Abstract
Schistosomiasis is a tropical, parasitic disease affecting humans and several animal species. The aim of this study was to identify proteins involved in the growth and survival of the parasitic forms inside a host. Schistosomula of Schistosoma japonicum were isolated from three different hosts: the susceptible BALB/c mice; the Wistar rats, which have a considerably lower susceptibility; and the resistant reed vole, Microtus fortis. Soluble proteins of the schistosomula collected from the above three hosts 10 days postinfection were subjected to two-dimensional difference gel electrophoresis. Comparative proteomic analyses revealed that 39, 21, and 25 protein spots were significantly differentially expressed between schistosomula from mice and rats, mice and reed voles, or rats and reed voles, respectively (ANCOVA, p < 0.05). Further, the protein spots were identified by liquid chromatography-tandem MS. Bioinformatics analysis showed that the differentially expressed proteins were essentially those involved in the metabolism of proteins, ribonucleotides, or carbohydrates, or in stress response or cellular movement. This study represents the first attempt at profiling S. japonicum living in different states and provides a basis for a better understanding of the molecular mechanisms in the development and survival of S. japonicum in different host environments.
Schistosomiasis is one of world's most prevalent tropical diseases caused by parasitic blood flukes. It is estimated that the disease afflicts more than 200 million individuals in 74 developing countries, with 200 thousand deaths per year and more than 600 million people facing a risk of infection (1, 2). Currently, the main strategy for schistosomiasis control relies on praziquantel-based morbidity control. However, chemotherapy does not prevent re-infection. Moreover, drug resistance has been noted as some isolates of Schistosoma mansoni showing resistance to high doses of praziquantel were found in Egypt (3). In addition, for individuals with a high worm burden, transition to severe and irreversible pathology cannot be prevented by chemotherapy at this late stage (4). Thus, a vaccine or a new drug against schistosomiasis is desirable as a strategic tool for the control of this disease (5), but none are currently available for practical use (6).
Proteomic analysis is a powerful tool to screen samples derived from pathogens and identify proteins that are possibly involved in pathogenesis. This technique had been applied to identification of schistosome proteins from complex samples or to study differential expression of schistosome proteins (7–9). This approach has uncovered an interesting and a wide array of schistosome antigens. For example, proteins expressed in different life cycle stages (8) and genders (7), tegument and secretion proteins etc. (10–12) have been identified. All the previous investigations focused on worms from rabbits, mice, or other susceptible hosts. Here we present a comparative analysis of protein expression in worms isolated from three different kinds of hosts: susceptible, unsusceptible, and resistant. The results of this analysis will be helpful in identifying the molecules which may be essential to the survival and development of the schistosomes, and may therefore be new candidates for a vaccine or drug targets for the control of schistosomiasis.
It was reported that in China S. japonicum can infect more than 40 kinds of mammals including cattle, sheep, goat, rabbit, and mice, which are susceptible to the infection, and some others such as water buffalo, pig, and rat, which are less susceptible as indicated by the observations that worms exhibit a low developmental rate and a smaller worm size in these hosts. The reed vole or Microtus fortis, is the only mammal reported so far in which the schistosome cannot develop and thus do not have any significant pathological effects (13–16). As reported earlier, growth and survival of the worms was extremely poor in M. fortis 15 days after a challenge with cercariae (14). This may be that M. fortis developed a stronger immune response and more severe pathological lesion in response to the schistosomes during the early phase of the infection (17). Different hosts provide a distinctive environment for the schistosome, which impacts the survival and development of the parasite. Protein expression variation in schistosome would be influenced directly by the development of worms in different hosts, and some of the proteins that are differentially expressed in worms from different hosts may be key molecules required for the survival of the worms, and the establishment of the parasite. In this study, schistosomula of S. japonicum were recovered 10 days postinfection from the susceptible host, mouse, or the unsusceptible host rat or the nonpermissive host M. fortis. Soluble proteins extracted from the schistosomula were compared by the robust and quantitative two-dimensional-differential gel electrophoresis (2D-DIGE)1 technique. Proteins that were highly and differentially expressed were selected for identification by mass spectrometry combined with bioinformatics. The purpose of this study was to identify proteins that may perform functions that may be important for the survival and development of the schistosome. A comparative proteomic analysis of schistosomula from rodent hosts differing in their susceptibility to infection is likely to provide valuable information for screening potential vaccine candidates or new drug targets for the control of the disease.
MATERIALS AND METHODS
Worm Collection
BALB/c mice (body weight, ∼25g each) and WISTAR rats (body weight, ∼150g each) were purchased from Shanghai laboratory animal center at the Chinese Academy of Sciences (Shanghai). Microtus fortis (body weight, ∼60g each) were purchased from Shanghai Xipu'er-bikai Experimental Animal Co., Ltd (Shanghai). They were raised in a sterilized room and feed sterilized food and water. The study protocol was approved by the Animal Care and Use Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
The S. japonicum (Chinese strain) life-forms were maintained in Oncomelania hupensis snails and New Zealand rabbits at our Institute. Cercariae were collected by exposing infected snails to light to induce shedding. Cercarial numbers and viability were determined by direct observation under a light microscope.
BALB/c mice, Wistar rats and M. fortis were infected with 200, 2000, or 3000 cercariae, respectively. Schistosomula were obtained by perfusion of infected mice, rats, and M. fortis 10 days postchallenge. The worms were manually washed in phosphate buffered saline at 37 °C in order to remove any residual host proteins. The worms obtained from Wistar rats, BALB/c mice and M. fortis were designated as A, B, and C, respectively. After collection they were snap frozen and stored in liquid nitrogen until use.
Protein Extraction
Lysis buffer (pH 8.5) containing 7 m urea, 2 m thiourea, 65 mm Tris, 2% dithiotreitol (DTT), 4% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS), 0.2% IPG buffer (GE Amersham Biosciences, now GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) and 0.1% v/v protease inhibitor mixture (Merck, Darmstadt, Germany) was added into the tubes containing frozen schistosomula. The worms were ground using Sample Grinding Kit (GE Healthcare). Cell disruption was achieved by sonication at 80 W for 10 s × 5 with intervals of 15 s, followed by the addition of lysis buffer to a final volume of 1 ml. Samples were maintained on ice during these procedures. The mixture was then centrifuged at 12,000 rpm for 40 min at 4 °C to remove insoluble material. The protein content in the supernatant was quantified by the Bradford method using Bio-Rad protein assay reagent. Samples of aliquots were stored at −80 °C until use in proteomic analysis.
Protein Labeling with DIGE Dyes
The three protein samples were labeled using fluorescent CyDyes™ (Cy2, Cy3, and Cy5) developed for DIGE (GE Healthcare). The dyes were designed to ensure that a given protein originating from different samples would have the same relative mobility regardless of the dye used to tag them. The rationale for using a pooled internal standard with DIGE to control for gel-to-gel variation has been described in detail elsewhere (18, 19).
The experimental design using the three-dye approach is illustrated in Table I. Fifty micrograms of an internal standard (IS) containing an equal amount of A, B, and C group proteins were labeled with 400 pmol Cy2. Fifty micrograms each of A, B, and C group protein samples were labeled with 400 pmol Cy3 or Cy5. Dye swap among A, B, and C protein samples was carried out to avoid artifacts due to preferential labeling.
Table I. Experimental design for protein labeling. Proteins of schistosomula recovered from Wistar rats (A), or BALB/c mice (B), or Microtus fortis (C) after 10 days of infection. IS, Internal Standard.
| Gel 1 | Gel 2 | Gel 3 | Gel 4 | Gel 5 | Gel 6 | |
|---|---|---|---|---|---|---|
| Cy2 | IS | IS | IS | IS | IS | IS |
| Cy3 | A | B | C | B | C | A |
| Cy5 | B | C | A | A | B | C |
Protein samples were maintained on ice and fluorescently labeled in the dark for 30 min. The reaction was then quenched by incubating with 1 μl of 10 mm l-lysine (GE Amersham Biosciences) on ice in the dark for 15min.
2D-DIGE
For each gel, Cy2-, Cy3-, or Cy5- labeled proteins (50 μg each) were pooled and an equal volume of rehydration buffer (8 m urea, 4% CHAPS, 2% DTT and 2% IPG buffer pH 3–10) was added so that the final concentrations of DTT and IPG buffer was 1%. The pooled protein samples were subjected to isoelectric focusing carried out on nonlinear IPG strips, 13 cm long, pH 3–10 (GE Healthcare), rehydrated at 30 V for 12 h at room temperature. isoelectric focusing was conducted at 500 V for 1 h, followed by 1000 V for 1 h, then 8000 V 10 h to reach a total of ∼60–80 Kvh using the Ettan II IPG-phor apparatus (GE Healthcare).
After the IEF run was completed, strips were equilibrated under gentle shaking in two steps of 10 min each, first in equilibration buffer I (1% DTT, 50 mm Tris-HCl, pH 6.8, 6 m Urea, 30% Glycerol, 2% SDS bromphenol blue) and then in equilibration buffer II (same as equilibration buffer I, except that DTT was increased to 2.5%,). Equilibrated strips were loaded onto a 12.5%, pH 8.8, 13 cm polyacrylamide gel with a 1 cm, 4%, pH 6.8 stacker gel. The second-dimension gels were cast between low fluorescent Pyrex glass plates (GE Healthcare) to minimize background fluorescence during scanning. The strips were overlaid with 1% agarose in SDS running buffer containing 5 mg of bromphenol blue. The gels were run at 15 mA for 15 min and then at 30 mA at 20 °C in dark until the bromphenol blue dye front had migrated out of the gels. A running buffer of 25 mm Tris, pH 8.3, 192 mm glycine, and 0.1% SDS was used.
Image Acquisition, Analysis, and Processing
The gels were scanned using a Typhoon 9400 Variable Mode Imager (Amersham Biosciences). Excitation and emission wavelengths for Cy2, Cy3, and Cy5 were 488/520, 532/580, and 633/670 nm, respectively. Gels were scanned at 100 μm resolution and the photo multiplier tube voltage was set to values ranging between 500 and 700 V to ensure maxima pixel intensity between 40,000 and 60,000 pixels for the three dyes. Images were cropped using Image-Quant™ version 5.2 to remove areas extraneous to the gel image.
Image gel analysis was carried out using the DeCyder software (Version 6.5, GE Healthcare). The experimental setup and relationship between samples were assigned using the DeCyder software. The protein spots on gels were codetected automatically as 2D-DIGE image pairs, which intrinsically link a sample to its in-gel standard. Matching between gels was performed using the in-gel standard from each image pair. The presence of a Cy2-labeled pooled internal standard on every gel allowed accurate relative quantitation of protein spot features across different gels. The estimated number of spots for each codetection procedure was set to 2500. The gel with the highest spot count was designated the master gel. A gel-to-gel matching of the standard spot maps from each gel, followed by calculation of normalized spot volumes and protein abundance and statistical analysis of protein abundance change between samples was performed using the DeCyder Differential In-gel Analysis and DeCyder Biological Variation Analysis software module, setting Student's t test to p < 0.05.
Two-dimensional Gel Excision, Tryptic Digestion, and Desalting
Protein extracts were separated on preparative gels and proteins of interest were recovered from the gels for identification. Proteins (800 μg) from A, B, or C samples were resolved on separate preparative polyacrylamide gels and were visualized by staining with Coomassie brilliant blue (Bio-Rad, Hercules, CA). All the differentially expressed spots were selected and excised manually from the three preparative gels. The excised gel pieces carrying the proteins of interest were placed in a tube, destained for 20 min in 100mmol/L NH4HCO3/30% acetonitrile and then lyophilized. All the lyophilized samples were digested overnight with 12.5 ng/μl trypsin in 0.1 m NH4HCO3 at 37 °C. The tryptic peptides were extracted three times with 50% acetonitrile, 0.1% trifluoroacetic acid and dried completely by centrifugal lyophilization.
Matrix-assisted Laser Desorption Ionization (MALDI)-Time-of-Flight (TOF) MS Analysis, Database Searches, and Bioinformatics Analysis
The tryptic peptide samples were sent to Shanghai GeneCore BioTechnologies Co. Ltd. for MALDI-TOF-MS peptide mass fingerprint analysis. Peptide mass fingerprint dates were submitted to MASCOT Sequence Query server (http://www.matrixscience.com) for identification against nonredundant NCBI database (http://www.ncbi.nlm.nih.gov/BLAST) and the S. japonicum sequence database (http://lifecenter.sgst.cn/main/index.jsp). Identification required a MASCOT confidence interval of 95%. The other search parameters were: cleavage enzyme, trypsin; fixed modification, carbamidomethyl (C); variable modifications, oxidation (M); max missed cleavage, 1. Peptide tolerance of 100 ppm; fragment mass tolerance of ± 0.5 Da; and a peptide charge of 1+ were considered significant. The criteria used to accept protein identifications were based on the data of peptide mass fingerprint, including the extent of sequence coverage, number of peptides matched and probability score. Only high-scoring proteins were accepted. Gene ontology (GO) annotations for identified proteins based on BLAST results were performed using Blast2GO (www.blast2go.de) and AmiGO (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi). The Blast2GO software was also used to complement multiple identified proteins within the Kyoto Encyclopedia of Genes and Genomes metabolic pathway maps (www.genome.jp/kegg/).
Quantitative Real-time PCR Verification
Total RNA was isolated from 10 day schistosomula obtained by perfusion of infected mice, rats, or M. fortis. All RNA samples were first treated with RNase-free DNase I (TaKaRa, Japan) to remove contaminating genomic DNA and then purified using the RNeasy mini Kit (Qiagen, Germany) according to the manufacturer's instructions. The samples were quantitated by spectrophotometry using Biophotometer (Eppendorf, Germany). Ten micrograms of purified mRNA of 10 day schistosomula from the three different hosts were used as a template in a reverse-transcription (RT) reaction performed with random hexamer primers and Superscript III reverse transcriptase (Invitrogen) according to standard protocols. The resulting cDNAs were used for quantitative PCR.
Quantitative real time PCR was conducted on triplicate samples using SYBR Green tag (Takara, Japan) in a Rotor-Gene 3000A Dual Channel Multiplexing System (Corbett Research, Australia). Primers for qPCR were designed using Real-Time PCR primer design tool (Beacon Designer 7) and the settings were adjusted to the highest possible stringency to generate 100–200 bp amplicons as recommended (Table II). Transcript levels were normalized against the corresponding transcript levels of the gene encoding NADH dehydrogenase in the 10 day schistosomula from the three different hosts.
Table II. Primer sequences and amplicon lengths of RT-PCR products of target genes. Transcript levels were normalized relative to those of NADH dehydrogenase.
| Spot ID | Target genes | Primers | Amplicon length (bp) |
|---|---|---|---|
| 435 | putative protein disulfide isomerase-associated 3 precursor | FP: 5′ GCTCCTTGGTGTGGTCATTG 3′ | 198 |
| RP: 5′ TTGGTAAAGCATGGGTGAAGAC 3′ | |||
| 522 | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | FP: 5′ GGACAAATGAATGAACCACCAG 3′ | 182 |
| RP: 5′ CCGACAGCACTAGGAATACG 3′ | |||
| 578 | Mitochondrial import receptor subunit TOM34 | FP: 5′ GCCGTCTCATGCCATAGC 3′ | 163 |
| RP: 5′ AGCATCGTCTGTGTAGTCTAAC 3′ | |||
| 790 | phosphoglycerate kinase 1 | FP: 5′ GCCTTTGGAACAGCACATC 3′ | 177 |
| RP: 5′ CAACTGGATCTTATCGCTAACC 3′ | |||
| 1212 | proteasome subunit beta type 4 | FP: 5′ ACTCATGTGGACAACCTTC 3′ | 182 |
| RP: 5′ ACACCAAGCATATTACAATACC 3′ | |||
| 1268 | Thioredoxin peroxidase | FP: 5′ GCTGGTGGATTAGGACAAATG 3′ | 200 |
| RP: 5′ AGAAGACGAATCGCCTCATC 3′ | |||
| NADH dehydrogenase | FP: 5′ ATCCAAGTTGACGGTGTTC 3′ | 174 | |
| RP: 5′ GCAGCTATCATTTCATCTTCAG 3′ |
Reaction conditions and cycling protocols were followed as described in the SYBR green tag kit to add the fluorescent tag during every final extension step. Negative (no template) controls were included in each PCR run. Five positive controls of known concentration were included in every run to confirm consistent amplification. Quantitation of relative differences in expression was calculated using the Rotor-Gene version 6.0.38 software (Corbett Research, Australia) (20, 21).
RESULTS
2D-DIGE Analysis of Differentially Expressed Proteins
Schistosomula were isolated from three different hosts after 10 days of infection with S. japonicum cercariae. Soluble proteins extracted from the schistosomula were compared by the 2D-DIGE technique. A total of 1721 spots were matched across four analytical gels and one preparative gel (Fig. 1A). The analysis of 2D-DIGE revealed 54 spots that showed a statistically significant (t test, p < 0.01) change in intensity of > 1.2. Thirty-nine protein spots were found to be differentially expressed between the 10 day schistosomula from mice or rats, of which 27 were up-regulated and 12 were down-regulated in schistosomula from mice (Fig. 1B). Twenty-one protein spots were found to be differentially expressed between 10 day schistosomula from mice or M. fortis, of which 13 were up-regulated and eight were down-regulated in schistosomula from mice (Fig. 1C). Six of the above proteins appeared to be similarly regulated in rats or reed voles compared with mice.
Fig. 1.
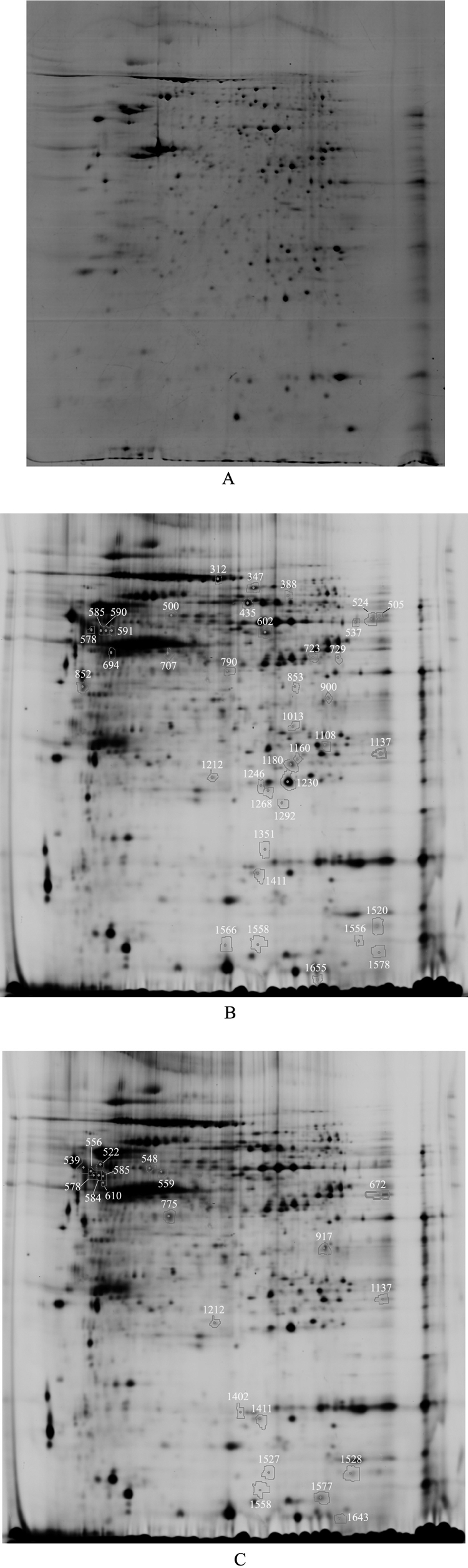
Separation of total proteins extracted from 10 d schistosomula of S. japonicum on two-dimensional electrophoresis gels over the pH range 3–10. A, proteins extracted from 10 day schistosomula were separated on preparative gel visualized by Coomassie brilliant blue staining. B, proteins differentially expressed in schistosomula recovered from rats compared with those from mice. C, proteins differentially expressed in schistosomula recovered from M. fortis compared with those from mice.
Identification of Differentially Expressed Proteins
The proteins selected as differentially expressed were separated on a preparative gel and visualized by staining with Coomassie blue. Gel pieces containing the proteins of interest were excised and subjected to digestion with trypsin. Thus, 44 protein spots were considered suitable for downstream analysis by MALDI MS/MS, based on the amount of protein, matching across gels, and the presence of the protein spot on the preparative gels. Out of these, 41 protein spots which represented 33 proteins were successfully identified by MS and are listed in Tables III and IV.
Table III. Identification of Schistosoma japonicum schistosomulum proteins differentially expressed between rats and mice: MALDI-TOF MS analyses. Schistosomula were recovered from the hosts 10 days after infection with cercariae. All the identified proteins matched with those in Schistosoma japonicum database.
| Spot IDa | Database ID no. | Identified protein name | MASCOT score | Sequence coverage (%)b | Theoretical Mr/pIc | Functional clusteringd | Fold changee |
|---|---|---|---|---|---|---|---|
| 312 | gi 56753427 | similar to heat shock 70kDa protein 9B | 625 | 43 | 71252/6.25 | Response to stimulus/stress | 1.25 |
| 388 | gi 56757978 | similar to Pkm2 protein | 182 | 27 | 61146/6.48 | carbohydrate metabolic process | 1.23 |
| 435 | gi 226468624 | putative protein disulfide isomerase-associated 3 precursor | 353 | 34 | 54671/6 | protein transport | 1.3 |
| 500 | gi 226471566 | similar to p50 immunophilin | 146 | 33 | 48551/5.65 | Response to stimulus/stress | 1.36 |
| 723/729 | gi 56754181 | pyruvate dehydrogenase E1 alpha 1 | 259/485 | 24/47 | 43940/8.44 | carbohydrate metabolic process | 1.31/1.45 |
| 1268 | gi 226489438 | Thioredoxin peroxidase | 233 | 28 | 21564/5.93 | regulation of gene expression | 1.64 |
| 1351 | gi 226475504 | putative cold shock domain protein A | 125 | 21 | 22381/10.63 | RNA metabolic process | 1.76 |
| 1558 | gi 76163073 | similar to cysteine protease inhibitor | 104 | 51 | 10026/8.53 | protein metabolic process | 1.26 |
| 1578 | gi 171473822 | similar to heat shock 10kD protein 1 | 191 | 67 | 12074/5.88 | protein metabolic process | 2.25 |
| 347 | gi 76154176 | similar to matricin | 255 | 32 | 24343/9.41 | protein metabolic process | −1.27 |
| 505 | gi 56759094 | similar to Heterogeneous nuclear ribonucleoprotein K | 80 | 23 | 42176/8.61 | RNA metabolic process | −1.62 |
| 524 | gi 462011 | Enolase | 130 | 17 | 47221/6.18 | carbohydrate metabolic process | −1.83 |
| 578/590 | gi 226473300 | Mitochondrial import receptor subunit TOM34 | 77/72 | 17/17 | 30253/5.02 | protein metabolic process | −3.03/−2.15 |
| 585 | gi 76156278 | similar to Mitochondrial import receptor subunit TOM34 | 82 | 14 | 27877/4.78 | protein metabolic process | −2.46 |
| 602 | gi 56756857 | similar to DEAD box ATP-dependent RNA helicase | 299 | 35 | 46113/6.16 | RNA metabolic process | −1.37 |
| 694 | gi 60688010 | similar to heat shock cognate protein | 206 | 52 | 9282/9.76 | −1.68 | |
| 707 | gi 226467798 | actin 5C | 156 | 26 | 41706/5.3 | cytoskeleton organization | −1.29 |
| 790 | gi 226475588 | phosphoglycerate kinase 1 | 197 | 25 | 44253/6.76 | carbohydrate metabolic process | −1.80 |
| 852 | gi 226478690 | Tropomyosin | 108 | 35 | 33006/4.63 | −1.84 | |
| 853/900 | gi 226473592 | aldo-keto reductase family 1, member B4 (aldose reductase) | 113/64 | 20/23 | 35612/6.87 | carbohydrate metabolic process | −1.53/−1.50 |
| 1013 | gi 226472458 | carbonyl reductase 1 | 199 | 54 | 30647/6.36 | carbohydrate metabolic process | −1.55 |
| 1180 | gi 62738608 | Chain A, Orthorhombic Glutathione S-Transferase | 329 | 53 | 26018/6.09 | protein metabolic process | −1.58 |
| 1212 | gi 226487138 | proteasome subunit beta type 4 | 96 | 46 | 27877/5.92 | protein metabolic process | −2.02 |
| 1230 | gi 226478722 | Tegument antigen (I(H)A) | 591 | 62 | 21679/6.45 | −1.29 | |
| 1246 | gi 226479412 | tryparedoxin peroxidase | 313 | 51 | 20776/6.42 | protein metabolic process | −1.25 |
| 1292 | gi 226472634 | similar to preprocathepsin C | 287 | 26 | 52787/8.76 | protein metabolic process | −1.66 |
a The spot ID was determined at the beginning of analysis of gels.
b The percentage coverage was defined as the ratio (%) of the protein sequence covered by the matched peptides.
c Theoretical Mr/pI was calculated using DNAStar software.
d The functions are classified by gene ontology (GO) annotations using Blast2GO and AmiGO.
e Fold change is calculated by DeCyder software (Version 6.5).
Table IV. Identification of Schistosoma japonicum schistosomulum proteins differentially expressed between M. forti and mice: MALDI-TOF MS analyses. Schistosomula were isolated from the hosts 10 days after infection with cercariae. All the identified proteins matched with those in Schistosoma japonicum database.
| Spot IDa | Database ID no. | Identified protein name | MASCOT score | Sequence coverage (%)b | Theoretical Mr/pIc | Functional clusteringd | Fold changee |
|---|---|---|---|---|---|---|---|
| 522 | gi 226487050 | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | 363 | 28 | 55763/5.21 | ribonucleotide metabolic process | 1.90 |
| 672 | gi 226479492 | Aldolase | 273 | 51 | 39580/6.56 | carbohydrate metabolic process | 1.54 |
| 1558 | gi 76163073 | similar to cysteine protease inhibitor | 104 | 37 | 10026/8.53 | protein metabolic process | 1.20 |
| 1577 | gi 226486682 | U6 snRNA-associated Sm-like protein LSm2 | 99 | 64 | 5049/7.79 | RNA metabolic process | 1.57 |
| 1643 | gi 226484011 | ubiquitin C | 318 | 35 | 17214/6.75 | protein metabolic process | 1.4 |
| 539/584/585 | gi 76156278 | similar to Mitochondrial import receptor subunit TOM34 | 67/81/82 | 10/17/14 | 27877/4.78 | protein metabolic process | −4.25/−2.49/−2.09 |
| 548 | gi 226472932 | Actin 5C | 111 | 25 | 40099/5.66 | cytoskeleton organization | −1.28 |
| 556 | gi 226471982 | putative Stress-induced-phosphoprotein 1 | 86 | 20 | 22352/4.55 | protein metabolic process | −6.49 |
| 559 | gi 226469026 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | 67 | 17 | 48028/5.46 | protein metabolic process | −1.24 |
| 578 | gi 226473300 | Mitochondrial import receptor subunit TOM34 | 77 | 17 | 30253/5.02 | protein metabolic process | −4.43 |
| 917 | gi 226472222 | Heterogeneous nuclear ribonucleoprotein A1, A2/B1 homolog | 201 | 25 | 32405/6.92 | RNA metabolic process | −1.41 |
| 1212 | gi 226487138 | proteasome subunit beta type 4 | 96 | 46 | 27877/5.92 | protein metabolic process | −1.80 |
| 1528 | gi 226480906 | nucleoside-diphosphate kinase | 236 | 59 | 16850/6.9 | RNA metabolic process | −2.02 |
a The spot ID was determined at the beginning of analysis of gels.
b The percentage coverage is defined as the ratio (%) of the protein sequence covered by the matched peptides.
c Theoretical Mr/pI was calculated using DNAStar software.
d The functions are classified by gene ontology (GO) annotations using Blast2GO and AmiGO.
e Fold change is calculated by DeCyder software (Version 6.5).
Quantitative Real-time PCR Analysis of Differentially Expressed Proteins
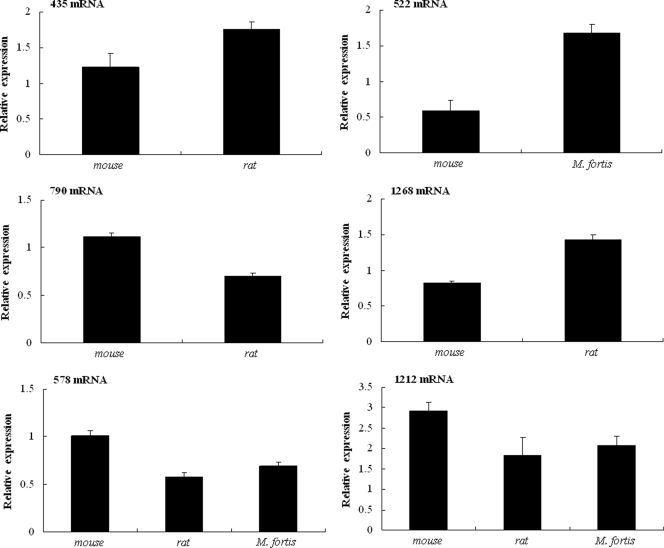
Six genes corresponding to the protein spots designated 435, 522, 578, 790, 1212, and 1268 were chosen for quantitative real-time PCR (qPCR) analysis to quantify their transcript levels (Fig. 2). Two of the schistosomula peptides appeared to be down-regulated in both rats and reed voles compared with mice, namely 578 and 1212, which had been identified as the mitochondrial import receptor subunit TOM34 and proteasome subunit beta type 4, respectively (Tables III and IV). In contrast, other spots were down-regulated in rats alone (790); or up-regulated in rats (435 and 1268) or in reed voles (522). The qPCR results were consistent with those of the DIGE studies, and suggested that these proteins identified as differentially expressed were regulated at transcriptional level.
Fig. 2.
Confirmation of transcriptional regulation of differentially expressed proteins by real-time RT-PCR. Transcript levels corresponding to six (spot IDs: 435, 522, 790, 1268, 1212, and 578) out of the 33 differentially expressed proteins in schistosomula isolated from mice, rats or reed voles (M. fortis) are presented relative to those corresponding to NADH dehydrogenase in the same host.
Gene Ontology Annotation
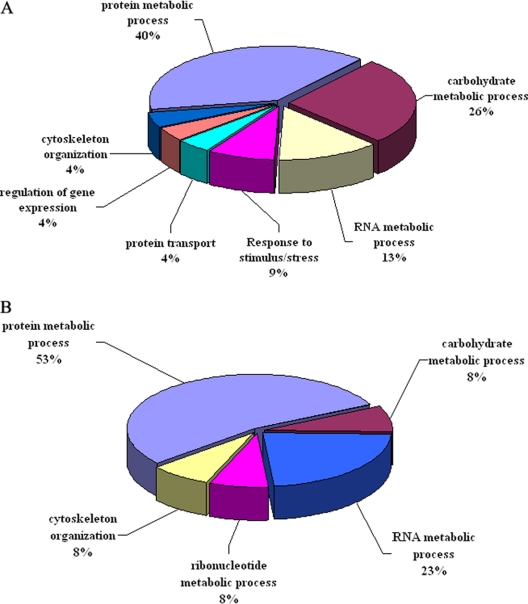
To better understand the biological functions of the 33 differentially expressed proteins, Gene Ontology Annotation was performed. The annotated S. japonicum proteins from the NCBI nonredundant database were analyzed by the Blast2GO and AmiGO servers initially. Each identified known protein was classified according to its GO functional annotation. These differentially expressed proteins were essentially those involved in metabolism of proteins, ribonucleotides or carbohydrates, or in stress response or cytoskeleton/movement (Fig. 3). Mitochondrial import receptor subunit TOM34 protein, proteasome subunits, enolase, heterogeneous nuclear ribonucleoprotein K, and actin 5C protein were highly expressed in the susceptible host, mice compared with the unsusceptible host, rats or the resistant host, M. fortis. In contrast, cysteine protease inhibitor, aldolase, and heat shock 70 kDa protein were relatively highly expressed in rats or M. fortis compared with mice. Most of the differentially expressed proteins were involved in important biological functions. The analysis also revealed that the proteins of 10 day schistosomula that were differentially expressed between rats and mice were mainly related to protein (40%) or carbohydrate metabolism (26%), while those differentially expressed between M. fortis and mice were mainly related to protein (53%) or ribonucleotide metabolism (23%).
Fig. 3.
Pie charts showing the gene ontology (GO) distribution of differentially expressed proteins of schistosomula from three host species, according to major biological process categories. A, proteins differentially expressed in schistosomula recovered from rats compared with those from mice. B, proteins differentially expressed in schistosomula recovered from M. fortis compared with those from mice.
DISCUSSION
Schistosomulum is a critical stage for the development and maturation of schistosome. The parasites in this stage undergo a number of morphological changes and physiological adaptations in their mammalian definitive hosts, and finally develop into mature adult worms. Several research groups have focused on the schistosomulum transcriptome or proteome with the purpose of identifying key molecules that are differentially expressed at this stage and which may therefore be essential for the survival and development of schistosome (8, 22–25). These studies have provided valuable information for screening vaccine candidates and new drug targets.
In recent years, the proteomic approach has been successfully applied to schistosome protein analysis (26), and has proved to be a powerful technique for the detection and identification of proteins specific to the schistosome stage of the parasite. In this study, we report for the first time a comparative proteomic analysis of proteins of 10 day schistosomula isolated from three different types of hosts: the susceptible host, BALB/c mice, unsusceptible host, Wistar rats, and resistant host, M. fortis. The proteins that were differentially expressed among the schistosomula from these hosts were selected for further identification by MALDI-MS/MS. GO analysis and other bioinformatic techniques were applied to predict the possible function of differentially expressed proteins. The analyses revealed that some of these proteins might possess important biologic functions affecting the living status of the schistosome and many were essential for the survival of schistosome. The distinctive expression of some of these molecules possibly influence the survival of the worms, such that they grow well in BALB/c mice, poorly in Wistar rats, or die within 2 weeks in M. fortis.
Proteomic maps showed that there were 39 schistosomulum proteins differentially expressed between mice and rats, and 21 between mice and M. fortis. GO analysis revealed that most of the differentially expressed proteins were mainly involved in five categories of cellular processes as discussed below.
The first category consists of proteins involved in metabolic processes. Successful development of schistosome in the final host requires a series of structural, biochemical, and physiological changes. These changes depend to a large degree on the synthesis and degradation of proteins, and are critical to the development and maturation of the schistosome.
Extensive studies with lower eukaryotes have led to the identification of many components of the protein import machinery, including proteins termed as translocases of the outer mitochondrial membrane (TOM proteins) (27). TOM34 is a cytosolic protein with chaperone-like activity that helps import certain preproteins to the mitochondria by keeping them in an unfolded, import-compatible state. TOM34 was found to be up-regulated frequently in colorectal tumors, suggesting that it has a role in the growth of cancer cell (28). Exogenous addition of the soluble domain of TOM34 and antibodies against this protein strongly inhibit in vitro mitochondrial import of several preproteins (29). In our study, Tom34 was down-regulated in schistosomula from both rats and M. fortis compared with mice. The down-regulation of TOM34 may result from an inability of the schistosome to grow and develop normally in unsusceptible hosts, rats and resistant hosts, M. forti, thereby affecting mitochondrial import of preproteins in the worms.
In eukaryotic cells, the turnover of intracellular proteins is mediated primarily by the ubiquitin-proteasome system (30). The 26S proteasome is a complex of ubiquitin-proteasome proteolytic pathway responsible for degradation of proteins that are naturally unfolded, mutated, or oxygen-damaged (31, 32). Many studies have indicated that the proteasome may play an important role in the development and survival of schistosome (33). In this study, the 26S proteasome subunit beta type 4 was found to be down-regulated in schistosomula from rats and M. fortis, and subunit ATPase 3 was also found to be down-regulated in schistosomula from M. fortis, compared with mice. Though down-regulation of subunit beta type 4 and subunit ATPase 3 seems unrelated to the protease activity provided by subunit beta types 1, 2, and 5 (30), we deduce that the expression of these active subunits are also likely down-regulated although were not identified in the present work. The down-regulation of the 26S proteasome might affect the development of schistosomes through influencing the cell cycle progression, transcriptional control, and other critical cellular processes in the worms (34).
The second category of differentially expressed proteins comprises of those related to carbohydrate metabolism. Like all parasitic helminths, the schistosomula as well as adult schistosomes depend largely on anaerobic energy metabolism for their growth and development (35). Schistosomes absorb copious amounts of host sugar, consuming glucose equivalent to their dry weight, every five hours (36). They seem to employ several enzymes in carbohydrate metabolism, and glycolysis leading to lactic acid production is an important energy pathway for schistosomes (35). Therefore, glycolytic enzymes are essential and important for the survival of the parasite. The results of this study revealed that pyruvate kinase (Pkm2) and pyruvate dehydrogenase family proteins were up-regulated, while enolase and phosphoglycerate kinase family molecules were down-regulated in 10 day schistosomula from rats. The up-regulation of pyruvate kinase and pyruvate dehydrogenase could enhance phosphoenolpyruvate conversion to pyruvate, and then to 2-hydroxyethyl-ThPP, lactate or other products. The down-regulation of phosphoglycerate kinase and enolase is deduced to suppress the 1.3–2-phosphoglycerate converting to phosphoenolpyruvate. The results suggested that the change of these gene expressions may effect the parasite survival and development by affecting the homeostasis of carbohydrate metabolism in the worms.
The third category corresponds to the molecules associated with the ribonucleotide metabolic processes and transcription and translation machinery. Heterogeneous nuclear ribonucleoprotein (hnRNP), one of the most important RNA-binding proteins, binds to mRNA precursors concomitant with transcription and forms ribonucleoprotein complexes essential for post-transcriptional events (37). ATP-dependent RNA helicases are implicated in all aspects of cellular RNA metabolism and promote unwinding of RNA during splicing and translation. Nucleoside diphosphate kinase (NDK) generates nucleoside triphosphates from respective diphosphates other than ATP. In the present study, heterogeneous nuclear ribonucleoprotein K and DEAD box ATP-dependent RNA helicases were down-regulated in schistosomula from rats compared with those from mice; while hnRNP A1 and NDK were down-regulated in schistosomula from M. fortis compared with those from mice. The down-regulation of these molecules in schistosomula in rats and M. fortis could adversely affect the ribonucleotide metabolism, transcription and translation processes of the worms in these two hosts, resulting in poor growth and survival or death.
The fourth category of differentially expressed proteins consists of stress-associated proteins. Stress responses in all organisms play an important role during cellular development. This is particularly true for a schistosome during the introduction of schistosomula into the immune-hostile environment of the mammalian host (22). In humans, heat-shock proteins are over-expressed when schistosomes are exposed to significant levels of stress (38). In the present study, HSPs, similar to HSP70 and HSP10 were up-regulated in schistosomula from rats. HSP70 protein is considered as the predominant HSP family and plays the key regulatory role in parasite development and pathogenesis (39). High expression of HSP70 in schistosomes might be associated with the acquisition of heat tolerance, or perhaps tolerance to other environmental stresses related to the host immune response (22). In lung schistosome and 24 h mechanically transformed schistosomula, HSP70 protein is overexpressed compared with that in adult schistosomes (24).
Rat probably provides a less suitable micro-environment for the schistosomes and is hence an unsusceptible host. Therefore, the up-regulation of HSP70 protein in 10 day schistosomula from rats is a reasonable and expected response. In recent research, the chaperone-like activity and the thermotolerance of recombinant SjHSP70 was demonstrated, and a recombinant SjHSP70 was able to elicit Th1-type bias immune response and induce partial immuno-protection against S. japonicum in mice as well (40). HSP10, also known as a relatively small stress protein, usually works in conjunction with a HSP60. The HSP10-HSP60 complex regulates the transport of proteins across the mitochondrial membrane (41, 42). HSP70 also assists a large variety of proteins in folding and or in the degradation of unstable proteins in the cell (43). Thus, both HSP70 and HSP10 take part not only in a stress response process but also in protein folding and metabolic processes (44).
The last category identified in our differential screening consists of cytoskeletal proteins and those required for cellular movement. Actin belongs to a highly conserved family of proteins found in all eukaryotes and is a critical component of the muscle system (45, 46). It was shown to be present on the surface of schistosomula, in the muscle, tegumental tubercles and spines of male and female adult worms (47–50). The movement of schistosomes is accomplished through well-developed muscular arrangements that are mainly facilitated by tubulin and actin molecules (51). The down-regulation of actin in schistosomula from both rats and M. fortis possibly change the relatively complex muscle organization within the body wall and have detrimental effects on movement of the schistosomes. We deduce that this process may influence the growth and development of schistosomes by inhibiting the muscular system movement and thus leading to an abnormal state resulting in the death of worms.
In the present study, we applied DIGE and MALDI MS/MS techniques to identify differentially expressed proteins of 10 day schistosomula from mice, rats and M. fortis. GO analysis revealed that some of the differentially expressed proteins had important functions that likely impact the survival and development of schistosomes. The identification of these molecules provides a new basis for a better understanding of the developmental mechanism in the schistosome and suggests new vaccine candidates or drug targets for the control of schistosomiasis.
Acknowledgments
We thank Hao Li and Ke Lu from Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences for technical assistance with parasite collection. We also thank Shanghai Applied Protein Technology Co. Ltd. for the technology support.
Footnotes
* This work was supported by National Basic Research Program of China (No. 2007CB513108), National Natural Science Foundation of China (No. 30671581), National High Technology Research and Development Program of China (No. 2006AA10A207-1) and Special Fund for Agro-scientific Research in the Public Interest (200903036).
1 The abbreviations used are:
- 2D-DIGE
- two-dimensional differential del electrophoresis
- DTT
- dithiotreitol
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate
- MALDI
- matrix-assisted laser desorption ionization
- TOM
- translocase of the outer mitochondrial membrane
- HSP
- heat shock protein.
REFERENCES
- 1. Bergquist N. R. (2002) Schistosomiasis: from risk assessment to control. Trends Parasitol. 18, 309–314 [DOI] [PubMed] [Google Scholar]
- 2. Engels D., Chitsulo L., Montresor A., Savioli L. (2002) The global epidemiological situation of schistosomiasis and new approaches to control and research. Acta Trop. 82, 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ismail M., Metwally A., Farghaly A., Bruce J., Tao L. F., Bennett J. L. (1996) Characterization of isolates of Schistosoma mansoni from Egyptian villagers that tolerate high doses of praziquantel. Am. J. Trop. Med. Hyg. 55, 214–218 [DOI] [PubMed] [Google Scholar]
- 4. Hu S., Law P. K., Fung M. C. (2009) Microarray analysis of genes highly expressed in cercarial stage of Schistosoma japonicum and the characterization of the antigen Sj20H8. Acta Trop. 112, 26–32 [DOI] [PubMed] [Google Scholar]
- 5. Bergquist N. R., Leonardo L. R., Mitchell G. F. (2005) Vaccine-linked chemotherapy: can schistosomiasis control benefit from an integrated approach? Trends Parasitol. 21, 112–117 [DOI] [PubMed] [Google Scholar]
- 6. DeMarco R., Verjovski-Almeida S. (2009) Schistosomes–proteomics studies for potential novel vaccines and drug targets. Drug Discov. Today 14, 472–478 [DOI] [PubMed] [Google Scholar]
- 7. Cheng G. F., Lin J. J., Feng X. G., Fu Z. Q., Jin Y. M., Yuan C. X., Zhou Y. C., Cai Y. M. (2005) Proteomic analysis of differentially expressed proteins between the male and female worm of Schistosoma japonicum after pairing. Proteomics 5, 511–521 [DOI] [PubMed] [Google Scholar]
- 8. Curwen R. S., Ashton P. D., Johnston D. A., Wilson R. A. (2004) The Schistosoma mansoni soluble proteome: a comparison across four life-cycle stages. Mol. Biochem. Parasitol. 138, 57–66 [DOI] [PubMed] [Google Scholar]
- 9. van Hellemond J. J., van Balkom B. W., Tielens A. G. (2007) Schistosome biology and proteomics: progress and challenges. Exp. Parasitol. 117, 267–274 [DOI] [PubMed] [Google Scholar]
- 10. Curwen R. S., Ashton P. D., Sundaralingam S., Wilson R. A. (2006) Identification of novel proteases and immunomodulators in the secretions of schistosome cercariae that facilitate host entry. Mol. Cell Proteomics 5, 835–844 [DOI] [PubMed] [Google Scholar]
- 11. Knudsen G. M., Medzihradszky K. F., Lim K. C., Hansell E., McKerrow J. H. (2005) Proteomic analysis of Schistosoma mansoni cercarial secretions. Mol. Cell Proteomics 4, 1862–1875 [DOI] [PubMed] [Google Scholar]
- 12. Perez-Sanchez R., Ramajo-Hernandez A., Ramajo-Martin V., Oleaga A. (2006) Proteomic analysis of the tegument and excretory-secretory products of adult Schistosoma bovis worms. Proteomics 6 Suppl 1, S226–236 [DOI] [PubMed] [Google Scholar]
- 13. He Y. X., Salafsky B., Ramaswamy K. (2001) Host–parasite relationships of Schistosoma japonicum in mammalian hosts. Trends Parasitol. 17, 320–324 [DOI] [PubMed] [Google Scholar]
- 14. Li H., He Y. Y., Lin J. J. (2000) The observation for the phenomenon of Microtus fortis aganisting Schistosoma japonicum. Chin. J. Vet. Parasitol. 8, 12–15 [Google Scholar]
- 15. Wu K. (1957) Schistosomiasis japonica among domestic and wild animals in China. Chin. Vet. J 3, 98–100 [Google Scholar]
- 16. Li S. K., Zhu Z. L., Jin B. R. (1965) Uninfectibility to Schistosoma japonicum of Microtus fortis. Acta Parasitol. 2, 103 [Google Scholar]
- 17. Jiang W., Hong Y., Peng J., Fu Z., Feng X., Liu J., Shi Y., Lin J. (2010) Study on differences in the pathology, T cell subsets and gene expression in susceptible and non-susceptible hosts infected with Schistosoma japonicum. PLoS One 5, e13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alban A., David S. O., Bjorkesten L., Andersson C., Sloge E., Lewis S., Currie I. (2003) A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3, 36–44 [DOI] [PubMed] [Google Scholar]
- 19. Tonge R., Shaw J., Middleton B., Rowlinson R., Rayner S., Young J., Pognan F., Hawkins E., Currie I., Davison M. (2001) Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 1, 377–396 [DOI] [PubMed] [Google Scholar]
- 20. Gobert G. N., Moertel L., Brindley P. J., McManus D. P. (2009) Developmental gene expression profiles of the human pathogen Schistosoma japonicum. BMC Genomics 10, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moertel L., McManus D. P., Piva T. J., Young L., McInnes R. L., Gobert G. N. (2006) Oligonucleotide microarray analysis of strain- and gender-associated gene expression in the human blood fluke, Schistosoma japonicum. Molecular and Cellular Probes 20, 280–289 [DOI] [PubMed] [Google Scholar]
- 22. Wang X., Gobert G. N., Feng X., Fu Z., Jin Y., Peng J., Lin J. (2009) Analysis of early hepatic stage schistosomula gene expression by subtractive expressed sequence tags library. Mol. Biochem. Parasitol. 166, 62–69 [DOI] [PubMed] [Google Scholar]
- 23. Farias L. P., Tararam C. A., Miyasato P. A., Nishiyama M. Y., Jr., Oliveira K. C., Kawano T., Verjovski-Almeida S., Leite L. C. (2010) Screening the Schistosoma mansoni transcriptome for genes differentially expressed in the schistosomulum stage in search for vaccine candidates. Parasitol. Res. 108, 123–135 [DOI] [PubMed] [Google Scholar]
- 24. Chai M., McManus D. P., McInnes R., Moertel L., Tran M., Loukas A., Jonesa M. K., Gobert G. N. (2006) Transcriptome profiling of lung schistosomula, in vitro cultured schistosomula and adult Schistosoma japonicum. Cell Mol. Life Sci. 63, 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu F., Lu J., Hu W., Wang S. Y., Cui S. J., Chi M., Yan Q., Wang X. R., Song H. D., Xu X. N., Wang J. J., Zhang X. L., Zhang X., Wang Z. Q., Xue C. L., Brindley P. J., McManus D. P., Yang P. Y., Feng Z., Chen Z., Han Z. G. (2006) New perspectives on host-parasite interplay by comparative transcriptomic and proteomic analyses of Schistosoma japonicum. PLoS Pathog. 2, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang L. L., Lv Z. Y., Hu S. M., He S. J., Li Z. Y., Zhang S. M., Zheng H. Q., Li M. T., Yu X. B., Fung M. C., Wu Z. D. (2009) Schistosoma japonicum: proteomics analysis of differentially expressed proteins from ultraviolet-attenuated cercariae compared to normal cercariae. Parasitol. Res. 105, 237–248 [DOI] [PubMed] [Google Scholar]
- 27. Mukhopadhyay A., Avramova L. V., Weiner H. (2002) Tom34 unlike Tom20 does not interact with the leader sequences of mitochondrial precursor proteins. Arch. Biochem. Biophys. 400, 97–104 [DOI] [PubMed] [Google Scholar]
- 28. Blesa J. R., Prieto-Ruiz J. A., Abraham B. A., Harrison B. L., Hegde A. A., Hernández-Yago J. (2008) NRF-1 is the major transcription factor regulating the expression of the human TOMM34 gene. Biochem. Cell Biol. 86, 46–56 [DOI] [PubMed] [Google Scholar]
- 29. Chewawiwat N., Yano M., Terada K., Hoogenraad N. J., Mori M. (1999) Characterization of the novel mitochondrial protein import component, Tom34, in mammalian cells. J. Biochem. 125, 721–727 [DOI] [PubMed] [Google Scholar]
- 30. Coux O., Tanaka K., Goldberg A. L. (1996) Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847 [DOI] [PubMed] [Google Scholar]
- 31. Orlowski M., Wilk S. (2003) Ubiquitin-independent proteolytic functions of the proteasome. Arch. Biochem. Biophys. 415, 1–5 [DOI] [PubMed] [Google Scholar]
- 32. Guerra-Sá R., Castro-Borges W., Evangelista E. A., Kettelhut I. C., Rodrigues V. (2005) Schistosoma mansoni: functional proteasomes are required for development in the vertebrate host. Exp. Parasitol. 109, 228–236 [DOI] [PubMed] [Google Scholar]
- 33. Hong Y., Han H., Peng J., Li Y., Shi Y., Fu Z., Liu J., Lin J., Li X. (2010) Schistosoma japonicum: cloning, expression and characterization of a gene encoding the alpha5-subunit of the proteasome. Exp. Parasitol. 126, 517–525 [DOI] [PubMed] [Google Scholar]
- 34. Nabhan J. F., El-Shehabi F., Patocka N., Ribeiro P. (2007) The 26S proteasome in Schistosoma mansoni: bioinformatics analysis, developmental expression, and RNA interference (RNAi) studies. Exp. Parasitol. 117, 337–347 [DOI] [PubMed] [Google Scholar]
- 35. Bueding E., Fisher J. (1982) Metabolic requirements of schistosomes. J. Parasitol. 68, 208–212 [PubMed] [Google Scholar]
- 36. Camacho M., Agnew A. (1995) Glucose uptake rates by Schistosoma mansoni, S. haematobium, and S. bovis adults using a flow in vitro culture system. J. Parasitol. 81, 637–640 [PubMed] [Google Scholar]
- 37. Xiong X., Feng Q., Xie L., Zhang R. (2007) Cloning and characterization of a heterogeneous nuclear ribonucleoprotein homolog from pearl oyster, Pinctada fucata. Acta Biochim. Biophys. Sin 39, 955–963 [DOI] [PubMed] [Google Scholar]
- 38. Maresca B., Carratùö L. (1992) The biology of the heat shock response in parasites. Parasitol. Today 8, 260–266 [DOI] [PubMed] [Google Scholar]
- 39. De Jong-Brink M. (1995) How schistosomes profit from the stress responses they elicit in their hosts. Adv. Parasitol. 35, 177–256 [DOI] [PubMed] [Google Scholar]
- 40. He S., Yang L., Lv Z., Hu W., Cao J., Wei J., Sun X., Yang J., Zheng H., Wu Z. (2010) Molecular and functional characterization of a mortalin-like protein from Schistosoma japonicum (SjMLP/hsp70) as a member of the HSP70 family. Parasitol. Res. 107, 955–966 [DOI] [PubMed] [Google Scholar]
- 41. Hickey R. W., Zhu R. L., Alexander H. L., Jin K. L., Stetler R. A., Chen J., Kochanek P. M., Graham S. H. (2000) 10 kD mitochondrial matrix heat shock protein mRNA is induced following global brain ischemia in the rat. Brain Res. Mol. Brain Res. 79, 169–173 [DOI] [PubMed] [Google Scholar]
- 42. Höhfeld J., Hartl F. U. (1994) Role of the chaperonin cofactor Hsp10 in protein folding and sorting in yeast mitochondria. J. Cell Biol. 126, 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mayer M. P., Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Walter S., Buchner J. (2002) Molecular chaperones–cellular machines for protein folding. Angew Chem. Int. Ed Engl. 41, 1098–1113 [DOI] [PubMed] [Google Scholar]
- 45. Sheterline P., Sparrow J. C. (1994) Actin. Protein Profile 1, 1–121 [PubMed] [Google Scholar]
- 46. Cooke R. (1986) The mechanism of muscle contraction. CRC Crit. Rev. Biochem. 21, 53–118 [DOI] [PubMed] [Google Scholar]
- 47. Kusel J. R., Gazzinelli G., Colley D. G., de Souza C. P., Cordeiro M. N. (1984) The formation of surface membrane vesicles from schistosomula of Schistosoma mansoni. Parasitology 89, 483–494 [DOI] [PubMed] [Google Scholar]
- 48. Davis A. H., Blanton R., Klich P. (1985) Stage and sex specific differences in actin gene expression in Schistosoma mansoni. Mol. Biochem. Parasitol. 17, 289–298 [DOI] [PubMed] [Google Scholar]
- 49. Abbas M. K., Cain G. D. (1987) Actin and intermediate-sized filaments of the spines and cytoskeleton of Schistosoma mansoni. Parasitol. Res. 73, 66–74 [DOI] [PubMed] [Google Scholar]
- 50. MacGregor A. N., Shore S. J. (1990) Immunocytochemistry of cytoskeletal proteins in adult Schistosoma mansoni. Int. J. Parasitol. 20, 279–284 [DOI] [PubMed] [Google Scholar]
- 51. Mair G. R., Maule A. G., Fried B., Day T. A., Halton D. W. (2003) Organization of the musculature of schistosome cercariae. J. Parasitol. 89, 623–625 [DOI] [PubMed] [Google Scholar]