Abstract
Ribosome biogenesis starts with transcription of the large ribosomal RNA precursor (47S pre-rRNA), which soon combines with numerous factors to form the 90S pre-ribosome in the nucleolus. Although the subsequent separation of the pre-90S particle into pre-40S and pre-60S particles is critical for the production process of mature small and large ribosomal subunits, its molecular mechanisms remain undetermined. Here, we present evidence that p32, fibrillarin (FBL), and Nop52 play key roles in this separation step. Mass-based analyses combined with immunoblotting showed that p32 associated with 155 proteins including 31 rRNA-processing factors (of which nine were components of small subunit processome, and six were those of RIX1 complex), 13 chromatin remodeling components, and six general transcription factors required for RNA polymerase III-mediated transcription. Of these, a late rRNA-processing factor Nop52 interacted directly with p32. Immunocytochemical analyses demonstrated that p32 colocalized with an early rRNA-processing factor FBL or Nop52 in the nucleolus and Cajal bodies, but was excluded from the nucleolus after actinomycin D treatment. p32 was present in the pre-ribosomal fractions prepared by cell fractionation or separated by ultracentrifugation of the nuclear extract. p32 also associated with pre-rRNAs including 47S/45S and 32S pre-rRNAs. Furthermore, knockdown of p32 with a small interfering RNA slowed the early processing from 47S/45S pre-rRNAs to 18S rRNA and 32S pre-rRNA. Finally, Nop52 was found to compete with FBL for binding to p32 probably in the nucleolus. Given the fact that FBL and Nop52 are associated with pre-ribosome particles distinctly different from each other, we suggest that p32 is a new rRNA maturation factor involved in the remodeling from pre-90S particles to pre-40S and pre-60S particles that requires the exchange of FBL for Nop52.
Human p32 (splicing factor 2-associated protein p32)1 contains 282 amino acid residues, 73 of which are in the N-terminal mitochondrial signal sequence. It associates with splicing factor ASF/SF2 (1) and regulates pre-mRNA splicing through its ability to inhibit both phosphorylation and binding of ASF/SF2 to pre-mRNA (2). p32 is also believed to be involved in alternative splicing events by regulating competitive binding of ASF/SF2 and heterogeneous nuclear ribonucleoprotein (hnRNP) A1 to the splicing complex (3–9). p32 has also been identified as a protein (C1QBP) that binds to complement component 1, q subcomponent, which recognizes and binds to the heavy chain of immunoglobulin G or M initiating the classical complement pathway (10), and as hyaluronic acid-binding protein, which is an inhibitor of Streptococcus pneumoniae hyaluronidase (11, 12). p32 also interacts with B subunit of the CCAAT-binding factor and inhibits specifically CCAAT-binding factor-mediated transcription activation (13). In addition, p32 binds the tumor suppressor ARF C terminus, is required for alternative reading frame protein (ARF) to localize to mitochondria, and induces apoptosis (14). Cancer-derived point mutations in the ARF C terminus disrupt simultaneously the ARF-p32 interaction and ARF's apoptotic function; thus, p32 is a critical mediator of ARF's apoptotic function in the mitochondria. p32 links the BH3-only protein Hrk to mitochondria and may be critically involved also in the regulation of Hrk-mediated apoptosis (15). So far, a dozen other intracellular proteins, including the lamin B receptor (16), mitogen-activated protein kinases, protein kinase C family proteins, and matrix metallopeptidase, have been found to associate with p32 (NCBI Entrez Gene, C1QBP). Furthermore, it interacts with several viral proteins, including herpes simplex virus 1 Orf-P protein (17), the adenovirus polypeptide V (18), Epstein-Barr virus EBNA 1 protein (19, 20), rubella virus capsid protein (21), and the human immunodeficiency virus 1 (HIV1) Rev (22, 23) and Tat proteins (24). Although most of the physiological functions of these interactions are not understood, p32 was found to inhibit the splicing of HIV pre-RNA transcripts in HIV-transfected cells (25). A single mutation in human p32 of Gly35 to Asp35, which is also found in murine p32, abrogates this effect. HIV uses the splicing inhibition function of human p32, but not of the mouse counterpart, in RNA splicing to escape from a mechanism underlying the host cell defense against viral infection.
Although p32 is predominantly localized in mitochondria and on the cell surface, we previously found that it interacts with a nucleolar protein, fibrillarin (FBL), in a subcomplex containing a minimal set of proteins including protein arginine methyltransferase 5 (PRMT5, Janus Kinase-binding protein 1), and PRMT1 (26). FBL is the most abundant protein in the dense fibrillar regions of the nucleolus where rRNA transcription and early pre-rRNA processing take place (27). FBL is a methyltransferase and a component of RNP complexes (small subunit prosessome) that contain U3, U8, U13, and other small nucleolar RNAs. FBL associates with Nop56, Nop5/58 and a 15.5-kDa protein (a counterpart of yeast Snu13p) to form box C/D snoRNP complexes that function in site-specific 2′-O-methylation of RNA (28–31), and is a component of the pre-90S ribosomal particle formed at the early stages of ribosome biogenesis in the nucleolus. Human FBL (∼36 kDa) comprises glycine- and arginine-rich (GAR) domain, RNA-binding domain (RBD) and methyltransferase-like domain (32). Both the FBL GAR domain and the subsequent spacer region 1 (residues 78–132) interact directly with p32 (26).
In this study, we performed a proteomic analysis of protein complexes pulled down from nuclear extract with p32 as bait and found that it associated with many probable rRNA processing factors. Among them, p32 was found to interact with Nop52 (NNP1/RRP1A) without any cofactors, as does it FBL. Nop52 is a human homolog of yeast Rrp1p, which is associated with several distinct 66S pre-ribosomal particles in yeast cells and is involved in late events related to the production of 60S ribosomal subunit (33). Nop52, which was originally identified with human autoantibodies, is a nucleolar protein that is excluded from rRNA transcription sites, accumulates in the granular external domain, and mainly co-localizes with nucleolar proteins such as B23 that are involved in the late processing step in the nucleoli (34, 35). The perinucleolar bodies sequentially recruit FBL, nucleolin, and Nop52 together with protein B23, in this order, during the formation of the nucleolus at the end of mitosis (34, 35). Our present study demonstrates that p32 is required for processing of pre-rRNAs at the early nucleolar stages of ribosome biogenesis.
EXPERIMENTAL PROCEDURES
Construction of Epitope-tagged Expression Plasmids
Reverse transcriptase (RT)-PCR was performed with the Super Script II kit (Invitrogen) according to the manufacturer's instructions using total mRNA that was prepared from 293EBNA cells (Invitrogen). C-terminally FLAG (DYKDDDDK)-tagged p32 (p32-FLAG) cDNA, C-terminally FLAG-tagged Guα (FLAG-Guα) cDNA, and N-terminally FLAG-tagged Guβ (FLAG-Guβ) cDNA were amplified by PCR using the primer sets 5′-ATTGAGCTAGCGCCACCATGCTGCCTCTGCTGCGCTGC-3′ and 5′-TCAGTGGATCCCTACTTGTCGTCGTCGTCCTTGTAGTCCTGGCTCTTGACAAAACTCTT-3′ for p32-FLAG, 5′-ATATAGCTAGCGCCACCATGCCGGGAAAACTCCGTAGTGACGCTGGT-3′ and 5′-ATATAGGATCCTTACTTGTCGTCGTCGTCCTTGTAGTCTTGACCAAATGCTTTACTGAAACTCCGCTT-3′ for FLAG-Guα, and 5′-TATAATGGATCCGCCACCATGGACTACAAGGACGACGACGACAAGATGCCTGGGAAACTCCTCTGG-3′ and 5′-CAACTCGAGTCAGTCAAAACTCCGTTTGTG-3′ for FLAG-Guβ. The PCR products were cloned into the NheI/BamHI sites (for p32-FLAG and FLAG-Guα) and into the BamHI/XhoI sites (for FLAG-Guβ) of the expression vector pcDNA3.1(+). Previous reports have described the construction of FLAG-tagged nucleolin (FLAG-NCL) (36), FLAG-tagged FBL (FLAG-FBL) (26), and FLAG-tagged Nop52 (FLAG-Nop52) (37). N-terminally HA (YPYDVPDYA)-tagged FBL (HA-FBL) was amplified from the expression vector containing the FLAG-tagged FBL-coded DNA fragment as the template by PCR using the primer sets 5′GAAGAAGGATCCGCCACCATGTACCCATACGACGTGCCTGACTATGCCAAGCCAGGATTCAGTCCCCGT-3′ and 5′-GAAGAAGAATTCTCAGTTCTTCACCTTGGGGGG-3′. The PCR product was cloned into the BamHI/EcoRI sites of the expression vector pcDNA3.1(+). To establish inducible FLAG-Nop52 expression cell lines, cDNA for FLAG-Nop52 was amplified by PCR using the forward primer 5′ATATCAAGCTTGCCAACCATGGACTACAAGGACGACGACGACAAGGTTTCGCGCGTGCAGCTCCCG-3′ and the reverse primers described previously (37). The PCR product was cloned into the HindIII/BamHI sites of the expression vector pcDNA5/FRT/TO. All constructs were verified by DNA sequencing.
RNA Interference (RNAi)
HeLa cells were cultured in 35-mm dishes until they reached 80% confluency, and transfected with 2.5 μl Lipofectamine 2000 and 50 nm stealth small interfering RNA (siRNA)615. The following stealth siRNA sequences were used: 5′-AUGACAGUCCAACACAAGGGCCUUC-3′ and 5′-GAAGGCCCUUGUGUUGGACUGUCAU-3′ for p32 knockdown and 5′-AUGCCAACUGCCAAACACGAGGUUC-3′ and 5′-GAACCUCGUGUUUGGCAGUUGGCAU-3′ for the negative control. The cells were transferred to 35-mm or 90-mm dishes 24 h after the first transfection and then transfected again with the same stealth siRNA 48 h after the first transfection. The cells were then cultured for 12 h and were washed once with PBS and cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) containing 10% FBS, streptomycin (0.1 mg/ml), penicillin G (100 U/ml), and 1000 mg/L glucose for additional 12 h.
Cell Fractionation
Subconfluent p32-FLAG-transfected 293EBNA, inducible FLAG-Nop52 expression (Flp-In T-REx 293) cells, 293EBNA cells, or HeLa cells were collected from four 90-mm dishes, lysed by vortexing for 10 s with 1 ml buffer A (16.7 mm Tris-HCl (pH 8.0), 50 mm NaCl, 1.67 mm MgCl2, 1 mm phenylmethylsulfonyl fluoride (PMSF)) containing 0.1% Triton X-100, incubated for 5 min on ice, and centrifuged at 1,000 × g for 5 min. The supernatant was collected as the cytoplasmic fraction. The nuclear extract (Nu), nucleoplasmic extract (NE) and nucleolar/Cajal bodies extract (NoE) were prepared from the remaining pellet as described (38) with the following modifications. To fully remove the cytoplasmic constituents, the pellet was suspended again with 1 ml buffer A and collected by centrifugation (the nuclear pellet). To prepare Nu, the nuclear pellet was sonicated in 500 μl of lysis buffer (50 mm Tris-HCl pH 8.0, 150 mm NaCl, 5 mm MgCl2, and 0.5% IGEPAL CA630) containing 1 mm PMSF and 20 U SUPERase·In (Ambion) three times for 20 s at 4 °C with a Bioruptor (Cosmo Bio) at the highest setting, and centrifuged for 15 min at 15,000 × g. The supernatant was used as the Nu. To prepare NE, the nuclear pellet was re-suspended with 500 μl of the lysis buffer, and sonicated ten times for 20 s at 4 °C with a Bioruptor at the highest setting, and then centrifuged for 30 min at 15,000 × g. The supernatant was used as the NE. The resulting pellet was resuspended with 500 μl NoE buffer (50 mm Tris-HCl pH 8.0, 150 mm NaCl, 10 mm EDTA, 10 mm dithiothreitol, 1 mm PMSF and 20 U Superasin), and sonicated ten times for 20 s at 4 °C, and then centrifuged for 30 min at 15,000 × g. The supernatant was used as the NoE. These extracts were examined for cross-contamination by immunoblot analysis using antibodies against GAPDH as a marker of the cytoplasm, lamin B as a nuclear marker, Nop58 as a nucleolar marker, and p80-coilin as a marker for Cajal bodies.
Immunoprecipitation and Immunoblot Analysis
After transfection for 48 h, FLAG-tagged protein- or FLAG-tagged mutant-transfected 293EBNA cells were harvested from two 90-mm dishes, washed with PBS and lysed by vigorous vortexing for 30 s in 1 ml lysis buffer, and incubated on ice for 30 min (40). The soluble fraction was obtained by centrifugation at 20,000 × g for 30 min at 4 °C. At least 5 mg of total cell lysate or the Nu described above was incubated with 10 μl anti-FLAG M2 agarose beads for 2 h at 4 °C. After washing the agarose beads with 1 ml lysis buffer five times and once with 50 mm Tris-HCl pH 8.0, containing 150 mm NaCl and 5 mm MgCl2, the complexes bound to the agarose beads were eluted with 500 μg/ml FLAG peptide. The eluted complexes were separated by SDS-PAGE and electrophoretically transferred to PVDF membranes. The membranes were blocked with 3% nonfat dried skim milk in phosphate-buffered saline (PBS) at 4 °C at least for 1 h, and incubated with the appropriate primary antibody overnight at 4 °C. The membranes were washed three times with PBST (PBS containing 0.1% (w/v) Tween 20) for 10 min, incubated with secondary antibody conjugated with alkaline phosphatase for 1 h at room temperature, washed three times with PBST for 10 min, and washed once with PBS for 5 min. Staining was performed in NBT (nitro-blue tetrazolium chloride)/BCIP (5-bromo-4-chloro-3′-Indolylphosphatase p-toluidine salt) solution, prepared by a (1:50) dilution of NBT/BCIP stock solution with alkaline phosphatase buffer (100 mm Tris-HCl pH 9.5, 100 mm NaCl, 50 mm MgCl2).
Ribonuclease Treatment of the Immunoprecipitated Protein Complexes
The complexes bound to the agarose beads described above were incubated with 50 mm Tris-HCl pH 8.0, 150 mm NaCl, 5 mm MgCl2 containing 10 μg/ml RNase A for 10 min at 37 °C, incubated for 30 min on ice, washed twice with lysis buffer and once with 50 mm Tris-HCl pH 8.0, 150 mm NaCl, and 5 mm MgCl2, and eluted with buffer containing the FLAG peptide as described above.
Sucrose Density Gradient Ultracentrifugation
Sucrose density gradient ultracentrifugation was performed as described (41). Briefly, after removing the cytoplasm by the method described above, the pellet was sonicated in sonication buffer (25 mm Tris-HCl pH 8.0, 100 mm KCl, 1 mm NaF, 2 mm EDTA, 1 mm dithiothreitol, 0.05% IGEPAL CA-630, 1 mm PMSF, and 20 U SUPERase·In) three times for 20 s at 4 °C with a Bioruptor at the highest setting, and centrifuged for 15 min at 15,000 × g. The prepared Nu (1 mg) was overlaid on 9.5 ml of a 10–30% (w/w) sucrose gradient in 25 mm Tris-HCl pH 8.0, containing 100 mm KCl, 2 mm EDTA, and 1 mm dithiothreitol, and centrifuged at 36,000 rpm (average 162,000 × g) for 3 h at 4 °C in a Hitachi P40ST rotor, and separated into 20 fractions (500 μl each). The absorbance at 254 nm in each fraction was measured by Nano-Drop spectrophotometer (Thermo SCIENTIFIC, Wilmington, USA), and graphed by Excel. Proteins from each fraction were precipitated by 10% trichloroacetic acid before SDS-PAGE and immunoblot analysis. RNAs were prepared by addition to an equal volume of denaturing buffer (7 m urea, 350 mm NaCl, 10 mm EDTA, 10 mm Tris-HCl pH 8.0, 1% SDS, 2% 2-mercaptoethanol), to which was added 1/10 volume of 2 m sodium acetate (pH 4.0). The samples were vortexed, extracted by acidic phenol-chloroform, and precipitated with isopropanol. Ribosomal RNAs (18S, 28S, and 32S) were visualized with 0.02% methylene blue after transffered to Hybond N+ membrane (GE Healthcare) in 50 mm sodium acetate buffer (pH 5.5) for 10 min.
Immunofluorescence Staining
293EBNA cells were grown on collagen-coated culture slides and transfected with expression plasmids by the calcium phosphate method. To visualize whole cells, the transfected and intact cells were washed with PBS followed by fixation with 3.7% formaldehyde in PBS for 10 min at room temperature. After washing twice with PBST (0.05% (w/v) Tween 20), the cells were permeabilized with PBS containing 0.5% (w/v) Triton X-100 for 5 min at room temperature and washed once with PBST. To visualize nuclei, the transfected and intact cells were washed with PBS followed by permeabilization with PBS containing 0.5% (w/v) Triton X-100 for 10 min at room temperature. After washing twice with PBST, the cells were fixed with 3.7% formaldehyde in PBS for 10 min at room temperature and washed twice with PBST. The cells were then blocked with 3% (w/v) nonfat dried milk in PBS for 1 h, and incubated with the appropriate primary antibody for 1 h at room temperature. The cells were washed three times in PBST for 10 min and then incubated with fluorochrome-conjugated secondary antibody for 1 h at room temperature. The cells were washed three times in PBST for 10 min and then mounted with Vectashield. The resulting cells were examined with an Axiovert 200 m microscope (Carl Zeiss, Germany).
Protein Identification by Liquid Chromatography-Tandem MS (LC-MS/MS) Analysis
Pulled-down proteins were separated on a gel by SDS-PAGE; the gel was cut into five pieces, each of which was subjected to in-gel digestion with trypsin (36), and the resulting peptides were analyzed using a nanoscale LC-MS/MS system with quadrupole-time-of-flight (Q-Tof)2 hybrid mass spectrometer (Q-Tof 2, Micromass, Wythenshawe, UK) as described (39, 42). The peptide mixture was applied to a Mightysil-RP-18 (3 μm particle, Kanto Chemical, Osaka, Japan) fritless column (45 mm × 0.150 mm i.d.) and separated using a 0–40% gradient of acetonitrile containing 0.1% formic acid over 80 min at a flow rate of 50 nl/min. Eluted peptides were sprayed directly into Q-Tof2. The peptides were detected in the MS mode to select a set of precursor ions for a data-dependent, collision-induced dissociation (CID) mass spectrometric (MS/MS) analysis, and every 4 s the four ions having the greatest signal intensity were subjected to the MS/MS analysis. The MS/MS signals were acquired by MassLynx version 3.5 (Micromass, UK) and converted to text files by ProteinLynx software (Micromass). The database search was performed by MASCOT version 2.2.1 (Matrix Science Ltd., London, UK) against the Uniprot human proteins databases Sprot 57.5, 471472 sequences, with the following parameters: fixed modification; carbamidemethyl (Cys), variable modifications: oxidation (Met), N-acetylation, pyroglutamine; maximum missed cleavages: 1; peptide mass tolerance: 200 ppm; MS/MS tolerance: 0.3 Da. The criteria were based on the vendor's definitions (Matrix Science, Ltd.). Furthermore, we set more strict criteria for protein assignment: (i) any peptide candidate with an MS/MS signal number of <2 was eliminated from the ‘hit’ candidates, regardless of the match score (total score minus threshold); and (ii) more than two peptides containing at least one peptide with ion score over 33 (p < 0.05) plus peptide(s) with ion score over 10 were considered as ‘hit’.
We also performed LC-MS/MS analysis with LTQ-Orbitrap hybrid mass spectrometer (model XL, Thermo Fisher Scientific, CA, USA). The mass spectrometer was operated in a data-dependent mode to automatically switch between Orbitrap-MS and linear ion trap-MS/MS acquisition. Survey full scan MS spectra (from m/z 450 to 1500) acquired in the Orbitrap with resolution r = 15,000 (after accumulation to a target value of 500,000 ions in the linear ion trap). The most intense ions (up to ten, depending on signal intensity) were sequentially isolated for fragmentation for every 0.2 s in the linear ion trap using CID at a target value of 30,000 ions. An MS scan and an MS/MS scan was one with centroid. The resulting fragment ions were recorded in the linear ion trap with a normal mode. Target ions selected for MS/MS were dynamically excluded for 30 s while the difference of 10 ppm. General mass spectrometric conditions were as follows: electrospray voltage, 1.6 kV; no lock mass option; with sheath and auxiliary gas flow; normalized collision energy, 35% for MS/MS. Ion selection threshold was 10 000 counts for MS/MS. An activation q-value of 0.25 and activation time of 30 ms was applied for MS/MS acquisitions.
The raw data acquired by Xcalibur version 2.0.7 (Thermo Fisher Scientific, USA) was converted to mgf file by Proteome Discoverer version 1.1 (Thermo Fisher Scientific). The database search was performed by MASCOT version 2.2.07 against the Uniprot human database version SwissProt_2010_05, 516603 sequences, with the following parameters: fixed modification; carbamidemethyl (Cys), variable modifications: oxidation (Met), N-acetylation, pyroglutamine; maximum missed cleavages: 1; peptide mass tolerance 10 ppm; MS/MS tolerance: 0.8 Da. We selected the candidate peptides with probability-based Mowse scores (total score) that exceed its threshold indicating a significant homology (p < 0.05; score over 20), and referred to them as “hits.” The criteria were based on the vendor's definitions. Furthermore, we set more strict criteria for protein assaignment: (1) any peptide candidate with an MS/MS signal number of <2 was eliminated from the ‘hits’ candidates, regardless of the match score (total score minus threshold); (2) proteins with match scores under 100 were also eliminated from the hits candidates. Peptides that are shared within a group of proteins were excluded from peptide assignment lists in supplemental Tables S1 and S2.
As a control, FLAG tag bound to anti-FLAG M2 agarose beads was also pulled down with the Nu. The proteins released from the M2 beads by eluting with FLAG peptide were separated by SDS-PAGE and subjected to in-gel trypsin digestion-LC-MS/MS analysis as described above, and were subtracted from the proteins identified in the total FLAG-tagged protein-associated complexes; thus, those proteins identified in the FLAG-tag eluate were not included in the FLAG-tagged protein-associated proteins unless the quantitative increase was confirmed.
Proliferation Assay
The siRNA615-transfected HeLa cells were counted 24 h after second transfection. Cells (5.0 × 103) were placed into new 24-well plates and incubated for various times. At proper time point, the cells were incubated with 0.1 μCi/ml [methyl-3H]Thymidine (PerkinElmer, NET 027E) for 1 h. After a brief rinse with ice-cold PBS, 10% trichloroacetic acid were added and incubated on ice for 30 min for DNA precipitation. The precipitated DNA were washed with 10% trichloroacetic acid, and incubated with 0.1 N NaOH, 1% SDS solution at 37 °C for 30 min for solubilization. The [methyl-3H]Thymidine incorporation into DNA was measured by scintillation counting with Insta-Gel Plus (PerkinElmer, Waltham, MA).
Metabolic Labeling and Analysis of RNA Transcripts
For [3H]uridine labeling, subconfluent siRNA-transfected HeLa cells in 35-mm dishes were incubated with 3 μCi/ml [5,6-3H]uridine (PerkinElmer, NET367) for 2 h. After a brief rinse with ice-cold PBS, total RNA was isolated using the RNAgent total RNA isolation system (Promega, Madison, WI) according to the manufacturer's instructions, and the incorporation of [3H]uridine was measured with Insta-Gel Plus by scintillation counting. RNA concentration was determined by Nano-Drop. An aliquot of RNA with an equal count (20,000 cpm) of the [3H]uridine incorporation from each sample was loaded on each lane of a 0.7% agarose/formaldehyde gel and electrophoresed in 3-(N-morpholino)propanesulfonic acid (MOPS) running buffer at 3.5 V/cm for 5 h for analysis of large rRNAs, and a 10% polyacrylamide/7.5 M urea gel in TBE running buffer at 15 V/cm for 3.5 h for small RNAs. Separated RNAs were transferred to a Hybond N+ membrane, which was subsequently dried and sprayed by EN3HANCE (PerkinElmer) and exposed to a Kodak BioMax MS film (Kodak) for 3 days in a deep freezer. For metabolic labeling of RNA with [3H]uridine, sub-confluent siRNA-transfected HeLa cells in a 35-mm dish were incubated for 30 min in medium containing [3H]uridine (5 μCi/ml). The cells were then chased in medium containing 0.5 mm nonradioactive uridine, after which RNA was isolated using RNAgent, and the RNAs were analyzed as described above. For [3H]uridine labeling of RNAs in the p32-associated complex, Flp-In T-REx 293 cells or those stably expressed p32-FLAG were incubated with 3 μCi/ml [5,6-3H]uridine for 2 h in 90-mm dishes, harvested from six dishes, and lysed as described above in 1 ml MgCl2-free lysis buffer containing 1 mm PMSF and 20 U SUPERase·In. The soluble fraction was obtained by centrifugation at 12,000 × g for 10 min at 4 °C, and was incubated with 10 μl anti-FLAG M2 agarose beads for 2 h at 4 °C. After washing the agarose beads with MgCl2-free lysis buffer, the RNAs in the complexes bound to the agarose beads or soluble cell lysates were extracted using the RNAgent, and were analyzed as described above.
Northern Blot Hybridization
RNAs were extracted from the p32-associated complex using the RNAgent, and were electrophoresed with agarose gel and transferred to a Hybond N+ membrane as described above. After stained with methylene blue, the membrane was hybridized to biotin-labeled probes at 50 °C overnight in pre-hybridization solution (5× SSC, 20 mm Na2HPO4 pH 7.2, 7% SDS, 2 X Denhardt's Solution, 1 mg/ml salmon sperm DNA). The membrane was washed with NonStringent wash solution (3× SSC, 25 mm NaH2PO4 (pH 7.5), 5% SDS) once for 10 min at 50 °C, twice for 30 min at 50 °C, and once with Stringent wash solution (1× SSC, 1% SDS) for 15 min at 50 °C. The hybridized RNA was detected by using Chemiluminescent Nucleic Acid Detection Module Kit (Thermo). The following oligonucleotides were used as probes to detect human pre-rRNAs in Northern blot hybridization (see Fig. 4D): 5′ ETS probe (5′- ACAGCGACGGAGGCAATACC-3′) to nt 1468 to 1487 of the 5′ ETS; ITS1 probe (5′-AACGCGCTAGGTACCTGGACGG-3′) to nt 586 to 607 of the ITS1; 5.8S probe (5′- AGACAGGCGTAGCCCCGGGAGGAA -3′) to nt 123 to 146 of the 5.8S; ITS2 probe (5′- ACGCCGCCGGGTCTGCGCTTA-3′) to nt 93 to 113 of the ITS2. Probes were 3′-end labeled using Biotin 3′ End DNA Labeling Kit (Thermo).
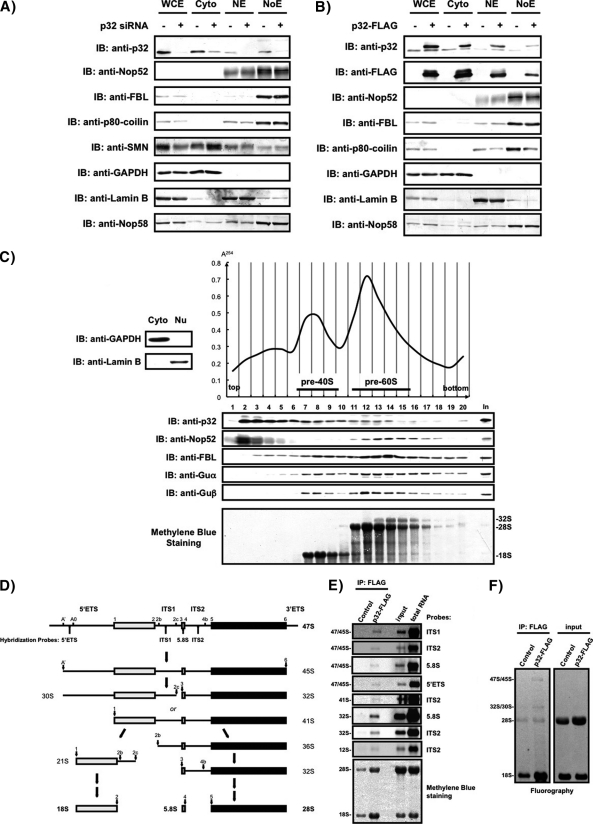
Fig. 4.
The presence of p32 in the nucleolus/Cajal body fraction prepared by cell fractionation and in the preribosome fractions separated by ultracentrifugation. Equivalent amounts of whole-cell extract (WCE), cytoplasmic fraction (Cyto), nucleoplasmic extract (NE), and nucleolar extract (NoE) prepared from 293EBNA cells transfected with (+) or without (–) a p32-specific siRNA (A), or transfected with (+) or without (–) p32-FLAG expression vector (B), were analyzed by immunoblotting with the antibodies indicated at the left. C, Cells were fractionated into nuclear (Nu) and cytoplasmic (Cyto) fractions and were analyzed by immunoblotting (IB) with the antibodies indicated (left, top). The Nu fraction that corresponded to the nuclear extract combining NE and NoE in (A) was separated on 10–30% sucrose density gradients and separated into 20 fractions. The absorbance at 254 nm was monitored and is shown on the top right. Proteins in each fraction were analyzed by immunoblotting with the antibodies indicated at the left (middle). RNAs were also extracted from each fraction, separated by agarose gel electrophoresis, transferred to a membrane, and stained with methylene blue (bottom). Input: 20 μg of protein from unfractionated Nu was loaded for immunoblotting; 2 μg of RNA from unfractionated Nu was loaded for agarose gel electrophoresis and methylene blue staining. D, Structure of the primary rRNA transcript (47S), outline of pre-rRNA processing in mammalian cells adapted from refs. 41, 52, 75, and oligonucleotide probes used for hybridization analysis are indicated. The main processing sites termed 1 to 6 correspond to the ends of mature 18S, 5.8S, and 28S rRNAs are also indicated. The external and internal transcribed spacers (ETS and ITS, respectively) contain additional cleavage sites designated A', A0, 2b, 2c, and 4b. E, RNAs extracted from p32-associated complex were detected by Northern blot analysis with the oligonucleotide probes indicated. Positions of pre-rRNAs and the probes used are indicated on the left and right, respectively. Input: 2 μg of RNA extracted from soluble cell lysates from control cells were loaded. Total RNA: 2 μg of total RNA were loaded. F, RNAs containing newly synthesized pre-rRNAs labeled with [3H]uridine were isolated from p32-associated complex, separated on an agarose gel, transferred to a membrane, and detected by fluorography. Positions of pre-rRNAs are indicated on the left. Input: RNAs (5000 cpm/lane) extracted from soluble cell lysates were loaded.
Purification of GST-Fusion Proteins and In Vitro Binding Assay
GST-fusion proteins were prepared based on the method described by Fujiyama-Nakamura and Yoshikawa et al. (39). Briefly, Escherichia coli BL21 (DE3) strain carrying a pGEX 4T plasmid encoding GST, GST-fused p32 (GST-p32), GST-thrombin cleavage site-FBL, or GST-thrombin cleavage site-FLAG-Nop52 was grown in 250 ml of Luria-Bertani medium, was treated with isopropyl β-D-thiogalactoside (final concentration 0.1 mm) for 4 h at 25 °C, and collected by centrifugation. The cells were suspended in 10 ml of PBS containing 2 mm EDTA, 0.1% 2-mercaptoethanol, and 0.5 mm PMSF, divided into 3 tubes, and sonicated for 20 s × 18 times at 4 °C with a Bioruptor at the high setting with a microtip. Cell lysate in each tube was centrifuged for 20 min at 20,000 × g. The supernatant (10 ml) was mixed with 200 μl of a 50% (v/v) slurry of glutathione-Sepharose-4B beads (GE Healthcare). The GST-protein-bound beads were washed with PBS containing 2 mm EDTA, 0.1% 2-mercaptoethanol, 0.5 mm PMSF, and 2% Tween 20 until absorbance at 280 nm in flow through became zero. FLAG-Nop52 or FBL was eluted from the beads by centrifugation after the cleavage with thrombin (final concentration 1%) for 2 h at 25 °C. GST or GST-p32 were mixed with the isolated FLAG-Nop52, or FBL, and incubated with glutathione-Sepharose-4B beads for 1 h at 4 °C. The beads were centrifuged, washed five times with 1 ml lysis buffer and once more with 50 mm Tris-HCl pH 8.0, containing 150 mm NaCl and 5 mm MgCl2. Proteins bound to glutathione-Sepharose-4B beads were separated by SDS-PAGE and detected by coomassie brilliant blue (CBB, quick-CBB WAKO) staining or by immunoblot analysis with anti-FLAG antibody, or anti-FBL antibody.
RESULTS
p32 Associates with Many Pre-rRNA Processing Factors Including Nop52
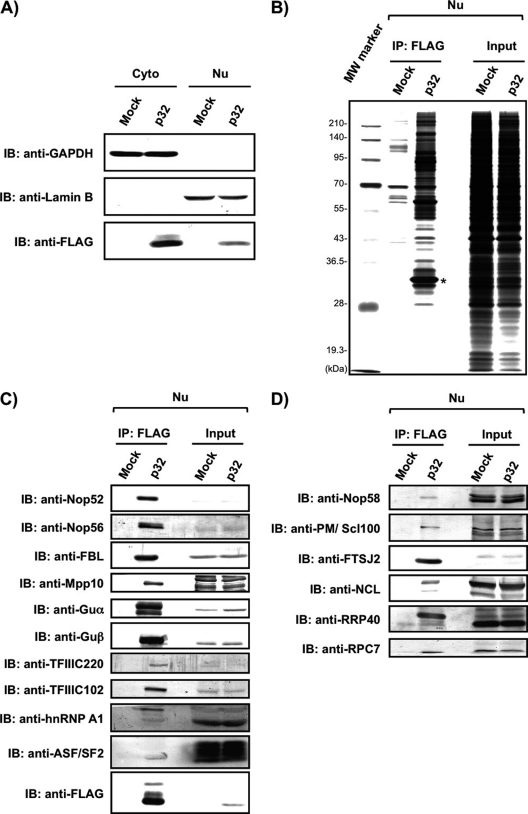
We first confirmed that p32-FLAG and endogenous p32 had the same subcellular localization (supplemental Fig. S1). We pulled down p32-FLAG-associated proteins from the Nu of 293EBNA cells (Figs. 1A and 1B), carried out nano-LC-MS/MS analyses of the p32-FLAG-associated proteins, and assigned the obtained MS/MS spectra to proteins using the Mascot search software (39) (supplemental Tables S1 and S2). We identified 149 proteins by these MS/MS analyses (Tables I and II and supplemental Tables S1 and S2). To confirm the mass-based identification, we also performed immunoblot analyses of the p32-FLAG-associated proteins using antibodies to the identified proteins against Nop52, Nop56, FBL, Mpp10, RNA helicases Guα and Guβ, TFIIIC220, TFIIIC102, and hnRNP A1, and detected all the proteins examined by the mass-based analysis (Fig. 1C). Mass-based analysis identified the six known nuclear p32-binding proteins (lamin B receptor, FBL, hnRNPA1, BAT2, RRP1B, and CDK13) (2, 3, 16, 26, 43, 44), and identified 143 proteins as new candidates that associate with p32. Although the mass-based analysis did not identify ASF/SF2, which is known as the nuclear p32-binding protein, we detected it among the p32-FLAG-associated proteins by immunoblotting using anti-ASF/SF2 (Fig. 1C). Among the proteins identified by mass-based analysis, 23 proteins, FBL, Nop56, IMP4, MPHOSPH10 (Mpp10), Noc4L, Nop14, UTP3, RRP7A, Guα (DDX21), DDX51, PELP1, TEX10, WDR18, LAS1L, MDN1, NOL9, SENP3, Nop52, DDX54, DDX56, NO66, and RRP12-like protein, and Guβ (DDX50), are involved or expected to be involved in human ribosome biogenesis (Table I) (45–57). In addition, three nucleolar proteins (MYBBP1A, NOM1, and RRP1B), and five subunits of the general transcription factors (3C polypeptides 1–5) required for RNA polymerase III-dependent transcription, were also identified (Tables I and II).
Fig. 1.
Association of p32 with several proteins involved in ribosome biogenesis. A, Equivalent amounts of cytoplasmic (Cyto) or nuclear (Nu) extract prepared from mock- or p32-FLAG (p32)-transfected 293EBNA cells were analyzed by immunoblotting (IB) with anti-GAPDH as a marker of the cytoplasm, anti-Lamin B as a marker of the nucleus, and anti-FLAG for the detection of the expressed p32-FLAG. B, Nuclear extracts prepared from 293EBNA cells transfected with a plasmid containing a FLAG- (mock) or p32-FLAG-encoded DNA fragment (p32) were subjected to immunoprecipitation with anti-FLAG-fixed beads. The isolated proteins as well as an aliquot of the nuclear extract of mock- or p32-FLAG-transfected cells (input, 5 μg) were analyzed by 10% SDS-PAGE and visualized by silver staining. The asterisk indicates p32-FLAG used as bait. The molecular masses (kDa) are indicated at the left side of the figure. C, Association of p32-FLAG with the proteins identified by LC-MS/MS analysis and presence of ASF/SF2 was confirmed by immunoblot analysis with the indicated antibodies. D, The presence of known pre-rRNA processing factors that were not identified by LC-MS/MS analysis was examined by immunoblot analysis of the isolated p32-FLAG-associated proteins with the indicated antibodies.
Table I. The Nuclear p32-associated proteins that are involved or expected to be involved in ribosome biogenesis. The nuclear p32-associated proteins involved or expected to be involved in ribosome biogenesis are indicated. Proteins are classified into functional groups based on SwissProt classification. Proteins were identified by LC-MS/MS (LTQ-Orbitrap XL or Q-Tof2) and/or immunoblot analysis (IB), as described in Supplementary Tables I and II, and Figs. 1C and 1D. Proteins detected in mock are not listed in this table. Uniprot KB accession numbers, entry names, protein names as well as gene symbols are shown. For proteins having yeast orthologs, the gene names are indicated (obtained by Blink analysis of the NCBI database). Cellular localization is also indicated: No, nucleoli; Nm, nuclear membrane; Np, nucleoplasm; Nu, nucleus; Cy, cytoplasm; CBs, Cajal bodies.
| Uniprot KB accession | Uniprot KB entry name | Protein name | Gene symbol | ID method |
Cellular localization | Yeast homologs | aFunction | |||
|---|---|---|---|---|---|---|---|---|---|---|
| LTQ-Orbtrap XL | Q-Tof2 | IB | ||||||||
| pre-rRNAs modification and processing | ||||||||||
| 1 | P22087 | FBRL_HUMAN | rRNA 2∼-O-methyltransferase fibrillarin (FBL)b | FBL | ○ | ○ | ○ | No/CBs | NOP1 | Pre-90S, pre-60S, boxC/D snoRNP, SSU processome, methyltransferase |
| 2 | O00567 | NOP56_HUMAN | Nucleolar protein 56 (Nop56) | NOP56 | ○ | ○ | No | NOP56 | Pre-90S, boxC/D snoRNP, SSU processome | |
| 3 | Q9Y2X3 | NOP58_HUMAN | Nucleolar protein 58 (Nop58) | NOP58 | ○ | No | NOP5 | Pre-90S, boxC/D snoRNP, SSU processome | ||
| 4 | Q96G21 | IMP4_HUMAN | U3 small nucleolar ribonucleoprotein protein IMP4 | IMP4 | ○ | No | IMP4 | Pre-90S, U3 snoRNP, SSU processome | ||
| 5 | O00566 | MPP10_HUMAN | U3 small nucleolar ribonucleoprotein protein MPP10 | MPHOSPH10 | ○ | ○ | No | MPP10 | Pre-90S, U3 snoRNP, SSU processome | |
| 6 | Q9BVI4 | NOC4L_HUMAN | Nucleolar complex protein 4 homolog | NOC4L | ○ | No | NOC4 | Pre-90S, SSU processome, Noc4-Nop14 complex function unknown in mammals | ||
| 7 | P78316 | NOP14_HUMAN | Nucleolar protein 14 | NOP14 | ○ | No | NOP14 | Pre-90S, SSU processome, Noc4-Nop14 complex function unknown in mammals | ||
| 8 | Q9NQZ2 | SAS10_HUMAN | Something about silencing protein 10 | UTP3 | ○ | Nu | UTP3 | Pre-90S, SSU processome, function unknown in mammals | ||
| 9 | Q9Y3A4 | RRP7A_HUMAN | Ribosomal RNA-processing protein 7 homolog A | RRP7A | ○ | - | RRP7 | Pre-90S, function unknown in mammals | ||
| 10 | P19338 | NUCL_HUMAN | Nucleolin (NCL) | NCL | ○ | No | NSR1 | Pre-90S, pre-60S | ||
| 11 | Q9NR30 | DDX21_HUMAN | Nucleolar RNA helicase 2 (Guα) | DDX21 | ○ | ○ | ○ | No | DBP1 | Pre-60S, DEAD box RNA helicase |
| 12 | Q8N8A6 | DDX51_HUMAN | ATP-dependent RNA helicase DDX51 | DDX51 | ○ | No | DBP6 | Pre-60S, DEAD box RNA helicase | ||
| 13 | Q8IZL8 | PELP1_HUMAN | Proline-, glutamic acid- and leucine-rich protein 1 | PELP1 | ○ | No/Nu/Cy | RIX1 | Pre-60S, RIX1 complex, SENP3-sensitive target of SUMO | ||
| 14 | Q9NXF1 | TEX10_HUMAN | Testis-expressed sequence 10 protein | TEX10 | ○ | No/Nu/Nm | IPI1 | Pre-60S, RIX1 complex | ||
| 15 | Q9BV38 | WDR18_HUMAN | WD repeat-containing protein 18 | WDR18 | ○ | - | IPI3 | pre-60S, RIX1 complex | ||
| 16 | Q9Y4W2 | LAS1L_HUMAN | Protein LAS1 homolog | LAS1L | ○ | ○ | No | LAS1 | Pre-60S, associated with human RIX1 complex, SENP3-sensitive target of SUMO | |
| 17 | Q9NU22 | MDN1_HUMAN | Midasin | MDN1 | ○ | Nu | REA1 | Pre-60S, associated with human RIX1 complex | ||
| 18 | Q9H4L4 | SENP3_HUMAN | Sentrin-specific protease 3 | SENP3 | ○ | No/Np | ULP1 | Pre-60S, associated with human RIX1 complex | ||
| 19 | Q5SY16 | NOL9_HUMAN | Nucleolar protein 9 | NOL9 | ○ | No | GRC3 | Pre-60S | ||
| 20 | P56182 | RRP1_HUMAN | Ribosomal RNA processing protein 1 homolog A (Nop52) | RRP1 | ○ | ○ | ○ | No | RRP1 | Pre-60S, function unknown in mammals |
| 21 | Q8TDD1 | DDX54_HUMAN | ATP-dependent RNA helicase DDX54 | DDX54 | ○ | No | DBP10 | Pre-60S, DEAD box RNA helicase, function unknown in mammals | ||
| 22 | Q9NY93 | DDX56_HUMAN | Probable ATP-dependent RNA helicase DDX56 | DDX56 | ○ | No | DBP9 | Present in 65S preribosomal particles, DEAD box RNA helicase | ||
| 23 | Q9H6W3 | NO66_HUMAN | Lysine-specific demethylase NO66 | C14orf169 | ○ | ○ | No/Np | - | Present in large preribosomal particles | |
| 24 | Q5JTH9 | RRP12_HUMAN | RRP12-like protein | RRP12 | ○ | ○ | No/Nm | RRP12 | Nuclear export of pre-40S and pre-60S, function unknown in mammals | |
| 25 | Q9BQ39 | DDX50_HUMAN | ATP-dependent RNA helicase DDX50 (Guβ) | DDX50 | ○ | ○ | ○ | No | DBP1 | DEAD box RNA helicase |
| 26 | Q01780 | EXOSX_HUMAN | Exosome component 10 (PM/Scl-100) | EXOSC10 | ○ | No/Cy | RRP6 | Exosome | ||
| 27 | Q9NQT5 | EXOS3_HUMAN | Exosome complex exonuclease RRP40 | EXOSC3 | ○ | No/Cy | RRP40 | Exosome | ||
| 28 | Q9UI43 | RRMJ2_HUMAN | Putative ribosomal RNA methyltransferase 2 (FTSJ2) | FTSJ2 | ○ | No | MRM2 | Methyltransferase | ||
| Nucleolar Protein | ||||||||||
| 29 | Q9BQG0 | MBB1A_HUMAN | Myb-binding protein 1A | MYBBP1A | ○ | No/Cy | POL5 | |||
| 30 | Q5C9Z4 | NOM1_HUMAN | Nucleolar MIF4G domain-containing protein 1 | NOM1 | ○ | ○ | No | SGD1 | ||
| 31 | Q14684 | RRP1B_HUMAN | Ribosomal RNA processing protein 1 homolog Bb | RRP1B | ○ | No | RRP1 | |||
a Known roles in ribosome biogenesis and/or associated pre-ribosome particles for yeast homologs or for human proteins are indicated.
b The proteins known to be associated with p32.
Table II. The Nuclear p32-associated proteins that are not involved in ribosome biogenesis. The nuclear p32-associated proteins are indicated. Proteins are classified into functional groups based on SwissProt classification. Proteins were identified by LC-MS/MS (LTQ-Orbitrap XL or Q-Tof2) and/or immunoblot analysis (IB), as described in Supplementary Tables I and II, and Figs. 1C and 1D. Proteins detected in mock are not listed in this table. Uniprot KB accession numbers, entry names, protein names as well as gene symbols are shown.
| Uniprot KB accession | Uniprot KB Entry name | Protein name | Gene symbol | ID method |
|||
|---|---|---|---|---|---|---|---|
| LTQ-Orbitrap XL | Q-Tof2 | IB | |||||
| Chromatin remodeling | |||||||
| 1 | Q969G3 | SMCE1_HUMAN | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 | SMARCE1 | ○ | ||
| 2 | Q12824 | SNF5_HUMAN | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | SMARCB1 | ○ | ||
| 3 | O60264 | SMCA5_HUMAN | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 | SMARCA5 | ○ | ||
| 4 | Q8TAQ2 | SMRC2_HUMAN | SWI/SNF complex subunit SMARCC2 | SMARCC2 | ○ | ○ | |
| 5 | Q92922 | SMRC1_HUMAN | SWI/SNF complex subunit SMARCC1 | SMARCC1 | ○ | ||
| 6 | Q86U86 | PB1_HUMAN | Protein polybromo-1 | PBRM1 | ○ | ||
| 7 | Q8WUB8 | PHF10_HUMAN | PHD finger protein 10 | PHF10 | ○ | ||
| 8 | P51610 | HCFC1_HUMAN | Host cell factor 1 | HCFC1 | ○ | ||
| 9 | Q6ZRS2 | SRCAP_HUMAN | Helicase SRCAP | SRCAP | ○ | ||
| 10 | Q9P2D1 | CHD7_HUMAN | Chromodomain-helicase-DNA-binding protein 7 | CHD7 | ○ | ||
| 11 | Q14839 | CHD4_HUMAN | Chromodomain-helicase-DNA-binding protein 4 | CHD4 | ○ | ||
| 12 | Q9NPI1 | BRD7_HUMAN | Bromodomain-containing protein 7 | BRD7 | ○ | ||
| 13 | Q68CP9 | ARID2_HUMAN | AT-rich interactive domain-containing protein 2 | ARID2 | ○ | ||
| RNA polymerase III transcription | |||||||
| 14 | Q12789 | TF3C1_HUMAN | General transcription factor 3C polypeptide 1 (TFIIIC220) | GTF3C1 | ○ | ○ | ○ |
| 15 | Q8WUA4 | TF3C2_HUMAN | General transcription factor 3C polypeptide 2 | GTF3C2 | ○ | ○ | |
| 16 | Q9Y5Q9 | TF3C3_HUMAN | General transcription factor 3C polypeptide 3 (TFIIIC102) | GTF3C3 | ○ | ○ | ○ |
| 17 | Q9UKN8 | TF3C4_HUMAN | General transcription factor 3C polypeptide 4 | GTF3C4 | ○ | ○ | |
| 18 | Q9Y5Q8 | TF3C5_HUMAN | General transcription factor 3C polypeptide 5 | GTF3C5 | ○ | ○ | |
| 19 | O15318 | RPC7_HUMAN | DNA-directed RNA polymerase III subunit RPC7 | POLR3G | ○ | ||
| Transcription | |||||||
| 20 | Q9H869 | YYAP1_HUMAN | YY1-associated protein 1 | YY1AP1 | ○ | ||
| 21 | Q8WXI9 | P66B_HUMAN | Transcriptional repressor p66-beta | GATAD2B | ○ | ||
| 22 | Q13263 | TIF1B_HUMAN | Transcription intermediary factor 1-beta | TRIM28 | ○ | ||
| 23 | P05412 | JUN_HUMAN | Transcription factor AP-1 | JUN | ○ | ||
| 24 | P51532 | SMCA4_HUMAN | Transcription activator BRG1 | SMARCA4 | ○ | ||
| 25 | Q14498 | RBM39_HUMAN | RNA-binding protein 39 | RBM39 | ○ | ||
| 26 | Q8N7H5 | PAF1_HUMAN | RNA polymerase II-associated factor 1 homolog | PAF1 | ○ | ||
| 27 | P30876 | RPB2_HUMAN | DNA-directed RNA polymerase II subunit RPB2 | POLR2B | ○ | ||
| 28 | P15336 | ATF2_HUMAN | Cyclic AMP-dependent transcription factor ATF-2 | ATF2 | ○ | ||
| 29 | Q7RTS1 | BHA15_HUMAN | Class A basic helix-loop-helix protein 15 | BHLHA15 | ○ | ||
| DNA binding/replication/repair | |||||||
| 30 | Q14683 | SMC1A_HUMAN | Structural maintenance of chromosomes protein 1A | SMC1A | ○ | ||
| 31 | P12004 | PCNA_HUMAN | Proliferating cell nuclear antigen | PCNA | ○ | ||
| 32 | Q9Y2S7 | PDIP2_HUMAN | Polymerase delta-interacting protein 2 | POLDIP2 | ○ | ||
| 33 | Q9UBB5 | MBD2_HUMAN | Methyl-CpG-binding domain protein 2 | MBD2 | ○ | ||
| 34 | O00255 | MEN1_HUMAN | Menin | MEN1 | ○ | ||
| 35 | P78527 | PRKDC_HUMAN | DNA-dependent protein kinase catalytic subunit | PRKDC | ○ | ||
| 36 | P26358 | DNMT1_HUMAN | DNA (cytosine-5)-methyltransferase 1 | DNMT1 | ○ | ||
| 37 | Q7Z7K6 | CENPV_HUMAN | Centromere protein V | CENPV | ○ | ||
| 38 | Q53HL2 | BOREA_HUMAN | Borealin | CDCA8 | ○ | ||
| mRNA splicing/stabilization | |||||||
| 39 | Q07955 | SRSF1_HUMAN | Serine/arginine-rich splicing factor 1 (ASF/SF2)a | SRSF1 | ○ | ||
| 40 | O95104 | SFR15_HUMAN | Splicing factor, arginine/serine-rich 15 | SFRS15 | ○ | ||
| 41 | Q99590 | SFRIP_HUMAN | SFRS2-interacting protein | SRSF2IP | ○ | ||
| 42 | Q86U06 | RBM23_HUMAN | Probable RNA-binding protein 23 | RBM23 | ○ | ||
| 43 | P09651 | ROA1_HUMAN | Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1)a | HNRNPA1 | ○ | ○ | |
| 44 | P48634 | BAT2_HUMAN | Large proline-rich protein BAT2a | BAT2 | ○ | ○ | |
| 45 | Q14004 | CDK13_HUMAN | Cell division protein kinase 13a | CDK13 | ○ | ||
| RNA binding protein | |||||||
| 46 | Q9NQ29 | LUC7L_HUMAN | Putative RNA-binding protein Luc7-like 1 | LUC7L | ○ | ||
| 47 | Q9UHX1 | PUF60_HUMAN | Poly(U)-binding-splicing factor PUF60 | PUF60 | ○ | ||
| 48 | P51116 | FXR2_HUMAN | Fragile X mental retardation syndrome-related protein 2 | FXR2 | ○ | ||
| 49 | Q06787 | FMR1_HUMAN | Fragile X mental retardation 1 protein | FMR1 | ○ | ||
| Telomere | |||||||
| 50 | Q9NYB0 | TE2IP_HUMAN | Telomeric repeat-binding factor 2-interacting protein 1 | TERF2IP | ○ | ||
| 51 | Q15554 | TERF2_HUMAN | Telomeric repeat-binding factor 2 | TERF2 | ○ | ||
| Nuclear envelop/pore/lamina | |||||||
| 52 | Q5SNT2 | TM201_HUMAN | Transmembrane protein 201 | TMEM201 | ○ | ||
| 53 | O94901 | SUN1_HUMAN | SUN domain-containing protein 1 | SUN1 | ○ | ||
| 54 | Q8N1F7 | NUP93_HUMAN | Nuclear pore complex protein Nup93 | NUP93 | ○ | ||
| 55 | Q92621 | NU205_HUMAN | Nuclear pore complex protein Nup205 | NUP205 | ○ | ||
| 56 | Q12769 | NU160_HUMAN | Nuclear pore complex protein Nup160 | NUP160 | ○ | ||
| 57 | Q8WUM0 | NU133_HUMAN | Nuclear pore complex protein Nup133 | NUP133 | ○ | ||
| 58 | P57740 | NU107_HUMAN | Nuclear pore complex protein Nup107 | NUP107 | ○ | ||
| 59 | Q96HA1 | P121A_HUMAN | Nuclear envelope pore membrane protein POM 121 | POM121 | ○ | ||
| 60 | Q14739 | LBR_HUMAN | Lamin-B receptora | LBR | ○ | ||
| Protein glycosylation | |||||||
| 61 | P39656 | OST48_HUMAN | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase 48 kDa subunit | DDOST | ○ | ||
| 62 | P04843 | RPN1_HUMAN | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit 1 | RPN1 | ○ | ||
| Importin | |||||||
| 63 | P52292 | IMA2_HUMAN | Importin subunit alpha-2 | KPNA2 | ○ | ||
| 64 | P52294 | IMA1_HUMAN | Importin subunit alpha-1 | KPNA1 | ○ | ||
| Mitochondorial protein | |||||||
| OXPHOS complex | |||||||
| 65 | P21912 | DHSB_HUMAN | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | SDHB | ○ | ||
| 66 | P31040 | DHSA_HUMAN | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | SDHA | ○ | ||
| 67 | O75306 | NDUS2_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial | NDUFS2 | ○ | ||
| 68 | P36542 | ATPG_HUMAN | ATP synthase subunit gamma, mitochondrial | ATP5C1 | ○ | ||
| Mitochondrial membrane protein | |||||||
| 69 | Q9Y277 | VDAC3_HUMAN | Voltage-dependent anion-selective channel protein 3 | VDAC3 | ○ | ||
| 70 | P45880 | VDAC2_HUMAN | Voltage-dependent anion-selective channel protein 2 | VDAC2 | ○ | ||
| 71 | P21796 | VDAC1_HUMAN | Voltage-dependent anion-selective channel protein 1 | VDAC1 | ○ | ○ | |
| 72 | Q9H9B4 | SFXN1_HUMAN | Sideroflexin-1 | SFXN1 | ○ | ||
| 73 | O96008 | TOM40_HUMAN | Mitochondrial import receptor subunit TOM40 homolog | TOMM40 | ○ | ||
| 74 | O75746 | CMC1_HUMAN | Calcium-binding mitochondrial carrier protein Aralar1 | SLC25A12 | ○ | ||
| 75 | Q9Y4W6 | AFG32_HUMAN | AFG3-like protein 2 | AFG3L2 | ○ | ||
| Lipid metabolism | |||||||
| 76 | P40939 | ECHA_HUMAN | Trifunctional enzyme subunit alpha, mitochondrial | HADHA | ○ | ||
| 77 | P55084 | ECHB_HUMAN | Trifunctional enzyme subunit beta, mitochondrial | HADHB | ○ | ||
| Others | |||||||
| 78 | P42704 | LPPRC_HUMAN | Leucine-rich PPR motif-containing protein, mitochondrial | LRPPRC | ○ | ||
| 79 | Q96EY1 | DNJA3_HUMAN | DnaJ homolog subfamily A member 3, mitochondrial | DNAJA3 | ○ | ||
| 80 | O00411 | RPOM_HUMAN | DNA-directed RNA polymerase, mitochondrial | POLRMT | ○ | ||
| 81 | Q96TA2 | YMEL1_HUMAN | ATP-dependent metalloprotease YME1L1 | YME1L1 | ○ | ||
| 82 | O76031 | CLPX_HUMAN | ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial | CLPX | ○ | ||
| 83 | O95831 | AIFM1_HUMAN | Apoptosis-inducing factor 1, mitochondrial | AIFM1 | ○ | ||
| ER protein | |||||||
| 84 | P43307 | SSRA_HUMAN | Translocon-associated protein subunit alpha | SSR1 | ○ | ||
| 85 | Q9UGP8 | SEC63_HUMAN | Translocation protein SEC63 homolog | SEC63 | ○ | ||
| 86 | P55072 | TERA_HUMAN | Transitional endoplasmic reticulum ATPase | VCP | ○ | ||
| 87 | Q9NZ01 | TECR_HUMAN | Trans-2,3-enoyl-CoA reductase | TECR | ○ | ||
| 88 | P16615 | AT2A2_HUMAN | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | ATP2A2 | ○ | ||
| 89 | Q13724 | MOGS_HUMAN | Mannosyl-oligosaccharide glucosidase | MOGS | ○ | ||
| Others | |||||||
| 90 | Q15836 | VAMP3_HUMAN | Vesicle-associated membrane protein 3 | VAMP3 | ○ | ||
| 91 | Q93009 | UBP7_HUMAN | Ubiquitin carboxyl-terminal hydrolase 7 | USP7 | ○ | ||
| 92 | P07437 | TBB5_HUMAN | Tubulin beta chain | TUBB | ○ | ||
| 93 | Q14232 | EI2BA_HUMAN | Translation initiation factor eIF-2B subunit alpha | EIF2B1 | ○ | ||
| 94 | Q07157 | ZO1_HUMAN | Tight junction protein ZO-1 | TJP1 | ○ | ||
| 95 | P48594 | SPB4_HUMAN | Serpin B4 | SERPIN B4 | ○ | ||
| 96 | P29508 | SPB3_HUMAN | Serpin B3 | SERPIN B3 | ○ | ||
| 97 | P48651 | PTSS1_HUMAN | Phosphatidylserine synthase 1 | PTDSS1 | ○ | ||
| 98 | Q15758 | AAAT_HUMAN | Neutral amino acid transporter B(0) | SLC1A5 | ○ | ||
| 99 | Q8NFW8 | NEUA_HUMAN | N-acylneuraminate cytidylyltransferase | CMAS | ○ | ||
| 100 | Q8WVX9 | FACR1_HUMAN | Fatty acyl-CoA reductase 1 | FAR1 | ○ | ||
| 101 | P35613 | BASI_HUMAN | Basigin | BSG | ○ | ||
| 102 | P06733 | ENOA_HUMAN | Alpha-enolase | ENO1 | ○ | ||
| Unknown | |||||||
| 103 | Q96JP5 | ZFP91_HUMAN | Zinc finger protein 91 homolog | ZFP91 | ○ | ○ | |
| 104 | Q9H5H4 | ZN768_HUMAN | Zinc finger protein 768 | ZNF768 | ○ | ||
| 105 | Q14966 | ZN638_HUMAN | Zinc finger protein 638 | ZNF638 | ○ | ||
| 106 | Q8NAF0 | ZN579_HUMAN | Zinc finger protein 579 | ZNF579 | ○ | ||
| 107 | Q5VZL5 | ZMYM4_HUMAN | Zinc finger MYM-type protein 4 | ZMYM4 | ○ | ||
| 108 | Q8N9Z2 | YG034_HUMAN | Uncharacterized protein FLJ36031 | FLJ36031 | ○ | ||
| 109 | Q9Y4P3 | TBL2_HUMAN | Transducin beta-like protein 2 | TBL2 | ○ | ||
| 110 | Q9UJZ1 | STML2_HUMAN | Stomatin-like protein 2 | STOML2 | ○ | ||
| 111 | Q9P2N5 | RBM27_HUMAN | RNA-binding protein 27 | RBM27 | ○ | ||
| 112 | Q5T8P6 | RBM26_HUMAN | RNA-binding protein 26 | RBM26 | ○ | ||
| 113 | Q9NWS8 | RMND1_HUMAN | Required for meiotic nuclear division protein 1 homolog | RMND1 | ○ | ||
| 114 | Q6P158 | DHX57_HUMAN | Putative ATP-dependent RNA helicase DHX57 | DHX57 | ○ | ||
| 115 | Q69YN4 | VIR_HUMAN | Protein virilizer homolog | KIAA1429 | ○ | ||
| 116 | Q9BX40 | LS14B_HUMAN | Protein LSM14 homolog B | LSM14B | ○ | ||
| 117 | Q9Y520 | BA2L2_HUMAN | Protein BAT2-like 2 | BAT2L2 | ○ | ||
| 118 | Q9ULL5 | PRR12_HUMAN | Proline-rich protein 12 | PRR12 | ○ | ||
| 119 | Q96DN6 | MBD6_HUMAN | Methyl-CpG-binding domain protein 6 | MBD6 | ○ | ||
| 120 | Q9UNF1 | MAGD2_HUMAN | Melanoma-associated antigen D2 | MAGED2 | ○ | ||
| 121 | Q9NXS2 | QPCTL_HUMAN | Glutaminyl-peptide cyclotransferase-like protein | QPCTL | ○ | ||
| 122 | Q96SL8 | FIZ1_HUMAN | Flt3-interacting zinc finger protein 1 | FIZ1 | ○ | ||
| 123 | Q68CQ4 | DEF_HUMAN | Digestive organ expansion factor homolog | C1orf107 | ○ | ||
| 124 | Q9H0W5 | CCDC8_HUMAN | Coiled-coil domain-containing protein 8 | CCDC8 | ○ | ||
a The proteins known to be associated with p32.
To examine the possible involvement of p32 in ribosome biogenesis, we first performed an additional immunoblot analysis of the p32-FLAG-associated proteins with 10 available antibodies against Nop58, PM/Scl100, PWP1, RRP40, RRP46, FTSJ2, nucleolin (NCL), B23, and NOC2L as well as the catalytic component of RNA polymerase III RPC7, which are involved or probably involved in ribosome biogenesis but were not identified by the mass-based analyses. Six of the proteins (Nop58, PM/Scl100, FTSJ2, NCL, RRP40, and RPC7) were detected by the analysis (Fig. 1D, and Table I); those that were not detected are excluded from Fig. 1D (data not shown).
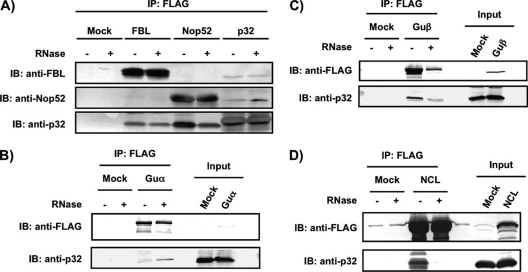
We next performed reverse pull-down analysis using Nop52, FBL, RNA helicases Guα and Guβ, and NCL, which are involved in ribosome biogenesis, as affinity bait to examine their reciprocal associations with p32 with and without RNase treatment (Figs. 2A, 2B, 2C, and 2D). Nop52, FBL, Guα, and Guβ remained associated with p32 even after RNase treatment (Figs. 2A, 2B, and 2C), whereas NCL was associated with p32 only in the presence of RNA (Fig. 2D). Consistent with the proteomic analysis reported (26), FLAG-Nop52 did not associate with FBL, and FLAG-FBL did not associate with Nop52 (Fig. 2A). However, p32-FLAG associated with both FBL and Nop52 (Fig. 2A), suggesting that the association of p32 with FBL was independent of its association with Nop52.
Fig. 2.
Reciprocal association of FBL, Nop52, Guα, Guβ, or NCL with p32. Proteins were immunoprecipitated with anti-FLAG from the whole-cell extract of 293EBNA cells expressing FLAG-tagged FBL (A), Nop52 (A), Guα (B), Guβ (C), or nucleolin (NCL) (D), and detected by immunoblotting (IB) with anti-FBL, anti-Nop52, anti-FLAG or anti-p32 before (–) and after (+) treatment with RNase A. Input: 30 μg of an aliquot of whole-cell extract.
p32 Is Localized in the Nucleolus and Is Present in Preribosomal Fractions
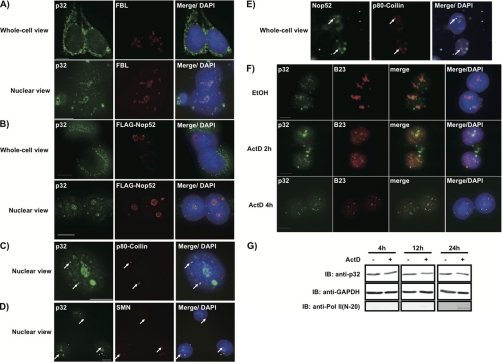
p32 is primarily localized in mitochondria and in the cytoplasm but can accumulate in the nucleus in response to leptomycin B or actinomycin D treatment of cells (58), whereas FBL and Nop52 are predominantly localized in the nucleoli during interphase (34). Therefore, we re-examined the localization of p32 in 293EBNA cells by immunocytochemical costaining. Although most p32 molecules were localized in mitochondria as reported (Fig. 3A, whole-cell view), they were dispersed as several minimum dots within the nucleus, some of which were present in high density and co-localized with FBL (Fig. 3A, nuclear view). By contrast, Nop52 was concentrated in the nucleolar periphery (Fig. 3B, nuclear view) and colocalized partly with p32. We also found that Nop52 co-localized with p80-coilin and that p32 colocalized with both p80-coilin and SMN, each of which is a marker of Cajal bodies (Figs. 3C, 3D, and 3E), suggesting that p32 and Nop52 also localize in Cajal bodies, as does FBL. In addition, upon treatment with actinomycin D, p32 was excluded from the nucleolus and was dispersed throughout the nucleoplasm in many dots (Fig. 3F), consistent with the relocalization of B23, a nucleolar marker that relocalizes to the nucleoplasm in response to actinomycin D (39); notably, the total amount of p32 in the cells was not affected (Fig. 3G). In concert, actinomycin D caused hyper-phosphorylation of the RNA polymerase II large subunit (Fig. 3G), as expected (59).
Fig. 3.
Co-localization of endogenous p32 with FBL and Nop52 in the nucleolus and Cajal bodies, and exclusion of p32 from the nucleolus by actinomycin D treatment. 293EBNA cells were fixed before (whole-cell view) or after (nuclear view) permeabilization and immunostained using an antibody against p32, FBL, FLAG, Nop52, p80-coilin, or SMN. p32, corresponding immunolocalization of endogenous p32. FBL, corresponding images stained for FBL indicating dense fibrillar component (DFC) of the nucleoli. FLAG-Nop52 and Nop52, corresponding images stained for FLAG-Nop52 and endogenous Nop52, respectively, indicating granular component (GC) of the nucleoli. p80-coilin, corresponding images stained for p80-coilin indicating Cajal bodies. SMN, corresponding images stained for SMN indicating Cajal bodies. Merge/DAPI for (A)∼(D), merge of p32 and DAPI, and FBL (A), or FLAG-Nop52 (B), or p80-coilin (C), or SMN (D). Merge/DAPI for (E), merge of Nop52 and DAPI, and p80-coilin. F, 293EBNA cells were treated with 2 μg/ml actinomycin D (ActD) for 2 h or 4 h, and stained for analysis by indirect immunofluorescence (p32 in green; B23 in red). Ethanol, treated with ethanol for 4 h. Merge/DAPI, merge of p32, B23, and DAPI. Bars indicate 10 μm. G, Total proteins from cells treated for longer periods with ActD analyzed by immunoblotting with the antibodies indicated at the left. Pol II (N-20), RNA polymerase II (antibody against hypo- and hyper-phosphorylated forms of Pol II).
We also detected p32 in the nucleolar/Cajal body fraction (NoE) (Figs. 4A and 4B). siRNA treatment reduced the level of p32 in the NoE (Fig. 4A), whereas transfection of a p32-FLAG-encoded plasmid increased p32 in the NoE (Fig. 4B). These results indicate that the antibody we used specifically reacted with p32 and strongly suggest that p32 is present in the NoE. Furthermore, p32 coeluted with Nop52 and FBL in pre-60S fractions and fractions with higher sedimentation velocity separated by sucrose gradient ultracentrifugation of the Nu (Fig. 4C). This ultracentrifugation separated pre-60S particles from the pre-40S particles almost completely, as indicated by methylene blue staining of their rRNA components; namely, 18S rRNA was detected exclusively in the pre-40S fractions, whereas 28S and 32S rRNAs were detected only in pre-60S fractions (Fig. 4C). p32 was also detected with FBL, but not with Nop52, in the pre-40S fractions; however, p32 separation between pre-40S and pre-60S peaks is less clear than that of either FBL or Nop52 (Fig. 4C). The RNA helicases Guα and Guβ, whose association with p32 was independent of the presence of RNA (Figs. 2B and 2C), also coeluted with p32 in the pre-40S and pre-60S fractions (Fig. 4C). In addition, p32 associated at least with 47S, 45S, 41S, 32S, and 12S pre-rRNAs as detected by Northern blot analysis (Figs. 4D and 4E). 47S/45S and 32S/30S pre-rRNA were also detected in the p32-associated complex that was pulse-labeled with [3H]uridine (Fig. 4F). Thus, p32 behaves in vivo as a component of preribosomal particles in all aspects that were examined.
p32 Is Involved in the Processing of pre-rRNA
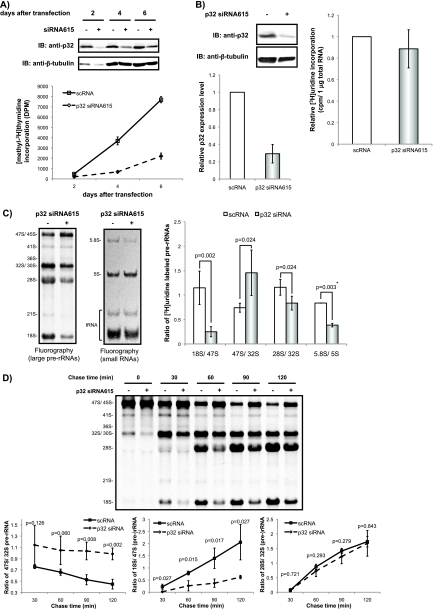
To clarify the involvement of p32 in ribosome biogenesis, we examined the effects of RNAi-mediated p32 knockdown on the production of rRNAs. The knockdown of p32 mRNA level of the cell with the stealth p32 siRNA615 reduced the expression of the protein when compared with the control siRNA treated cells and suppressed significantly the cell growth as reported (60) (Fig. 5A). The p32 siRNA615 caused only a marginal reduction in cellular uptake of [3H]uridine (Fig. 5B). The knockdown reduced significantly the production of 18S rRNA and 32S pre-rRNAs, whereas it accumulated 47S/45S pre-rRNA (Fig. 5C). Pulse-chase analysis using [3H]uridine confirmed the reduction of the early processing of pre-rRNAs from 47S/45S to 18S rRNA and 32S pre-rRNA (Figs. 4D and 5D).
Fig. 5.
Suppression of 47S/45S pre-rRNA processing by knockdown of p32. A, Immunoblotting (IB) was used to determine the p32 expression level in the cells used for the proliferation assay. Whole-cell extracts prepared from cells transfected with scRNA (–) or p32 siRNA615 (+) were analyzed by immunoblotting with anti-p32 and anti-β-tubulin. The proliferation of siRNA615-transfected cells was monitored by measuring [methyl-3H]Thymidine incorporation into DNA with Insta-Gel Plus. The y axis is shown as [3H] incorporation disintegration per minute (DPM). The values indicated are averages (±S.D.) of six independent experiments. B, Immunoblotting was used to determine the p32 expression level in whole-cell extracts prepared from HeLa cells transfected with scRNA (–) or p32 siRNA615 (+) for 72 h. Newly synthesized RNA prepared from the cells transfected as in left was measured by [3H]uridine incorporation into total RNA by scintillation counting (cpm) and normalized to 1 μg of total RNA. The values indicated are averages (±S.D.) of three independent experiments. C, RNA containing newly synthesized pre-rRNAs labeled with [3H]uridine was isolated, and 20,000 cpm per sample were resolved on an agarose gel and transferred to a membrane (left) or on a polyacrylamide gel and transferred to a membrane (middle), and detected by fluorography. Positions of rRNAs are indicated on the left of each fluorogram. Ratio of labeled (pre-)rRNAs in each lane was compared between scRNA and siRNA615 treated cells. The values indicated in the graph are averages (±S.D.) of four independent experiments or two independent experiments (*). p values are also indicated in each comparison. D, Cells transfected as in (B) were pulse-labeled with [3H]uridine for 30 min and cultured in nonradioactive medium for the time indicated. Total RNA (20,000 cpm/lane) was resolved on an agarose gel and transferred to a membrane and detected by fluorography (upper left). Positions of rRNAs and major precursors are indicated on the left of the picture. Ratio of 18S/47S (lower left), 47S/32S (upper right), or 28S/32S (lower right) in the staining intensity in the fluorography (upper left) is plotted against the times indicated. The values are averages (±S.D.) of three independent experiments.
Nop52 Competes with FBL for Binding to p32
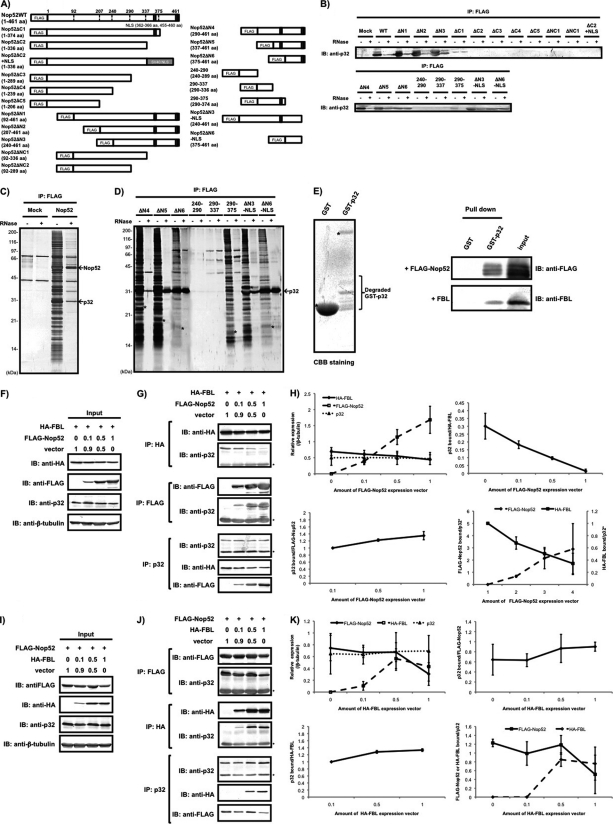
Given the result that the association of p32 with FBL was independent of its association with Nop52, we postulated that p32 was involved in ribosome biogenesis by mediating competitive binding of FBL and Nop52 to pre-ribosome particles. To first ascertain a direct association between Nop52 and p32, we constructed expression vectors for 19 FLAG-tagged-Nop52 truncated mutants to identify the region in Nop52 that was responsible for binding to p32 (Fig. 6A). These truncated mutants were constructed based on the exon-intron boundaries of the Nop52 gene, the domain structure, and the presence of two probable nuclear localization signals at the C-terminal region of Nop52 (http://www.ncbi.nlm.nih.gov/sites/entrez?Db = gene&Cmd = retrieve&dopt = full_reporttwo nuc&list_uids = 8568&log$ = databasead&logdbfrom = nuccore#refseq). The expression and cellular localization of the truncated proteins were examined by SDS-PAGE and immunocytochemistry, respectively (supplemental Fig. S2A and S2B). All truncated mutants were expressed in the cell (supplemental Fig. S2B) and showed the expected molecular sizes as estimated from their amino acid sequences, with the exception of mutant 240–290, which was detected by immunocytochemical analysis but not by the SDS-PAGE (supplemental Fig. S2A). Immunoblotting with anti-p32 showed that domain mutants Nop52ΔN1, ΔN2, ΔN3, ΔN4, ΔN5, and ΔN6 were associated with p32 (Fig. 6B). In addition, Nop52ΔN3 and ΔN6 mutants lacking the C-terminal NLS (Nop52ΔN3-NLS and Nop52ΔN6-NLS) also associated with p32 (Fig. 6B). All of these mutants primarily localized in the nucleolus (supplemental Fig. S2B), suggesting that the interaction between Nop52 and p32 occurs in the nucleolus. On the other hand, all other mutants lacking the region 375–454 (except Nop52ΔC1 and Nop52 290–375) did not associate with p32 (Fig. 6B). These results clearly indicate that the minimal region of Nop52 corresponding to ΔN6-NLS (375–454) is sufficient to interact with p32. Silver staining of SDS-PAGE gels showed that the ΔN5, ΔN6, and ΔN6-NLS mutants were associated almost exclusively with p32 after RNase treatment, whereas wild-type Nop52 associated with several proteins, including p32, independently of the presence of RNA (Figs. 6C and 6D). To examine further the direct interaction between p32 and Nop52, we synthesized recombinant GST-p32 and GST-thrombin cleavage site-FLAG-Nop52 using E. coli DL21 and isolated GST-p32 by affinity chromatography and FLAG-Nop52 by thrombin cleavage from GST-thrombin cleavage site-FLAG-Nop52 bound glutathione-fixed beads (39). We mixed the isolated GST-p32 with FLAG-Nop52, pulled down with glutathione-fixed beads, and confirmed the interaction between GST-p32 and FLAG-Nop52 by immunoblotting with anti-FLAG antibody (Fig. 6E). These results indicate that Nop52 associates directly with p32 at the C-terminal region of Nop52 and that the binding does not require cofactors.
Fig. 6.
Competitive binding of FBL and Nop52 to p32. A, Schematic representation of FLAG-tagged wild-type Nop52 (WT) and its truncation mutants used in this study. Abbreviations used are indicated in the text. Proteins were immunoprecipitated (IP) with anti-FLAG from whole-cell extracts of 293EBNA cells transfected with FLAG-Nop52 or its truncation mutants (see text for descriptions) and detected by immunoblotting (IB) with anti-p32 before (–) and after (+) treatment with RNase A (B) or by silver staining of a 12% SDS-PAGE gel (C, FLAG-Nop52; D, its truncation mutants). Asterisk indicates the position of its corresponding domain mutant migrated. The arrows indicate p32 and FLAG-Nop52 (Nop52). E,The isolated GST-p32 or GST was mixed with purified FLAG-Nop52 or FBL, pulled down with glutathione-fixed beads, and detected by immunoblotting with anti-FLAG antibody or anti-FBL antibody. The CBB-stained SDS-PAGE gel showed that the used-GST-p32 contained several degraded fragments that were detected by immunoblotting with anti-GST antibody (Degraded GST-p32). The isolated FLAG-Nop52 had several forms that were probably produced during thrombin cleavage (see Materials and Methods). (F, G) The expression vector encoding FLAG-Nop52 was mixed with pcDNA3.1(+) empty vector in the ratios indicated and then cotransfected with a constant amount of the HA-FBL expression vector. HA-FBL, FLAG-Nop52, p32 or β-tubulin was detected by immunoblotting with the corresponding antibodies in the cell extract (input) (F) or in the complex isolated by immunoprecipitation with anti-HA, anti-FLAG, or anti-p32 from the co-transfected cells (G). Asterisks in F and G indicate the light chain of the antibody detected by immunoblotting. (H) Statistical evaluation of the results obtained in Figs. 6F and 6G. The horizontal axis is shown as the amount of FLAG-Nop52 expression vector mixed. Vertical axis is shown as: each protein expression detected with the indicated antibody shown in Fig. 6F is normalized by that of β-tubulin in the same lane (upper left); p32 bound/HA-FBL, amount of p32 bound to HA-FBL shown in Fig. 6G (IP:HA) is divided by that of HA-FBL (upper right); p32 bound/FLAG-Nop52, amount of p32 bound to FLAG-Nop52 shown in Fig. 6G (IP:FLAG) is divided by that of FLAG-Nop52 (lower left); FLAG-Nop52 bound/p32, amount of FLAG-Nop52 bound to p32 shown in Fig. 6G (IP:p32) is divided by that of p32, vertical axis right-amount of HA-FBL bound to p32 is divided by that of p32 (IP:p32) (lower right). The values indicated in the graph are averages (±S.D.) of three independent experiments or two independent experiments (*). (I, J) Transfection and immunoprecipitation of the mixture of the HA-FBL expression vector and pcDNA3.1(+) empty vector in different ratios over a constant amount of the FLAG-Nop52 expression vector was also performed. Asterisks in I and J indicate the light chain of the antibody detected by immunoblotting. (K) Statistical evaluation of the results obtained in Figs. 6I and 6J. The horizontal axis is shown as the amount of HA-FBL expression vector mixed. Vertical axis is shown as: each protein expression detected with the indicated antibody shown in Fig. 6I is normalized by that of β-tubulin in the same lane (upper left); p32 bound/FLAG-Nop52, amount of p32 bound to FLAG-Nop52 shown in Fig. 6J (IP:FLAG) is divided by that of FLAG-Nop52 (upper right); p32 bound/HA-FBL, amount of p32 bound to HA-FBL shown in Fig. 6J (IP:HA) is divided by that of HA-FBL (lower left,); FLAG-Nop52 or HA-FBLbound/p32, amount of FLAG-Nop52 or HA-FBL bound to p32 shown in Fig. 6J (IP:p32) is divided by that of p32 (lower right). The values indicated in the graph are averages (±S.D.) of three independent experiments.
To examine the possible competition between FBL and Nop52 for binding to p32, we transfected the HA-FBL plasmid with a mixture of pcDNA3.1 vector and FLAG-Nop52 plasmid at different ratios, pulled down proteins with anti-HA, anti-FLAG, or anti-p32 immobilized on beads, and detected p32, HA-FBL, or FLAG-Nop52 among the pulled-down proteins by immunoblotting. The expression of FLAG-Nop52 reduced FBL-associated p32 in a dose-dependent manner, whereas the amount of Nop52-associated p32 increased with an increase in Nop52 expression (Figs. 6F-6H). In addition, p32-associated HA-FBL decreased with an increased association of FLAG-Nop52 in the protein complex isolated with anti-p32 antibody (Figs. 6G and 6H). On the other hand, when the expression of HA-FBL was increased whereas Nop52 expression remained constant, the amount of FBL-associated p32 increased some extent with an increase in FBL expression (Figs. 6I–6K); however, the decrease of the amount of FLAG-Nop52-associated p32 was not as evident as that of the amount of HA-FBL-associated p32 when the expression of FLAG-Nop52 was increased whereas FBL expression remained constant (Figs. 6F–6K). These results suggest that Nop52, but not the other way can replace FBL bound to p32.
The Association of FBL with p32 Takes Place in the Nucleolus
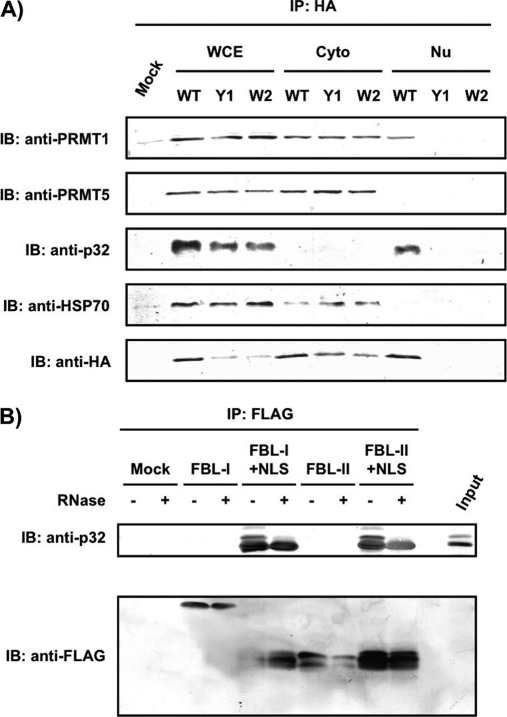
We next examined where in the cell the competition between FBL and Nop52 for binding to p32 takes place. We first used two FBL mutants that localized predominantly in the cytoplasm: Gly-Ala (GA) at residues 167–168 and Ala-Ala (AA) at residues 168–169 were replaced by Tyr-Tyr (YY; Y1) and Trp-Tyr (WY; W2), respectively (supplemental Fig. 3A and 3B). Each of the two mutants was able to associate with p32 in whole-cell extracts (Fig. 7A, WCE); however, wild-type FBL or the mutants did not associate with p32 in the cytoplasmic fraction. On the other hand, wild-type FBL associated with p32 in the nuclear fraction (Fig. 7A, Nu), indicating that the interaction between p32 and FBL takes place in the nucleus. As a reference, we also examined co-precipitation with PRMT1, PRMT5, and HSP70, which associate with FBL, and found that wild-type FBL associated with PRMT1 in both the nucleus and the cytoplasm, whereas it associated with PRMT5 and HSP70 only in the cytoplasm (Fig. 7A). Next we used two domain mutants without a NLS fusion—FBL-I corresponding to the GAR domain (residues 1–77), and FBL-II corresponding to the spacer region 1 (residues 78–132)—which were excluded from the nucleolus and localized in the nucleoplasm and the cytoplasm, respectively (supplemental Figs. S3C and S3D). We found that neither mutant associated with p32 (Fig. 7B). We observed that the expressed FBL-I had a molecular weight two fold greater than that predicted from its amino acid sequence; however, at present we do not know the cause of this discrepancy. However, when the NLS was fused to these mutants, they localized in the nucleolus but not in Cajal bodies (supplemental Figs. S3C and S3D), and they were able to associate with p32 (Fig. 7B) (26). These results suggest that nucleolar localization is required for the association of FBL with p32, and imply that p32 changes its interaction partner from FBL to Nop52 in the nucleolus.
Fig. 7.
The association of FBL with p32 takes place in the nucleolus. A, Proteins were immunoprecipitated (IP) with anti-HA from whole-cell extract (WCE), cytoplasmic fraction (Cyto), and nuclear fraction (Nu), which were prepared from 293EBNA cells transfected with an expression vector encoding HA-FBL, HA-Y1, or HA-W2. They were analyzed by immunoblotting (IB) with the antibodies indicated at the left. B, FLAG-FBL truncation mutant-associated protein complexes were immunoprecipitated with anti-FLAG after expression of various truncation mutants (FBL-l, FBL-l+NLS, FBL-ll, or FBL-ll+NLS) in 293EBNA cells, and analyzed by immunoblotting with anti-p32 before (–) and after (+) treatment with RNase A. Input: 30 μg of an aliquot of whole-cell extract was loaded.
p32 Is Associated with Nop52 in Pre-60S Particles
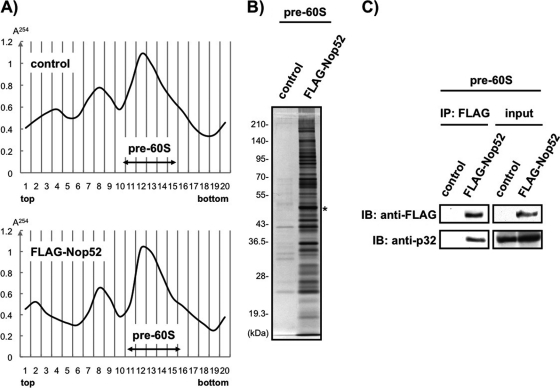
To confirm further the association of p32 with pre-60S particle, we used the pull-down method to isolate FLAG-Nop52-associated pre-60S particles from pre-60S fractions separated by sucrose gradient ultracentrifugation of the Nu (Figs. 8A and 8B). Immunoblotting with anti-p32 unambiguously detected p32 in isolated FLAG-Nop52-associated pre-60S particles (Fig. 8C). Thus, p32 is recruited indeed to Nop52-associated pre-60S ribosomal particles.
Fig. 8.
p32 is present in the Nop52-associated pre-60S particles. A, The nuclear fraction prepared from control cells (control) that stably expressed hygromycin-resistant protein constructed by the transfection of pcDNA5/FRT/TO vector into Flp-In T-REx 293 cells or the inducible FLAG-Nop52 expression cells (FLAG-Nop52) was separated into 20 fractions by sucrose gradient ultracentrifugation. The absorbance at 254 nm was monitored to generate the profile indicated. The pre-60S fractions 11–15 were combined for each of the profiles independently and used for immunoprecipitation with anti-FLAG-fixed beads. B, The isolated proteins were analyzed by 10% SDS-PAGE and visualized by silver staining. Asterisk indicates FLAG-Nop52 as bait. The molecular masses (kDa) are indicated at the left. C, Association of p32 with Nop52 in isolated pre-60S ribosomal particles was confirmed by immunoblot (IB) analysis with anti-p32. An aliquot (5%) of the pre-60S combined fractions prepared from control or FLAG-Nop52 was used as input. IP: Immunoprecipitation.
DISCUSSION
In this study, we showed that 1) p32 associates with 31 proteins known or expected to be involved in rRNA processing (of which 10 proteins are classified as components of pre-90S particle, and 11 proteins as pre-60S particle, Table I), at least four of which are found to associate independently with p32; 2) p32 colocalizes with FBL or Nop52 in the nucleolus and is excluded from the nucleolus upon inhibition of transcription with actinomycin D; 3) p32 is present in the preribosomal fraction prepared by cell fractionation; 4) p32 coelutes with FBL or Nop52 in the pre-ribosomal fractions separated by ultracentrifugation; 5) p32 associates with pre-rRNAs, 6) p32 is present in the isolated Nop52-associated pre-60S ribosomal particles; and 7) p32 knockdown slows down the early processing of pre-rRNAs. We conclude that p32 is a de facto component of pre-ribosomal particles and is involved in early nucleolar steps of ribosome biogenesis.
Our hypothesis for p32 involvement in ribosome biogenesis is that p32 exchanges its binding partner, i.e. p32 binds Nop52 in place of FBL, in preribosomal particles in the nucleolus. The p32-Nop52 complex then mediates transformation of FBL-associated pre-90S ribosomal particles formed in the dense fibrillar component (DFC) to Nop52-associated pre-60S ribosomal particles in the granular component (GC). This hypothesis is based on our results that 1) p32 association with Nop52 is independent of that with FBL; 2) p32 interacts with Nop52 without any cofactors, just as with FBL; 3) the association between FBL and p32 takes place in the nucleolus; 4) the association between Nop52 and p32 possibly occurs in the nucleolus, too; 5) p32 is required for the early processing of pre-rRNA from 47S/45S pre-rRNA to 18S rRNA and 32S pre-rRNA, 6) p32 associates with pre-rRNAs including 47S/45S and 32S pre-rRNAs, 7) p32 is present in Nop52-associated pre-60S ribosomal particles isolated from the nucleus; and 8) Nop52 competes with FBL for association with p32. We previously demonstrated that FBL associates with U3 snoRNP components (Nop56, Nop58, and a 15.5-kDa protein) and at least 27 pre-rRNA processing factors, but not with Nop52 (26, 57); we proposed that FBL functions in the pre-ribosomal particles formed from the early to middle stages of human ribosome biogenesis (26, 42, 61). We have also used mass-based proteomics to examine the constituents of the Nop52-associated preribosomal particles and identified 20 proteins involved or expected to be involved in ribosome biogenesis, but did not detect FBL as well as the other U3 snoRNP components (42, 62). These results indicate that FBL-associated preribosomal particles are distinct from those associated with Nop52. It is well established that FBL and Nop52 are involved in different stages of ribosome biogenesis in the nucleolus; FBL participates in early processing of rRNA, i.e. the processing of the 5′-external transcribed spacer, and localizes in the nucleolar DFC (46, 47, 63), whereas Nop52 is associated with late events related to the production of 60S ribosomal particle and localizes in the nucleolar GC (33–35). These observations support our hypothesis.
We propose two scenarios for this hypothesis. In the first scenario, p32 associates with pre-90S ribosomal particles formed in the early stages of ribosome biogenesis through FBL in the DFC. Subsequently, Nop52 targets p32 on the FBL-associated pre-90S ribosomal particles, thereby replacing FBL and initiating the formation of Nop52-associated pre-60S ribosomal particles, which accelerates the maturation of 28S rRNA in the later stages of ribosome biogenesis in the GC. In this scenario, preribosomal particles formed in the early stages would contain not only FBL but also p32. In fact, p32 was detected in fractions with a sedimentation coefficient larger than 60S (i.e. fractions 16–20 in Fig. 4C) and was associated with at least 10 proteins classified as components of pre-90S particle (Table I) as well as 47S/45S pre-rRNA (Figs. 4E and 4F). The presence of Nop52 in these fractions may indicate a transient preribosomal complex containing FBL, p32, and Nop52. This transit complex may be unstable and difficult to be isolated by the pull-down method we used. As p32 forms trimer with a doughnut-shaped quaternary structure with an unusually asymmetric charge distribution on the surface (64), it is possible that p32 acts as a bridge between FBL and Nop52 on the transient pre-ribosomal complex. After this exchange event, the Nop52-p32 complex in pre-60S ribosomal particle accelerates the processing of 32S pre-rRNA to 28S rRNA. The fact that one molecule of FBL binds to two molecules of p32—one in the GAR domain and another in spacer region 1 (26)—may explain the presence of p32 in FBL-associated pre-40S ribosome particles as well; i.e. only one of the p32 molecules is involved in competitive binding with Nop52, leaving the other p32 molecule associated with FBL in pre-40S ribosomal particles after the remodeling of preribosomal particles.
In the second scenario, FBL and Nop52 are both associated with preribosomal particles in a small population formed at a certain point in the early stages of ribosome biogenesis, but these intermediate preribosomal particles associated with FBL and Nop52 may be too unstable to isolate by the pull-down method. Then, a posttranslational modification of p32 and/or other proteins, or perhaps the entry of p32 and/or another protein(s) into the nucleolus, triggers remodeling of the FBL-Nop52-associated preribosomal particles to FBL-p32- and Nop52-p32-associated preribosomal particles formed at later stages of ribosome biogenesis. This scenario can also explain some of our results (i.e. Fig. 4, Fig. 5, and Fig. 8), such that p32 was required for the early processing of 47S/45S pre-rRNA, and that it was detected in pre-40S and pre-60S particles and in fractions with a sedimentation coefficient larger than 60S fractions. Both scenarios suggest that p32 is required to distinguish Nop52-associated pre-60S ribosomal particles from those associated with FBL. p32 may function to validate the proper assembly of FBL-associated pre-90S ribosomal particles formed at early stages of ribosome biogenesis before carrying out processes such as the cleavage of the internal transcribed spacer 2 in Nop52-associated pre-60S ribosomal particles at later stages.
We also detected p32, FBL, and Nop52 in fractions with sedimentation velocity smaller than 40S separated by sucrose gradient ultracentrifugation (Fig. 4C). Although no direct evidence for the involvement of these protein populations in ribosome biogenesis is currently available, the relative amounts and interactions among these proteins may determine the supply of each protein to the sites of ribosome biogenesis. In this context, their localization in Cajal bodies may be related to a mechanism underlying this level of regulation.
It has been reported that yeast Mam33p (mitochondrial acidic matrix protein) and the putative Caenorhabditis elegans gene product F59A-2.3 are functional homologs of human p32 (65). The two homologs are synthesized as precursors with an N-terminal mitochondrial targeting sequence that is cleaved upon import, and they are located mainly in mitochondria. Those proteins also have conserved structural features, including the prevalence of acidic amino acids and the occurrence in homo-oligomeric complexes (64). To re-examine possible evolutionary conservation of the p32 amino acid sequence in organisms lower than yeast (prokaryotic organisms), we searched for protein sequences that align with the entire human p32 sequence (UniProt: C1QBP_HUMAN) in the database containing nonfragmented sequences of bacteria (5,338,921 entries) and archaebacteria (182,058 entries) extracted from UniProt (SwissProt, release 57.4 and TrEMBL, release 40.4) using BlastP (E-value < 0.1 with SEG filtering) and PSI-BLAST (E-value < 0.1, 10 iterations). No homologs were detected by PSI-BLAST, and only one hit, B8K7P_VIBPA, was found by BlastP. Although this protein has 28% sequence identity with human p32, it is unlikely to be a functional homolog, as it has neither the sequence motif (Pfam02330) nor the structural unit (SCOP d.25.1.1) of human p32 (64), which adopts a novel fold with seven consecutive anti-parallel β-strands flanked by one N-terminal and two C-terminal α-helices. By contrast, yeast homologs of human p32, MAM33_YEAST (Saccharomyces cerevisiae) and MAM33_SCHPO (Schizosaccharomyces pombe) were identified as homologs of human p32 at E-values < 0.1 by BlastP and PSI-BLAST, and both have Pfam02330 and SCOP d.25.1.1 domains. These results suggest that p32 evolved in eukaryotes rather than prokaryotes. Although the involvement of Mam33p in ribosome biogenesis has not been reported, large-scale proteomic analyses have demonstrated that Mam33p associates with Nop7p and Mrt4p, which are involved in rRNA processing and are components of yeast pre-66S ribosome particles (66, 67). By large-scale proteomic analyses Mam33p was also shown to be associated with several mitochondrial ribosome proteins in yeast (67). Thus, it is likely that the involvement of p32 in ribosome biogenesis is conserved among eukaryotes from yeast to human. However, Mam33p may be involved in ribosome biogenesis by a mechanism different from that in humans, as yeast Rrp1p does not have much sequence similarity in the region of Nop52 that binds to p32 (33).
p32 also interacts with herpes simplex virus 1 Orf-P protein (17), HIV-1 Tat (24), and Epstein-Barr virus EBNA 1 protein (20). Ribosomal proteins are synthesized up to the late stage of herpes simplex virus 1 infection and efficiently assemble into mature ribosomes, whereas there is a severe shutoff of the synthesis of other cellular proteins (68). Upon infection, this virus also alters the nuclear transport of preribosomal particles (69). In the case of HIV1, Tat protein affects processing of pre-rRNA, probably through the interaction with FBL (70). Regarding Epstein-Barr virus, the yeast homolog of EBNA1-binding protein is essential for biogenesis of the 60S subunit (71). These reports suggest that these virus proteins influence ribosome biogenesis by interrupting or modifying the interaction of p32 with FBL or Nop52. It has also been established that several viruses and viral proteins enter the nucleolus; viral proteins co-localize with factors such as nucleolin, B23, and FBL, and can cause their redistribution during infection (72, 73). In the case of hepatitis C virus, its infection even modifies transcription of rDNA (74); thus, the nucleolar processes of ribosome biogenesis seem to be common targets for viral infection. As such, a more detailed understanding of viral protein interactions with the ribosome-generating machinery may lead to novel antiviral therapies.
Acknowledgments
We would like to express our deepest condolences to the death of Miss Miyuki Kawasaki who initiated this study, and thank Dr. D. Hernandez-Verdun for the generous gift of human anti-Nop52 (NNP1), Dr. A. R. Krainer for mouse monoclonal anti-p32, and Dr. W. C. Russell for rabbit polyclonal anti-p32.
Footnotes
* This work was supported in part by a grant for promotion from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), the Japan Health Science Foundation (H18-Soyaku-Ippan-001), the Core Research for Evolutional Science and Technology (CREST), Japan Science and Technology Agency (JST), and the Targeted Proteins Research Program (TPRP) from the MEXT.
 This article contains supplemental Figs. S1 to S3 and Tables S1 and S2.
This article contains supplemental Figs. S1 to S3 and Tables S1 and S2.
1 The abbreviations used are:
- p32
- splicing factor 2–associated protein p32
- FBL
- fibrillarin
- PRMT
- protein arginine methyltransferase
- HIV
- human immunodeficiency virus
- GAR
- glycine- and arginine-rich
- RBD
- RNA-binding domain
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- MW
- molecular weight
- MS/MS
- tandem mass spectrometer
- pre-rRNP
- preribosomal ribonucleoprotein
- siRNA
- small interfering RNA
- NLS
- nuclear localization signal.
REFERENCES
- 1. Krainer A. R., Mayeda A., Kozak D., Binns G. (1991) Functional expression of cloned human splicing factor SF2: homology to RNA binding protein VI 70K, and Drosophila splicing regulators. Cell 66, 383–394 [DOI] [PubMed] [Google Scholar]
- 2. Petersen-Mahrt S. K., Estmer C., Ohrmalm C., Matthews D. A., Russell W. C., Akusjävi G. (1999) The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 18, 1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mayeda A., Krainer A. R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68, 365–375 [DOI] [PubMed] [Google Scholar]
- 4. Mayeda A., Helfman D. M., Krainer A. R. (1993) Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein-A1 and premessenger RNA splicing factor SF2/ASF. Mol. Cell. Biol. 13, 2993–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sun Q., Mayeda A., Hampson R. K., Krainer A. R., Rottman F. M. (1993) General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 7, 2598–2608 [DOI] [PubMed] [Google Scholar]
- 6. Yang X., Bani M. R., Lu S. J., Rowan S., Ben-David Y., Chabot B. (1994) The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl. Acad. Sci. U.S.A. 91, 6924–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chabot B., Blanchette M., Lapierre I., La Branche H. (1997) An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol. Cell. Biol. 17, 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hanamura A., Cáceres J. F., Mayeda A., Franza B. R., Jr., Krainer A. R. (1998) Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4, 430–444 [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang Z. H., Zhang W. J., Rao Y., Wu J. Y. (1998) Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc. Natl. Acad. Sci. U.S.A. 95, 9155–9160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghebrehiwet B., Lim B. L., Peerschke E. I., Willis A. C., Reid K. B. (1994) Isolation, cDNA cloning, and overexpression of a 33-kD cell surface glycoprotein that binds to the globular ‘heads’ of C1q. J. Exp. Med. 179, 1809–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yadav G., Prasad R. L., Jha B. K., Rai V., Bhakuni V., Datta K. (2009) Evidence for inhibitory interaction of hyaluronan-binding protein 1 (HABP1/p32/gC1qR) with Streptococcus pneumoniae hyaluronidase. J. Biol. Chem. 284, 3897–3905 [DOI] [PubMed] [Google Scholar]
- 12. Deb T. B., Datta K. (1996) Molecular cloning of human fibroblast hyaluronic acid-binding protein confirms its identity with P-32, a protein co-purified with splicing factor SF2. Hyaluronic acid-binding protein as P-32 protein, co-purified with splicing factor SF2. J. Biol. Chem. 271, 2206–2212 [DOI] [PubMed] [Google Scholar]
- 13. Chattopadhyay C., Hawke D., Kobayashi R., Maity S. N. (2004) Human p32, interacts with B subunit of the CCAAT-binding factor, CBF/NF-Y, and inhibits CBF-mediated transcription activation in vitro. Nucleic Acids Res. 32, 3632–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Itahana K., Zhang Y. (2008) Mitochondrial p32 Is a Critical Mediator of ARF-Induced Apoptosis. Cancer Cell 13, 542–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sunayama J., Ando Y., Itoh N., Tomiyama A., Sakurada K., Sugiyama A., Kang D., Tashiro F., Gotoh Y., Kuchino Y., Kitanaka C. (2004) Physical and functional interaction between BH3-only protein Hrk and mitochondrial pore-forming protein p32. Cell Death Differ. 11, 771–781 [DOI] [PubMed] [Google Scholar]
- 16. Simos G., Georgatos S. D. (1994) The lamin B receptor-associated protein p34 shares sequence homology and antigenic determinants with the splicing factor 2-associated protein p32. FEBS Lett. 346, 225–228 [DOI] [PubMed] [Google Scholar]
- 17. Bruni R., Roizman B. (1996) Open reading frame P-a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc. Natl. Acad. Sci. U.S.A. 93, 10423–10427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matthews D. A., Russell W. C. (1998) Adenovirus core protein V interacts with p32-a protein, which is associated with both the mitochondria and the nucleus. J. Gen. Virol. 79, 1677–1685 [DOI] [PubMed] [Google Scholar]
- 19. Wang Y., Finan J. E., Middeldrop J. M., Hayward S. D. (1997) p32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 236, 18–29 [DOI] [PubMed] [Google Scholar]
- 20. Chen M. R., Yang J. F., Wu C. W., Middeldorp J. M., Chen J. Y. (1998) Physical association between the EBV protein EBNA-1 and p32/TAP/hyaluronectin. J. Biomed. Sci. 5, 173–179 [DOI] [PubMed] [Google Scholar]
- 21. Beatch M. D., Hobman T. C. (2000) Rubella virus capsid associates with host cell protein p32 and localizes to mitochondria. J. Virol. 74, 5569–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luo Y., Yu H., Peterlin B. M. (1994) Cellular protein modulates effects of human immunodeficiency virus type 1 Rev. J. Virol. 68, 3850–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tange T. O., Jensen T. H., Kjems J. (1996) In vitro interaction between human immunodeficiency virus type 1 rev protein and splicing factor ASF/SF2-associated protein p32. J. Biol. Chem. 271, 10066–10072 [DOI] [PubMed] [Google Scholar]
- 24. Yu L., Loewenstein P. M., Zhang Z., Green M. (1995) In vitro interaction of the human immunodeficiency virus type 1 Tat transactivator and the general transcription factor TFIIB with the cellular protein TAP. J. Virol. 69, 3017–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng Y. H., Yu H. F., Peterlin B. M. (2003) Human p32 protein relieves a post-transcriptional block to HIV replication in murine cells. Nat. Cell Biol. 5, 611–618 [DOI] [PubMed] [Google Scholar]
- 26. Yanagida M., Hayano T., Yamauchi Y., Shinkawa T., Natsume T., Isobe T., Takahashi N. (2004) Human fibrillarin forms a sub-complex with splicing factor 2 associated p32, protein/arginine methyltransferases, tubulin α3 and β1, which is Independent of its association with preribosomal ribonucleoprotein complexes. J. Biol. Chem. 279, 1607–1614 [DOI] [PubMed] [Google Scholar]
- 27. Warner J. R. (1990) The nucleolus and ribosome formation. Curr. Opin. Cell Biol. 2, 521–527 [DOI] [PubMed] [Google Scholar]
- 28. Kiss-László Z., Henry Y., Bachellerie J. P., Caizergues-Ferrer M., Kiss T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 29. Tycowski K. T., Smith C. M., Shu M. D., Steitz J. A. (1996) A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl. Acad. Sci. U.S.A. 93, 14480–14485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tyc K., Steitz J. A. (1989) U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 8, 3113–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baserga S. J., Yang X. D., Steitz J. A. (1991) An intact Box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 10, 2645–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aris J. P., Blobel G. (1991) cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc. Natl. Acad. Sci. U.S.A. 88, 931–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Horsey E. W., Jakovljevic J., Miles T. D., Harnpicharnchai P., Woolford J. L., Jr. (2004) Role of the yeast Rrp1 protein in the dynamics of preribosome maturation. RNA 10, 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Savino T. M., Bastos R., Jansen E., Hernandez-Verdun D. (1999) The nucleolar antigen Nop52p, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleogenesis. J. Cell Sci. 112, 1889–1900 [DOI] [PubMed] [Google Scholar]
- 35. Savino T. M., Gébrane-Younès J., De Mey J., Sibarita J. B., Hernandez-Verdun D. (2001) Nucleolar Assembly of the rRNA Processing Machinery in Living Cells. J. Cell Biol. 153, 1097–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yanagida M., Shimamoto A., Nishikawa K., Furuichi Y., Isobe T., Takahashi N. (2001) Isolation and proteomic characterization of the major proteins of the nucleolin-associating ribonucleoprotein complexes. Proteomics 1, 1390–1404 [DOI] [PubMed] [Google Scholar]
- 37. Stavreva D. A., Kawasaki M., Dundr M., Koberna K., Müller W. G., Tsujimura-Takahashi T., Komatsu W., Hayano T., Isobe T., Raska I., Misteli T., Takahashi N., McNally J. G. (2006) Potential roles for ubiquitin and the proteasome during ribosome biogenesis. Mol. Cell. Biol. 26, 5131–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watkins N. J., Lemm I., Ingelfinger D., Schneider C., Hossbach M., Urlaub H., Lührmann R. (2004) Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell 16, 789–798 [DOI] [PubMed] [Google Scholar]
- 39. Fujiyama-Nakamura S., Yoshikawa H., Homma K., Hayano T., Tsujimura-Takahashi T., Izumikawa K., Ishikawa H., Miyazawa N., Yanagida M., Miura M., Shinkawa T., Yamauchi Y., Isobe T., Takahashi N. (2009) Parvulin (Par14), a peptidyl prolyl cis-trans isomerase, is a novel rRNA-processing factor that evolved in the metazoan lineage. Mol. Cell. Proteomics 8, 1552–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hayano T., Yanagida M., Yamauchi Y., Shinkawa T., Isobe T., Takahashi N. (2003) Proteomic analysis of human Nop56p-associated preribosomal ribonucleoprotein complexes: Possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J. Biol. Chem. 278, 34309–34319 [DOI] [PubMed] [Google Scholar]
- 41. Pestov D. G., Lapik Y. R., Lau L. F. (2008) Assays for ribosomal RNA processing and ribosome assembly. Curr. Protoc. Cell Biol. 22, Unit 22.11 [DOI] [PubMed] [Google Scholar]
- 42. Takahashi N., Isobe T. in Proteomic Biology using LC/MS, Wiley Interscience, John Wiley and Sons Inc., Hoboken NJ U.S.A. (2007) pp. 1–254 [Google Scholar]
- 43. Even Y., Durieux S., Escande M. L., Lozano J. C., Peaucellier G., Weil D., Genevière A. M. (2006) CDC2L5, a Cdk-like kinase with RS domain, interacts with the ASF/SF2-associated protein p32 and affects splicing in vivo, J. Cell. Biochem. 99, 890–904 [DOI] [PubMed] [Google Scholar]
- 44. Crawford N. P., Yang H., Mattaini K. R., Hunter K. W. (2009) The metastasis efficiency modifier ribosomal RNA processing 1 homolog B (RRP1B) is a chromatin-associated factor. J. Biol. Chem. 284, 28660–28673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tycowski K. T., Smith C. M., Shu M. D., Steitz J. A. (1996) A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl. Acad. Sci. U.S.A. 93, 14480–14485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jansen R. P., Hurt E. C., Kern H., Lehtonen H., Carmo-Fonseca M., Lapeyre B., Tollervey D. (1991) Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J. Cell Biol. 113, 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Turner A. J., Knox A. A., Prieto J. L., McStay B., Watkins N. J. (2009) A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol. Cell. Biol. 29, 3007–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Granneman S., Vogelzangs J., Lührmann R., van Venrooij W. J., Pruijn G. J., Watkins N. J. (2004) Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP. Mol. Cell. Biol. 24, 8600–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ginisty H., Amalric F., Bouvet P. (1998) NCL functions in the first step of ribosomal RNA processing. EMBO J. 17, 1476–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Henning D., So R. B., Jin R., Lau L. F., Valdez B. C. (2003) Silencing of RNA helicase II/Guα inhibits mammalian ribosomal RNA production. J. Biol. Chem. 278, 52307–52314 [DOI] [PubMed] [Google Scholar]
- 51. Castle C. D., Cassimere E. K., Lee J., Denicourt C. (2010) Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis. Mol. Cell. Biol. 30, 4404–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Srivastava L., Lapik Y. R., Wang M., Pestov D. G. (2010) Mammalian DEAD box protein Ddx51 acts in 3′ End maturation of 28S rRNA by promoting the release of U8 snoRNA. Mol. Cell. Biol. 30, 2947–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eilbracht J., Reichenzeller M., Hergt M., Schnölzer M., Heid H., Stöhr M., Franke W. W., Schmidt-Zachmann M. S. (2004) NO66, a highly conserved dual location protein in the nucleolus and in a special type of synchronously replicating chromatin. Mol. Biol. Cell 15, 1816–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zirwes R. F., Eilbracht J., Kneissel S., Schmidt-Zachmann M. S. (2000) A novel helicase-type protein in the nucleolus: protein NOH61. Mol. Biol. Cell 11, 1153–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heindl K., Martinez J. (2010) Nol9 is a novel polynucleotide 5′-kinase involved in ribosomal RNA processing. EMBO J. 29, 4161–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Haindl M., Harasim T., Eick D., Muller S. (2008) The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 9, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Finkbeiner E., Haindl M., Muller S. (2011) The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 30, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brokstad K. A., Kalland K. H., Russell W. C., Matthews D. A. (2001) Mitochondrial protein p32 can accumulate in the nucleus. Biochem. Biophys. Res. Comm. 281, 1161–1169 [DOI] [PubMed] [Google Scholar]
- 59. Izumikawa K., Yanagida M., Hayano T., Tachikawa H., Komatsu W., Shimamoto A., Futami K., Furuichi Y., Shinkawa T., Yamauchi Y., Isobe T., Takahashi N. (2008) Association of human DNA helicase RecQ5β with RNA polymerase II and its possible role in transcription. Biochem. J. 413, 505–516 [DOI] [PubMed] [Google Scholar]
- 60. Fogal V., Richardson A. D., Karmali P. P., Scheffler I. E., Smith J. W., Ruoslahti E. (2010) Mitochondrial p32 Protein Is a Critical Regulator of Tumor Metabolism via Maintenance of Oxidative Phosphorylation. Mol. Cell. Biol. 30, 1303–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Takahashi N., Yanagida M., Fujiyama S., Hayano T., Isobe T. (2003) Proteomic snapshot analysis preribosomal ribonucleoprotein complexes formed at various stages of ribosome biogenesis in yeast and mammalian cells. Mass Spectrom. Rev. 22, 287–317 [DOI] [PubMed] [Google Scholar]
- 62. Kawasaki M. (2004) Master's thesis (Isolation and proteomic characterization of the preribosomal ribonucleoprotein complex associated with a novel nucleolar protein-1 that is ubiquitously expressed in human tissue), Graduate School of Agriculture, Tokyo University of Agriculture & Technology [Google Scholar]
- 63. Snaar S., Wiesmeijer K., Jochemsen A. G., Tanke H. J., Dirks R. W. (2000) Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol. 151, 653–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jiang J., Zhang Y., Krainer A. R., Xu R. M. (1999) Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc. Natl. Acad. Sci. U.S.A. 96, 3572–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Seytter T., Lottspeich F., Neupert W., Schwarz E. (1998) Mam33p, an oligomeric, acidic protein in the mitochondrial matrix of Saccharomyces cerevisiae is related to the human complement receptor gC1q-R. Yeast 14, 303–310 [DOI] [PubMed] [Google Scholar]
- 66. Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., Meagher D., Imai B., Guo Y., Brame C. J., Shabanowitz J., Hunt D. F., Woolford J. L., Jr. (2001) Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8, 505–515 [DOI] [PubMed] [Google Scholar]
- 67. Ho Y., Gruhler A, Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford. J., Poulsen V., Sørensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 68. Simonin D., Diaz J. J., Massé T., Madjar J. J. (1997) Persistence of ribosomal protein synthesis after infection of HeLa cells by herpes simplex virus type 1. J. Gen. Virol. 78, 435–443 [DOI] [PubMed] [Google Scholar]
- 69. Besse S., Puvion-Dutilleul F. (1996) Intranuclear retention of ribosomal RNAs in response to herpes simplex virus type 1 infection. J. Cell Sci. 109, 119–129 [DOI] [PubMed] [Google Scholar]
- 70. Ponti D., Troiano M., Bellenchi G. C., Battaglia P. A., Gigliani F. (2008) The HIV Tat protein affects processing of ribosomal RNA precursor. BMC Cell Biol. 9, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shirai C., Takai T., Nariai M., Horigome C., Mizuta K. (2004) Ebp2p, the yeast homolog of Epstein-Barr virus nuclear antigen 1-binding protein 2, interacts with factors of both the 60 S and the 40 S ribosomal subunit assembly. J. Biol. Chem. 279, 25353–25358 [DOI] [PubMed] [Google Scholar]
- 72. Hiscox J. A. (2002) The nucleolus-a gateway to viral infection? Arch. Virol. 14, 1077–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hiscox J. A. (2007) RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5, 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Raychaudhuri S., Fontanes V., Barat B., Dasgupta A. (2009) Activation of ribosomal RNA transcription by hepatitis C virus involves upstream binding factor phosphorylation via induction of cyclin D1. Cancer Res. 69, 2057–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hadjiolova K. V., Nicoloso M., Mazan S., Hadjiolov A. A., Bachellerie J. P. (1993) Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 212, 211–215 [DOI] [PubMed] [Google Scholar]