Abstract
Tyrosine nitration is a low-abundance post-translational protein modification that requires appropriate enrichment techniques to enable proteomic analyses. We report a simple yet highly specific method to enrich nitropeptides by chemoprecipitation involving only two straightforward chemical modifications of the nitropeptides before capturing the obtained derivatives with a strategically designed solid-phase active ester reagent. Specifically, capping of the aliphatic amines in the peptides is done first by reductive methylation to preserve the charge state of peptides for electrospray ionization mass spectrometric analysis, followed by reduction of nitrotyrosines to the corresponding aminotyrosines. These peptides are then immobilized on the solid-phase active ester reagent, whereas other peptides carrying no free amino groups are separated from the immobilized species by thoroughly washing the beads from which the tagged peptide derivatives can easily be released by acid-catalyzed hydrolysis at room temperature. The benefits of selective enrichment from a matrix of unmodified peptides for liquid chromatography-tandem mass spectrometry are demonstrated on three synthetic nitropeptides that are nitrated fragments of biologically relevant proteins. Identification of several in vitro nitrated human plasma proteins, also implicated under various pathological processes, by database searches from the enriched and tagged tryptic nitropeptides is presented as a practical application. We also show that converting the nitro-group to the small 4-formylbenzoylamido tag does not significantly alter fragmentation properties upon collision-induced dissociation compared with those of the native nitropeptides, and at the same time this derivatization actually improves electron capture dissociation due to conversion of the electron-predator nitro-group to this novel tag.
Tyrosine (Tyr, Y) nitration is a low-abundance posttranslational protein modification associated with significant changes in protein functions and implicated in a variety of biological processes and diseases (1, 2). 3-Nitrotyrosine has been considered the biomarker of nitrative stress (3). Nitration of a protein Tyr decreases the pKa of the neighboring phenolic hydroxyl group, rendering it mostly deprotonated at physiological pH that ultimately results in altered protein binding and protein interactions (4). Identifications of protein targets for posttranslational nitration should be considered only putative, when two-dimensional polyacrylamide gel electrophoresis (2D-PAGE)1 in combination with Western blot detection of nitrotyrosine immunoreactive spots is used (5) because these studies usually do not seek or cannot obtain direct information about specific nitration sites (6), although there have been reports on MS/MS-based localizations of tyrosine nitration from peptides after digestion of proteins in relevant 2D-PAGE spots (7). On the other hand, immunoprecipitation and gel electrophoresis apparently suffer from serious limitations, when they are used as front-end procedures to enable the exploration of the nitroproteome in vivo by liquid chromatography-tandem MS (LC-MS/MS) (8). Since MS/MS-based misidentifications of nitropeptides have been revealed (9, 10), appropriate enrichment of the usually very scarce nitrated species (1) from an overwhelming “matrix” of unmodified proteins or peptides before LC-MS/MS analyses should be considered crucial for discovery-driven investigations of this posttranslational modification on a proteome-wide scale (11–15). In addition, critical analysis of MS/MS data is also required, when instruments furnishing low mass resolution and accuracy are used (9, 10).
Chemical labeling of nitrotyrosines is carried out after conversion to the corresponding aminotyrosines (4), because the amino group offers a convenient chemical handle for covalent derivatization (“tagging”) that enables the enrichment of nitropeptides from the matrix of unmodified species. Reduction to aminotyrosines also introduces a chromatographic shift that has been proposed as a basis to distinguish nitropeptides from unmodified matrix species (16), although unreliable fragmentation of the reduced peptides (17), time-consuming procedure because of the need for meticulous offline fractionation, and potential dynamic range issues are, among others, of concern with this strategy. Biotin tagging and subsequent avidin immunoaffinity-based enrichment have been reported for nitropeptides after converting nitrotyrosines to aminotyrosines (11).
Because aliphatic amino groups abundantly present in the proteolytic digest can also be partially tagged even at acidic pH, capping by acetylation before reducing the nitrotyrosines to aminotyrosines has been introduced (12). With acylation, the accompanying decrease in functional groups responsible for the ionization of peptides may, however, result in an unwanted loss of peptide ion signals, when ESI is the method of choice to enable online data-dependent LC-MS/MS analysis (13). Another disadvantage of acylation is that removal of O-acyl intermediates requires treatment with a large quantity of an additional chemical (18) or harsh reaction conditions (15). A more selective method than biotin tagging for avidin immunoaffinity enrichment to capture aminotyrosine peptides and facilitate mass spectrometric analysis utilizes solid-phase chemistry (“chemoprecipitation”) on thiopropyl Sepharose beads after a four-step derivatization resulting in acetylated sulfhydryl peptides (12). In general, newer enrichment strategies (12–15) tend to involve multiple-step chemical conversions to obtain the corresponding tagged peptides from a complex biological medium, which may not only result in low overall efficiency making unequivocal identification of nitropeptides difficult or ambiguous, but also requires cumbersome and time-consuming sample preparation (12–14).
To overcome these shortcomings, our goal was to develop a simple yet highly specific method to identify nitration sites in peptides/proteins by significantly decreasing the number of chemical modifications required for the selective capture of nitropeptides from biological matrices by solid-phase chemistry. Before reducing the nitropeptides to the corresponding aminopeptides, we used reductive methylation to block aliphatic amines inherently present in proteolytic peptides and to avoid potential loss of peptide signals (13). Selective chemoprecipitation was performed by a novel, strategically designed solid-phase active ester reagent (SPAER) on glass beads from which the tagged peptides could be easily recovered via acid-catalyzed hydrolysis at room temperature.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
All chemicals and reagents were purchased from Sigma-Aldrich unless otherwise specified. Succinimidyl-4-formylbenzoate was obtained from Pierce (Rockford, IL). Human plasma was obtained from Innovative Research, Inc. (Novi, MI). Trypsin was purchased from Applied Biosystems Inc. (Foster City, CA). VSFLSALEEY*TK, GDY*DLNAVR, IINEPTAAAIAY*GLDK, and EGYY*GYTGAFR (where Y* denotes nitrotyrosine) were custom-synthesized by Synthetic Biomolecules (San Diego, CA), and they were used for the experiments without further purification.
Preparation of the Solid-phase Active Ester Reagent (SPAER)
Aminopropyl glass beads (0.2 mmol/g loading) were converted to solid-phase hydrazide (SPH) beads as reported before (19). The SPH beads (250 mg) were then reacted with 120 mg (0.48 mmol) of succinimidyl-4-formylbenzoate in 5 ml of acetonitrile/H2O, 7:3 (v/v), containing 0.2% acetic acid (pH 4) at room temperature for 5 h with continuous shaking. The obtained SPAER beads were filtered off and washed thoroughly with 3 × 3 ml of reaction medium, 3 × 3 ml of acetonitrile and 2 × 2 ml of dichloromethane, respectively. The glass beads were then dried in vacuo and stored under nitrogen at −20 °C.
Nitration of Human Plasma
Human plasma (200 μl) was nitrated with tetranitromethane (0.5 μl) for 5 min at room temperature (20). Plasma proteins were immediately precipitated by the addition of 4 sample volumes of cold (−20 °C) acetone (Pierce technical note http://www.piercenet.com/files/TR0049-Acetone-precipitation.pdf). The sample was then vortexed and incubated at −20 °C for 2 h. After centrifugation at 13000g for 10 min, the supernatant was removed and discarded. The pellet was washed with cold acetone and the sample was centrifuged again. The supernatant was removed and the pellet was carefully dried with a gentle stream of nitrogen. The pellet was then resuspended in 100 mm ammonium bicarbonate buffer containing 8 m urea (pH 8, freshly prepared using electrophoresis-grade urea). Protein concentration was estimated by the BCA protein assay kit (Pierce, Rockford, IL). Before digestion, the sample (∼5 mg of protein) was reduced with 5 mm dithiothreitol at 56 °C for 30 min and then alkylated with 10 mm iodoacetamide for 30 min at room temperature in the dark. Afterward, the mixture was diluted with 4 volumes of 100-mm ammonium bicarbonate buffer, and trypsin (1:50 trypsin to protein, w/w) was added for overnight digestion at 37 °C.
Preparation of Nitropeptide-containing Matrices for Solid-phase Enrichment
Synthetic model nitropeptides (VSFLSALEEY*TK, GDY*DLNAVR, IINEPTAAAIAY*GLDK, approx. 1 nmol each) in BSA digest (0.25 mg, 1 mg/ml) or nitrated human plasma digest obtained above was first undertaken to reductive methylation with 0.5% formaldehyde and 50 mm sodium cyanoborohydride in 100 mm sodium acetate buffer (pH 5.5) at 37 °C for 1 h (21). Excess formaldehyde was quenched with 0.5% NH4OH (22). Subsequently, solid-phase extraction (SPE, C18 Sep-Pak, Waters, Milford, MA) was done. The eluted peptides were dried by speedvac and nitrotyrosines were reduced to the corresponding aminotyrosines with 500-fold molar excess of Na2S2O4 in 100 mm phosphate buffer (pH = 8) at room temperature for 30 min in a customary manner (4). Excess Na2S2O4 was decomposed by acidifying the solution (with acetic acid, pH 3) for 30 min.
Alternatively, we have also used immobilized reagents to develop a “one-pot” derivatization method, without the need of sample clean up to obtain dimethylated aminotyrosines for the enrichment process by SPAER. Briefly, formaldehyde (final concentration 0.5% v/v) was added to 250 μl of peptide mixture in bovine serum albumin (BSA) tryptic digest, and the sample was incubated with 20 mg of immobilized cyanoborohydride polymer (∼2 mmol/g loading, Sigma) by end-over-end rotation for 2 h at room temperature (LC-MS/MS analysis showed a complete dimethylation similarly to that of solution-phase cyanoborohydiride method). The cyanoborohydride polymer was removed by centrifugation at 11000 rpm for 30 s, and the resin was washed with 50 μl of sodium acetate buffer. The pH of the combined solutions was adjusted to 5.5 and 250 mg of aminopropyl glass beads (19) were added to quench excess formaldehyde by end-over-end rotation for 4 h, at room temperature. The beads were then removed by centrifugation at 11,000 rpm for 30 s and washed with the reaction medium (100 μl). After adjusting the pH of the combined solutions to 8, nitrotyrosines were reduced to aminotyrosines, as described above. Aliquots of the solution (400 μl) were then used for the enrichment process without further purification.
Enrichment of Nitropeptides Using SPAER
The pH of the above solution containing the dimethylated and reduced peptides was adjusted to 5.5 followed by the addition of 10 mg of SPAER. The resultant slurry was rotated end-over-end at room temperature for 4 h. SPAER was then pelleted by centrifugation, and the supernatant was removed. The beads were thoroughly washed with 400 μl of solvent each time in the following order to remove non-aminotryrosine containing peptides and impurities: H2O (4×), 80% acetonitrile/H2O containing 0.1% acetic acid (4×), acetonitrile (3x) and, then, H2O. The captured aminotyrosines were released from the glass beads with 200 μl of 95% (v/v) trifluoroacetic acid/H2O at room temperature within 1 h. The solvent was evaporated under a nitrogen stream, and the 4-formylbenzoylamido-tagged peptides were desalted by solid-phase extraction (bonded C18 pipette Ziptips, Millipore, Billerica, MA) before online data-dependent reverse phase (RP) LC-MS/MS analysis.
Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry
An Eksigent nano-LC-2D system (Dublin, CA) with a 15 cm × 75 μm PepMap C18 (LC Packings, Dionex, San Francisco, CA) nanoflow column was used for the analyses. Mobile phases containing solvent A [0.1% acetic acid and 99.9% water (v/v)] and solvent B [0.1% acetic acid and 99.9% acetonitrile (v/v)] were at a constant flow rate of 250 nL/min. Five microliters of peptide mixture in 4.8% acetonitrile, 95.1% water, and 0.1% acetic acid were automatically loaded onto the IntegraFritTM sample trap (2.5 cm × 75 μm i.d.) (New Objective, Woburn, MA) for sample concentration and desalting, at a flow rate of 1.5 μl/min in a loading solvent of 0.1% (v/v) acetic acid and 5% (v/v) acetonitrile in 94.9% (v/v) water. Separations were performed on the C18 column and equilibrated for 5 min at 4.8% solvent B, followed by a 45-min gradient for the BSA digest spiked with three nitropeptides or a 90-min gradient for the nitrated plasma to 40% solvent B. Solvent B was then held at 40% for 5 min, increasing up to 90% for the next 5 min and finally at 4.8% within 10 min. LC-ESI-MS/MS analysis was performed using a hybrid linear ion trap-Fourier transform ion cyclotron resonance (7-Tesla) mass spectrometer (LTQ-FT, Thermo Fisher, San Jose, CA) equipped with the manufacturer's nanoelectrospray ionization source and operated with the Xcalibur (version 2.0) and Tune Plus (version 2.2) data acquisition software.
Data-dependent mode of acquisition was used in which an accurate m/z survey scan was performed in the Fourier transform (FT)-ion cyclotron resonance (ICR) cell followed by parallel MS/MS linear ion trap analysis of the top five most intense precursor ions. FT-ICR full-scan mass spectra were acquired at 50000 mass resolving power (m/z 400) from m/z 350 to 1500 using the automatic gain control mode of ion trapping. Collision-induced dissociation (CID) was performed in the linear ion trap using a 4.0-Th isolation width and 35% normalized collision energy with helium as the collision gas. Singly charged precursors and unassigned charge states were excluded from the precursor selection. Also, the precursor ion that had been selected for CID was dynamically excluded from further MS/MS analysis for 60 s.
Electron Capture Dissociation-Fourier-Transform Ion Cyclotron Resonance Mass Spectrometry
EGYY*GYTGAFR or #EGYYΔGYTGAFR (prepared from EGYY*GYTGAFR according to the procedure described above; viz. reductive methylation followed by reduction of Y* to aminotyrosine, then capture by and release from SPAER) was prepared in methanol/water (50:50, v/v/) containing 1% acetic acid in a concentration of 3 pmol/μl. The sample was electrosprayed at a flow rate of 3 μl/min. Precursor ions were isolated in the linear ion trap of linear trap quadrupole (LTQ)-FT and, then, transferred into the ICR cell for electron capture dissociation (ECD). The isolation width was 5 Da. Electrons were produced by an indirectly heated (with current typically around 1.1 A) barium-tungsten cylindrical dispenser cathode, and ions trapped in the ICR cell were irradiated for 200 ms at −5 V cathode potential. ECD mass spectra were averaged over 2 min of data acquisition.
Data Analysis
Bioworks (version 3.3, Thermo) was used to generate peak lists for database search. Mascot (Matrix Science, London, UK; version 2.2) and Sequest (Thermo; version 27, rev. 12) were used to identify peptides by searching the International Protein Index (IPI) human database (version 3.71, 86745 entries). Trypsin was selected as the digesting enzyme and one missed cleavage was allowed. X! Tandem (The GPM, thegpm.org; version 2007.01.01.1) was set up to search a subset of the ipi_HUMAN_v3.71 database. Mascot and Sequest were searched with parent-ion and fragment-ion mass tolerances of 25 ppm and 0.80 Da respectively. Carbamidomethylation of cysteine (57.0215 Da) dimethylation of N terminus (28.0313 Da), and dimethylation of lysine (28.0313 Da) were specified as fixed modifications. Oxidation of methionine (15.99492 Da) and tagging of tyrosine (see Fig. 1, 147.0320 Da) were set as variable modifications in Mascot and Sequest searches. Scaffold (version Scaffold_3_00_01, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted at greater than 95% probability as determined by the peptide prophet algorithm (23). Protein identifications were evaluated by the Protein Prophet algorithm (24). MS/MS spectra from enriched peptides were manually annotated using theoretical CID-MS/MS fragment ion masses obtained from the MS-Product program available through Protein Prospector (prospector.ucsf.edu). To discard potential false positives, validation was performed according to the flowchart in our earlier report (9). The data associated with this manuscript may be downloaded from the ProteomeCommons.org Tranche network using the following hash: Yu23mxL2Xfo6HJfXZjmn8CSFPqhQSCsX2nj2X/s9WoHunq0w3owJ6YGaYjgBJZxcB3LQiXYccXFD5n36P623zn2e3CkAAAAAAAADEg==.
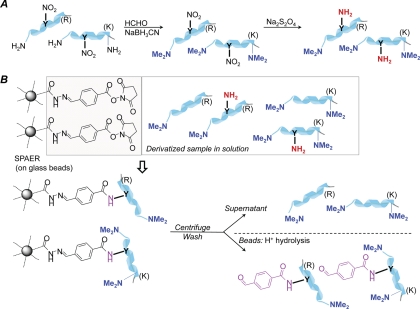
Fig. 1.
A, Schematic representation of the two-step derivatization procedure before chemoprecipitation of aminotyrosines by SPAER. B, Enrichment of nitropeptides by solid-phase active ester reagent (SPAER).
RESULTS AND DISCUSSION
Our simple enrichment method involves only two chemical modifications on the nitropeptides to allow subsequent chemoprecipitation (Fig. 1). First blocking the aliphatic amines by reductive methylation (21) is performed to preserve the charge state of peptides for ESI-MS analysis (13), which proceeds with practically quantitative yield, in agreement with previous reports (22, 25) and also shown (supplemental Fig. S1) by LC-ESI-MS/MS of a mixture containing the derivatized IINEPTAAAIAY*GLDK, VSFLSALEEY*TK and GDY*DLNAVR. Reduction of nitrotyrosines to the corresponding aminotyrosines is carried out by a well-established protocol that documented about 98% yield for proteomic analysis (12). Our actual enrichment technique illustrated in Fig. 1 relies on the immobilization of aminotyrosines on SPAER. Unmodified peptides that did not yield aminotyrosine upon precapture derivatization are removed by thorough washing of the beads from which the 4-formylbenzoyl-tagged peptide derivatives can easily be released for LC-MS analysis by acid-catalyzed hydrolysis at room temperature. We derived SPAER from an SPH reagent (19) that our laboratory has been successfully using to enrich peptide carbonyls (26, 27) by reacting SPH with the commercially available 4-formylbenzoic acid N-succinimidyl ester. We believe that other active esters (28) may also be used that can be synthesized either a priori or in situ on the glass beads. Efficiency of chemoprecipitation by SPAER was confirmed by showing that a single enrichment step using 10 mg of beads for 0.4 ml sample solution already immobilized all the modified peptides, since essentially no more target peptides were captured when the procedure was repeated with a fresh batch of SPAER.
The simplicity and high specificity of our method have been demonstrated by selectively enriching three synthetic nitropeptides from a biological matrix for LC-MS/MS analysis. These nitropeptides, IINEPTAAAIAY*GLDK, VSFLSALEEY*TK, and GDY*DLNAVR are the modified fragments of the heat shock protein implicated in the autoimmune inflammatory disorder of the central nervous system (29), apolipoprotein A-I associated with the development of atherosclerosis (30), and the NF-κB p65 subunit that causes the inactivation of this transcription factor (31), respectively.
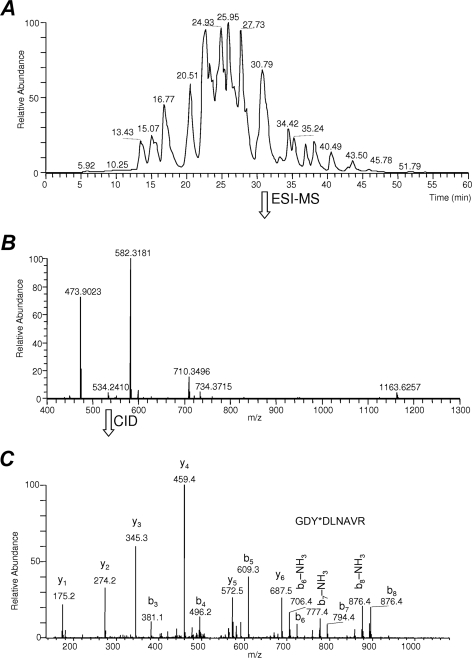
To mimic a biological matrix such as plasma proteins, BSA tryptic digest was spiked with VSFLSALEEY*TK, GDY*DLNAVR, and IINEPTAAAIAY*GLDK, in >200-fold excess in molar ratio of unmodified peptides versus nitropeptides (considering 1201 Da as an average molecular weight of BSA tryptic peptides according to an in silico digestion performed by the MS-Digest module of Protein Prospector, http://prospector.ucsf.edu, assuming no missed cleavages and carbamidomethylation of Cys). Fig. 2 shows the data-dependent online RP-LC-ESI-MS/MS analysis of this peptide mixture in BSA digest obtained by a linear ion trap-Fourier transform ion cyclotron resonance hybrid mass spectrometer coupled with a nano-LC system. As an example, we show that, in the full-scan ESI mass spectrum recorded at retention time (tR) of 30.79 min, GDY*DLNAVR ([M+2H]+ at m/z 534.2410, Fig. 2B) is overwhelmed by BSA tryptic peptides SLHTLFGDELC@K (where C@ denotes carbamidomethylated Cys; [M+3H]3+ at m/z 473.9023 and [M+2H]2+ at m/z 710.3496) and DLVNELTEFAK ([M+2H]2+ at m/z 582.3181). It is, therefore, apparent from this model experiment that low-abundance nitropeptides may not be chosen during data-dependent acquisitions for subsequent MS/MS scans from a peptide matrix represented by the proteolytic digest of a complex biological fluid such as plasma. For reference, Fig. 2C also shows the MS/MS product-ion spectrum of GDY*DLNAVR obtained by CID of the doubly protonated precursor ion at m/z 534.2 (MS/MS product-ion spectra of VSFLSALEEY*TK and IINEPTAAAIAY*GLDK obtained by CID of their [M+H]2+ precursor ions are available in supplemental Fig. S2A and S3A, respectively).
Fig. 2.
A, Base peak chromatogram of IINEPTAAAIAY*GLDK, VSFLSALEEY*TK and GDY*DLNAVR (with Y* denoting 3-nitrotyrosine) added to BSA tryptic digest indicating that unmodified peptides may bias against low-abundance nitropeptides upon data-dependent LC-ESI-MS/MS analysis because of potential undersampling (i. e. the modified peptides may elude selection for tandem mass spectrometry and, thus, preclude identification) and/or suppression by overwhelming matrix components (26, 27). We selected at tR of 30.79 min GDY*DLNAVR ([M+2H]+ at m/z 534.2410) as a representative example to show: B, The full-scan ESI mass spectrum where the nitropeptide was detected; C, CID-MS/MS product-ion spectrum recorded from m/z 534.2 as precursor.
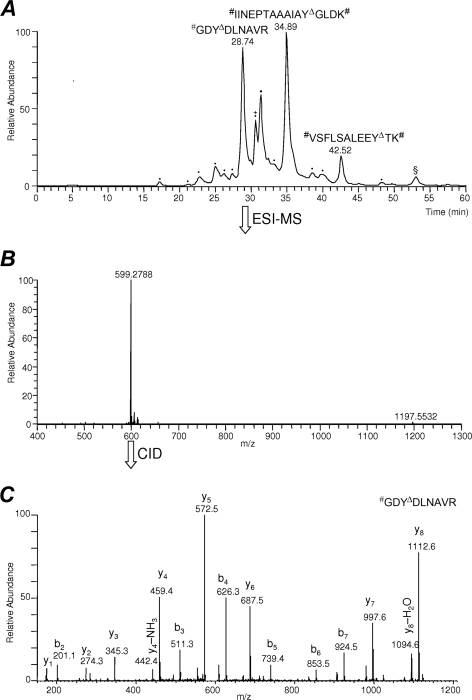
Fig. 3 summarizes data-dependent online LC-MS/MS analysis of the sample obtained from nitropeptides spiked into the BSA tryptic digest after completing the procedure shown in Fig. 1. The full-scan mass spectrum recorded at tR of 28.74 min (Fig. 3B) in this initial proof-of-concept study demonstrates that the 4-formylbenzoyl-tagged derivative of the dimethylated and reduced GDY*DLNAVR eluted without interference from the non-nitrated peptides of BSA tryptic digest whose presence was minimized to a few contaminating species that were not completely removed from the enriched sample. However, the introduction of SPAER immobilized on the surface of glass beads for chemoprecipitation would allow for a more aggressive washing with organic solvents to remove adsorbed but not covalently immobilized peptides and other synthetic impurities than the one used here in the procedure, which will be exploited for future improvement of the reported method in this regard. Additional method refinements may include optimization of SPAER quantity necessary for the consistent performance of chemoprecipitation; however, a procedure reported recently for phosphopeptide enrichment (32) used TiO2-beads in quantity identical to that of SPEAR beads in our nitropeptide enrichment method, which indicated that handling in this scale (i.e. 10 mg of beads) might be the most convenient for routine use in batch mode. Pipette-tips filled with beads may also be developed as an alternative (33), especially to handle small sample volumes.
Fig. 3.
A, Base peak chromatogram of SPAER-enriched low-abundance nitropeptides #IINEPTAAAIAYΔGLDK#, #VSFLSALEEYΔTK# and #GDYΔDLNAVR (where # indicates dimethylation of the amino terminus and YΔ denotes conversion of 3-nitrotyrosine to the 4-formylbenzoylamido derivative) showing interference-free data-dependent LC-ESI-MS/MS analysis of the tagged species. B, Full-scan ESI mass spectrum at tR = 28.74 min of #GDYΔDLNAVR ([M+2H]+ at m/z 599.2788) is shown as an example for derivatized and tagged nitropeptide, C, CID-MS/MS product-ion spectrum recorded from m/z 599.2788 as precursor. Peaks marked with dots were non-peptidic impurities, whereas the peak with the § was singly charged species excluded from MS/MS analysis upon data-dependent acquisition. As a minor constituent in the peak also marked with the + sign, the fully dimethylated BSA tryptic peptide #LFTFHADIC@TLPDTEK# ([M+2H]+ at m/z 655.33; @: carbamidomethylated Cys) was identified as the only non-modified matrix species present.
As shown in Fig. 3C, CID-MS/MS spectra of the derivatized and tagged peptide, #GDYΔDLNAVR [where YΔ represents 3-(4-formylbenzamido)tyrosine and # indicates dimethylation of the amino terminus], show fragmentation properties similar to the parent nitropeptide GDY*DLNAVR that permitted its unequivocal identification by MS/MS. Specifically, although the doubly charged GDY*DLNAVR gave adjoining y1 to y6 and b3 to b8 series (Fig. 2C), [M+2H]2+ of #GDYΔDLNAVR displayed, without giving irregular fragmentation patterns reported for 3-aminotyrosine-containing peptides (17), contiguous y1 to y8 and b2 to b7 series of fragment ions upon CID in the linear ion trap. Similar features were observed for the two additional nitropeptides (IINEPTAAAIAY*GLDK and VSFLSALEEY*TK) and their corresponding tagged peptides enriched from BSA tryptic digest (see supplemental Figs. S2B and S3B, respectively) by the procedure summarized in Fig. 1.
Next, the fidelity of our method to explore the nitroproteome has been demonstrated by making convincing identifications of nitroproteins from a highly complex and biologically relevant matrix, specifically from nitrated human plasma. Plasma protein nitration has been implicated in aging (34) and a wide variety of pathological processes, including inflammation (35), thrombosis (36), kidney (37), lung (38, 39), and heart diseases (40). In particular, nitration of fibrinogen has been observed in these diseases (41). In addition, nitrated albumin and serotransferrin were also identified in the blood of patients undergoing hemodialysis (37). Detection of the nitration of these proteins was done by immunoassays or immunoprecipitation followed by Western blotting.
To make the nitrated human plasma, we relied on a procedure frequently applied to the constituents of the plasma proteome (16, 42, 43), but made some with an important adjustment. Specifically, reaction time was shortened significantly [to 5 min from 20 min (43) and 1 h (16, 42), respectively] to limit the extent of protein modification. For comparison, light and heavy chains of IgG1 monoclonal antibody appeared to undergo nitration in about 20% and >40%, respectively, when the reaction was conducted for 20 min (43) and using reagent to protein ratio somewhat higher than those reported here in the Experimental Procedures section [The extent of protein nitration was not reported in and could not be inferred from references (16) and (42)]. Overall, nitroproteins in our treated plasma sample represented about 14% of the proteome based on estimation through spectral counts (261 for identified nitroproteins out of 1815 for all identified proteins, when processed by Scaffold) after shotgun LC-MS/MS analyses, albeit the latter method appeared to overestimate the actual degree of tyrosine nitration. In addition, the latter varied significantly within and among proteins of the human plasma, as illustrated in supplemental Fig. S4. For example, we obtained about 0.7 and 60% for the relative abundance of Y*ICENQDSISSK and QNCELFEQLGEY*K, respectively, in albumin, and 2% for the abundance of FNWY*VDGVEVHNAK in the IGHG_1 protein based on the extracted-ion chromatograms of the modified and unmodified tryptic peptides in the sample. These observations also support the hypothesis about the selectivity of protein nitration in a complex proteome (1, 2).
Our results for in vitro protein nitration of human plasma using SPAER for chemoprecipitation and database searches relying on CID-MS/MS spectra obtained through data-dependent acquisition during online nano-RPLC separation are summarized in Table I. Noteworthy, that nitration of several lower-abundance proteins (e.g. α-2 macroglobulin, fibronectin, haptoglobin, and hemopexin) could not be identified without enrichment. Only eight untagged peptides from abundant plasma proteins were identified possible because of carryover from incomplete rinsing of the beads that, again, will be addressed in a future study aimed at improving the protocol by introducing additional washing steps involving organic solvents such as dimethylformamide. Annotated MS/MS spectra of the identified nitropeptide derivatives are given in supplemental Fig. S5. We found that one hit not reaching p < 0.05 threshold for identification by Mascot (#LSITGTYΔDLK# Table I) was actually above the threshold required for identification by Sequest (44) and also passed validation according to our recently published procedure (9). On the other hand, Mascot search convincingly identified five peptides nitrated at internal Tyr residues, yet they eluded identification by Sequest. Therefore, we recommend combining results from these complementary search algorithms upon application of our enrichment procedure for the exploration of the nitroproteome. We also showed that the CID-MS/MS spectrum of #YΔLQEIYNSNNQK#, indicating the nitration of Tyr-135 of the gamma-B isoform of fibrinogen γ-chain (41) was identical to that of the derivatized and tagged synthetic peptide (supplemental Fig. S6A in the Supporting Information online). Similarly, we confirmed the identification of #EGYYΔGYTGAFR (Table I) indicating Tyr-534 nitration in serotransferrin, a modification that has been found in the blood of patients with kidney failure (37), by comparing the CID-MS/MS of the plasma-derived species to that of the authentic synthetic peptide (see supplemental Fig. S6B).
Table I. Peptides identified from nitrated human plasma after trypsin digestion and completing the derivatization and enrichment procedure summarized in Fig. 1. # Represents dimethylation of N-terminal amino-group and ϵ-amino-group of Lys (K); @ Denotes carbamidomethylation of Cys (C); Δ Designates the final tag (C8H6NO2-) on the tyrosine residues of enriched peptides. Peptide identifications were accepted at <10 ppm precursor-ion mass accuracy and greater than 95% probability as determined by the Peptide Prophet algorithm. All MS/MS spectra of identified proteins were manually annotated using the peptide fragmentation program MS-Product through Protein Prospector (University of California, San Francisco). Protein identifications assigned by the Protein Prophet algorithm were mostly at >99%. α-1-acid glycoprotein 2 was identified at 73% protein probability by Mascot and 94% by Sequest; isoform 1 of fibronectin was at 73% protein probability by Mascot and 94% by Sequest; α-1-acid glycoprotein 1 and IGHG1 protein probabilities were 73% and 60%, respectively. Only few (8) untagged peptides from abundant plasma proteins were identified (possible due to carryover from incomplete washing of the beads): #ALVLIAFAQYLQQC@PFEDHVK#, #HPDYSVVLLLR, #RHPDYSVVLLLR, #RHPYFYAPELLFFAK#, #SLHTLFGDK#LC@TVATLR, #DVFLGMFLYEYAR, #EFNAETFTFHADIC@TLSEK#, and #FNK#PFVFLMIEQNTK# by both Mascot and Sequest.
| IPI No. | Protein name | Peptide sequence | Nitration site(s) | Charge (+) | Mascot score (threshold, p < 0.05) | Sequest Xcorr (threshold for ID:a 2.5 for 2+; 3.8 for 3+) |
|---|---|---|---|---|---|---|
| IPI00022434 | Albumin | #YΔLYEIAR | 172 | 2 | 39.5 (34.1) | 2.42 |
| IPI00022434 | Albumin | #RHPYFYΔAPELLFFAK# | 174 | 2 | 44.9 (33.9) | 5.81 |
| IPI00022434 | Albumin | #RHPYΔFYΔAPELLFFAK# | 172, 174 | 2 | 48.8 (33.9) | 5.05 |
| IPI00022434 | Albumin | #YΔK#AAFTEC@C@QAADK# | 186 | 2 | 58.6 (31.1) | 4.54 |
| IPI00022434 | Albumin | #YΔIC@ENQDSISSK# | 287 | 2 | 49.9 (30.5) | 3.48 |
| IPI00022434 | Albumin | #DVFLGMFLYEYΔAR | 358 | 2 | 74.4 (33.3) | 4.31 |
| IPI00022434 | Albumin | #QNC@ELFEQLGEYΔK# | 425 | 2 | 63.5 (31.1) | 4.01 |
| IPI00022434 | Albumin | #MPC@AEDYΔLSVVLNQLC@VLHEK# | 476 | 2 | 78.0 (33.2) | 5.48 |
| IPI00022429 | α-1-acid glycoprotein 1 | #YΔVGGQEHFAHLLILR | 109 | 2 | 75.5 (34.1) | 5.07 |
| IPI00020091 | α-1-acid glycoprotein 2 | #EQLGEFYΔEALDC@LC@IPR | 160 | 2 | 69.8 (30.9) | 5.77 |
| IPI00553177 | α-1-antitrypsin | #ELDRDTVFALVNYΔIFFK# | 211 | 3 | 48.3 (33.4) | 4.52 |
| IPI00553177 | α-1-antitrypsin | #LSITGTYΔDLK# | 321 | 2 | 30.7 (34.3) | 3.06 |
| IPI00478003 | α-2-macroglobulin | #LLIYΔAVLPTGDVIGDSAK# | 543 | 2 | 46.5 (31.8) | 3.78 |
| IPI00478003 | α-2-macroglobulin | #VGFYΔESDVMGR | 708 | 2 | 49.4 (29.6) | 3.14 |
| IPI00478003 | α-2-macroglobulin | #AFQPFFVELTMPYΔSVIR | 800 | 2 | 63.2 (34.4) | 4.11 |
| IPI00478003 | α-2-macroglobulin | #AIGYΔLNTGYQR | 1007 | 2 | 47.5 (33.9) | N/A |
| IPI00021841 | Apolipoprotein A-I | #DYΔVSQFEGSALGK# | 53 | 2 | 53.9 (33.0) | 4.06 |
| IPI00021841 | Apolipoprotein A-I | #VSFLSALEEYΔTK# | 260 | 2 | 80.9 (34.7) | 4.36 |
| IPI00021854 | Apolipoprotein A-II | #EPC@VESLVSQYΔFQTVTDYGK#DLMEK# | 37 | 3 | 56.7 (32.1) | N/A |
| IPI00021854 | Apolipoprotein A-II | #EPC@VESLVSQYFQTVTDYΔGK# | 44 | 2 | 66.4 (32.9) | N/A |
| IPI00783987 | Complement C3 | #AAVYΔHHFISDGVR | 219 | 2 | 81.9 (33.6) | 3.45 |
| IPI00022418 | Fibronectin | #NTFAEVTGLSPGVTYYΔFK# | 974 | 2 | 60.6 (34.3) | 5.25 |
| IPI00021891 | Fibrinogen γ chain | #FGSYC@PTTC@GIADFLSTYΔQTK# | 58 | 3 | 75.3 (31.2) | 5.22 |
| IPI00021891 | Fibrinogen γ chain | #YΔLQEIYNSNNQK# | 135 | 2 | 40.4 (33.6) | 4.35 |
| IPI00641737 | Haptoglobin | #DIAPTLTLYΔVGK#K# | 230 | 2 | 54.2 (30.3) | 3.05 |
| IPI00641737 | Haptoglobin | #YΔVMLPVADQDQC@IR | 311 | 2 | 42.9 (32.5) | 3.58 |
| IPI00641737 | Haptoglobin | #VMPLC@LPSK#DYΔAEVGR | 286 | 2 | 44.7 (34.0) | 3.64 |
| IPI00022488 | Hemopexin | #DYΔFMPC@PGR | 227 | 2 | 32.2 (24.3) | 2.83 |
| IPI00022488 | Hemopexin | #GGYΔTLVSGYPK# | 335 | 2 | 35.7 (34.6) | N/A |
| IPI00022463 | Serotransferrin | #MYLGYΔEYVTAIR | 336 | 2 | 54.1 (33.9) | 4.18 |
| IPI00022463 | Serotransferrin | #EGYYΔGYTGAFR | 534 | 2 | 40.0 (33.9) | 3.65 |
| IPI00448938 | IGHG1 protein | #FNWYΔVDGVEVHNAK# | 313 | 2 | 81.7 (33.5) | N/A |
a See ref. (44).
Complementary to the CID method of ion dissociation, the electron-based ECD and ETD have been increasingly used for MS/MS characterization of posttranslationally modified peptides (45, 46). However, the electron predator effect of the NO2-group (47) has been noted, which limits the capacity of ECD to obtain sequence information on nitropeptides (48). Therefore, an additional benefit of replacing the nitro- with the 4-formylbenzoylamido-group in the modified peptide is that this conversion has permitted the recording of ECD mass spectra that are devoid of strong hydroxyl-radical (HO•), water and ammonia losses observed for nitropeptides (48) and, hence, significantly improved sequence coverage (see supplemental Fig. S7). The chemical conversion abolishing electron predator effect will also be beneficial for the ETD method of ion dissociation, that is an MS/MS technique related to ECD and increasingly used for analyses of posttranslational modifications upon coupling with online LC separation (49).
CONCLUSIONS
In summary, we have developed a novel and simple chemoprecipitation method for the highly selective enrichment of nitrotyrosine-containing peptides from complex biological matrices involving only two straightforward derivatization steps on the nitropeptides before their covalent capture from the matrix by a strategically designed SPAER on glass beads. After converting nitrotyrosines having N-dimethylated aliphatic amino groups to the corresponding aminotyrosines for their immobilization on SPAER, their tagged forms are released by acid-catalyzed hydrolysis at room temperature for LC-MS/MS analysis or subsequent additional enrichment (e.g. by SPH chemistry) (19, 26, 27), if necessary. The simplicity and efficiency of the SPEAR approach has been demonstrated by enriching three synthetic nitropeptides derived from biologically important proteins (29–31) from BSA tryptic digest. We have also demonstrated that the small 4-formylbenzoylamido tag on the released peptides preserves the fragmentation properties of those of the parent nitropeptides making their MS/MS-based identification unambiguous. Identification of protein nitration in human plasma by database searches from enriched and tagged tryptic nitropeptides was also presented as an example for practical application of SPEAR for the exploration of the nitroproteome. Several nitrated proteins we found in the human plasma (e.g. fibrinogen, serotransferrin, albumin, apoprotein A-1) by SPAER enrichment have also been identified in the plasma of patients having various pathological conditions (34–41).
Acknowledgments
We thank Navin Rauniyar for his help in data analysis. Laszlo Prokai is the Robert A. Welch Chair in Biochemistry at the University of North Texas Health Science Center (endowment number BK-0031).
Footnotes
* This research was supported by the National Institutes of Health, grant number AG025384.
 This article contains supplemental Figs. S1 to S7.
This article contains supplemental Figs. S1 to S7.
1 The abbreviations used are:
- 2D-PAGE
- two-dimensional polyacrylamide gel electrophoresis
- SPAER
- solid-phase active ester reagent
- SPH
- solid-phase hydrazide
- ESI
- electrospray ionization
- SPE
- solid phase extraction
- BSA
- bovine serum albumin
- RPLC
- reverse-phase liquid chromatography
- LTQ
- linear trap quadrupole
- FT
- Fourier transform
- ECD
- electron capture dissociation
- CID
- collision-induced dissociation.
REFERENCES
- 1. Radi R. (2004) Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. U.S.A. 101, 4003–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abello N., Kerstjens H. A., Postma D. S., Bischoff R. (2009) Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteome Res. 8, 3222–3238 [DOI] [PubMed] [Google Scholar]
- 3. Tsikas D. (2010) Analytical methods for 3-nitrotyrosine quantification in biological samples: The unique role of tandem mass spectrometry. Amino Acids DOI 10.1007/s00726-010-0604-5 [DOI] [PubMed] [Google Scholar]
- 4. Sokolovsky M., Riordan J. F., Vallee B. L. (1967) Conversion of 3-nitrotyrosine to 3-aminotyrosine in peptides and proteins. Biochem. Biophys. Res. Commun. 27, 20–25 [DOI] [PubMed] [Google Scholar]
- 5. Amoresano A., Chiappetta G., Pucci P., D'Ischia M., Marino G. (2007) Bidimensional tandem mass spectrometry for selective identification of nitration sites in proteins. Anal. Chem. 79, 2109–2117 [DOI] [PubMed] [Google Scholar]
- 6. Butterfield D. A., Sultana R. (2008) Identification of 3-nitrotyrosine-modified brain proteins by redox proteomics. Methods Enzymol. 440, 295–308 [DOI] [PubMed] [Google Scholar]
- 7. Zhan X., Desiderio D. M. (2004) The human pituitary nitroproteome: detection of nitrotyrosyl-proteins with two-dimensional Western blotting, and amino acid sequence determination with mass spectrometry. Biochem. Biophys. Res. Commun. 325, 1180–1186 [DOI] [PubMed] [Google Scholar]
- 8. Bigelow D. J., Qian W. J. (2008) Quantitative proteome mapping of nitrotyrosines. Methods Enzymol. 440, 191–205 [DOI] [PubMed] [Google Scholar]
- 9. Stevens S. M., Jr., Prokai-Tatrai K., Prokai L. (2008) Factors that contribute to the misidentification of tyrosine nitration by shotgun proteomics. Mol. Cell. Proteomics 7, 2442–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prokai L. (2009) Misidentification of nitrated peptides: Comments on Hong, S.J., Gokulrangan, G., Schöneich, C., (2007) Proteomic analysis of age-dependent nitration of rat cardiac proteins by solution isoelectric focusing coupled to nanoHPLC tandem mass spectrometry. Exp. Gerontol. 44, 367–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nikov G., Bhat V., Wishnok J. S., Tannenbaum S. R. (2003) Analysis of nitrated proteins by nitrotyrosine-specific affinity probes and mass spectrometry. Anal. Biochem. 320, 214–222 [DOI] [PubMed] [Google Scholar]
- 12. Zhang Q., Qian W. J., Knyushko T. V., Clauss T. R., Purvine S. O., Moore R. J., Sacksteder C. A., Chin M. H., Smith D. J., Camp D. G., 2nd, Bigelow D. J., Smith R. D. (2007) A method for selective enrichment and analysis of nitrotyrosine-containing peptides in complex proteome samples. J. Proteome Res. 6, 2257–2268 [DOI] [PubMed] [Google Scholar]
- 13. Nuriel T., Deeb R. S., Hajjar D. P., Gross S. S. (2008) Protein 3-nitrotyrosine in complex biological samples: quantification by high-pressure liquid chromatography/electrochemical detection and emergence of proteomic approaches for unbiased identification of modification sites. Methods Enzymol. 441, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee J. R., Lee S. J., Kim T. W., Kim J. K., Park H. S., Kim D. E., Kim K. P., Yeo W. S. (2009) Chemical approach for specific enrichment and mass analysis of nitrated peptides. Anal. Chem. 81, 6620–6626 [DOI] [PubMed] [Google Scholar]
- 15. Abello N., Barroso B., Kerstjens H. A., Postma D. S., Bischoff R. (2010) Chemical labeling and enrichment of nitrotyrosine-containing peptides. Talanta 80, 1503–1512 [DOI] [PubMed] [Google Scholar]
- 16. Ghesquière B., Colaert N., Helsens K., Dejager L., Vanhaute C., Verleysen K., Kas K., Timmerman E., Goethals M., Libert C., Vandekerckhove J., Gevaert K. (2009) In vitro and in vivo protein-bound tyrosine nitration characterized by diagonal chromatography. Mol. Cell. Proteomics 8, 2642–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghesquière B., Goethals M., Van Damme J., Staes A., Timmerman E., Vandekerckhove J., Gevaert K. (2006) Improved tandem mass spectrometric characterization of 3-nitrotyrosine sites in peptides. Rapid Commun. Mass Spectrom. 20, 2885–2893 [DOI] [PubMed] [Google Scholar]
- 18. Miller B. T., Rogers M. E., Smith J. S., Kurosky A. (1994) Identification and characterization of O-biotinylated hydroxy amino acid residues in peptides. Anal. Biochem. 219, 240–248 [DOI] [PubMed] [Google Scholar]
- 19. Roe M. R., Xie H., Bandhakavi S., Griffin T. J. (2007) (2007) Proteomic mapping of 4-hydroxynonenal protein modification sites by solid-phase hydrazide chemistry and mass spectrometry. Anal. Chem. 79, 3747–3756 [DOI] [PubMed] [Google Scholar]
- 20. Sokolovsky M., Riordan J. F., Vallee B. L. (1966) Tetranitromethane. A reagent for the nitration of tyrosyl residues in proteins. Biochemistry 5, 3582–3589 [DOI] [PubMed] [Google Scholar]
- 21. Dottavio-Martin D., Ravel J. M. (1978) Radiolabeling of proteins by reductive alkylation with [14C]formaldehyde and sodium cyanoborohydride. Anal. Biochem. 87, 562–565 [DOI] [PubMed] [Google Scholar]
- 22. Hsu J. L., Chen S. H., Li D. T., Shi F. K. (2007) Enhanced a1 fragmentation for dimethylated proteins and its applications for N-terminal identification and comparative protein quantitation. J. Proteome Res. 6, 2376–2383 [DOI] [PubMed] [Google Scholar]
- 23. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 24. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 25. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J. R. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 26. Rauniyar N., Stevens S. M., Prokai-Tatrai K., Prokai L. (2009) Characterization of 4-hydroxy-2-nonenal-modified peptides by liquid chromatography-tandem mass spectrometry using data-dependent acquisition: neutral loss-driven MS3 versus neutral loss-driven electron capture dissociation. Anal. Chem. 81, 782–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rauniyar N., Prokai-Tatrai K., Prokai L. (2010) Identification of carbonylation sites in apomyoglobin after exposure to 4-hydroxy-2-nonenal by solid-phase enrichment and liquid chromatography-electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 45, 398–410 [DOI] [PubMed] [Google Scholar]
- 28. Prokai-Tatrai K., Prokai L., Bodor N. (1996) Brain-targeted delivery of a leucine-enkephalin analogue by retrometabolic design. J. Med.Chem. 39, 4775–4782 [DOI] [PubMed] [Google Scholar]
- 29. Qi X., Lewin A. S., Sun L., Hauswirth W. W., Guy J. (2006) Mitochondrial protein nitration primes neurodegeneration in experimental autoimmune encephalomyelitis. J. Biol. Chem. 281, 31950–31962 [DOI] [PubMed] [Google Scholar]
- 30. Shao B., Bergt C., Fu X., Green P., Voss J. C., Oda M. N., Oram J. F., Heinecke J. W. (2005) Tyrosine 192 in apolipoprotein A-I is the major site of nitration and chlorination by myeloperoxidase, but only chlorination markedly impairs ABCA1-dependent cholesterol transport. J. Biol. Chem. 280, 5983–5993 [DOI] [PubMed] [Google Scholar]
- 31. Park S. W., Huq M. D., Hu X., Wei L. N. (2005) Tyrosine nitration on p65: a novel mechanism to rapidly inactivate nuclear factor-kappaB. Mol. Cell. Proteomics 4, 300–309 [DOI] [PubMed] [Google Scholar]
- 32. Gnad F., Forner F., Zielinska D. F., Birney E., Gunawardena J., Mann M. (2010) Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria. Mol. Cell. Proteomics 9, 2642–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stevens S. M., Jr., Chung A. Y., Chow M. C., McClung S. H., Strachan C. N., Harmon A. C., Denslow N. D., Prokai L. (2005) Enhancement of phosphoprotein analysis using a fluorescent affinity tag and mass spectrometry. Rapid Commun Mass Spectrom. 19, 2157–2162 [DOI] [PubMed] [Google Scholar]
- 34. Kim C. H., Zou Y., Kim D. H., Kim N. D., Yu B. P., Chung H. Y. (2006) Proteomic analysis of nitrated and 4-hydroxy-2-nonenal-modified serum proteins during aging. J. Gerontol. A –Biol. Sci. Med. Sci. 61, 332–338 [DOI] [PubMed] [Google Scholar]
- 35. Ischiropoulos H. (1998) Biological tyrosine nitration: A pathophysiological function of nitric oxide and reactive oxygen species. Arch. Biochem. Biophys. 356, 1–11 [DOI] [PubMed] [Google Scholar]
- 36. Nowak P., Zbikowska H. M., Ponczek M., Kolodziejczyk J., Wachowicz B. (2007) Different vulnerability of fibrinogen subunits to oxidative/nitrative modifications induced by peroxynitrite: Functional consequences. Thromb. Res. 121, 163–174 [DOI] [PubMed] [Google Scholar]
- 37. Mitrogianni Z., Barbouti A., Galaris D., Siamopoulos K. C. (2004) Tyrosine nitration in plasma proteins from patients undergoing hemodialysis. Am. J. Kidney Disease 44, 286–292 [DOI] [PubMed] [Google Scholar]
- 38. Pignatelli B., Li C. Q., Boffetta P., Chen Q., Ahrens W., Nyberg F., Mukeria A., Bruske-Hohlfeld I., Fortes C., Constantinescu V., Ischiropoulos H., Ohshima H. (2001) Nitrated and oxidized plasma proteins in smokers and lung cancer patients. Cancer Res. 61, 778–784 [PubMed] [Google Scholar]
- 39. Gole M. D., Souza J. M., Choi I., Hertkorn C., Malcolm S., Foust R. F., 3rd, Finkel B., Lanken P. N., Ischiropoulos H. (2000) Plasma proteins modified by tyrosine nitration in acute respiratory distress syndrome. Am. J. Physiol. Lung Cell. Mol. Physiol. 278, L961–L967, [DOI] [PubMed] [Google Scholar]
- 40. Bakillah A. (2009) Nitrated apolipoprotein A-I, a potential new cardiovascular marker, is markedly increased in low high-density lipoprotein cholesterol subjects. Clin. Chem. Lab. Med. 47, 60–69 [DOI] [PubMed] [Google Scholar]
- 41. Tang Z., Wu H., Du D., Wang J., Wang H., Qian W. J., Bigelow D. J., Pounds J. G., Smith R. D., Lin Y. (2010) Sensitive immunoassays of nitrated fibrinogen in human biofluids. Talanta 81, 1662–1669 [DOI] [PubMed] [Google Scholar]
- 42. Walcher W., Franze T., Weller M.G., Pöschl U., Huber C. G. (2003) Liquid- and gas-phase nitration of bovine serum albumin studied by LC-MS and LC-MS/MS using monolithic columns. J. Proteome Res. 2, 534–542 [DOI] [PubMed] [Google Scholar]
- 43. Liu H., Gaza-Bulseco G., Chumsae C., Radziejewski C. H. (2008) Mass spectrometry analysis of in vitro nitration of a recombinant human IgG1 monoclonal antibody. Rapid Commun. Mass Spectrom. 22, 1–10 [DOI] [PubMed] [Google Scholar]
- 44. Sadygov R. G., Liu H., Yates J. R. (2004) Statistical models for protein validation using tandem mass spectral data and protein amino acid sequence databases. Anal. Chem. 76, 1664–1671 [DOI] [PubMed] [Google Scholar]
- 45. Guan Z., Yates N. A., Bakhtiar R. (2003) Detection and characterization of methionine oxidation in peptides by collision-induced dissociation and electron capture dissociation. J. Am. Soc. Mass Spectrom. 14, 605–613 [DOI] [PubMed] [Google Scholar]
- 46. Alley W. R., Jr., Mechref Y., Novotny M. V. (2009) Characterization of glycopeptides by combining collision-induced dissociation and electron-transfer dissociation mass spectrometry data. Rapid Commun. Mass Spectrom. 23, 161–170 [DOI] [PubMed] [Google Scholar]
- 47. Sohn C. H., Chung C. K., Yin S., Ramachandran R., Loo J. A., Beauchamp J. L. (2009) Probing the mechanism of electron capture and electron transfer dissociation using tags with variable electron affinity. J. Am. Chem. Soc. 131, 5444–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jones A. W., Mikhailov V. A., Iniesta J., Cooper H. J. (2010) Electron capture dissociation mass spectrometry of tyrosine nitrated peptides. J. Am. Soc. Mass Spectrom. 21, 268–277 [DOI] [PubMed] [Google Scholar]
- 49. Udeshi N. D., Compton P. D., Shabanowitz J., Hunt D. F., Rose K. L. (2008) Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry. Nat. Protoc. 3, 1709–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]