t034 isolates were resistant to trimethoprim and susceptible to sulphonamides.
Different antibiotic resistance profiles were also found among the less frequent isolates, ST398-t011 and ST398-t1451. They all displayed resistance to ß-lactams [mecA] and tetracycline [tet(K), tet(M)]; however, resistance to MLSB antibiotics [erm(C)], tiamulin [vga(A)v], streptomycin [str], and spectinomycin varied from strain to strain (Table 2). The ST1-t2279-IVc isolate was only detected once and showed resistance to ß-lactams [mecA, blaZ], tetracycline [tet(M)], MLSB antibiotics [erm(A)], and spectinomycin [ant(9)-Ia]; additionally, a resistance to ciprofloxacin was detected (Table 2). The resistance mechanism to fluoroquinolones which may be due to mutations in the topoisomerase genes [27] was not determined. In contrast, MRSA isolates from 2009 and 2010 were susceptible to vancomycin, gentamicin, kanamycin, chloramphenicol, quinupristin/dalfopristin, fusidic acid, mupirocin, rifampicin, and linezolid; importantly, none of the isolates carried the Panton-Valentine Leukocidin toxin.
Discussion
Molecular typing and epidemiology
This study was the first to reveal the presence of MRSA ST49 in pigs. MRSA ST49 was already present in 2009, but its prevalence increased in 2010. The presence of the new clonal lineage ST49-t208, which is unique to MRSA from pigs in Switzerland, suggests that the selection occurred within the Swiss pig population. This idea is supported by the fact that MRSA ST49 was detected in seven different pigs from seven different farms slaughtered in three different slaughterhouses, and that ST49 isolates displayed different resistance profiles mainly due to the acquisition of an additional erm(A) or erm(C) gene (Table 2). The spa type t208, which is associated with ST49, was previously found in seven out of 48 methicillin-susceptible S. aureus MSSA from pigs in Switzerland [20], suggesting a possible acquisition of the SCCmec element. ST49 is rarely mentioned in the literature; it was not detected among 2890 MSSA and MRSA from human infections in the 26 EU countries, the 133 S. aureus colonising healthy adults in Switzerland nor within the 572 MSSA isolates from Swiss children [28-30]. ST49 has only been associated with MSSA involved in one human infection in the United Kingdom and in three cases of skin lesions and laryngeal ulceration in wild squirrels [31,32]. In the latter three cases, the authors mentioned close contact with persons who fed the squirrels as a possible source of infection. Further screening of humans, including people working with food-producing animals, would be necessary to determine if humans also play a role in the dissemination of MRSA ST49, as demonstrated with ST398. Human carriage of ST398 has been widely documented in other European countries, and the exchange of ST398 between humans and pigs has been reported in the Netherlands and Denmark [13,33,34]. Furthermore, ST398-t034 was the most predominant lineage found in Swiss pigs, and has been detected in one out of 133 veterinarians in Switzerland [22]. In our study, ST1 was only detected once, although it has been associated with MRSA infections in humans in the United Kingdom and Spain, and in horses in Austria [31,35,36]. ST1 represented, however, 15.6% of the MRSA-positive pig finishing holdings in Italy [37]. Cases of transmission of ST1 between humans and cows have also been reported [38].
Transmission of MRSA among pigs during animal transport and at slaughterhouse has been described [39]. In our study, no direct association of STs to a specific abattoir has been observed, except for slaughterhouse A, where ST398-t034 has been predominantly detected in the samples. Transmission which may have occurred at slaughterhouse may not be excluded for this abattoir. Otherwise, MRSA ST398 and ST49 were found in different slaughterhouses at different sampling dates and the pigs originated from different farms.
Increase of MRSA prevalence
This study is also the first to describe MRSA prevalence in slaughter pigs in Switzerland using a sampling plan which was representative of the Swiss slaughter pig population and which was evenly distributed over time. This strategy allowed us to obtain statistically significant and representative data for the evaluation of the chronological distribution of MRSA. Such representative sampling criteria were not considered in the two previous studies reporting MRSA in pigs in Switzerland, resulting in a less accurate estimation of the prevalence over the entire country [20,22]. However, these studies indicated that the prevalence of MRSA was low among the proportion of pigs analysed before 2009. Our study also showed that the prevalence of MRSA in 2009 was low, and indicated an approximate three-fold increase within one year. The increased diversity of MRSA and the emergence of the clonal lineage ST49 unique to MRSA from Swiss pigs suggested that MRSA-positive pigs imported into Switzerland have not introduced this MRSA. However, Switzerland sporadically imports a low number of breeding pigs, from which MRSA ST398 may have been introduced. In 2010, for example, Switzerland imported pigs from Denmark (n = 10), The Netherlands (n = 4), and Germany (n = 1), where the prevalence of MRSA ST398 is high (Federal Veterinary Office, pers. communication). Like Switzerland, the predominant clone in Denmark and Germany is ST398-t034, whereas MRSA ST398-t011 is the predominant in The Netherlands [40-42].
Conclusions
Two predominant clonal lineages, ST398-t034 and ST49-t208, are spreading within the Swiss slaughter pig population. Focus should be stressed on prevention of the transmission of these multi-resistant zoonotic bacteria from the farms into the community. Human carriage of MRSA represents a new challenge for public health because people that are in the vicinity of animals have been shown to have a higher risk of developing a MRSA infection when hospitalised. Targeted screening for MRSA in at-risk people, that is, farmers, veterinarians, and other people with close contact to animals, as recommended by Harbarth et al. (2011) [43], should be taken into account upon hospital admission. A study on MRSA decolonisation in humans with close contact to pigs showed a low efficacy due to continuous re-colonisation [44]. Therefore, effective hygiene measures into the entire pig production chain should be maintained as proposed by the Scientific Advisory Group on Antimicrobials [8]. Nevertheless, periodic monitoring of the MRSA in livestock animals at slaughterhouses is now established in Switzerland at the National Reference Laboratory for Antimicrobial Resistance (Centre of Zoonoses, Bacterial Animal Diseases and Antimicrobial Resistance (ZOBA), Institute of Veterinary Bacteriology, Vetsuisse Faculty, University of Bern), for the surveillance of MRSA in the livestock population.
Methods
Sampling
Representative samples were taken in accordance with the framework of a national monitoring program on antimicrobial resistance in food animals [23]. A minimum required sample size of 382 randomly selected animals was calculated with the assumptions of an infinite population size, a prevalence of 5%, a desired confidence level of 97.5% and an accuracy of 2.5%. The samples were randomly taken at the nine biggest abattoirs (A to I), where over 85% of the pigs in Switzerland were slaughtered. Only one sample was taken per animal holding. The number of samples in the sampling frame collected from each slaughterhouse was proportional to the number of pigs slaughtered at each establishment per year. One to 8 samples were taken per sampling day and abattoir. The samples collected on Mondays and Tuesdays were equally distributed over the years. Based on these data, random sampling plans were conducted, resulting in a total of 405 nasal swabs of fattening pigs in 2009 and 392 swabs from pigs in 2010.
Isolation of MRSA
Samples were taken using transport swabs (Oxoid Ltd, Basingstoke, England) from the nares of the pigs subsequent to stunning by officials of the Swiss abattoir authorities and immediately after sampling, were transported to the laboratory without cooling. After arrival, swabs were transferred into tubes containing 10 ml Mueller Hinton Broth supplemented with 6.5% NaCl and incubated aerobically at 37°C for 24 h under agitation. One ml from this pre-enrichment was inoculated into 9 ml tryptone soy broth containing 3.5 mg/L cefoxitin and 75 mg/L aztreonam, and further incubated aerobically at 37°C for 24 h. A loopful was then spread onto MRSA selective agar plates (BBL ™ CHROMagar ™ MRSA; Becton Dickinson, Franklin Lakes, NJ), which were incubated at 37°C for 24 h. Pink to mauve-colored colonies were regarded as suspicious and five presumptive colonies were cultivated onto tryptone soy agar plates containing 5% sheep blood (TSA-SB) (Oxoid Ltd, Basingstoke, England) at 37°C for 24 h. S. aureus was identified using Vitek 2 with Gram-Positive (GP) cards (BioMérieux, Mary l'Etoile, France) following manufacturer's recommendations.
Molecular typing and antibiotic resistance
The minimal inhibitory concentration (MIC) of the antibiotics was determined by broth microdilution in Müller-Hinton using the Sensititre susceptibility plate EUST (Trek Diagnostics Systems, East Grinstead, England; MCS Diagnostics BV, Swalmen, The Netherlands), except for spectinomycin, which was tested using homemade microbroth dilution plates. The MIC was interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines http://www.eucast.org using clinical resistance breakpoints for cefoxitin (> 4 mg/L), chloramphenicol (> 8 mg/L), clindamycin (> 0.5 mg/L), ciprofloxacin (> 1 mg/L), erythromycin (> 2 mg/L), fusidic acid (> 1 mg/L), gentamicin (> 1 mg/L), linezolid (> 4 mg/L), penicillin (> 0.125 mg/L), quinupristin/dalfopristin (> 2 mg/L), rifampicin (> 0.5 mg/L), tetracycline (> 2 mg/L), trimethoprim (> 4 mg/L), and vancomycin (> 2 mg/L). Otherwise, resistance breakpoints were tentatively derived from epidemiological MIC cut off values from EUCAST for kanamycin (> 8 mg/L), mupirocin (> 1 mg/L), streptomycin (> 16 mg/L), spectinomycin (> 128 mg/L), sulfamethoxazole (> 128 mg/L), and tiamulin (> 2 mg/L). Antibiotic resistance genes, including mecA, were detected using a microarray [45]. SCCmec types were determined by multiplex Polymerase Chain Reaction PCR assays [46]. Sequence types (ST) were determined using multilocus sequence typing (MLST) [31]. Spa type was determined as previously described and analysed using the Ridom StaphType software (Ridom StaphType, Ridom GmbH, Würzburg, Germany) [47]. The presence of the Panton-Valentine Leukocidin operon lukS-lukF was determined by PCR as described previously [48].
Statistical Analysis
Sampling calculations were performed using the Win Episcope 2.0 software http://www.clive.ed.ac.uk/winepiscope. The difference in the prevalence between the years was analysed by Chi-square test. The level of significance was 5%. All statistics were performed using the NCSS 2007 statistical software (Hintze J. NCSS (2008) Version 07.1.8. Kaysville, UT, USA).
Authors' contributions
GO wrote the manuscript, supervised the analyses for the detection and identification of MRSA and contributed to the conception of the study. SB designed the sampling plan, organised the sampling at the slaughterhouses and performed statistical analyses. AR carried out the molecular genetic studies. VP designed and coordinated the study, and helped to write the manuscript. All authors read and approved the final manuscript.
Contributor Information
Gudrun Overesch, Email: gudrun.overesch@vetsuisse.unibe.ch.
Sabina Büttner, Email: sabina.buettner@bvet.admin.ch.
Alexandra Rossano, Email: alexandra.rossano@vetsuisse.unibe.ch.
Vincent Perreten, Email: vincent.perreten@vetsuisse.unibe.ch.
Acknowledgements
We thank the participating slaughterhouses and their personnel for the sampling and acquisition of data. We also thank E. Lüthi, I. Brodard, and M. Binggeli for their technical assistance. The Swiss Federal Veterinary Office and the Canton of Bern financed this study.
References
- Cuny C, Friedrich A, Kozytska S, Layer F, Nubel U, Ohlsen K. et al. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int J Med Microbiol. 2010;300:109–117. doi: 10.1016/j.ijmm.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Weese JS. Methicillin-resistant Staphylococcus aureus in animals. ILAR J. 2010;51:233–244. doi: 10.1093/ilar.51.3.233. [DOI] [PubMed] [Google Scholar]
- Vanderhaeghen W, Hermans K, Haesebrouck F, Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol Infect. 2010;138:606–625. doi: 10.1017/S0950268809991567. [DOI] [PubMed] [Google Scholar]
- Smith TC, Pearson N. The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis. 2011;11:327–339. doi: 10.1089/vbz.2010.0072. [DOI] [PubMed] [Google Scholar]
- Wagenaar JA, Yue H, Pritchard J, Broekhuizen-Stins M, Huijsdens X, Mevius DJ. et al. Unexpected sequence types in livestock associated methicillin-resistant Staphylococcus aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China. Vet Microbiol. 2009;139:405–409. doi: 10.1016/j.vetmic.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Guardabassi L, O'Donoghue M, Moodley A, Ho J, Boost M. Novel lineage of methicillin-resistant Staphylococcus aureus, Hong Kong. Emerg Infect Dis. 2009;15:1998–2000. doi: 10.3201/eid1512.090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhaeghen W, Cerpentier T, Adriaensen C, Vicca J, Hermans K, Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet Microbiol. 2010;144:166–171. doi: 10.1016/j.vetmic.2009.12.044. [DOI] [PubMed] [Google Scholar]
- Catry B, van Duijkeren E, Pomba MC, Greko C, Moreno MA, Pyorala S. et al. Reflection paper on MRSA in food-producing and companion animals: epidemiology and control options for human and animal health. Epidemiol Infect. 2010;138:626–644. doi: 10.1017/S0950268810000014. [DOI] [PubMed] [Google Scholar]
- Wulf M, Voss A. MRSA in livestock animals-an epidemic waiting to happen? Clin Microbiol Infect. 2008;14:519–521. doi: 10.1111/j.1469-0691.2008.01970.x. [DOI] [PubMed] [Google Scholar]
- van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D. et al. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis. 2007;13:1834–1839. doi: 10.3201/eid1312.070384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders MN, Haenen AP, Geenen PL, Vesseur PC, Poldervaart ES, Bosch T. et al. Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in The Netherlands. Epidemiol Infect. 2010;138:743–755. doi: 10.1017/S0950268810000075. [DOI] [PubMed] [Google Scholar]
- Khanna T, Friendship R, Dewey C, Weese JS. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet Microbiol. 2008;128:298–303. doi: 10.1016/j.vetmic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- van Belkum A, Melles DC, Peeters JK, van Leeuwen WB, van Duijkeren E, Huijsdens XW. et al. Methicillin-resistant and-susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg Infect Dis. 2008;14:479–483. doi: 10.3201/eid1403.0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TC, Male MJ, Harper AL, Kroeger JS, Tinkler GP, Moritz ED. et al. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS ONE. 2009;4:e4258. doi: 10.1371/journal.pone.0004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijen MML, van Keulen PH, Kluytmans JA. Increase in a Dutch Hospital of methicillin-resistant Staphylococcus aureus related to animal farming. Clin Infect Dis. 2008;46:261–263. doi: 10.1086/524672. [DOI] [PubMed] [Google Scholar]
- van Rijen MM, Bosch T, Heck ME, Kluytmans JA. Meticillin-resistant Staphylococcus aureus epidemiology and transmission in a Dutch hospital. J Hosp Infect. 2009;72:299–306. doi: 10.1016/j.jhin.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Witte W, Strommenger B, Stanek C, Cuny C. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg Infect Dis. 2007;13:255–258. doi: 10.3201/eid1302.060924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potel C, Alvarez-Fernandez M, Constenla L, Alvarez P, Perez S. First human isolates of methicillin-resistant Staphylococcus aureus sequence type 398 in Spain. Eur J Clin Microbiol Infect Dis. 2010;29:351–352. doi: 10.1007/s10096-009-0860-z. [DOI] [PubMed] [Google Scholar]
- European Food Safety Authority. Analysis of the baseline survey on the prevalence of methicillin resistant Staphylococcus aureus (MRSA) in holding with breeding pigs, in the EU, 2008, Part A: MRSA prevalence estimates. EFSA J. 2009;7:1376. [Google Scholar]
- Riesen A, Perreten V. Antibiotic resistance and genetic diversity in Staphylococcus aureus from slaughter pigs in Switzerland. Schweiz Arch Tierheilk. 2009;151:425–431. doi: 10.1024/0036-7281.151.9.425. [DOI] [PubMed] [Google Scholar]
- Nitzsche S, Zweifel C, Stephan R. Phenotypic and genotypic traits of Staphylococcus aureus strains isolated from pig carcasses. Vet Microbiol. 2007;120:292–299. doi: 10.1016/j.vetmic.2006.10.027. [DOI] [PubMed] [Google Scholar]
- Huber H, Koller S, Giezendanner N, Stephan R, Zweifel C. Prevalence and characteristics of meticillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Euro Surveill. 2010;15:1–4. [PubMed] [Google Scholar]
- Büttner S, Kuhn M. Antibiotikaresistenzmonitoring-Jahresbericht. 2008. http://www.bvet.admin.ch/themen
- Jung YH, Shin ES, Kim O, Yoo JS, Lee KM, Yoo JI. et al. Characterization of two newly identified genes, vgaD and vatG, conferring resistance to streptogramin A in Enterococcus faecium. Antimicrob Agents Chemother. 2010;54:4744–4749. doi: 10.1128/AAC.00798-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K, Dunsmore CJ, Fishwick CW, Chopra I. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob Agents Chemother. 2008;52:1737–1742. doi: 10.1128/AAC.01015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis. 2000;31(Suppl 2):S24–S28. doi: 10.1086/314056. [DOI] [PubMed] [Google Scholar]
- Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakwinska O, Kuhn G, Balmelli C, Francioli P, Giddey M, Perreten V. et al. Genetic diversity and ecological success of Staphylococcus aureus strains colonizing humans. Appl Environ Microbiol. 2009;75:175–183. doi: 10.1128/AEM.01860-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megevand C, Gervaix A, Heininger U, Berger C, Aebi C, Vaudaux B. et al. Molecular epidemiology of the nasal colonization by methicillin-susceptible Staphylococcus aureus in Swiss children. Clin Microbiol Infect. 2010;16:1414–1420. doi: 10.1111/j.1469-0691.2009.03090.x. [DOI] [PubMed] [Google Scholar]
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson V, Davison N, Hudson L, Whatmore AM. Staphylococcus aureus ST49 infection in red squirrels. Vet Rec. 2010;167:69. doi: 10.1136/vr.c3625. [DOI] [PubMed] [Google Scholar]
- Otter JA, French GL. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Europe. Lancet Infect Dis. 2010;10:227–239. doi: 10.1016/S1473-3099(10)70053-0. [DOI] [PubMed] [Google Scholar]
- Lewis HC, Mølbak K, Reese C, Aarestrup FM, Selchau M, Sørum M. et al. Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg Infect Dis. 2008;14:1383–1389. doi: 10.3201/eid1409.071576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspiroz C, Lozano C, Vindel A, Lasarte JJ, Zarazaga M, Torres C. Skin lesion caused by ST398 and ST1 MRSA, Spain. Emerg Infect Dis. 2010;16:157–159. doi: 10.3201/eid1601.090694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny C, Strommenger B, Witte W, Stanek C. Clusters of infections in horses with MRSA ST1, ST254, and ST398 in a veterinary hospital. Microb Drug Resist. 2008;14:307–310. doi: 10.1089/mdr.2008.0845. [DOI] [PubMed] [Google Scholar]
- Battisti A, Franco A, Merialdi G, Hasman H, Iurescia M, Lorenzetti R. et al. Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet Microbiol. 2010;142:361–366. doi: 10.1016/j.vetmic.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Juhasz-Kaszanyitzky E, Janosi S, Somogyi P, Dan A, van der Graaf-van Bloois, van Duijkeren E. et al. MRSA transmission between cows and humans. Emerg Infect Dis. 2007;13:630–632. doi: 10.3201/eid1304.060833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broens EM, Graat EAM, van der Wolf PJ, van de Giessen AW, De Jong MCM. Transmission of methicillin resistant Staphylococcus aureus among pigs during transportation from farm to abattoir. Vet J. 2010. [DOI] [PubMed]
- de Neeling AJ, van den Broek MJM, Spalburg EC, van Santen-Verheuvel MG, Dam-Deisz WDC, Boshuizen HC. et al. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet Microbiol. 2007;122:366–372. doi: 10.1016/j.vetmic.2007.01.027. [DOI] [PubMed] [Google Scholar]
- Hasman H, Moodley A, Guardabassi L, Stegger M, Skov RL, Aarestrup FM. Spa type distribution in Staphylococcus aureus originating from pigs, cattle and poultry. Vet Microbiol. 2010;141:326–331. doi: 10.1016/j.vetmic.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Tenhagen BA, Fetsch A, Stuhrenberg B, Schleuter G, Guerra B, Hammerl JA. et al. Prevalence of MRSA types in slaughter pigs in different German abattoirs. Vet Rec. 2009;165:589–593. doi: 10.1136/vr.165.20.589. [DOI] [PubMed] [Google Scholar]
- Harbarth S, Hawkey PM, Tenover F, Stefani S, Pantosti A, Struelens MJ. Update on screening and clinical diagnosis of meticillin-resistant Staphylococcus aureus (MRSA) Int J Antimicrob Agents. 2011;37:110–117. doi: 10.1016/j.ijantimicag.2010.10.022. [DOI] [PubMed] [Google Scholar]
- Lozano C, Aspiroz C, Lasarte JJ, Gomez-Sanz E, Zarazaga M, Torres C. Dynamic of nasal colonization by methicillin-resistant Staphylococcus aureus ST398 and ST1 after mupirocin treatment in a family in close contact with pigs. Comp Immunol Microbiol Infect Dis. 2011;34:e1–e7. doi: 10.1016/j.cimid.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Perreten V, Vorlet-Fawer L, Slickers P, Ehricht R, Kuhnert P, Frey J. Microarray-based detection of 90 antibiotic resistance genes of gram-positive bacteria. J Clin Microbiol. 2005;43:2291–2302. doi: 10.1128/JCM.43.5.2291-2302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J. et al. Combination of multiplex PCRs for Staphylococcal Cassette Chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D. et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter M-O, Gauduchon V. et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]