Abstract
In-depth MS-based proteomics has necessitated fractionation of either proteins or peptides or both, often requiring considerable analysis time. Here we employ long liquid chromatography runs with high resolution coupled to an instrument with fast sequencing speed to investigate how much of the proteome is directly accessible to liquid chromatography-tandem MS characterization without any prefractionation steps. Triplicate single-run analyses identified 2990 yeast proteins, 68% of the total measured in a comprehensive yeast proteome. Among them, we covered the enzymes of the glycolysis and gluconeogenesis pathway targeted in a recent multiple reaction monitoring study. In a mammalian cell line, we identified 5376 proteins in a triplicate run, including representatives of 173 out of 200 KEGG metabolic and signaling pathways. Remarkably, the majority of proteins could be detected in the samples at sub-femtomole amounts and many in the low attomole range, in agreement with absolute abundance estimation done in previous works (Picotti et al. Cell, 138, 795–806, 2009). Our results imply an unexpectedly large dynamic range of the MS signal and sensitivity for liquid chromatography-tandem MS alone. With further development, single-run analysis has the potential to radically simplify many proteomic studies while maintaining a systems-wide view of the proteome.
MS-based proteomics has proven to be an indispensable technology for the unbiased analysis of large numbers of proteins. It is routinely applied to study the composition and dynamics of subcellular organelles, protein complexes, interactions, modifications, and the mechanisms of cell signaling (1–5). Although most of these applications do not require the analysis of entire proteomes, even comprehensive “expression proteomics” is becoming a realistic proposition—at least in the sense of quantifying peptides for all the gene products expressed in a given cellular state (6).
Notwithstanding these successes, an intrinsic challenge in MS-based proteomics remains the large “dynamic range” of protein abundance levels; at least four orders of magnitude in yeast (7, 8) and even larger in human cells. In the standard “shotgun” proteomics strategy the enzymatic digestion of proteins to peptides followed by liquid chromatography tandem mass spectrometry (LC MS/MS)1 further compounds the complexity and dynamic range challenges (9, 10). For in-depth analysis of very complex mixtures such as those represented in total cell lysates, at least one step of protein or peptide fractionation is therefore always employed before LC MS/MS. However, each additional fractionation step is accompanied by corresponding increases in the required starting material and in the required measurement time. Furthermore, because of the very high sensitivity of modern mass spectrometers, peptides and proteins can easily be found in several adjacent biochemical fractions, diminishing the contribution of classical biochemical fractionation to achieving deep coverage of the proteome. In contrast, LC is in principle capable of very high separation power (11). Joergensen and coworkers pioneered the use of small, one micrometer-sized chromatographic particles, which increase chromatographic resolution (12). However, the backpressure in LC strongly depends on the size of these particles and these small particle sizes required ultrahigh pressure LC systems. Smith and coworkers similarly constructed very high pressure systems and coupled them to three-dimensional ion traps as well as toFourier transform-ion cyclotron resonance instruments with very high field strength (13, 14). Using columns up to 2 m in length, they reported identification of about 2000 proteins of Shewanella oneidensis in 12h gradients and demonstrated 15 attomole sensitivity for bovine serum albumin. Waters Corporation, along with several other companies, have commercially introduced high-pressure LC systems (UPLC for ultra high pressure chromatography). They reported that UPLC enabled the use of small (sub two-micrometer) beads and extended column lengths, which increased chromatographic resolution (15). Yates and coworkers described an LC/LC peptide separation system with extended column length of 50 cm, which led to 30% increase in protein identification compared with the previous set-up (16).
Monolithic columns offer a somewhat different approach to obtain high separation capacity, which does not necessitate as high a backpressure. Very recently, Ishihama and coworkers measured the E. coli proteome in triplicate 41 h gradients on a 350 cm monolithic column and identified more than 2500 proteins (17). Remarkably, this number slightly exceeded the transcriptome detected on microarrays in the same system, suggesting that very high coverage of the proteome had been achieved. Furthermore, these researchers reported a fivefold enhanced total peptide signal compared with standard columns typically used in shotgun proteomics, which they attributed to reduced peptide suppression in electrospray in their system.
Most of the above reports used very specialized equipment not routinely employed in proteomics. Furthermore, in just the last few years the resolution, mass accuracy and sequencing speed of modern mass spectrometers have increased dramatically (18, 19) and there have been corresponding advances in computational proteomics. We therefore set out to investigate the combined capabilities of a high resolution chromatographic system with a state of the art MS and computational proteomics workflow. We employed small particles, long columns and long and shallow gradients using standard HPLC pumps to answer the conceptual question of whether or not extensive fractionation was necessary to characterize a large part of the proteome. We used the yeast model system as well as a human cell line to judge the depth and the usefulness of the achieved proteome coverage against the comprehensive yeast proteome (6), a recent study designed to identify yeast proteins expressed at very low levels (20) and, in a bioinformatic approach, by the coverage of cellular pathways and processes.
EXPERIMENTAL PROCEDURES
Yeast Cell Culture and Protein Extraction
A S288C WT strain (TWY70) was grown in YNB medium to a final OD600 of 0.7. Cells were harvested by centrifugation for 5 min at 4000 × g at 4 °C. Cells were resuspended in 8 m urea and total protein was extracted by bead milling.
Human Cell Culture and SILAC Labeling
HEK-293 cell were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and 1% penicillin/streptomycin (Invitrogen). For stable isotope labeling with amino acids in cell culture (SILAC) experiments, HEK293 cells were grown in Dulbecco's modified Eagle's medium containing heavy isotope labeled amino acids, l-13C615N4-arginine (Arg10) and l-13C615N2-lysine (Lys8) instead of the natural amino acids, or the same concentrations of the light amino acids. Cells were cultured for approximately eight doublings in the SILAC medium to reach complete labeling. Subconfluent cultures were trypsinized and the cell pellets were lysed in a buffer containing 4% SDS and 100 mm dithiotreitol in 100 mm tris-HCl pH 7.6. Lysates were incubated at 95 °C for 5 min and briefly sonicated. After centrifugation, the protein concentration was measured using fluorescence emission at 350 nm using an excitation wavelength of 295 nm. The measurements were performed in 8 m urea using tryptophan as the standard. Proteins were then precipitated using methanol-chloroform and resuspended in a buffer containing 8 m urea in 0.1 m Tris HCl pH 8.5.
In-Solution Digestion
For digestion in solution proteins were reduced with 1 mm dithiotreitol and then alkylated with iodoacetamide. Yeast proteins were digested overnight with LysC at a concentration of 1:50 (w/w) at room temperature. HEK293 proteins were digested with LysC for 3 h at a concentration of 1:50 (w/w) followed by overnight digestion with trypsin at a concentration of 1:50 (w/w). After digestion, peptides were acidified with trifluoroacetic acid, and loaded on StageTips as described previously (21) (5 μg per StageTip). Peptides were eluted from the StageTips before LC MS/MS using buffer B (80% acetonitrile, 0.5% acetic acid).
Column Packing
Chromatographic separation power increases with the length of column and with decreasing size of packing materials (12). We therefore packed columns of 50 cm length and 75 μm inner diameter with 1.8 μm C18 beads (Reprosil-AQ Pur, Dr. Maisch). Uniformity of packing was achieved by using chloroform for packing instead of methanol. Columns were packed using the High Pressure Column Packer system (Proxeon Biosystems, Odense, Denmark, now part of Thermo Fisher Scientific), followed by further bead condensation on the EASY-nLC system (Proxeon Biosystems, Odense, Denmark). Nanospray tips were generated using a P-2000 Laser Based Micropipette Puller (Sutter Instruments).
Column Oven
Long columns combined with small beads increase the backpressure generated by the chromatographic pump, which can then be counteracted by elevating the column temperature. To facilitate the usage of long chromatography columns on our standard Proxeon-NanoESI pumps we therefore constructed a column oven (supplemental Fig. S1). It is based on cascaded Peltier elements (2 layers) driven by an electronic controller. The mechanical design of the oven makes it possible to use columns of different lengths (from 12 to 100 cm) by coiling them up around a central core. Longer high performance liquid chromatography (HPLC)-columns can be heated up to a temperature of maximum 60 °C, to reduce the backpressure on the HPLC.
LC-MS Analysis
Peptides were loaded on a 50-cm column (described above) with buffer A (0.5% acetic acid), and were separated with a linear gradient from 5% to 35% buffer B (0.5% acetic acid and 80% acetonitrile) at a flow rate of 75 nl/min followed by a wash reaching 90% buffer B. Gradient lengths were either 140 min or 480 min, as described in the main text. The LC system was directly coupled in-line with a linear trap quadrupole (LTQ)-Orbitrap Velos instrument (Thermo Fisher Scientific) via the Proxeon Biosystems nanoelectrospray source. The source was operated at 2.1 kV, with no sheath gas flow and with the ion transfer tube at 200 °C. The mass spectrometer was programmed to acquire in a data dependent mode. The survey scans were acquired in the Orbitrap mass analyzer with resolution 60,000 at m/z 400 with lock mass option enabled for the 445.120024 ion (22). Up to the 25 most intense peaks with charge state ≥2 and above an intensity threshold of 500 were selected for sequencing and fragmented in the ion trap by collision induced dissociation with normalized collision energy of 40%, activation q = 0.25, activation time of 10 ms and one microscan. For all sequencing events dynamic exclusion was enabled to minimize repeated sequencing. Peaks selected for fragmentation more than once within 30 s were excluded from selection (10 p.p.m. window) for 90 s. The maximum number of excluded peaks was 500.
Data Processing and Analysis
The raw data acquired were processed with the MaxQuant software version 1.1.0.39 according to the standard workflow (23). Database search was performed in MaxQuant with the Andromeda search engine (24) against International Protein Index Human version 3.68 database (87,083 entries in the forward database, including common contaminants) or against the translation of all ORFs in SGD (Saccharomyces Genome Database, 6752 entries in the forward database, including common contaminants) version from 5 Jan 2010, with initial precursor mass tolerance of 7 ppm and fragment mass deviation of 0.5 Da. The search included cysteine carbamidomethylation as a fixed modification and N-acetylation of protein and oxidation of methionine as variable modifications. Up to two missed cleavages were allowed for protease digestion. For trypsin digested proteins, peptides had to be fully tryptic and for LysC, the peptides had to fully match LysC digestion specificity. The “identify” module in MaxQuant was used to filter identifications at 1% false discovery rate on the peptide and protein level using a reverse database in which the lysines and arginines were swapped with the preceding amino acid (25). Only peptides with minimum six amino acid length were considered for identification. For SILAC analysis, two ratio counts were set as a minimum for quantification. The lists of identified proteins were filtered to eliminate reverse hits and known contaminants. Inspection of peptide scores revealed that the minimum Andromeda score in the human nonlabeled cell line data was 61 (Note that Andromeda scores are on average threefold higher than Mascot scores).
Estimation of Sensitivity of Single-run Analysis
For estimation of protein amounts, summed peptide intensities were taken as proxies. To account for the fact that larger proteins produce more peptides at the same copy number, we normalized the summed peptide intensities to the molecular weight of the protein. Likewise, to obtain the total number of molecules from the total amount of protein, we divided the protein amount loaded on the column (2 μg) by the average molecular weight (60 kDa). These two quantities must be equal to each other, which allowed us to estimate the number of molecules for each protein from its normalized MS signal.
RESULTS AND DISCUSSION
Evaluation of the Chromatographic Set-up
To investigate the dynamic range in LC MS/MS analysis of complex mixtures without prefractionation, we first wanted to improve chromatographic resolution and to provide more time for MS/MS events. We employed 50-cm columns packed with 1.8 μm C18 particle sizes and operated them with long gradients (Experimental Procedures). Construction of a column oven allowed operation of this system with a normal pressure HPLC pump (supplemental Fig. S1). The contour plot of an LC-MS/MS run visualizes that an optimized linear gradient spread the complex peptide mixtures as evenly across the increased elution time as a standard gradient (Fig. 1A, B). The LC system was on-line coupled via electrospray ionization to MS analysis on a linear ion trap Orbitrap instrument (LTQ-Orbitrap Velos). Because of the very high sequencing speed of this instrument (26), we chose a method in which high resolution MS scans were followed by up to 25 fragmentation events in the linear ion trap. All data evaluation was performed in the MaxQuant environment (25), which improved the absolute average peptide mass deviations in these experiments to 590 p.p.b. while providing individualized search tolerance windows for all peptide precursor masses (supplemental Table S1).
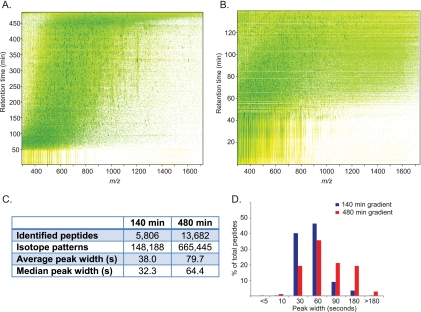
Fig. 1.
Chromatographic performance at long gradient times. A, Yeast peptides separated using a 480 min gradient on a 50 cm column packed with 1.8 μm C18 beads. Contour plot from MaxQuant shows uniform peptide separation along the retention time and the m/z range. B, Same as (A), but using a 140 min gradient. C, Comparison of the number of peptides, number of isotope patterns and peak widths in two different gradient lengths. D, Comparison of the distribution of peak widths between the two gradients.
We first characterized this single-run analysis system with respect to the elution gradient length and for this purpose performed triplicate analysis of yeast peptides in 140 min and 480 min gradients. Average and median peak widths for eluting peptides only increased twofold when increasing gradient time about 3.5-fold, indicating that chromatographic resolution was still improving at the longest gradient. As a consequence, the number of detected isotope patterns was increased more than fourfold and the number of identified peptides more than doubled (Fig. 1C and supplemental Table S2). Interestingly, in the 480 min gradient LC peak lengths are distributed normally, whereas the 140 min gradient has most peak widths concentrated at the narrower elution times (Fig. 1D). Plotting the cumulative number of identifications as a function of retention time shows the expected increase for peptide identifications. Protein identifications also continue to increase substantially over the entire gradient but more slowly than peptide identifications, implying an increase in sequence coverage per protein (supplemental Fig. S2). Comparison to standard conditions used in our laboratory demonstrated a 2.5-fold increase in the number of identifications for the single-run analysis (supplemental Table S3).
Evaluation in the Yeast Model Systems
The yeast proteome is a good model system for MS-based proteomics because protein expression has been measured by genome wide tagging experiments (7, 8). Furthermore, yeast has served as a model for two alternative approaches in MS-based proteomics: comprehensive analysis of the proteome by extensive fractionation (6) and recently using MRM-based targeting (20). This provides the opportunity to evaluate the single-run approach against these two established alternatives.
Applying our single-run LC MS/MS system to the yeast proteome, we detected 2990 yeast proteins in triplicate analysis with about 1 day of measurement time (supplemental Tables S2, S4). Each single run by itself identified more than 2400 proteins and each additional replicate added a decreasing number of new identifications (13% for the first replicate and 6% for a third replicate). In each single run, more peptides are present than are selected for fragmentation by the instrument. This limits the observed dynamic range of the fragmented and identified peptides and confounds the issue of how much of the proteome is available for analysis in LC MS/MS without fractionation. Therefore, to get a better estimate of the true dynamic range of the single-run analysis method, we matched peptide identities from triplicate analysis against one single run. The very high mass accuracy achieved in the analysis of the high resolution Orbitrap data facilitated this transfer of peptide identities (performed by the “match between runs” feature in MaxQuant). Including the comparison between different runs revealed that each single run contained enough information for the identification of over 2700 proteins. This demonstrates that each single run has sufficient dynamic range for the detection of the total number of proteins that are identified in the triplicate analysis.
Either triplicate single 480-min runs or one single 480-min run employing “match between runs” covered 68% of proteins in our previous comprehensive yeast proteome measurement (6). That study included a subcellular fractionation approach, which identified 3639 proteins. This is only 21.5% more than obtained here at drastically reduced measurement time and sample consumption. Using summed MS peptide intensity as proxy for absolute protein abundance (6, 27), we found that the more abundant half of yeast proteins were covered almost completely in addition to a substantial fraction of the low abundance proteins (supplemental Fig. S3).
Coverage of Functional Categories
For systems biology applications as well as for globally studying alteration of pathways it may not be necessary to monitor each pathway member (28). To test how well our dataset covers known cellular pathways, we overlaid our triplicate single-run yeast proteome data onto pathway database of the Kyoto Encyclopedia of Genes and Genome (KEGG) (29). Remarkably, 48 of 91 KEGG pathways were represented with over 80% of their members and 76 of 91 with over half of them (supplemental Table S5 and supplemental Fig. S4). This demonstrates that almost all KEGG pathways can be monitored within the dynamic range of LC MS/MS alone. These also include classical signaling pathways such as MAPK signaling cascades (26 of 45 proteins) as well as specialized activities such as DNA mismatch repair (11 of 15 proteins). Note that these numbers are only a lower limit of the actual coverage because not all pathway members are expressed in yeast cells growing under standard laboratory conditions (see also below). In contrast, coverage of the proteins acting in the cell cycle was relatively low (29 of 106 proteins). This likely because of their low abundance combined with brief expression period during cell division, which effectively dilutes these proteins in the total yeast proteome.
Comparison to Multiple-reaction Monitoring in Yeast
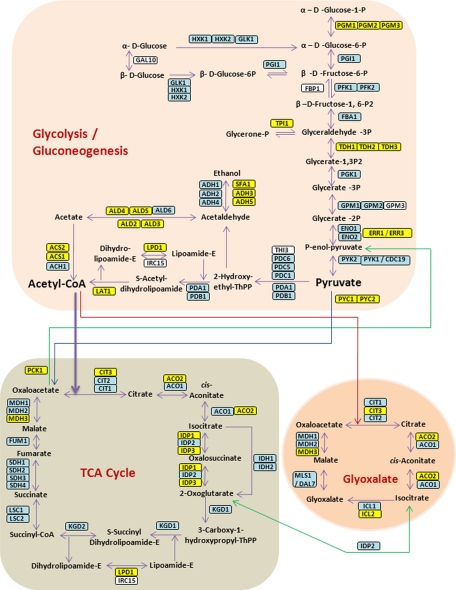
In the multiple-reaction monitoring (MRM) strategy, specific peptides of interest are targeted and a triple quadrupole mass spectrometer selectively records precursor mass and fragment mass transitions during these chromatographic runs (30). The MRM approach has recently been evaluated on the glycolysis and gluconeogenesis pathway in yeast (20). Of the 45 proteins targeted for MRM our triplicate data set contains 43, identified with a median of 14 peptides (Fig. 2 and supplemental Table S6). Annotated spectra of all discussed yeast proteins that have less than five identified unique peptides are shown in supplemental Fig. S5. One missing protein FBP1, was found in the comprehensive yeast MS-derived proteome (6) and was only identified in the MRM study after shifting to another nutrient source (20). Three other proteins (MLS1, IDP2, and ICL1) likewise only became detectable by MRM after nutrient shift but they were clearly identified by single-run LC MS/MS analysis with 14, 8, and 4 unique peptides, respectively. In addition, our data set contains 28 proteins in this pathway that were not targeted in the MRM study, often isozymes of the targeted proteins (Fig. 2). Taking an expanded view of the glycolysis and gluconeogenesis and TCA-related pathways that also covers pentose phosphate pathway and pyruvate metabolism, we detected 97 out of 103 proteins (supplemental Fig. S6).
Fig. 2.
Single-run analysis of the yeast proteome by LC MS/MS. Schematic representation of proteins involved in glycolysis/gluconeogenesis, in the citric acid cycle (TCA cycle) and in the glyoxylate pathway in yeast. The scheme is based on ref (20) and extra proteins were added based on the KEGG pathway database. Proteins that were identified in the single-run analysis and were not targeted in ref (20) are in yellow and proteins that were found by both strategies are in blue. Gal10 was not targeted and not found in the single-run analysis (white).
The same MRM study also exemplarily targeted proteins from each yeast protein abundance class to evaluate the dynamic range of that technique (20). Of these 127 proteins we identified 113 in single-run analysis, including all in the “below 50 copies per cell” category (supplemental Table S6; annotated spectra of proteins identified by less than five unique peptides are shown in supplemental Fig. S7). As they were identified with a median of 21 peptides it is unlikely that they were expressed at extremely low levels and their abundance was instead likely misclassified in the genome tagging studies (7, 8). Supporting this view, they were outliers in the correlation plot between MS signal and copy numbers determined by Western blotting in the comprehensive yeast MS-based proteome study (6). The three proteins with absolute abundance of 103 copies per cell or less, as determined with isotope labeled peptides (20), were all present in our single-run data set: YNR067C (identified with four unique peptides), YGL006W (10 unique peptides) as well as YKR031C (two unique peptide). For several proteins it had not been possible to devise MRM assays but they nevertheless appear in our data set. Some of them are highly phosphorylated or glycosylated (20) such as YMR173W and YIL162W, highlighting the ability of nontargeted analysis to identify a protein even if many peptides are modified.
Evaluation of Single LC-MS/MS Runs on a Human Cell Line
To test our single-run analysis pipeline on the more complex mammalian proteome, we applied it to a human embryonic kidney cell line (HEK293). We again performed triplicate analysis (about 1 day of MS measurement time in total), which yielded identification of 4695 proteins on average in single runs and 5376 proteins in the combined triplicate dataset (Table I and supplemental Table S7). In total, 35,155 sequence unique peptides were identified, with an average of six peptides per protein and average sequence coverage of 18%. Furthermore, matching the peptide identification of three reference runs to any single one identified more than 5000 proteins on average in each individual run.
Table I. Protein identification in replicate runs. The number of proteins is given for each single 480 min run analyzed alone and with transfer of identifications using the “match between runs” feature in MaxQuant. Matching was performed for two samples, three samples, and for each of the experiments with the three others.
| Protein in a single run | Proteins in two runs | Proteins in three runs | Matched runs | |
|---|---|---|---|---|
| Experiment 1 | 4529 | 5120 | 5348 | 5272 |
| Experiment 2 | 4622 | 5292 | ||
| Experiment 3 | 4542 | 5328 | ||
| Matched experiment | 4654 | 5286 |
Sensitivity in Single-run Analysis
Detection of 5000 proteins from about 2 μg of starting material implies very high detection sensitivity. Dividing 2 μg by 5000 proteins of an average molecular weight of 60 kDa results in an average sensitivity of less than 10 fmole per protein. To determine the sensitivity more accurately, we used the MS signal of the peptides of each protein, adjusted for protein length, and calculated the protein abundance based on total MS signal (Experimental Procedures). Protein signals estimated in this way ranged over six orders of magnitude (Fig. 3), which is about 10-fold more than in the yeast experiments (supplemental Fig. 8). The median protein amount was 0.6 fmole and remarkably, about a thousand proteins were detected at an estimated level of 100 attomoles or less. This very high sensitivity is a consequence of the absence of losses because of fractionation as well as the highly sensitive MS detection methods employed. Note that our experiments do not provide accurate copy numbers for each the 5500 measured proteins as this would require isotope labeled standards. However, even if the quantification of individual proteins is not very accurate, we do not expect any global deviation from these values. Furthermore, at the very low abundance levels the relationship between MS signal and absolute protein amount may be much less accurate and we may be underestimating the abundance of these proteins. That said, in our yeast data set 76% of the proteins are identified with three or more peptides (supplemental Table S2). The intensities of only these proteins range over five orders of magnitude, showing that the estimation of dynamic range is not overly skewed by a low number of peptides.
Fig. 3.
Dynamic range of single-run analysis of a human cell line. Ranking of HEK293 proteins according to their absolute amounts. Quantification is based on added peptide intensities of the proteins as described in the EXPERIMENTAL PROCEDURES section.
Pathway Coverage in Single LC-MS/MS Runs
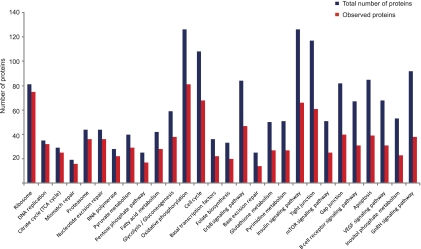
The triple single-run analysis of the HEK293 cells covered 76 of 202 KEGG pathways by at least half their members (supplemental Table S8). This is substantially less than in the case of yeast and likely reflects the higher complexity of mammalian proteomes as well as the fact that many pathways are cell type or state specific. Nevertheless, almost all known pathways (170 of 200) were represented by at least two proteins in the single-run analysis. Apart from major metabolic processes such as glycolysis and basic “cellular machines” such as the ribosome, signaling pathways were also well represented (Fig. 4). For example, we identified 47 of 84 members of the ErbB signaling pathway and 66 of 126 members of the insulin signaling pathway. Furthermore, regulatory protein families such as kinases and transcription factors were present as expected from their relative proportion in the genome (4.1% and 4.4%, respectively).
Fig. 4.
KEGG pathways in HEK293 cells. Analysis of the representation of KEGG pathways in a triplicate of single-run analyses compared with the total number of proteins in the pathway according to the KEGG database.
Compatibility with SILAC-based Quantification
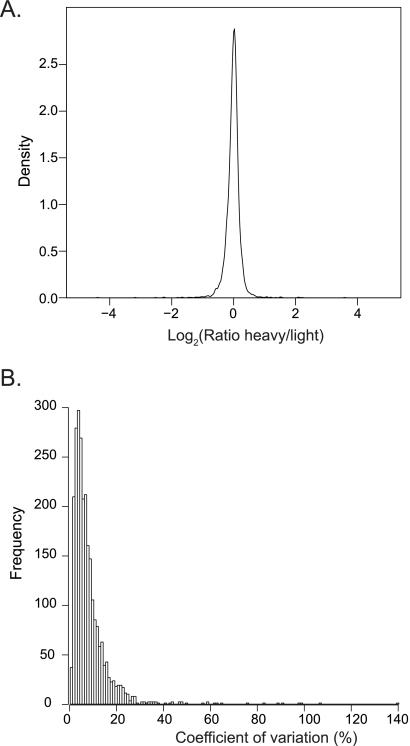
To test if the single-run analysis technology was compatible with accurate quantification, we SILAC-labeled the HEK293 cells with light and heavy arginine and lysine (Experimental Procedures). Triplicate analysis identified 4335 proteins of which 93% were quantified (supplemental Table S9). The somewhat lower number of identified proteins compared with non-SILAC conditions is because of the two-fold higher complexity of the peptide mixture introduced by labeling. Determination of ratios in MaxQuant revealed a very narrow ratio distribution with 66% of proteins within plus or minus 10% fold-change (Fig. 5A). Mean coefficient of variation of 8% with 99% of proteins within an apparent twofold variation between the two SILAC conditions (Fig. 5).
Fig. 5.
SILAC-based quantification of HEK293 proteins. HEK293 cells were labeled with heavy or light amino acids and quantified using single-run analysis. A, A density plot shows the ratio distribution of proteins from a triplicate single-run analysis. B, Histogram of the coefficient of variation of each of the quantified proteins.
Conclusion and Outlook
At the outset of this project we asked the question whether or not LC-MS/MS by itself—without pre-fractionation—could cover a large part of the proteome. The answer to this question was by no means self-evident. Indeed we had expected that there was a definite limit set by the combined dynamic range capabilities of LC, of MS and of MS/MS that would make it very difficult to move beyond a certain number of identified proteins and beyond a certain coverage. Surprisingly, we found that the absence of any prefractionation did not impose such a limit. Triplicate LC MS/MS with mainstream instrumentation was capable of identifying about 70% of the yeast proteome and more than 5500 proteins in a mammalian proteome.
Already at its current state of development, “single-run proteomics” has some very attractive features. First of all, many applications only require the analysis of less than a few thousand proteins making single-run proteomics directly applicable. Compared with extensive fractionation for the analysis of very complex proteomes on the one hand it involves drastically reduced sample preparation, sample consumption, and MS measurement time. Compared with the MRM strategy on the other hand its main appeal is that it retains the discovery and systems-wide character implicit in proteomics. It is also completely generic and does not require the development of specific assays for specific peptides. To be sure, single-run proteomics also has definitive weaknesses compared with these other strategies: At least for now, it cannot rival the coverage of LC-MS/MS combined with prefractionation. Gradient times per measurement condition are currently three- to eight-fold longer than they are in MRM. For these reasons, we suggest that single-run proteomics will be a useful complement rather than a competitor to the two established alternatives mentioned above.
The very high sensitivity achievable with single-run proteomics may be one of its most exciting features. Using a state of the art MS and computational proteomics workflow and a few μg protein starting material, we were able to identify about 1000 of a total of 5000 proteins at an estimated amount per protein of less than 100 attomoles. To put this number in perspective, a single 10-cm cell culture dish of HeLa cells yields sufficient protein material for several hundred single-run analyses. Furthermore, subfemtomole sensitivity for single protein analysis was considered cutting edge performance not many years ago. We imagine that the workflow described here could be combined with RePlay chromatographic analysis, in which the chromatographic effluent of a column is split into two flows. One is analyzed directly and one is analyzed after first being diverted into a delay line (31). This RePlay technology does not require additional sample, thus in the context of single-run proteomic analysis it may allow online targeted re-analysis or it may make replicate analysis dispensable, further increasing effective sensitivity.
Several considerations suggest that the identification and coverage numbers in single-run analysis can be further increased in the future. A dedicated UPLC pump (which was not available to us) would further enhance chromatographic resolution and performance of MS instruments is continuously improving as well. For example, Orbitrap analyzers with even higher resolution are possible, which would automatically increase the MS dynamic range (32). Even within the dynamic range that we were able to observe here, most peptides remained unfragmented and hence unidentified (Fig. 1C). Therefore the actual dynamic range of LC MS is much larger than what we have described here. Together these speculations lead us to predict that it may be eventually possible to cover essentially the complete yeast proteome by LC-MS/MS without any prefractionation.
Acknowledgments
We thank other members of our laboratory for fruitful discussions a nd help, Nadin Neuhauser for providing annotated spectra, Marlis Zeiler for help with protein abundance analysis and Tobias Walther for critical reading of the manuscript. T. G. is supported by the Humboldt Foundation.
Accession information. All described nanoLC-MS/MS data (Raw files, MaxQuant output for peptide tables and annotated spectra) may be downloaded from ProteomeCommons.org (http://proteomecommons.org/tranche/) using the following hash codes:
Yeast raw files: STV2ToZgz62Q9w1hg9EFujfjWzbl3kKZA8ZuSiaal5diVtQKzHKYx1mUzRlnOkxni9g19NM4EwQWLonzk2OlShxur2kAAAAAAAAEVg==
HEK293 raw files: 7q7SzUnNwc1AbvCefHcdfRP3B9cda11AAoL+5LbfRXlQYbIz7u5jkT1NOnseCFIdoNnIs2zhw86SbbRplbJSNE3wjJUAAAAAAAADDQ==
Yeast raw files- standard conditions 3 μm beads, 15 cm column length, 140 min gradient: du5Yy/m9eyAXTHhAOvlOqphDCQSumqbohog09LNqO9AGsTXkDGTzbCJeAMONtHgKs4CK4B2TM0zo+qYAi1C/j4jyF2QAAAAAAAAC6w==
SILAC labeled HEK293 cells raw files: Wt4Hmuj9DEa+1/CQ2fv9jG1B8V5FkaMSorgp35a3HMjLIKKxPn8EJsnHFZSsyufOvIph2aiPpb3s8wABOtZGJlr/rCUAAAAAAAAC+w==
Peptide annotated spectra- proteins identified with a single peptide: RIld9e1lJ+FgRL/DnxnKQusqqaDc55TTResKvhFl0Gxc/ca/ReuDHDAZDVoMAxQ3S8hURN9M96j58hCp8SZSHvXb4EQAAAAAAAADrA==
supplemental Tables S10 and S11: The encryption code: lGHc3aO7NuGlyPxRczu4
The Hash code: OiLa1q6gXfZYWgjRE96sElwwcoL+xrSRP4Nw3ZGkBrm89YGbBplKS0Ro5sqgHDUf7oUYPALK5OsOK+FaIwIvCvUVEd4AAAAAAAADWQ==
Footnotes
* This project was supported by the European Commission's 7th Framework Program PROteomics SPECification in Time and Space (PROSPECTS, HEALTH-F4-2008-021,648).
 This article contains supplemental Figs. S1 to S8 and Tables S1 to S11.
This article contains supplemental Figs. S1 to S8 and Tables S1 to S11.
1 The abbreviations used are:
- UHPLC
- ultra high pressure liquid chromatography
- LTQ
- linear trap quadrupole
- SILAC
- stable isotope labeling by amino acids in cell culture
- MS/MS
- tandem mass spectrometry
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- MRM
- multiple-reaction monitoring
- LC
- liquid chromatography.
REFERENCES
- 1. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 2. Yates J. R., 3rd, Gilchrist A., Howell K. E., Bergeron J. J. (2005) Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell Biol. 6, 702–714 [DOI] [PubMed] [Google Scholar]
- 3. Cravatt B. F., Simon G. M., Yates J. R., 3rd (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 4. Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C., Reader V., Sweetman G., Bauer A., Bouwmeester T., Hopf C., Kruse U., Neubauer G., Ramsden N., Rick J., Kuster B., Drewes G. (2007) Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol 25, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 5. Choudhary C., Mann M. (2010) Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 11, 427–439 [DOI] [PubMed] [Google Scholar]
- 6. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 7. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 8. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 9. Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd (1999) Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682 [DOI] [PubMed] [Google Scholar]
- 10. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 11. Guiochon G. (2006) The limits of the separation power of unidimensional column liquid chromatography. J. Chromatogr. A 1126, 6–49 [DOI] [PubMed] [Google Scholar]
- 12. MacNair J. E., Lewis K. C., Jorgenson J. W. (1997) Ultrahigh-pressure reversed-phase liquid chromatography in packed capillary columns. Anal. Chem. 69, 983–989 [DOI] [PubMed] [Google Scholar]
- 13. Shen Y., Zhao R., Belov M. E., Conrads T. P., Anderson G. A., Tang K., Pasa-Tolić L., Veenstra T. D., Lipton M. S., Udseth H. R., Smith R. D. (2001) Packed capillary reversed-phase liquid chromatography with high-performance electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry for proteomics. Anal. Chem. 73, 1766–1775 [DOI] [PubMed] [Google Scholar]
- 14. Luo Q., Shen Y., Hixson K. K., Zhao R., Yang F., Moore R. J., Mottaz H. M., Smith R. D. (2005) Preparation of 20-microm-i.d. silica-based monolithic columns and their performance for proteomics analyses. Anal. Chem. 77, 5028–5035 [DOI] [PubMed] [Google Scholar]
- 15. Liu H., Finch J. W., Lavallee M. J., Collamati R. A., Benevides C. C., Gebler J. C. (2007) Effects of column length, particle size, gradient length and flow rate on peak capacity of nano-scale liquid chromatography for peptide separations. J. Chromatogr. A 1147, 30–36 [DOI] [PubMed] [Google Scholar]
- 16. Motoyama A., Venable J. D., Ruse C. I., Yates J. R., 3rd (2006) Automated ultra-high-pressure multidimensional protein identification technology (UHP-MudPIT) for improved peptide identification of proteomic samples. Anal. Chem. 78, 5109–5118 [DOI] [PubMed] [Google Scholar]
- 17. Iwasaki M., Miwa S., Ikegami T., Tomita M., Tanaka N., Ishihama Y. (2010) One-dimensional capillary liquid chromatographic separation coupled with tandem mass spectrometry unveils the Escherichia coli proteome on a microarray scale. Anal. Chem. 82, 2616–2620 [DOI] [PubMed] [Google Scholar]
- 18. Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 19. Mann M., Kelleher N. L. (2008) Precision proteomics: the case for high resolution and high mass accuracy. Proc. Natl. Acad. Sci. U.S.A. 105, 18132–18138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 22. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 23. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protocols 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 24. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 25. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 26. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap orbitrap instrument with very high sequencing speed. Mol. Cell Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malmström J., Beck M., Schmidt A., Lange V., Deutsch E. W., Aebersold R. (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zubarev R. A., Nielsen M. L., Fung E. M., Savitski M. M., Kel-Margoulis O., Wingender E., Kel A. (2008) Identification of dominant signaling pathways from proteomics expression data. J. Proteomics 71, 89–96 [DOI] [PubMed] [Google Scholar]
- 29. Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waanders L. F., Almeida R., Prosser S., Cox J., Eikel D., Allen M. H., Schultz G. A., Mann M. (2008) A novel chromatographic method allows on-line reanalysis of the proteome. Mol. Cell Proteomics 7, 1452–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Makarov A., Denisov E., Lange O. (2009) Performance evaluation of a high-field Orbitrap mass analyzer. J. Am. Soc. Mass Spectrom. 20, 1391–1396 [DOI] [PubMed] [Google Scholar]