Abstract
The dependence of Plasmodium vivax on invasion mediated by Duffy binding protein (DBP) makes this protein a prime candidate for development of a vaccine. However, the development of a DBP-based vaccine might be hampered by the high variability of the protein ligand (DBPII), known to bias the immune response toward a specific DBP variant. Here, the hypothesis being investigated is that the analysis of the worldwide DBPII sequences will allow us to determine the minimum number of haplotypes (MNH) to be included in a DBP-based vaccine of broad coverage. For that, all DBPII sequences available were compiled and MNH was based on the most frequent nonsynonymous single nucleotide polymorphisms, the majority mapped on B and T cell epitopes. A preliminary analysis of DBPII genetic diversity from eight malaria-endemic countries estimated that a number between two to six DBP haplotypes (17 in total) would target at least 50% of parasite population circulating in each endemic region. Aiming to avoid region-specific haplotypes, we next analyzed the MNH that broadly cover worldwide parasite population. The results demonstrated that seven haplotypes would be required to cover around 60% of DBPII sequences available. Trying to validate these selected haplotypes per country, we found that five out of the eight countries will be covered by the MNH (67% of parasite populations, range 48–84%). In addition, to identify related subgroups of DBPII sequences we used a Bayesian clustering algorithm. The algorithm grouped all DBPII sequences in six populations that were independent of geographic origin, with ancestral populations present in different proportions in each country. In conclusion, in this first attempt to undertake a global analysis about DBPII variability, the results suggest that the development of DBP-based vaccine should consider multi-haplotype strategies; otherwise a putative P. vivax vaccine may not target some parasite populations.
Introduction
After more than a century of development of malaria control measures, Plasmodium vivax remains more widely distributed than Plasmodium falciparum and it is a potential cause of morbidity and mortality. Around 2.85 billion people live at risk of infection by P. vivax, with the greatest burden occurring in Middle East, Asia, the Western Pacific, Central and South America [1], [2]. Although neglected, P. vivax causes important socioeconomic loss, with the overall global cost of vivax infection estimated as being between US$1.4–4.0 billion per year [3]. The recent emergence of drug-resistant strains and severe (sometimes fatal) disease challenges the traditional view of P. vivax malaria as a benign infection [4], [5]. Consequently, the malaria eradication research agenda (MalERA) placed the P. vivax in the top list of priorities [www.ploscollections.org/malERA2011].
The complex life cycle of the Plasmodium includes an erythrocytic phase that is responsible for clinical symptoms of human malaria. While P. falciparum invades mature as well immature red blood cells (RBC) through multiple invasion pathways, P. vivax invades preferentially reticulocytes [6] and requires mainly the interaction of parasite ligand to Duffy antigen/receptor for chemokines (DARC) on RBC membrane [7]. The P. vivax ligand is a 140 kDa micronemal type I membrane protein, called the Duffy binding protein (DBP), and gene-deletion experiment showed that DBP plays an important role in the irreversible junction of the merozoite with host erythrocytes, a key step of human infection [8]. Cysteine-rich region II of the DBP (DBPII) comprises erythrocyte binding motif known as Duffy-binding-like domain (DBL) [9], which is also found in other erythrocyte binding proteins (erythrocyte binding antigen 175 - EBA-175, EBA-140 and EBA-181) and in cytoadherent proteins (Plasmodium falciparum erythrocyte membrane protein 1) [10]. The crystal structure of the orthologous DBP ligand domain of the simian malaria Plasmodium knowlesi provided insight into the molecular basis for receptor recognition of the PvDBP. The proposed DARC-recognition site of DBP lies in a solvent-accessible groove on a fairly flat surface and exposed site for DARC recognition in subdomain 2 of DBPII [11]. Recently, Bolton and Garry (2011) demonstrated an additional region on subdomain 1 of DBPII that might be also necessary for DARC binding [12].
DBPII is an important anti-P.vivax vaccine candidate since antibodies against this molecule: (i) inhibit in vitro their binding to DARC; (ii) reduce merozoite invasion of human erythrocytes; and (iii) confer protection against blood-stage infection [13], [14], [15], [16]. The important role of DBP-DARC interaction is reinforced by previous studies that showed individuals without DARC on their erythrocytes surface are highly resistant to P. vivax invasion [17], [18]. In addition, studies developed in Brazil and Papua New Guinea showed reduced susceptibility to P. vivax infection in heterozygous carriers of one DARC-negative allele compared to two DARC-positive allele carriers [19], [20]. However, the paradigm of the absolute dependence on the presence of Duffy on the red cell for P. vivax infection has been recently questioned for some findings indicating that P. vivax can infect and cause disease in Duffy-negative people [21], [22], [23]. Nevertheless, this situation seems to occur in specific areas and/or a small proportion of the populations, thus the epidemiological importance of this alternative pathway seem to be restricted to specific endemic areas, such as Madagascar [23].
Analysis of genetic variability from field parasites showed that the DBP binding domain (region II) is highly polymorphic [24], [25], [26], [27], [28], [29], [30], [31], therefore it might hamper the vaccine development as some variable residues alter immune recognition of protein [32], [33], [34]. The excess of non-synonymous substitutions observed in DBPII is consistent with the hypothesis of positive selective pressure acting on this protein domain, and suggests allelic variation as a mechanism of immune evasion [35]. As antigenic variation presents a major limitation in successful vaccine design, we analyzed all DBPII sequences recorded in GenBank in order to identify the main haplotypes shared among P. vivax isolates from different malaria-endemic areas and undertake a detailed analysis of the nucleotide diversity of the dbpII gene. Here we show the need to include a minimum number of DBPII haplotypes in a DBP-based vaccine; otherwise no broad coverage against worldwide P. vivax isolates might be reached.
Materials and Methods
P. vivax isolates
The following 511 Duffy binding protein gene sequences of P. vivax deposited in GenBank (last update on 13th April 2011) were downloaded for genetic analyses: the sequence from a reference strain Sal-I (access number: NC_009911); 113 sequences from Papua New Guinea isolates (PNG) (DQ156519; Wosera area in East Sepik Province: AF289480–AF289483, AF289635–AF289653 and AF291096; Madang town and rural villages within Madang Province: AY970837–AY970925, AF469515–AF469602); 17 sequences from Colombia isolates (COL) obtained from the villages of Villavicencio and Tumaco (U50575–U50590, DQ156513); 15 sequences from South Korea isolates (SK) (DQ156515, DQ156522–DQ156523, AF215737–AF215738, AF220657, AF220659–AF220667); 102 sequences from different parts of India (IND) (FJ491142–FJ491241); 30 sequences from Thailand (THAI) (EF219451, EF368159–EF368180, EF379127–EF379132, EF379134); 100 sequences from Sri Lanka (SLK) (GU143914–GU144013); 123 sequences from different parts of Brazil (BRA) (DQ156520, EU812839–EU812960) and eleven sequences from Iran (IRA) (EU860428–EU860438). Only unique haplotypes for each individual were analyzed.
Data Analysis
Genetic diversity analysis
DBPII sequences were aligned and compared using the Clustal W multiple alignment algorithm in BioEdit Sequence Alignment editor [36] to identify the single nucleotide polymorphisms (SNPs). Gaps were removed from alignments because indels (insertions/deletions) and repeats evolve by different mechanisms than SNPs and might result in false estimates of biologically significant diversity. The number of segregating sites (S), haplotypes (H), nucleotide diversity (π), haplotype diversity (Hd), and the corresponding standard deviations (SD) were estimated using DnaSP 5.10 software [37]. Between-population differentiation using the pairwise fixation index F ST was measured with Arlequin 3.5 software [38]. Haplotype construction. Haplotypes (combinations of nucleotides with no particular weight placed upon any position) were constructed by using DnaSP 5.10. We removed synonymous SNPs to focus the analysis only on the protein diversity. Cluster (population) analysis. To determine whether our sample could be grouped into genetic clusters and to infer the number of clusters (K) that best fit the data, we used the Bayesian clustering method implemented in the Structure 2.3 software [39], [40]. Structure was run 10 times for K = 1–10 for 30,000 Monte Carlo Markov Chain (MCMC) iterations after burn-in period of 10,000 using the admixture model and correlated allele frequencies. We did not use prior information about population origin for each individual (USEPOPINFO = 0). The mean log probability of the data (Ln P[D]) and its standard deviation was plotted to predict the optimal value for K. Graphs of Structure results were produced by using the DISTRUCT program [41].
Results
Polymorphism and genetic differentiation
We compiled 511 sequences from GenBank for the gene fragment encoding Duffy binding protein region II (DBPII). The population dataset included sequences from the natural parasite populations of eight countries (Table 1): Brazil [35], Colombia [25], India and Iran [30], Papua New Guinea [27], [42], [43], South Korea [28], Sri Lanka [31] and Thailand [44]. The average of sample was 64 sequences (ranged from 11 to 123) per country. By analyzing a region of the DBPII of 676 bp that is available for the overall dataset, 127 polymorphic sites were identified (ranged from 16 to 73 per country) with a nucleotide diversity (π) varying between 0.006 and 0.0109 (South Korea and Thailand, respectively). Most of these SNPs (55%) are singletons, i.e. observed only in one sequence. Despite the wide range of number of haplotypes per country (ranged from 9 to 73, mean of 24), the levels of haplotype diversity (Hd) among them were equally high and quite similar (ranged from 0.922 to 0.993 in Sri Lanka and Colombia, respectively). In order to remove most potential sequencing errors that could interfere with the analysis and interpretation of the results, additional analyses were performed excluding singleton polymorphisms (Table S1). By comparing both analyses a significant bias could be detected in the number of segregating sites and haplotypes in South Korea and PNG samples. However, nucleotide (π) and haplotype diversity (Hd) were not significantly affected by this further analysis because these diversity parameters exclude polymorphisms that are present at low frequencies.
Table 1. Estimates of diversity and genetic differentiation for PvDBPII encoding gene among P. vivax isolates.
| Population (N) | S | π (SD) | H | Hd (SD) | F ST | ||||||
| BRA | COL | PNG | SK | THAI | IRA | SLK | |||||
| BRA (123) | 21 | 0.0082 (0.0003) | 35 | 0.935 (0.012) | - | ||||||
| COL (17) | 16 | 0.0085 (0.0007) | 16 | 0.993 (0.023) | 0.163* | - | |||||
| PNG (113) | 73 | 0.0106 (0.0004) | 73 | 0.981 (0.005) | 0.117* | 0.196* | - | ||||
| SK (15) | 18 | 0.0060 (0.0013) | 10 | 0.924 (0.053) | 0.197* | 0.384* | 0.234* | - | |||
| THAI (30) | 29 | 0.0109 (0.0005) | 24 | 0.982 (0.014) | 0.083* | 0.234* | 0.127* | 0.177* | - | ||
| IRA (11) | 17 | 0.0094 (0.0016) | 9 | 0.964 (0.051) | −0.011 | 0.136* | 0.063* | 0.169* | 0.040 | - | |
| SLK (100) | 27 | 0.0097 (0.0005) | 39 | 0.922 (0.014) | 0.029* | 0.186* | 0.129* | 0.200* | 0.101* | 0.018 | - |
| IND (102) | 38 | 0.0088 (0.0005) | 36 | 0.923 (0.016) | 0.011* | 0.183* | 0.120* | 0.200* | 0.077* | 0.004 | 0.018* |
| All (511) | 127 | 0.0101 (0.0002) | 193 | 0.970 (0.004) | |||||||
N: number of isolates; S: number of segregating sites; π: average number of nucleotide substitutions per 1000 sites between pairs of sequences (standard deviation, SD); Hd: haplotype diversity (SD); H: number of haplotypes; FST: Fixation index, a measure of genetic differentiation between populations;
*: F ST values with P<0.05. BRA – Brazil, COL – Colombia, IRA – Iran, SK – South Korea, SLK – Sri Lanka, THAI – Thailand, IND – India, PNG – Papua new Guinea.
To determine how the observed diversity was distributed among geographic regions, population structure was inferred by measuring genetic differentiation among countries (F ST). The highest differentiation was identified between Colombia and South Korea (F ST = 0.384) and the lowest differentiation was detected between Iran and Brazil (F ST = −0.011) (Table 1). We repeated the analyses for F ST using the dataset in which singletons were excluded and no significant differences were observed for F ST values among countries (Table S1).
Haplotype diversity
To focus the analysis on the putative antigenic diversity (i.e. polymorphisms that change protein sequence), the nonsynonymous single nucleotide polymorphism (nsSNP) haplotypes were derived for DBPII sequences. In total 46 nsSNPs were identified among all sequences with most of them being rare (19 nsSNPs with frequency ≤1% and 17 with frequency between 1–10%). Only 10 nsSNPs showed allele frequency above 10% in the whole world (R308S, K371E, G384D, E385K, K386N, H390R, N417K, L424I, W437R, and I503K). All but one nsSNP were found at the eight countries with DBP genetic diversity data available, except for the SNP R308S that was absent in Colombia and South Korea (Table 2). Moreover we investigated if these polymorphisms were localized in regions previously identified or predicted as T- or B-cell epitopes. All but one nsSNP (K371E) are in regions predicted or experimentally identified as epitope in the DBPII (Table 2).
Table 2. Description of the most frequent nonsynonymous (nsSNPs) polymorphisms used to construct DBPII haplotypes, their frequencies in each geographic population and presence in described epitopes.
| nsSNPa | Frequency | |||||||||
| BRA | COL | IRA | SK | SLK | THAI | IND | PNG | World | Epitopeb | |
| R308S | 0.073 | 0.000 | 0.182 | 0.000 | 0.130 | 0.267 | 0.098 | 0.690 | 0.235 | H1, 5 |
| K371E | 0.260 | 0.176 | 0.182 | 0.467 | 0.340 | 0.200 | 0.324 | 0.115 | 0.254 | - |
| G384D | 0.187 | 0.412 | 0.455 | 0.467 | 0.060 | 0.233 | 0.137 | 0.345 | 0.211 | H3, 45 |
| E385K | 0.203 | 0.176 | 0.182 | 0.067 | 0.200 | 0.467 | 0.304 | 0.097 | 0.209 | H3, 45, 48 |
| K386N | 0.228 | 0.176 | 0.182 | 0.067 | 0.200 | 0.400 | 0.304 | 0.097 | 0.211 | H3, 45, 48 |
| H390R | 0.504 | 0.941 | 0.636 | 0.533 | 0.340 | 0.433 | 0.353 | 0.504 | 0.456 | H3, 45, 48 |
| N417K | 0.398 | 0.412 | 0.364 | 0.933 | 0.360 | 0.400 | 0.373 | 0.336 | 0.387 | M2, Ia * |
| L424I | 0.480 | 0.412 | 0.545 | 1.000 | 0.490 | 0.867 | 0.451 | 0.681 | 0.558 | M2, 66, Ia |
| W437R | 0.488 | 0.118 | 0.455 | 1.000 | 0.370 | 0.633 | 0.373 | 0.327 | 0.417 | M3 * |
| I503K | 0.431 | 0.059 | 0.636 | 1.000 | 0.550 | 0.567 | 0.569 | 0.425 | 0.497 | Id, 1638* |
: Amino acid numbers according to SAL-1 sequence [39] with the first letter representing the amino acid present in Sal-1 sequence and the latter representing the polymorphism;
: H1, H3, M2 and M3 are B-cell epitopes [51]; 5 – T and B-cell epitope; 45 and 48 are B-cell epitopes; 66 – T-cell epitope [52], [53]; Ia and Id – MHC class I in silico predicted promiscuous epitopes [35]; 1638 – MHC class II universal epitope [54];
*- polymorphisms that alter the antigenic character of DBPII [32].
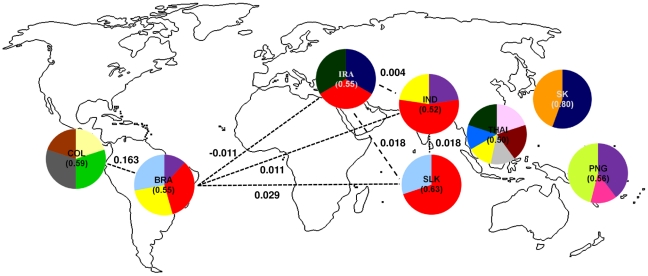
In further analysis, those 10 most frequent polymorphisms were used to build predominant DBPII haplotypes circulating in malaria-endemic areas. Seventy-three haplotypes were defined for the whole dataset, ranging from 7 to 29 haplotypes per country (Table S2). In order to determine a minimum number of DBPII haplotypes (MNH) required to be included in anti-P. vivax vaccine, we next sought to identify the number of haplotypes per country able to cover at least 50% of local parasite population. In the whole dataset, 17 out of 73 haplotypes fitted this criterion: two haplotypes in South Korea and Sri Lanka; three in Papua New Guinea, Iran and India; four haplotypes in Brazil and Colombia and six haplotypes in Thailand (Figure 1 and Table S2). Together, the 17 haplotypes covers about 70% of worldwide parasite population. Among them, seven were shared between two or more areas, being one haplotype (Hap 23, colored in red) found in high frequency in four countries from two different continents (America and Asia). This result agrees with those from F ST analysis, which estimated low genetic differentiation between DBPII sequences from Brazil and Iran, India or Sri Lanka (Table 1, Figure 1 and Table S2). The reference strain Sal-1 sequence, which has being used to develop a DBPII–based vaccine, covers only 10% of worldwide samples (Hap 12, colored in purple in Figure 1). So far, Sal-1 DBPII was detected only in three endemic areas (Brazil, India and PNG), being in very low frequency in other countries (Figure 1 and Table S2).
Figure 1. Frequency by country of the DBPII haplotypes that cover at least fifty percent of local parasite population.
Among the 73 haplotypes defined using the 10 most prevalent nonsynonymous single nucleotide polymorphisms (nsSNPs), 17 haplotypes (each haplotype was represented by a color) were able to cover at least 50% of each country parasite population: 2 in South Korea (Hap 4 – dark blue and Hap 40 - orange) and in Sri Lanka (Hap 23 - red and Hap 44 – light blue); 3 in Iran (Hap 4, Hap 23 and Hap 59 – dark green), in India (Hap 12 - purple, Hap 23 and Hap 24 - yellow) and in PNG (Hap 12, Hap 55 – pink and Hap 64 – light green); 4 in Colombia (Hap 9 – light yellow, Hap 27 – dark gray, Hap 30 - green and Hap 37 - brown) and 4 in Brazil (Hap 12, Hap 23, Hap 24 and Hap 44); 6 in Thailand (Hap 8 – dark brown, Hap 15 – light pink, Hap 21 - blue, Hap 24, Hap 36 – light gray and Hap 59). The haplotype from Sal-1 reference strain was represented by purple (Hap 12), for haplotype sequences please see Table S2. The number in parentheses indicates the frequency of the selected haplotypes in the respective country (range from 0.52 to 0.63). Some F ST values among countries are showed and represented by dashed lines.
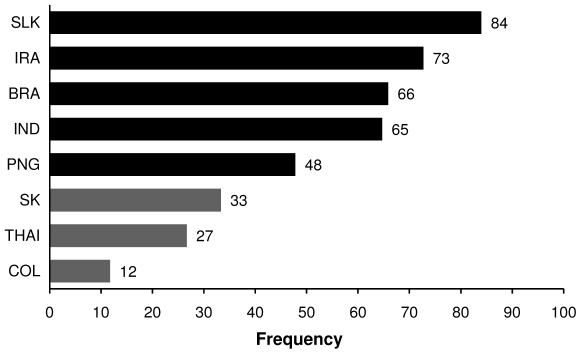
Concerning a more global approach for DBP-based vaccine, we further sought to determine the MNH that will be able to cover the majority (∼50%) of worldwide parasite population independent of the region of origin. Seven haplotypes fitted this criterion and were found in 60% of 511 DBPII sequences of P. vivax deposited in GenBank (Haplotypes 4, 12, 23, 24, 44, 59, 64 of Table S2). Considering the distribution of these 7 selected haplotypes by locality, it was possible to categorize those localities in two groups (Figure 2). The first group includes Sri Lanka, Iran, Brazil, India and PNG, where around 50% of the parasites population will be covered by these haplotypes. The second group includes South Korea, Thailand and Colombia, where about only 24% (range 12–33%) of parasite isolates will be covered by these seven selected haplotypes.
Figure 2. Frequency of the seven nsSNP haplotypes of DBPII that cover at least fifty percent of DBPII sequences deposited in GenBank.
Coverage of the selected haplotypes in each country: countries that at least 50% of parasites are covered by the selected haplotypes are represented by black bars and countries that less than 50% of the parasites are covered by the selected haplotypes are represented by grey bars. The selected haplotypes were: 4, 12, 23, 24, 44, 59, 64 (for haplotype sequences see Table S2).
Clustering
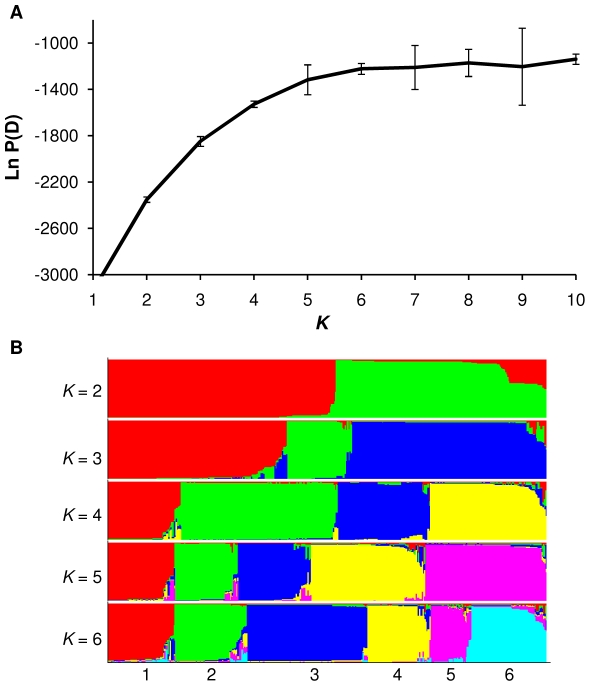
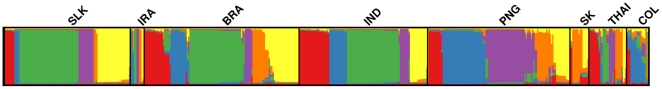
Aiming to identify related subgroups of DBPII sequences among parasites circulating in the study countries, individual samples were clustered to population on the basis of their genotypes, independent of their geographic location. For that, we used the clustering method that uses departures from Hardy-Weinberg equilibrium to detect population structure. The algorithm groups related individuals into a predefined number of clusters (K), herein K = 1–10. A Bayesian approach is taken to infer the K value that provides the best fit to the data as measured by the log-likelihood score. Each individual is then assigned a membership coefficient (Q) to each of the clusters with majority of the haplotypes being assigned to only one cluster at “true” K. The estimated log probability of our data [Ln P(D)] plateaued between K = 4–6 (Figure 3A). Simulation studies have shown that once the real K has been reached, Ln P(D) will typically plateau or continue to increase slightly, indicating that K = 6 provides the best fit to our data. We show clustering results for K values of 2–6, being each individual represented by a vertical line, and each ancestral population in a different color (Figure 3B). To determine whether the above-defined subgroups were geographically restricted we plotted the average Q for each country. For all countries but South Korea the analysis supported low levels of differentiation among geographical regions, with three to six ancestral populations present in each country (Figure 4).
Figure 3. Population structure of the gene encoding DBPII based on Bayesian cluster analysis using Structure 2.3 software.
(A) Plot of the log probability of the data [Ln P(D)] and its standard deviation (vertical bars) given values for K of 1–10. (B) Population clustering for K = 2–6. Each individual is represented by a thin vertical color. Each color represents an ancestral population (pop), and the color of individual represents their proportional membership in the different populations. Ancestral populations: red, pop1; green, pop2; blue, pop3; yellow, pop4; pink, pop5 and light blue, pop6. The figure shown for a given K is based on the highest probability run at that K.
Figure 4. Population structure for P. vivax sequences of DBPII at K = 6.
Graphic of Structure results were produced by using the DISTRUCT program. Individual P. vivax isolate are represented by a thin vertical line from each geographic population showed above the graphic and separated by a thin black line.
Discussion
The major pathway used by P. vivax to invade human reticulocytes depends on the interaction of the DBP with its cognate receptor, which makes DBP a high priority anti-P. vivax vaccine candidate. A significant challenge for the development of DBP-based vaccines is its highly polymorphic nature, which can biases antibodies response toward a specific DBP variant [32], [33], [34]. Engineering vaccines that combine all potential haplotypes that might be circulating in malaria-endemic areas could circumvent polymorphism's limitation; however, it is not feasible for a highly polymorphic antigen like DBP. Therefore, the purpose of this study was to identify predominant DBPII haplotypes that could be included in a putative vaccine, with potential to induce an immune response against P. vivax circulating around the world.
By comparing the DBPII diversity found in different countries worldwide, we demonstrated high levels of haplotype diversity among P. vivax isolates. The profile of DBPII genetic diversity was not related with the levels of malaria endemicity, since similar pattern was observed from areas with low and unstable malaria transmission, such as Brazil, as well as from highly endemic areas such as Papua New Guinea. These findings suggested that recombination plays an important role in determining the haplotype structure of DBPII, as we recently demonstrated [35]. Genetic recombination of parasites takes place in the vector, as part of the Plasmodium life cycle, and is likely facilitated by multiplicity of infections, i.e. the simultaneous infection of a host by more than one parasite variant. For P. falciparum, the levels of multiplicity of infection are partially correlated with the levels of transmission intensities [45]. Nevertheless for P. vivax, strikingly high values of multiplicity of infection are reported even in regions of low endemicity such as Brazil and Thailand. Thus, the proportion of multiple-clone P. vivax infections, estimated by using microsatellites analyses, range from 49–57% in Brazil [46], [47], 10–47% in India [48], [49], 9–60% in Sri Lanka [50] and 52–63% in Thailand [48], [49]. This difference in pattern of multiplicity of infections between P. vivax and P. falciparum could be related with specific biological features of the P. vivax parasite, such as earlier gametocytogenesis [51] and relapse [52]. Early gametocytogenesis might allow for a more efficient transmission to the mosquito vector before symptoms appear and, thus, before drug treatment is initiated, while relapses would also enhance transmission and increase the probability of detecting mixed infections as further inoculations occur.
Besides the role of recombination we have recently demonstrated that natural selection has an important role in the generation and maintenance of the genetic diversity of DBPII [35]. The maintenance of this variability by diversifying selection presumably helps parasite to evade the host immune recognition and favors a low-medium frequency of distinct haplotypes. As we can expect due differences in the endemicity spectrum and immune response profile, both recombination and selection seem to be acting differentially among distinct geographical areas. If no recombination is assumed, we would expect that the number of described polymorphic sites observed would be arranged into a maximum of n+1 haplotypes. Here, this profile was found in the majority of studied countries, some of them with the number of haplotypes lower than the number of segregating sites (South Korea and Iran). However, it is important to highlight that the small number of DBPII sequences available for those two countries might biased the number of haplotypes observed for those regions; specially, because there was a significant correlation between the number of haplotypes and sample size (Spearman correlation coefficient: rs = 0.6628, P = 0.0139). Interestingly, in two countries, Brazil and Sri Lanka, the number of haplotypes was higher than the number of segregating sites, suggesting a major role of recombination in these areas. At this time, it is not possible to conclude if the differences on haplotype number could be consequence of different recombination rates or due to the different levels of selective pressure.
For the purpose of rationalizing vaccine design will be necessary to define polymorphisms that represent antigenically distinct haplotypes. Here, we selected the most frequent nonsynonymous single nucleotide polymorphisms (nsSNPs) to derive DBPII haplotypes. Nine out of 10 nsSNPs lay in regions of DBPII that are immunologically relevant, mapping on previously defined T- and B-cell epitopes [32], [35], [53], [54], [55], [56]. Based on these nsSNPs, we identified a number between two and six DBPII haplotypes, which will be required to cover 50% of parasite population in each studied country. Unfortunately, most of those haplotypes were geographically restricted and we could not find a single high-frequency haplotype covering all endemic areas. Of note, the DBPII Sal-1 variant that is currently being used to develop a DBPII–based vaccine was found so far in low frequency (10%) and it seems to be restricted to specific geographic areas. Aiming to avoid geographically restrict DBPII haplotypes, in the next analysis we determined the MNH that covers at least fifty percent of DBPII sequences available in GenBank independent of the region of origin. By using this approach, seven frequent haplotypes were identified, that broadly cover parasite populations from 5 out of 8 endemic-countries, i.e., Sri Lanka, Iran, Brazil, India and PNG. These same selected haplotypes were not able to cover parasite populations from South Korea, Colombia and Thailand; however, the results for South Korea and Colombia must be interpreted with caution because of the limited number of sequences available. Together, these results show that the development of DBP-based vaccine should considered multi-haplotype strategies. Similar strategy has been proposed to another polymorphic malaria vaccine candidate, the P. falciparum Apical Membrane Antigen 1 (PfAMA-1) [57]. By using a similar approach to that used here, the authors demonstrated that a worldwide collection of PfAMA1 haplotypes could be clustered into six populations that were independent of geographic location. Because continuous culture of P. falciparum has been successfully automated for over 30 years [58], the same authors were able to demonstrate that immunization with one member of PfAMA-1 elicited antibodies that block in vitro parasite invasion against the same subgroup [57]. Altogether, these two studies provide an important proof of concept that vaccine against malaria polymorphic antigens should consider multi-haplotype strategies. Now it remains to be determined whether those DBPII haplotypes described here could elicit broadly immune response. Although essential, those studies will be a difficult task due to limitation on P. vivax culture. Recent progress in short-term culture of P. vivax field isolates [59], [60] and in vitro DBPII-DARC binding assays [15], [61] present new opportunities to improve understanding of the ability of DBPII haplotypes to elicit cross-reactive responses against those that are genetically similar. In addition, these assays might help to define which polymorphisms determine antigenically different DBPII haplotypes.
An additional consideration in the malaria vaccines design is the geographic structuring of the parasite populations because significant variation among regions would suggest a need for vaccines to be tailored accordingly. Overall, we detected low to medium differentiation between countries, except to sequences from Colombia and South Korea, which showed high differentiation compared to the other regions. Remarkably, the four haplotypes that covers more than half Colombian parasites were not shared with any other parasite population, even with the Brazilian, another Latin America parasite population. The contrast among-region structuring was confirmed by F ST analysis with whole data set and clustering analysis using the STRUCTURE software with the ten selected nsSNPs. The Bayesian clustering tool identified six subgroups of related DBPII sequences in the worldwide parasite population. Nevertheless, clustering was not related to the geographical origin of P. vivax isolates and all subgroups were present in the eight endemic countries studied, although in different proportions. Moreover, the large majority of isolates (>75%) was formed predominantly by only one ancestral population. Similar pattern was described for P. falciparum merozoite antigens in a meta-population analysis [62]. In that study, the merozoite antigens generally had lower levels of differentiation among countries. In addition, cluster analysis suggested that haplotypes formed subgroups independent of geographic origin with high levels of diversity within population. Thus, to improve probability of broad efficacy, vaccines based on merozoite antigens as DBPII may incorporate predominant haplotypes from different regions. Further analysis will be required to define the stability of the haplotypes in a region over time, as natural fluctuations can result from frequency dependent selection or new haplotypes can appear as result of recombination. For DBP, a single study so far has investigated the dynamic of asymptomatic P. vivax infections in a cohort of PNG children [63]. By following those children over six-month period, it was possible to demonstrate that the DBPII dominant haplotypes remained relatively stable throughout the study. However, it is difficult to determine the period of time necessary to observe such changes.
The present study comprises the first attempt to undertake a global analysis about DBPII variability and provides new insights on future design of a broad-spectrum P. vivax vaccine. Overcome the limitation of DBP diversity may require the inclusion of representative haplotypes otherwise a large proportion of the P. vivax population will be not target.
Supporting Information
Estimates of genetic diversity and differentiation for PvDBPII encoding gene among P. vivax isolates in the absence of singleton sequences.
(DOC)
Characterization of DBPII haplotypes present in the eight endemic countries studied.
(DOC)
Acknowledgments
We are grateful to all of the researchers whose publically available sequences made this study possible. We also thank the Program for Technological Development in Tools for Health - PDTIS platform (FIOCRUZ) for DNA sequencing facilities.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Research Foundation of Minas Gerais State (FAPEMIG), The Brazilian National Research Council (CNPq), and Oswaldo Cruz Foundation (FIOCRUZ, PAPES V) and Malaria Network/Support Program for Centers of Excellence - Pronex (MS/DECIT, CNPq, FAPEMIG, FAPEMAT and FAPERJ). Scholarships from CNPq (TNS, LHC and CFAB) are also acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 2.Guerra CA, Howes RE, Patil AP, Gething PW, Van Boeckel TP, et al. The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl Trop Dis. 2010;4:e774. doi: 10.1371/journal.pntd.0000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, et al. Vivax malaria: Neglected and not benign. Am J Trop Med Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- 4.Price RN, Douglas NM, Anstey NM. New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr Opin Infect Dis. 2009;22:430–435. doi: 10.1097/QCO.0b013e32832f14c1. [DOI] [PubMed] [Google Scholar]
- 5.Alexandre MA, Ferreira CO, Siqueira AM, Magalhães BL, Mourão MP, et al. Severe Plasmodium vivax malaria, Brazilian Amazon. Emerg Infect Dis. 2010;16:1611–1614. doi: 10.3201/eid1610.100685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mons B, Collins WE, Skinner JC, Vanderstar W, Croon J, et al. Plasmodium vivax - in vitro growth and reinvasion in red blood-cells of aotus-nancymai. Exp Parasitol. 1988;66:183–188. doi: 10.1016/0014-4894(88)90089-6. [DOI] [PubMed] [Google Scholar]
- 7.Horuk R, Chitnis C, Darbonne W, Colby T, Rybicki A, et al. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science. 1993;261:1182–1184. doi: 10.1126/science.7689250. [DOI] [PubMed] [Google Scholar]
- 8.Singh AP, Ozwara H, Kocken CHM, Puri SK, Thomas AW, et al. Targeted deletion of Plasmodium knowlesi Duffy binding protein confirms its role in junction formation during invasion. Mol Microbiol. 2005;55:1925–1934. doi: 10.1111/j.1365-2958.2005.04523.x. [DOI] [PubMed] [Google Scholar]
- 9.Adams JH, Sim BKL, Dolan SA, Fang XD, Kaslow DC, et al. A family of erythrocyte binding-proteins of malaria parasites. Proc Natl Acad Sci U S A. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michon P, Stevens JR, Kaneko O, Adams JH. Evolutionary relationships of conserved cysteine-rich motifs in adhesive molecules of malaria parasites. Mol Biol Evol. 2002;19:1128–1142. doi: 10.1093/oxfordjournals.molbev.a004171. [DOI] [PubMed] [Google Scholar]
- 11.Singh S, Hora R, Belrhali H, Chitnis C, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–744. doi: 10.1038/nature04443. [DOI] [PubMed] [Google Scholar]
- 12.Bolton MJ, Garry RF. Sequence similarity between the erythrocyte binding domain 1 of the Plasmodium vivax Duffy binding protein and the V3 loop of HIV-1 strain MN reveals binding residues for the Duffy Antigen Receptor for Chemokines. Virol J. 2011;8:45. doi: 10.1186/1743-422X-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceravolo IP, Souza-Silva FA, Fontes CJF, Braga EM, Madureira AP, et al. Inhibitory properties of the antibody response to Plasmodium vivax Duffy binding protein in an area with unstable malaria transmission. Scand J Immunol. 2008;67:270–278. doi: 10.1111/j.1365-3083.2007.02059.x. [DOI] [PubMed] [Google Scholar]
- 14.Grimberg BT, Udomsangpetch R, Xainli J, McHenry A, Panichakul T, et al. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med. 2007;4:1940–1948. doi: 10.1371/journal.pmed.0040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michon P, Fraser T, Adams JH. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect Immun. 2000;68:3164–3171. doi: 10.1128/iai.68.6.3164-3171.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King C, Michon P, Shakri A, Marcotty A, Stanisic D, et al. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci U S A. 2008;105:8363–8368. doi: 10.1073/pnas.0800371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller L, Mason S, Clyde D, McGinniss M. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 18.Welch SG, McGregor IA, Williams K. The Duffy blood group and malaria prevalence in Gambian West Africans. Trans R Soc Trop Med Hyg. 1977;71:295–296. doi: 10.1016/0035-9203(77)90102-x. [DOI] [PubMed] [Google Scholar]
- 19.Sousa TN, Sanchez BAM, Ceravolo IP, Carvalho LH, Brito CFA. Real-time multiplex allele-specific polymerase chain reaction for genotyping of the Duffy antigen, the Plasmodium vivax invasion receptor. Vox Sang. 2007;92:373–380. doi: 10.1111/j.1423-0410.2007.00902.x. [DOI] [PubMed] [Google Scholar]
- 20.Kasehagen L, Mueller I, Kiniboro B, Bockarie M, Reeder J, et al. Reduced Plasmodium vivax erythrocyte infection in PNG Duffy-negative heterozygotes. PLoS One. 2007;2:e336. doi: 10.1371/journal.pone.0000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan JR, Stoute JA, Amon J, Dunton RF, Mtalib R, et al. Evidence for transmission of Plasmodium vivax among a Duffy antigen negative population in western Kenya. Am J Trop Med Hyg. 2006;75:575–581. [PubMed] [Google Scholar]
- 22.Cavasini CE, de Mattos LC, Couto AAD, Bonini-Omingos CR, Valencia SH, et al. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception? Trans R Soc Trop Med Hyg. 2007;101:1042–1044. doi: 10.1016/j.trstmh.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Menard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci U S A. 2010;107:5967–5971. doi: 10.1073/pnas.0912496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sousa TN, Ceravolo IP, Fontes CJF, Couto A, Carvalho LH, et al. The pattern of major polymorphisms in the Duffy binding protein ligand domain among Plasmodium vivax isolates from the Brazilian Amazon area. Mol Biochem Parasitol. 2006;146:251–254. doi: 10.1016/j.molbiopara.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Ampudia E, Patarroyo MA, Patarroyo ME, Murillo LA. Genetic polymorphism of the Duffy receptor binding domain of Plasmodium vivax in Colombian wild isolates. Mol Biochem Parasitol. 1996;78:269–272. doi: 10.1016/s0166-6851(96)02611-4. [DOI] [PubMed] [Google Scholar]
- 26.Tsuboi T, Kappe SH, Alyaman F, Prickett MD, Alpers M, et al. Natural variation within the principal adhesion domain of the Plasmodium vivax Duffy binding-protein. Infect Immun. 1994;62:5581–5586. doi: 10.1128/iai.62.12.5581-5586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xainli J, Adams J, King C. The erythrocyte binding motif of Plasmodium vivax duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol Biochem Parasitol. 2000;111:253–260. doi: 10.1016/s0166-6851(00)00315-7. [DOI] [PubMed] [Google Scholar]
- 28.Kho W, Chung J, Sim E, Kim D, Chung W. Analysis of polymorphic regions of Plasmodium vivax Duffy binding protein of Korean isolates. Korean J Parasitol. 2001;39:143–150. doi: 10.3347/kjp.2001.39.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gosi P, Fukuda M, Schaecher K, Lanar D, Khusmith S, et al. Polymorphisms of Plasmodium vivax duffy binding protein in isolates from Thai patients. Am J Trop Med Hyg. 2007;77:247–247. [Google Scholar]
- 30.Babaeekho L, Zakeri S, Djadid ND. Genetic mapping of the duffy binding protein (DBP) ligand domain of Plasmodium vivax from unstable malaria region in the Middle East. Am J Trop Med Hyg. 2009;80:112–118. [PubMed] [Google Scholar]
- 31.Premaratne PH, Aravinda BR, Escalante AA, Udagama-Randeniya PV. Genetic diversity of Plasmodium vivax Duffy Binding Protein II (PvDBPII) under unstable transmission and low intensity malaria in Sri Lanka. Infect Genet Evol. 2011 doi: 10.1016/j.meegid.2011.04.023. Apr 28. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 32.VanBuskirk KM, Cole-Tobian JL, Baisor M, Sevova ES, Bockarie M, et al. Antigenic drift in the ligand domain of Plasmodium vivax Duffy binding protein confers resistance to inhibitory antibodies. J Infect Dis. 2004;190:1556–1562. doi: 10.1086/424852. [DOI] [PubMed] [Google Scholar]
- 33.Ceravolo IP, Sanchez BA, Sousa TN, Guerra BM, Soares IS, et al. Naturally acquired inhibitory antibodies to Plasmodium vivax Duffy binding protein are short-lived and allele-specific following a single malaria infection. Clin Exp Immunol. 2009;156:502–510. doi: 10.1111/j.1365-2249.2009.03931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cole-Tobian JL, Michon P, Biasor M, Richards JS, Beeson JG, et al. Strain-specific duffy binding protein antibodies correlate with protection against infection with homologous compared to heterologous Plasmodium vivax strains in Papua New Guinean children. Infect Immun. 2009;77:4009–4017. doi: 10.1128/IAI.00158-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sousa TN, Tarazona-Santos EM, Wilson DJ, Madureira AP, Falcao PR, et al. Genetic variability and natural selection at the ligand domain of the Duffy binding protein in Brazilian Plasmodium vivax populations. Malar J. 2010;9:334. doi: 10.1186/1475-2875-9-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 37.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 38.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 39.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg N. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- 42.Gosi P, Khusmith S, Khalambaheti T, Lanar D, Schaecher K, et al. Polymorphism patterns in Duffy-binding protein among Thai Plasmodium vivax isolates. Malar J. 2008;7:112. doi: 10.1186/1475-2875-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole-Tobian JL, Cortes A, Baisor M, Kastens W, Xainli J, Bockarie M, Adams JH, King CL. Age-acquired immunity to a Plasmodium vivax invasion ligand, the duffy binding protein. J Infect Dis. 2002;186:531–539. doi: 10.1086/341776. [DOI] [PubMed] [Google Scholar]
- 44.Cole-Tobian JL, Biasor M, King CL. High complexity of Plasmodium vivax infections in Papua New Guinean children. Am J Trop Med Hyg. 2005;73:626–633. [PubMed] [Google Scholar]
- 45.Anderson T, Haubold B, Williams J, Estrada-Franco J, Richardson L, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 46.Rezende A, Tarazona-Santos E, Fontes C, Souza J, Couto A, et al. Microsatellite loci: determining the genetic variability of Plasmodium vivax. Trop Med Int Health. 2010;15:718–726. doi: 10.1111/j.1365-3156.2010.02535.x. [DOI] [PubMed] [Google Scholar]
- 47.Ferreira MU, Karunaweera ND, da Silva-Nunes M, da Silva NS, Wirth DF, et al. Population structure and transmission dynamics of plasmodium vivax in rural amazonia. J Infect Dis. 2007;195:1218–1226. doi: 10.1086/512685. [DOI] [PubMed] [Google Scholar]
- 48.Imwong M, Nair S, Pukrittayakamee S, Sudimack D, Williams JT, et al. Contrasting genetic structure in Plasmodium vivax populations from Asia and south America. Int J Parasitol. 2007;37:1013–1022. doi: 10.1016/j.ijpara.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 49.Imwong M, Snounou G, Pukrittayakamee S, Tanomsing N, Kim JR, et al. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J Infect Dis. 2007;195:927–933. doi: 10.1086/512241. [DOI] [PubMed] [Google Scholar]
- 50.Karunaweera ND, Ferreira MU, Hartl DL, Wirth DF. Fourteen polymorphic microsatellite DNA markers for the human malaria parasite Plasmodium vivax. Mol Ecol Notes. 2007;7:172–175. [Google Scholar]
- 51.McKenzie FE, Jeffery GM, Collins WE. Gametocytemia and fever in human malaria infections. J Parasitol. 2007;93:627–633. doi: 10.1645/GE-1052R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krotoski WA. The hypnozoite and malarial relapse. Prog Clin Parasitol. 1989;1:1–19. [PubMed] [Google Scholar]
- 53.Chootong P, Ntumngia F, Vanbuskirk K, Xainli J, Cole-Tobian J, et al. Mapping Epitopes of the Plasmodium vivax Duffy Binding Protein with Naturally Acquired Inhibitory Antibodies. Infect Immun. 2010;78:1089–1095. doi: 10.1128/IAI.01036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole-Tobian J, King CL. Diversity and natural selection in Plasmodium vivax Duffy binding protein gene. Mol Biochem Parasitol. 2003;127:121–132. doi: 10.1016/s0166-6851(02)00327-4. [DOI] [PubMed] [Google Scholar]
- 55.Xainli J, Baisor M, Kastens W, Bockarie M, Adams J, et al. Age-dependent cellular immune responses to Plasmodium vivax Duffy binding protein in humans. J Immunol. 2002;169:3200–3207. doi: 10.4049/jimmunol.169.6.3200. [DOI] [PubMed] [Google Scholar]
- 56.Saravia C, Martinez P, Granados D, Lopez C, Reyes C, et al. Identification and evaluation of universal epitopes in Plasmodium vivax Duffy binding protein. Biochem Biophys Res Commun. 2008;377:1279–1283. doi: 10.1016/j.bbrc.2008.10.153. [DOI] [PubMed] [Google Scholar]
- 57.Duan J, Mu J, Thera MA, Joy D, Kosakovsky Pond SL, et al. Population structure of the genes encoding the polymorphic Plasmodium falciparum apical membrane antigen 1: implications for vaccine design. Proc Natl Acad Sci U S A. 2008;105:7857–7862. doi: 10.1073/pnas.0802328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 59.Russell BM, Udomsangpetch R, Rieckmann KH, Kotecka BM, Coleman RE, et al. Simple in vitro assay for determining the sensitivity of Plasmodium vivax isolates from fresh human blood to antimalarials in areas where P. vivax is endemic. Antimicrob Agents Chemother. 2003;47:170–173. doi: 10.1128/AAC.47.1.170-173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Udomsangpetch R, Somsri S, Panichakul T, Chotivanich K, Sirichaisinthop J, et al. Short-term in vitro culture of field isolates of Plasmodium vivax using umbilical cord blood. Parasitol Int. 2007;56:65–69. doi: 10.1016/j.parint.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 61.Hans D, Pattnaik P, Bhattacharyya A, Shakri AR, Yazdani SS, et al. Mapping binding residues in the Plasmodium vivax domain that binds Duffy antigen during red cell invasion. Mol Microbiol. 2005;55:1423–1434. doi: 10.1111/j.1365-2958.2005.04484.x. [DOI] [PubMed] [Google Scholar]
- 62.Barry AE, Schultz L, Buckee CO, Reeder JC. Contrasting population structures of the genes encoding ten leading vaccine-candidate antigens of the human malaria parasite, Plasmodium falciparum. PLoS One. 2009;4:e8497. doi: 10.1371/journal.pone.0008497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cole-Tobian JL, Michon P, Dabod E, Mueller I, King CL. Dynamics of asymptomatic Plasmodium vivax infections and Duffy binding protein Polymorphisms in relation to Parasitemia levels in Papua new guinean children. Am J Trop Med Hyg. 2007;77:955–962. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Estimates of genetic diversity and differentiation for PvDBPII encoding gene among P. vivax isolates in the absence of singleton sequences.
(DOC)
Characterization of DBPII haplotypes present in the eight endemic countries studied.
(DOC)