Abstract
Abdominal aortic aneurysms (AAA) are characterized by pathological remodeling of the aortic extracellular matrix (ECM). However, besides the well-characterized elastolysis and collagenolysis little is known about changes in other ECM proteins. Previous proteomics studies on AAA focused on cellular changes without emphasis on the ECM. In the present study, ECM proteins and their degradation products were selectively extracted from aneurysmal and control aortas using a solubility-based subfractionation methodology and analyzed by gel-liquid chromatography-tandem MS and label-free quantitation. The proteomics analysis revealed novel changes in the ECM of AAA, including increased expression as well as degradation of collagen XII, thrombospondin 2, aortic carboxypeptidase-like protein, periostin, fibronectin and tenascin. Proteomics also confirmed the accumulation of macrophage metalloelastase (MMP-12). Incubation of control aortic tissue with recombinant MMP-12 resulted in the extensive fragmentation of these glycoproteins, most of which are novel substrates of MMP-12. In conclusion, our proteomics methodology allowed the first detailed analysis of the ECM in AAA and identified markers of pathological ECM remodeling related to MMP-12 activity.
Abdominal aortic aneurysms (AAA)1 affect 2–10% of the elderly population (1, 2) and are histopathologically characterized by the extensive degeneration of the aortic extracellular matrix (ECM) (3). The breakdown of structural proteins, in particular medial elastin and collagen I and III, is responsible for the inability of the aortic wall to withstand the hemodynamic forces (4). Transmural inflammatory infiltrates (5) combined with a reduction in the number of medial smooth muscle cells trigger the destruction of the aortic connective tissue via unregulated proteinase activity (6, 7). However, apart from the well-established elastolysis and collagenolysis, little is known about other changes in ECM composition and turnover in AAA. Recent proteomics studies on aneurysms have used whole tissue lysates (8, 9) and reported significant changes in cellular proteins but provided limited insights into matrix remodeling. Regarding the ECM these approaches had several limitations: first, whole tissue lysates are rich in cellular proteins, which mask the identification of scarce extracellular components, including proteinases and proteinase inhibitors. Second, ECM proteins are difficult to solubilize and analyze by proteomics because of their unique biochemical properties (aggregation, cross-linking, and glycosylation). Third, during pathological processes, such as aneurysm formation, the ECM undergoes extensive turnover with continuous degradation as well as deposition of newly synthesized matrix proteins. In analysis of whole tissue lysates, newly synthesized ECM proteins cannot be readily discerned from existing matrix proteins and only abundant degradation products are detected (9). To overcome these shortcomings we have recently developed a solubility-based, protein subfractionation procedure, which facilitates the selective extraction of newly synthesized ECM proteins and degradation products, reduces the contamination with cellular proteins, and ensures the solubilization of the interstitial ECM and its associated proteins, thereby improving their analysis by proteomics (10). In this study, we applied this subfractionation method to AAA. Our aim was to characterize the changes in ECM and associated proteins involved in the pathological remodeling process. We achieved the most detailed proteomics characterization of the aneurysmal ECM to date and revealed major differences between healthy and AAA tissues including the increased deposition and fragmentation of collagen XII, thrombospondin 2, aortic carboxypeptidase-like protein (ACLP), periostin, and fibronectin. In addition, we were able to show that these glycoproteins are novel proteolytic targets of MMP-12. Thus, our proteomics approach can serve as a platform for the identification of candidate markers of pathological remodeling in clinical samples (11).
EXPERIMENTAL PROCEDURES
Source of Tissue
This study was approved by the Research Ethics Committees of King's College London and St. George's Hospital London. All patients gave written informed consent. Advanced, asymptomatic, infrarenal AAA specimens were collected from four male patients >65 yr of age during open aneurysm repair surgery based on aneurysm size exceeding 5.5 cm in diameter (representative hematoxylin and eosin stained sections are shown in supplemental Fig. S1). Four control samples from patients without connective tissue disorder (one female and three males aged 20 to 55) were obtained upon aortotomy performed during routine aortic valve replacement from positions of the ascending aorta that were free of macroscopically evident vascular pathology (supplemental Fig. S1). There were no known potential confounders during sampling of clinical specimens and tissues were immediately snap-frozen and kept in liquid nitrogen for later use.
Tissue Preparation
Before extraction the tissue specimens were partially thawed and weighed. Approximately 150 mg of tissue per aortic sample were placed in ice-cold phosphate buffered saline to remove plasma contaminants. The buffer was supplemented with 1% v/v proteinase inhibitor mixture (Sigma-Aldrich), which contains AEBSF, aprotinin, bestatin, E-64, leupeptin, and pepstatin, with a broad specificity for serine, cysteine, and acid proteinases, and aminopeptidases and 1% v/v phosphatase inhibitor mixture (Sigma-Aldrich), which contains, sodium orthovanadate, cantharidin, imidazole, and calyculin A, to inhibit a broad range of phosphatases. In addition, 25 mm EDTA were added to ensure inhibition of metalloproteinases. While the tissue samples were immersed in the cold saline, they were diced with a scalpel into 8–10 smaller pieces to facilitate the removal of plasma contaminants and the effective extraction of extracellular proteins as described below.
Three-step Extraction Methodology
For the proteomics methodology we used three healthy aortic specimens (CON) and four AAA specimens. The diced samples were first incubated in a 0.5 m NaCl, 10 mm Tris, pH 7.5 buffer supplemented with proteinase and phosphatase inhibitor cocktails as above and 25 mm EDTA. The volume of the buffer was adjusted to 10:1 of the tissue weight (i.e. 100 mg in 1 ml). The samples were mildly vortexed for 4 h at room temperature. The NaCl extracts were then desalted with centrifugation using desalting columns (Zeba Spin, Pierce Biotechnology, Cramlington, Northumberland, UK). After desalting the extracts were mixed with 100% acetone (5:1 volume ratio) at −20 °C for 16 h. Proteins were precipitated with centrifugation (16,000 × g for 45 min) and the pellets were vacuum-dried and redissolved in deglycosylation buffer (see below). Subsequently, the aortic samples were incubated with 0.08% SDS (10:1 buffer volume to tissue weight), including proteinase and phosphatase inhibitor cocktails and 25 mm EDTA. The samples were mildly vortexed, to minimize mechanical disruption of the ECM, for 4 h at room temperature. The SDS solution was then removed and stored frozen for later use. Finally, the samples were incubated in a 4 m guanidine-HCl, 50 mm sodium acetate pH 5.8 buffer (5:1 buffer volume to tissue weight), plus proteinase and phosphatase inhibitor cocktails and 25 mm EDTA. pH was adjusted to 5.8 to improve the extractability of vascular proteoglycans (12). The samples were incubated for 48 h at room temperature and vortexed vigorously to enhance the mechanical disruption of matrix proteins. After the denaturing extraction guanidine was removed by mixing the samples with 100% ethanol (5:1 volume ratio) at −20 °C for 16 h. Proteins were then precipitated with centrifugation (16,000 × g for 45 min) and the pellets were washed with 90% ethanol, dried, and redissolved in deglycosylation buffer.
Deglycosylation
Deglycosylation (removal of glycosaminoglycan side chains) of the different extracts was achieved in a 150 mm NaCl, 50 mm sodium acetate pH 6.8 buffer supplemented with proteinase inhibitors and 10 mm EDTA for 16 h at 37 °C. The deglycosylation enzymes (0.05 U) were: 1) chondroitinase ABC from Proteus vulgaris (catalyzes the removal of polysaccharides containing 1⃗4-β-d-hexosaminyl and 1⃗3-β-d-glucuronosyl or 1⃗3-α-l-iduronosyl linkages to disaccharides containing 4-deoxy-β-d-gluc-4-enuronosyl groups. It acts on chondroitin 4-sulfate, chondroitin 6-sulfate, and dermatan sulfate glycosaminoglycan side chains). One unit will liberate 1.0 μm of 2-acetamido-2-deoxy-3-O-(β-d-gluc-4-ene-pyranosyluronic acid)-4-O-sulfo-d-galactose from chondroitin sulfate A or 1.0 μm of 2-acetamido-2-deoxy-3-O-(β-d-gluc-4-ene-pyranosyluronic acid)-6-O-sulfo-d-galactose from chondroitin sulfate C per minute at pH 8.0 at 37 °C. 2) keratanase from Bacteroides fragillis (cleaves internal 1⃗4-β-galactose linkages in unbranched, repeating poly-N-acetyl-lactosamine and keratan sulfate). One unit will release 1.0 μm of reducing sugar per minute at 37 °C and pH 5.8 from bovine corneal keratan sulfate. 3) heparatinase II Flavobacterium heparinum (cleaves heparan sulfate). One unit forms 0.1 μm of unsaturated uronic acid per hour at pH 7.0 at 37 °C. All enzymes were purchased from Sigma-Aldrich. After, deglycosylation the solutions were clarified with centrifugation (16.000 × g for 10 min) to ensure that the samples were free of turbidity and protein concentration was estimated by UV absorbance at 280 nm. Because of the expected contamination with nucleic acids that remain in the tissue after decellularization, in the 4 m guanidine extracts we used the Groves equation [(A280 × 1.55) – (A260 × 0.76)] to account for nucleic acid interference (13).
One-dimensional Electrophoresis
The 0.5 m NaCl and 4 m guanidine extracts were denatured and reduced in sample buffer containing 100 mm Tris, pH 6.8, 40% glycerol, 0.2% SDS, 2% β-mercaptoethanol and 0.02% bromphenol blue and boiled at 96 °C for 10 min. Thirty-five micrograms of protein per sample were loaded and separated on Bis-Tris discontinuous 4–12% polyacrylamide gradient gels (NuPage, Invitrogen). Protein standards were run along side the samples (prestained All Blue, Precision Plus, Bio-Rad Laboratories, Cramlington, Northumberland, UK).
Nanoflow LC-MS/MS
After electrophoresis, gels were stained using the PlusOne Silver staining Kit (GE Healthcare). Silver staining was used for band staining to avoid cross-contamination with fainter gel bands (14) All gel bands were excised in identical parallel positions across lanes and no empty gel pieces were left behind. Subsequently, all gel bands were subjected to in-gel tryptic digestion using an Investigator ProGest (Genomic Solutions) robotic digestion system. Tryptic peptides were separated on a nanoflow LC system (Dionex UltiMate 3000, UK) and eluted with a 10 min gradient (10–25% B in 35 min, 25–40% B in 5 min, 90% B in 10 min, and 2% B in 30 min where A is 2% acetonitrile, 0.1% formic acid in high performance liquid chromatography (HPLC) H2O and B is 90% acetonitrile, 0.1% formic acid in HPLC H2O). The column (Dionex PepMap C18, 25 cm length, 75 μm internal diameter, 3 μm particle size) was coupled to a nanospray source (Picoview) using RePlay (Advion). After the direct liquid chromatography (LC)-MS run, the flow was switched and the portion stored in the capillary of the RePlay device re-analyzed (“replay run”) (15). Spectra were collected from a high-mass accuracy analyzer (LTQ Orbitrap XL, ThermoFisher Scientific) using full ion scan mode over the mass-to-charge (m/z) range 450–1600. Tandem MS (MS/MS) was performed on the top six ions in each MS scan using the data-dependent acquisition mode with dynamic exclusion enabled. MS/MS peaklists were generated by extract_msn.exe and matched to human database (UniProtKB/Swiss-Prot Release 14.6, 20333 protein entries) using SEQUEST v.28 (rev. 13), (Bioworks Browser 3.3.1 SP1, ThermoFisher Scientific) and X! Tandem, (Version 2007.01.01.2). Carboxyamidomethylation of cysteine was chosen as fixed modification and oxidation of methionine as variable modification. The mass tolerance was set at 1.5 AMU for the precursor ions and at 1.0 AMU for fragment ions. Two missed cleavages were allowed. Scaffold (version 2.0.6, Proteome Software Inc., Portland, OR) was used to calculate the normalized spectral counts, and to validate peptide and protein identifications (16, 17). According to the default values in the Scaffold software, peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (16). Only tryptic peptides were included in the analysis. Protein identifications were accepted if they could be established at greater than 99.0% probability (17) with at least two independent peptides and a mass accuracy of ≤10ppm of the precursor ion. ECM proteins identified for the first time by proteomics in the vasculature as well as ECM proteins with low spectral counts were further examined to ensure the quality of the identified spectra.
Western Blotting
Aliquots of the NaCl, guanidine, and SDS extracts were mixed with denaturing sample buffer and boiled. Thirty micrograms of protein per sample were loaded and separated on 4–12% gradient gels as above. Proteins were then transferred on nitrocellulose membranes. Membranes were blocked in 5% fat-free milk powder in PBS and probed for 16 h at 4 °C with primary antibodies to: CD68 (sc-70761, Santa Cruz Biotechnology, Santa Cruz, CA) and smooth muscle actin (A5228, Sigma-Aldrich) (0.08% SDS extracts). Collagen XII (sc-166020), thrombospondin 2 (sc-12313), fibronectin (sc-56391), ACLP (sc-32919), periostin (sc-67233), and MMP-12 (sc-133151) all from Santa Cruz Biotechnology, and tenascin (ab6393) and TIMP-1 (ab38978) both from Abcam (Cambridge, UK) (0.5 m NaCl extracts). The 4 m guanidine extracts were immunoblotted for eSOD (SOD-105) from Stressgen (Victoria, BC, Canada). All antibodies were used at 1:500 dilution in 5% bovine serum albumin. The membranes were treated with the appropriate secondary, horseradish peroxidase (HRP) conjugated antibodies (Dako) at a 1:2000 dilution. Finally, the blots were imaged using enhanced chemilluminenscence (ECL, GE Healthcare) and films were developed on an Xograph processor. The densitometry for the lanes from developed films for collagen XII, tenascin, thrombospondin 2, fibronectin, ACLP, periostin, eSOD, MMP-12, and TIMP-1 was measured using the ImageJ software (v1.42q, NIH, USA).
In Vitro MMP Digestion Assay
Normal aortic specimens taken upon aortotomy (as described above) from patients without connective tissue disorder, were dissected into equal blocks (≈30 mg), and were washed five times in ice-cold, 1× PBS. The tissue blocks were then mixed with 10 volumes MMP reaction buffer (10 mm CaCl2, 120 mm NaCl, 50 mm Trizma base, and 0.05%w/v Brij-35, pH 7.5), which was either used plain or supplemented with human recombinant MMP-9 or human recombinant MMP-12, both derived from murine myeloma cell lines (R&D Systems, Minneapolis, MN). MMP-9 was activated by incubation with 0.5 mm APMA (Aminophenylmercuric acetate) (Sigma-Aldrich) in MMP reaction buffer at 37 °C for 24 h, while MMP-12 was auto-activated in MMP reaction buffer at 37 °C for 30 h, both according to manufacturer's instructions. APMA was removed by desalting through a Zeba Spin (Thermo Scientific) desalting column (MMP-12 was also spun through a desalting column for comparison). The mixtures were supplemented with the proteinase inhibitor mixture to inhibit non-MMP proteinase activity. After the incubation of the aortic tissue with the activated MMPs, the tissue supernatants were deglycosylated, and then immunoblotted using antibodies to collagen XII, thrombospondin 2, tenascin, fibronectin ACLP, and periostin. Activated MMP-12 was also used on human recombinant periostin and thrombospondin 2 both derived from murine myeloma cell lines (R&D Systems) and on purified human fibronectin (Sigma-Aldrich). Ten micrograms of each protein were incubated in MMP reaction buffer, supplemented with proteinase inhibitor mixture, at 37 °C for 16 h with or without MMP-12.
Histochemistry
Small representative pieces from AAA and control aortic samples were placed in OCT compound (BD Healthcare) and used to generate consecutive 7 μm thick sections on a Leica cryostat. The sections were air-dried (30 min) and fixed in 100% acetone for 20 min. Subsequently, sections were either stained with hematoxylin and eosin (H&E) or incubated with primary antibodies. Before immunohistochemistry, the internal peroxidase activity was quenched using a 3% hydrogen peroxide solution in methanol for 15 min. Sections were probed with antibodies to CD68 (sc-70761), ACLP (sc-32919), MMP-12 (sc-133151) from Santa Cruz Biotechnology. Primary antibodies were used at 2 μg/ml. Staining was produced using appropriate HRP-conjugated secondary antibodies, developed with diaminobenzidine purchased from Dako. Negative controls were generated with mouse and rabbit isotype IgG antibodies (Sigma-Aldrich, 2 μg/ml). Nuclei were counterstained blue using Mayer's hematoxylin solution (Sigma-Aldrich) for 5 min. Images were taken by a Zeiss Axioplan 2ie microscope interfaced to AxioVision software (version 3.0.6).
Statistical Analysis
For each clinical sample (CON, n = 3 and AAA, n = 4) and biochemical fraction (0.5 m NaCl and 4 m guanidine), we obtained a technical LC-MS/MS replicate using the RePlay device (Advion). The value reported as spectral count for each protein was calculated as the average ±S.E. of all samples and technical replicates (i.e. CON, n = 6 and AAA, n = 8). All assigned spectra for each protein were used for spectral counting. Before statistical analysis protein spectral counts were normalized using the Scaffold software (version 2.0.6, Proteome Software Inc., Portland, OR) by calculating and averaging the number of identified spectra in each sample, then multiplying the number of spectra assigned to each protein by the ratio of the average spectral count and the number of total spectra in that sample. Protein ambiguity issues were handled using tools available in the Scaffold software and peptides assigned to more than one protein are highlighted in supplemental Table S1. The difference in normalized spectral counts for each identified extracellular protein between control and AAA was computed using two-tailed, unpaired t test (GraphPad Prism 5). A p value of ≤0.05 was considered significant. Only proteins identified with at least two unique peptides were included in the analysis and no outliers were removed. Proteins with a significant difference in normalized spectral counts (p ≤ 0.05) between control and AAA samples were progressively joined into clusters. Pairwise similarity in spectral counts between different proteins (rows) and different samples (columns) was computed using Pearson Correlation Coefficient. The dendrograms and quantitative heat maps were produced using the GenePattern software (version 3.2.3) (18). The relative difference in normalized spectral counts is represented by a color gradient. No outliers were removed. The correlation between the spectral counts and the densitometry values of immunoblots was calculated using Spearman's rank correlation coefficient (GraphPad Prism 5). A p value of ≤0.05 was considered significant.
RESULTS
Characterization of AAA and ECM Extraction
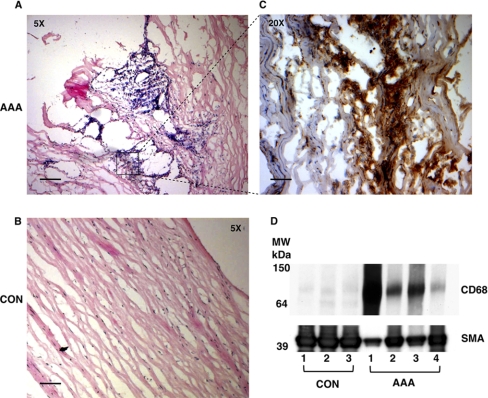
Before proteomics, all tissue specimens were examined histologically. Extensive medial degeneration with few intact elastic laminae, inflammatory infiltrates (indicated by CD68 staining) and loss of smooth muscle cells (SMCs) were evident in AAA. Representative images are shown in Fig. 1A–C. The macrophage marker CD68 was not detected in normal aortic specimens (Fig. 1D). CD68 positivity in AAA inversely correlated to smooth muscle actin (SMA) staining (Fig. 1D). The ECM and its associated proteins were extracted from four AAA and three control (CON) aortas following our established, procedure (10). The proteomics workflow is summarized in Fig. 2. Briefly, in the first step the ionic strength of the 0.5 m NaCl buffer is sufficient to solubilize extracellular proteins that are weakly bound to the ECM (19), including newly synthesized ECM proteins (20), which are not heavily cross-linked, degradation products and other proteins present in the extracellular space. In the second step the samples are decellularized using 0.08% SDS to reduce the highly abundant cellular proteins. Finally, in the third step the extraction of the mature, heavily cross-linked ECM (proteoglycans, glycoproteins, and collagens) is achieved using 4 m guanidine, a strongly denaturing agent.
Fig. 1.
Characterization of clinical specimens. Representative sections of AAA (A) and control aortas (B) were stained with hematoxylin & eosin as well as antibodies against the monocyte/macrophage marker CD68 (C). Immunoreactivity appears in brown and nuclei are counterstained with Mayer's hematoxylin. Scale bar, 5x: 100 μm and 20x: 25 μm. D, In the cellular extracts (0.08% SDS) of AAA, smooth muscle actin (SMA) was inversely correlated to CD68.
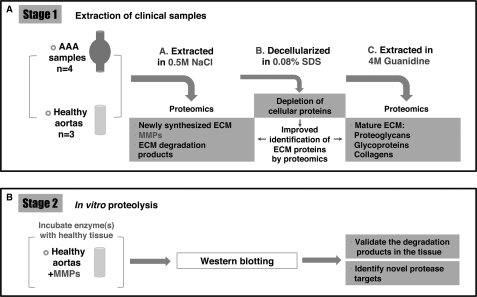
Fig. 2.
Proteomics workflow. The methodology is based on a three-step subfractionation of the vascular proteome (A), which enriches ECM, its associated proteins and degradation products. First, specimens are extracted in 0.5 m NaCl. This buffer solubilizes proteins or protein fragments that are weakly bound to the ECM, including newly synthesized ECM proteins and degradation products. In addition, secreted proteinases such as MMPs are also extracted. Second, specimens are decellularized with 0.08% SDS to deplete cellular proteins. Finally, the salt-insoluble ECM proteins are extracted using 4 m guanidine to solubilize the tightly interacting, cross-linked matrix. In the second stage (B), the identified MMPs are further investigated by incubating healthy tissue with recombinant enzymes to identify novel proteolytic targets and characterize their proteolytic activity in disease.
Proteomics Analysis of the 0.5 m NaCl and 4 m Guanidine Extracts
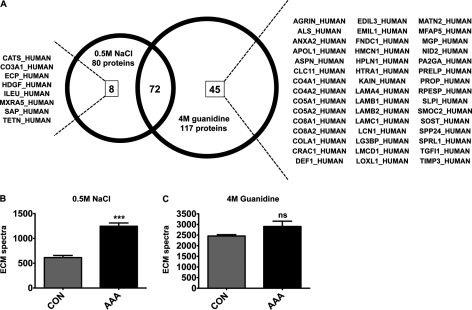
After deglycosylation, proteins in the 0.5 m NaCl and 4 m guanidine extracts were separated by one-dimensional SDS-PAGE (supplemental Fig. S2), subjected to in-gel tryptic digestion, and analyzed by LC-MS/MS using a high mass accuracy ion trap (LTQ-Orbitrap XL, Thermo Fisher Scientific) interfaced to the RePlay device (Advion) to obtain a technical replicate for each biological sample. The entire MS/MS dataset has been deposited in PRIDE, a public repository for mass spectrometry data (http://www.ebi.ac.uk/pride/, accession numbers: 16940–17107). Only proteins passing stringent identification criteria are listed in supplemental Tables S2 and S3 (0.5 m NaCl and 4 m guanidine extracts, respectively). According to the Gene Ontology Annotation database, 80 nonredundant proteins in the 0.5 m NaCl (Table I) and 117 nonredundant proteins in the 4 m guanidine (Table II) fractions were either ECM proteins (proteoglycans, glycoproteins, and collagens) or proteins associated with the ECM such as secreted proteinases, extracellular enzymes, growth factors, cytokines, and lipoproteins. Among the identified extracellular proteins eight were only found in the 0.5 m NaCl extracts whereas 45 proteins were unique to the 4 m guanidine extracts (Fig. 3A). As expected, most unique proteins in the 0.5 m NaCl extracts were secreted factors such as cathepsin S, eosinophil cationic protein, hepatoma-derived growth factor, leukocyte elastase inhibitor, and proactivator polypeptide, whereas the proteins exclusive to the 4 m guanidine extracts were predominantly matrix proteins such as asporin, cartilage acidic protein 1, link protein, prolargin, laminins, matrix Gla protein, sclerostin, and collagen V. Interestingly, spectral counts for ECM proteins were significantly increased in the salt-soluble protein fraction of AAA (p < 0.0001, Fig. 3B). In contrast, in the 4 m guanidine extracts the spectral counts for ECM proteins (Fig. 3C) as well as the number of total spectra in either the 0.5 m NaCl or 4 m guanidine extracts (data not shown) were not significantly different between control and AAA.
Table I. Extracellular matrix and associated proteins identified by proteomics in the 0.5 m NaCl extracts. aThree biological samples performed with two technical replicates each (n = 6). bFour biological samples performed with two technical replicates each (n = 8). cIndicates proteins identified for the first time by proteomics in the vasculature. ND: none detected.
| Protein name | Swiss Prot Accession | MW (kDa) | Mean spectra±S.E. CONa | Mean spectra±S.E. AAAb | p value (t-test) |
|---|---|---|---|---|---|
| Proteoglycans and glycoproteins | |||||
| Thrombospondin-1 | TSP1_HUMAN | 129 | 8 ± 0 | 238 ± 13 | 0.0004 |
| Fibronectin | FINC_HUMAN | 263 | 26 ± 2 | 144 ± 4 | ≥0.0001 |
| Lumican | LUM_HUMAN | 38 | 55 ± 1 | 67 ± 1 | 0.1000 |
| Aortic carboxypeptidase-like protein | AEBP1_HUMAN | 131 | 38 ± 1 | 53 ± 1 | 0.0500 |
| EGF-containing fibulin-like ECM protein 1 | FBLN3_HUMAN | 55 | 34 ± 1 | 50 ± 3 | 0.1200 |
| Tenascin-X | TENX_HUMAN | 464 | 47 ± 3 | 32 ± 1 | 0.1200 |
| Versican | CSPG2_HUMAN | 373 | 20 ± 1 | 20 ± 1 | 0.7600 |
| Tenascin | TENA_HUMAN | 241 | 1 ± 0 | 24 ± 1 | 0.0004 |
| Mimecan | MIME_HUMAN | 34 | 19 ± 0 | 10 ± 0 | 0.0001 |
| Perlecan | PGBM_HUMAN | 469 | 14 ± 3 | 8 ± 2 | 0.0840 |
| Thrombospondin-2 | TSP2_HUMAN | 130 | ND | 19 ± 1 | 0.0003 |
| Fibulin-1 | FBLN1_HUMAN | 77 | 9 ± 0 | 11 ± 0 | 0.4800 |
| Fibrillin-1 | FBN1_HUMAN | 312 | 5 ± 0 | 11 ± 2 | 0.4600 |
| Vitronectin | VTNC_HUMAN | 54 | ND | 15 ± 1 | 0.0034 |
| Fibromodulin | FMOD_HUMAN | 43 | 9 ± 1 | 7 ± 0 | 0.2300 |
| Decorin | PGS2_HUMAN | 40 | 2 ± 0 | 5 ± 0 | 0.0066 |
| Galectin-1 | LEG1_HUMAN | 15 | 5 ± 0 | 6 ± 0 | 0.3800 |
| TGFβ-induced protein ig-h3 | BGH3_HUMAN | 75 | ND | 5 ± 0 | 0.0019 |
| Podocan | PODN_HUMAN | 69 | 3 ± 0 | 2 ± 0 | 0.1700 |
| Cartilage oligomeric matrix protein | COMP_HUMAN | 83 | 1 ± 0 | 3 ± 0 | 0.2500 |
| Biglycan | PGS1_HUMAN | 42 | 1 ± 0 | 2 ± 0 | 0.1600 |
| Fibulin-5 | FBLN5_HUMAN | 50 | 3 ± 0 | 1 ± 0 | 0.0620 |
| Aggrecan | PGCA_HUMAN | 250 | 2 ± 0 | 1 ± 0 | 0.0580 |
| Lactadherin | MFGM_HUMAN | 43 | 1 ± 0 | 2 ± 0 | 0.8300 |
| Galectin-3 | LEG3_HUMAN | 26 | ND | 2 ± 0 | 0.2500 |
| Microfibril-associated glycoprotein 4 | MFAP4_HUMAN | 29 | 2 ± 0 | 1 ± 0 | 0.4600 |
| Periostin | POSTN_HUMAN | 93 | ND | 2 ± 0 | 0.2300 |
| Adlicanc | MXRA5_HUMAN | 312 | ND | 1 ± 0 | 0.0600 |
| Laminin subunit alpha-5 | LAMA5_HUMAN | 400 | 1 ± 0 | 1 ± 0 | 0.7100 |
| Spondin-1 | SPON1_HUMAN | 91 | 2 ± 0 | ND | 0.0390 |
| Nidogen-1 | NID1_HUMAN | 136 | 1 ± 0 | 1 ± 0 | 0.4100 |
| Dermatopontin | DERM_HUMAN | 24 | 1 ± 0 | 1 ± 0 | 0.6300 |
| ECM-associated proteins | |||||
| Extracellular superoxide dismutase | SODE_HUMAN | 26 | 60 ± 3 | 19 ± 2 | 0.0011 |
| Serum amyloid P-component | SAMP_HUMAN | 25 | 14 ± 0 | 38 ± 2 | 0.0190 |
| Pigment epithelium-derived factor | PEDF_HUMAN | 46 | 17 ± 0 | 29 ± 0 | 0.0003 |
| Apolipoprotein A-I | APOA1_HUMAN | 31 | 7 ± 1 | 30 ± 1 | 0.0006 |
| Myeloperoxidase | PERM_HUMAN | 84 | 2 ± 0 | 25 ± 2 | 0.0045 |
| Latent-transforming growth factor beta-binding protein 2 | LTBP2_HUMAN | 195 | 9 ± 1 | 12 ± 1 | 0.1500 |
| Apolipoprotein E | APOE_HUMAN | 36 | 3 ± 0 | 17 ± 1 | 0.0230 |
| Tetranectin | TETN_HUMAN | 23 | 10 ± 0 | 12 ± 0 | 0.3500 |
| Procollagen C-endopeptidase enhancer 1 | PCOC1_HUMAN | 48 | 8 ± 0 | 12 ± 1 | 0.1500 |
| Cathepsin D | CATD_HUMAN | 45 | 5 ± 0 | 9 ± 1 | 0.4200 |
| Ceruloplasmin | CERU_HUMAN | 122 | 2 ± 0 | 12 ± 1 | 0.0018 |
| Tryptase beta-1 | TRYB1_HUMAN | 31 | 7 ± 1 | 7 ± 1 | 0.8600 |
| Clusterin | CLUS_HUMAN | 52 | 2 ± 0 | 12 ± 1 | 0.0003 |
| Insulin-like growth factor-binding protein 7 | IBP7_HUMAN | 29 | 11 ± 0 | 4 ± 0 | 0.0004 |
| Apolipoprotein D | APOD_HUMAN | 21 | 2 ± 0 | 9 ± 1 | 0.0270 |
| Protein S100-A8c | S10A8_HUMAN | 11 | 1 ± 0 | 9 ± 1 | 0.0110 |
| Secreted frizzled-related protein 3 | SFRP3_HUMAN | 36 | 6 ± 0 | 3 ± 0 | 0.1800 |
| Calreticulin | CALR_HUMAN | 48 | 5 ± 0 | 5 ± 0 | 0.9300 |
| Protein S100-A9c | S10A9_HUMAN | 13 | ND | 6 ± 1 | 0.0300 |
| Apolipoprotein A-IV | APOA4_HUMAN | 45 | 4 ± 0 | 4 ± 0 | 0.8900 |
| Metalloproteinase inhibitor 1 | TIMP1_HUMAN | 23 | 1 ± 0 | 4 ± 0 | 0.0019 |
| Latent-TGFβ-binding protein, isoform 1S | LTB1S_HUMAN | 153 | 2 ± 0 | 3 ± 0 | 0.4800 |
| Cathepsin G | CATG_HUMAN | 29 | 1 ± 0 | 2 ± 0 | 0.6500 |
| Apolipoprotein B-100 | APOB_HUMAN | 516 | 1 ± 0 | 3 ± 0 | 0.0220 |
| Leukocyte elastase inhibitor | ILEU_HUMAN | 43 | 2 ± 0 | 2 ± 0 | 0.7900 |
| Target of Nesh-SH3 | TARSH_HUMAN | 119 | 2 ± 0 | 3 ± 0 | 0.4100 |
| Carboxypeptidase-like protein X2 | CPXM2_HUMAN | 86 | 2 ± 0 | 1 ± 0 | 0.9700 |
| Hepatoma-derived growth factor | HDGF_HUMAN | 27 | 2 ± 0 | 1 ± 0 | 0.0590 |
| Macrophage metalloelastasec | MMP12_HUMAN | 54 | ND | 3 ± 0 | 0.0500 |
| Cathepsin Z | CATZ_HUMAN | 34 | 3 ± 0 | 1 ± 0 | 0.0250 |
| Proactivator polypeptide | SAP_HUMAN | 58 | 3 ± 0 | ND | 0.0067 |
| Latent-TGFβ-binding protein 4 | LTBP4_HUMAN | 173 | 2 ± 0 | ND | 0.0600 |
| Secreted frizzled-related protein 1 | SFRP1_HUMAN | 35 | 2 ± 0 | ND | 0.0830 |
| Cathepsin S | CATS_HUMAN | 37 | ND | 1 ± 0 | 0.2300 |
| 72 type IV collagenase | MMP2_HUMAN | 74 | 1 ± 0 | ND | 0.1300 |
| Glia-derived nexin | GDN_HUMAN | 44 | ND | 1 ± 0 | 0.2900 |
| Chymase | CMA1_HUMAN | 27 | ND | 1 ± 0 | 0.3000 |
| Eosinophil cationic proteinc | ECP_HUMAN | 18 | ND | 1 ± 0 | 0.5300 |
| Collagens | |||||
| Collagen alpha-1(XIV) | COEA1_HUMAN | 194 | 71 ± 1 | 50 ± 2 | 0.0009 |
| Collagen alpha-1(XII) | COCA1_HUMAN | 333 | 13 ± 1 | 81 ± 5 | 0.0014 |
| Collagen alpha-3(VI) | CO6A3_HUMAN | 344 | 13 ± 1 | 36 ± 3 | 0.0430 |
| Collagen alpha-1(VI) | CO6A1_HUMAN | 109 | 4 ± 0 | 8 ± 1 | 0.2000 |
| Collagen alpha-1(XVIII) | COIA1_HUMAN | 178 | 5 ± 0 | 7 ± 0 | 0.1700 |
| Collagen alpha-2(VI) | CO6A2_HUMAN | 109 | 2 ± 0 | 3 ± 0 | 0.6100 |
| Collagen alpha-1(I) | CO1A1_HUMAN | 139 | 1 ± 0 | 3 ± 0 | 0.1900 |
| Collagen alpha-1(XV) | COFA1_HUMAN | 142 | 2 ± 0 | 3 ± 0 | 0.5600 |
| Collagen alpha-2(I) | CO1A2_HUMAN | 129 | 1 ± 0 | 3 ± 0 | 0.0130 |
| Collagen alpha-1(III) | CO3A1_HUMAN | 139 | ND | 1 ± 0 | 0.3900 |
Table II. Extracellular matrix and associated proteins identified by proteomics in the 4 m guanidine extracts. aThree biological samples performed with two technical replicates each (n = 6). bFour biological samples performed with two technical replicates each (n = 8). cIndicates proteins identified for the first time by proteomics in the vasculature. ND: none detected.
| Protein name | Swiss Prot Accession | MW (kDa) | Mean spectra±S.E. CONa | Mean spectra±S.E. AAAb | p value (t-test) |
|---|---|---|---|---|---|
| Proteoglycans and glycoproteins | |||||
| Fibronectin | FINC_HUMAN | 263 | 210 ± 15 | 336 ± 16 | ≥0.0001 |
| Perlecan | PGBM_HUMAN | 469 | 323 ± 13 | 176 ± 43 | 0.0280 |
| Versican | CSPG2_HUMAN | 373 | 190 ± 5 | 84 ± 18 | 0.0010 |
| Aggrecan | PGCA_HUMAN | 250 | 154 ± 23 | 28 ± 11 | 0.0003 |
| Prolargin | PRELP_HUMAN | 44 | 76 ± 8 | 86 ± 8 | 0.3100 |
| Tenascin | TENA_HUMAN | 241 | 11 ± 1 | 144 ± 21 | ≥0.0001 |
| Biglycan | PGS1_HUMAN | 42 | 74 ± 4 | 78 ± 10 | 0.4400 |
| Vitronectin | VTNC_HUMAN | 54 | 26 ± 2 | 117 ± 12 | ≥0.0001 |
| Decorin | PGS2_HUMAN | 40 | 35 ± 7 | 70 ± 14 | 0.0430 |
| Clusterin | CLUS_HUMAN | 52 | 34 ± 2 | 69 ± 7 | ≥0.0001 |
| Periostin | POSTN_HUMAN | 93 | 56 ± 6 | 47 ± 6 | 0.6400 |
| Lactadherin | MFGM_HUMAN | 43 | 60 ± 12 | 39 ± 11 | 0.2400 |
| Lumican | LUM_HUMAN | 38 | 41 ± 2 | 51 ± 6 | 0.1400 |
| Thrombospondin-1 | TSP1_HUMAN | 129 | 2 ± 1 | 90 ± 12 | ≥0.0001 |
| Tenascin-X | TENX_HUMAN | 464 | 31 ± 1 | 39 ± 10 | 0.3600 |
| Nidogen-1 | NID1_HUMAN | 136 | 29 ± 4 | 24 ± 7 | 0.6700 |
| Aortic carboxypeptidase-like protein | AEBP1_HUMAN | 131 | 21 ± 2 | 28 ± 6 | 0.2900 |
| Laminin subunit gamma 1 | LAMC1_HUMAN | 178 | 26 ± 6 | 16 ± 6 | 0.3300 |
| Mimecan | MIME_HUMAN | 34 | 15 ± 2 | 23 ± 8 | 0.3200 |
| Laminin subunit beta 2 | LAMB2_HUMAN | 196 | 23 ± 4 | 12 ± 4 | 0.0960 |
| Dermatopontin | DERM_HUMAN | 24 | 17 ± 5 | 16 ± 3 | 0.8100 |
| Annexin A2 | ANXA2_HUMAN | 39 | 13 ± 2 | 19 ± 5 | 0.2600 |
| Hyaluronan and proteoglycan link protein 1 | HPLN1_HUMAN | 40 | 25 ± 9 | 6 ± 2 | 0.0510 |
| Fibrillin-1 | FBN1_HUMAN | 312 | 4 ± 1 | 26 ± 4 | ≥0.0001 |
| Laminin subunit alpha 5 | LAMA5_HUMAN | 400 | 19 ± 4 | 6 ± 2 | 0.0060 |
| Fibulin-5 | FBLN5_HUMAN | 50 | 14 ± 3 | 3 ± 1 | 0.0110 |
| Metalloproteinase inhibitor 3 | TIMP3_HUMAN | 24 | 3 ± 1 | 14 ± 3 | 0.0067 |
| Galectin-1 | LEG1_HUMAN | 15 | 9 ± 3 | 8 ± 2 | 0.9900 |
| Asporin | ASPN_HUMAN | 43 | 2 ± 1 | 12 ± 5 | 0.0800 |
| EMILIN-1 | EMIL1_HUMAN | 107 | 8 ± 3 | 3 ± 1 | 0.1700 |
| Microfibril-associated glycoprotein 4 | MFAP4_HUMAN | 29 | 9 ± 2 | 2 ± 1 | 0.0120 |
| Nidogen-2 | NID2_HUMAN | 151 | 2 ± 0 | 7 ± 3 | 0.0630 |
| Fibromodulin | FMOD_HUMAN | 43 | 5 ± 2 | 3 ± 0 | 0.3700 |
| Fibulin-1 | FBLN1_HUMAN | 77 | 6 ± 2 | 1 ± 0 | 0.0110 |
| Agrin | AGRIN_HUMAN | 215 | 4 ± 2 | 3 ± 1 | 0.5700 |
| Thrombospondin-2 | TSP2_HUMAN | 130 | ND | 7 ± 1 | ≥0.0001 |
| Microfibrillar-associated protein 5 | MFAP5_HUMAN | 20 | 3 ± 1 | 3 ± 1 | 0.5400 |
| Glia-derived nexin | GDN_HUMAN | 44 | ND | 4 ± 1 | 0.0014 |
| Laminin subunit beta 1 | LAMB1_HUMAN | 198 | ND | 4 ± 1 | 0.0110 |
| RPE-spondin | RPESP_HUMAN | 30 | 4 ± 2 | 1 ± 1 | 0.0330 |
| Podocan | PODN_HUMAN | 69 | ND | 3 ± 1 | 0.0700 |
| Matrix Gla proteinc | MGP_HUMAN | 12 | 1 ± 0 | 1 ± 1 | 0.7500 |
| Sclerostin | SOST_HUMAN | 24 | 1 ± 0 | ND | 0.1800 |
| SPARC-like protein 1c | SPRL1_HUMAN | 75 | 1 ± 1 | ND | 0.1100 |
| Cartilage oligomeric matrix protein | COMP_HUMAN | 83 | 1 ± 1 | ND | 0.0930 |
| Matrilin-2 | MATN2_HUMAN | 107 | 1 ± 0 | 1 ± 1 | 0.7900 |
| Galectin-3 | LEG3_HUMAN | 26 | 1 ± 1 | ND | 0.2700 |
| Spondin-1 | SPON1_HUMAN | 91 | 1 ± 1 | ND | 0.1300 |
| Cartilage acidic protein 1c | CRAC1_HUMAN | 71 | ND | 1 ± 1 | 0.2100 |
| SPARC-related modular calcium-binding protein 2 | SMOC2_HUMAN | 50 | 1 ± 0 | ND | 0.3200 |
| Fibronectin type III domain-containing protein 1 | FNDC1_HUMAN | 205 | 1 ± 0 | ND | 0.2300 |
| Laminin subunit alpha 4 | LAMA4_HUMAN | 203 | 1 ± 0 | ND | 0.3000 |
| Hemicentin-1 | HMCN1_HUMAN | 613 | ND | 1 ± 0 | 0.4100 |
| ECM-associated proteins | |||||
| Serum amyloid P component | SAMP_HUMAN | 25 | 39 ± 6 | 91 ± 20 | 0.0220 |
| Apolipoprotein B-100 | APOB_HUMAN | 516 | ND | 96 ± 34 | 0.0470 |
| Apolipoprotein A-I | APOA1_HUMAN | 31 | ND | 50 ± 16 | 0.0230 |
| Serine protease HTRA1 | HTRA1_HUMAN | 51 | 16 ± 4 | 19 ± 7 | 0.7600 |
| TGF-beta induced protein ig-h3 | BGH3_HUMAN | 75 | 8 ± 3 | 27 ± 4 | ≥0.0001 |
| Insulin-like growth factor-binding protein 7 | IBP7_HUMAN | 29 | 20 ± 2 | 9 ± 3 | 0.0430 |
| EGF-containing fibulin-like extracellular matrix protein 1 | FBLN3_HUMAN | 55 | 13 ± 2 | 12 ± 5 | 0.9900 |
| Cathepsin D | CATD_HUMAN | 22 | 8 ± 2 | 14 ± 4 | 0.1090 |
| Extracellular superoxide dismutase | SODE_HUMAN | 26 | 18 ± 3 | 5 ± 2 | 0.0020 |
| Target of Nesh-SH3 | TARSH_HUMAN | 119 | 20 ± 4 | 3 ± 1 | 0.0002 |
| Metalloproteinase inhibitor 3 | TIMP3_HUMAN | 24 | 3 ± 1 | 14 ± 3 | 0.0067 |
| Apolipoprotein E | APOE_HUMAN | 36 | ND | 15 ± 4 | 0.0180 |
| Macrophage metalloelastasec | MMP12_HUMAN | 54 | ND | 15 ± 3 | 0.0033 |
| Apolipoprotein D | APOD_HUMAN | 21 | ND | 13 ± 2 | 0.0035 |
| Latent-transforming growth factor beta-binding protein 2 | LTBP2_HUMAN | 195 | 9 ± 2 | 4 ± 1 | 0.0570 |
| Latent-TGF-beta binding protein, isoform 1S | LTB1S_HUMAN | 153 | 8 ± 2 | 4 ± 2 | 0.3000 |
| Ceruloplasmin | CERU_HUMAN | 122 | ND | 12 ± 5 | 0.0650 |
| Transforming growth factor beta-1-induced transcript 1 protein | TGFI1_HUMAN | 50 | 10 ± 2 | 1 ± 1 | 0.0003 |
| Cathepsin G | CATG_HUMAN | 29 | 3 ± 2 | 7 ± 2 | 0.0450 |
| Chymase | CMA1_HUMAN | 27 | 7 ± 1 | 3 ± 2 | 0.2700 |
| Myeloperoxidase | PERM_HUMAN | 84 | ND | 10 ± 3 | 0.0210 |
| Neutrophil defensin 1 | DEF1_HUMAN | 10 | 1 ± 1 | 9 ± 1 | 0.0003 |
| LIM and cysteine-rich domains protein 1 | LMCD1_HUMAN | 41 | 9 ± 2 | ND | 0.0067 |
| Lysyl oxidase homolog 1 | LOXL1_HUMAN | 63 | 5 ± 2 | 3 ± 1 | 0.2900 |
| Galectin-3-binding protein | LG3BP_HUMAN | 65 | 6 ± 3 | 2 ± 0 | 0.1000 |
| Tryptase beta-1 | TRYB1_HUMAN | 31 | 4 ± 1 | 3 ± 2 | 0.9900 |
| Metalloproteinase inhibitor-3 | TIMP1_HUMAN | 23 | ND | 6 ± 2 | 0.0270 |
| Pigment epithelium-derived factor | PEDF_HUMAN | 46 | 2 ± 1 | 3 ± 1 | 0.6100 |
| Carboxypeptidase-like protein X2 | CPXM2_HUMAN | 86 | 4 ± 2 | ND | 0.1600 |
| Apolipoprotein A-IV | APOA4_HUMAN | 45 | ND | 4 ± 3 | 0.1900 |
| Phospholipase A2, membrane associated | PA2GA_HUMAN | 16 | 2 ± 1 | 2 ± 1 | 0.9300 |
| Apolipoprotein L1 | APOL1_HUMAN | 44 | ND | 4 ± 2 | 0.1900 |
| Protein S100-A9c | S10A9_HUMAN | 13 | ND | 4 ± 1 | 0.0290 |
| Latent-transforming growth factor beta-binding protein 4 | LTBP4_HUMAN | 173 | 3 ± 1 | ND | 0.0550 |
| Procollagen C-endopeptidase enhancer 1 | PCOC1_HUMAN | 48 | 2 ± 1 | ND | 0.1700 |
| Kallistatin | KAIN_HUMAN | 49 | ND | 2 ± 2 | 0.3300 |
| Properdin | PROP_HUMAN | 51 | 1 ± 1 | 1 ± 0 | 0.7500 |
| Calreticulin | CALR_HUMAN | 48 | 1 ± 1 | 1 ± 0 | 0.2900 |
| Antileukoproteinase | SLPI_HUMAN | 14 | 1 ± 0 | ND | 0.0980 |
| Cathepsin Z | CATZ_HUMAN | 34 | 1 ± 1 | ND | 0.4800 |
| Secreted frizzled-related protein 1 | SFRP1_HUMAN | 35 | 2 ± 1 | ND | 0.1200 |
| SPARC-like protein 1 | SPRL1_HUMAN | 75 | 1 ± 1 | ND | 0.1100 |
| Protein S100-A8c | S10A8_HUMAN | 11 | 1 ± 0 | 1 ± 0 | 0.4200 |
| Secreted frizzled-related protein 3 | SFRP3_HUMAN | 36 | 1 ± 1 | ND | 0.1400 |
| C-type lectin domain family 11 member Ac | CLC11_HUMAN | 36 | 1 ± 0 | 1 ± 0 | 0.9500 |
| Lipocalin 1c | LCN1_HUMAN | 19 | ND | 1 ± 0 | 0.1800 |
| Secreted phosphoprotein 24 | SPP24_HUMAN | 24 | ND | 1 ± 1 | 0.1600 |
| EGF-like repeat and discoidin I-like domain- | |||||
| containing protein 3 | EDIL3_HUMAN | 54 | ND | 1 ± 1 | 0.2400 |
| Insulin-like growth-factor binding protein complex acid labilec | ALS_HUMAN | 66 | ND | 1 ± 0 | 0.2900 |
| Collagens | |||||
| Collagen alpha 3(VI) | CO6A3_HUMAN | 344 | 194 ± 35 | 343 ± 37 | 0.0025 |
| Collagen alpha 1(VI) | CO6A1_HUMAN | 109 | 74 ± 10 | 95 ± 16 | 0.1100 |
| Collagen alpha 1(XIV) | COEA1_HUMAN | 194 | 112 ± 17 | 40 ± 9 | 0.0005 |
| Collagen alpha 1(XVIII) | COIA1_HUMAN | 178 | 69 ± 6 | 13 ± 4 | ≥0.0001 |
| Collagen alpha 1(I) | CO1A1_HUMAN | 139 | 36 ± 5 | 41 ± 2 | 0.0580 |
| Collagen alpha 1(XII) | COCA1_HUMAN | 333 | 17 ± 3 | 60 ± 10 | 0.0006 |
| Collagen alpha 2(VI) | CO6A2_HUMAN | 109 | 35 ± 6 | 40 ± 7 | 0.2400 |
| Collagen alpha 2(I) | CO1A2_HUMAN | 129 | 34 ± 2 | 36 ± 2 | 0.0580 |
| Collagen alpha 2(IV) | CO4A2_HUMAN | 168 | 37 ± 6 | 31 ± 12 | 0.7700 |
| Collagen alpha 1(XV) | COFA1_HUMAN | 142 | 24 ± 6 | 17 ± 5 | 0.4300 |
| Collagen alpha 1(V) | CO5A1_HUMAN | 184 | 8 ± 1 | 11 ± 2 | 0.0300 |
| Collagen alpha 1(VIII) | CO8A1_HUMAN | 73 | 7 ± 2 | 9 ± 3 | 0.6200 |
| Collagen alpha 2(VIII) | CO8A2_HUMAN | 67 | 2 ± 1 | 3 ± 1 | 0.3100 |
| Collagen alpha 1(IV) | CO4A1_HUMAN | 161 | 1 ± 1 | 2 ± 1 | 0.4700 |
| Collagen alpha 1(XXI) | COLA1_HUMAN | 99 | 1 ± 0 | 2 ± 1 | 0.2100 |
| Collagen alpha 2(V) | CO5A2_HUMAN | 145 | ND | 1 ± 0 | 0.0820 |
Fig. 3.
Differential solubility of extracellular proteins. A, The Venn diagram displays the number of ECM and associated proteins extracted with either 0.5 m NaCl or 4 m guanidine. The SwissProt entry codes of uniquely identified proteins are given for each extract. All identifications are listed in Tables I and II. B, In the 0.5 m NaCl extracts the spectral counts for ECM proteins were significantly increased in the AAA samples in comparison to control aortas (CON). The total number of identified spectra was similar. C, In the 4 m guanidine extracts neither the spectral counts for ECM proteins nor the total number of spectra were significantly different between controls and AAA. Values are mean ± S.E. *** p ≤ 0.001. Statistical analysis was performed using 2-tailed, unpaired t test.
ECM and Associated Proteins in AAA
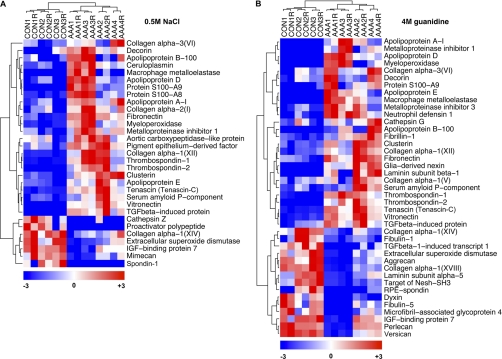
Differences in relative protein abundance between control (CON) and AAA were estimated using normalized spectral counts (Table I and Table II). No outliers were removed. Details can be found in Experimental Procedures. Hierarchical clustering of the differentially expressed extracellular proteins in both the NaCl and guanidine extracts clearly separated the control and AAA samples and highlighted major differences in their ECM composition (Fig. 4). Increased levels of collagens I and VI and the fibril-associated collagen XII, fibronectin, vitronectin, tenascin (alternative name: tenascin-C), thrombospondins 1 and 2, and ACLP were observed in the 0.5 m NaCl extracts of AAA (Fig. 4A, Table I). These proteins, excluding ACLP, were also more abundant in the 4 m guanidine extracts of AAA (Fig. 4B, Table II) indicating that they are components of the mature, salt-insoluble aneurysmal ECM. We also noted a decrease in the spectral counts of the large proteoglycans aggrecan, versican, and perlecan in AAA (Fig. 4B, Table II), perhaps because of proteolysis at an earlier stage of the disease. The reduction in perlecan could explain the loss of extracellular superoxide dismutase (eSOD) (Fig. 4A and 4B, Table I and II) because this antioxidant enzyme is attached to perlecan and other heparan sulfate proteoglycans in extracellular space (21). There was also a reduction in another fibril-associated collagen, type XIV, and the poorly characterized extracellular proteins, retinal pigment epithelium spondin (RPE-spondin), dyxin and target of Nesh-SH3 (Fig. 4B, Table II). Moreover, proteomics identified scarce extracellular proteins including the calgranulins S100A8 and A9 (Figs. 4A and 4B, Table I and II), calcium-binding neutrophil proteins that are involved in macrophage adhesion and transmigration (22), the tissue inhibitor of metalloproteinases 1 (TIMP-1) and macrophage metalloelastase, MMP-12 (Figs. 4A and 4B Table I and Table II). Representative MS/MS spectra from extracellular proteins identified for the first time by proteomics in the vasculature are shown in supplemental Fig. S3.
Fig. 4.
Hierarchical clustering of extracellular proteins. The dendrograms display hierarchical clustering analysis (Pearson correlation coefficient) of the differentially expressed extracellular proteins in the 0.5 m NaCl extracts (A) and in the 4 m guanidine extracts (B). “R” denotes the technical replicate obtained for each clinical sample using the RePlay device. Differences in spectral counts are color-coded as indicated.
Inflammatory Markers in AAA
Besides a clear distinction between control and AAA samples, the hierarchical clustering revealed notable differences among the four AAA specimens (Figs. 4A and 4B). Proteins involved in inflammation, such as apolipoprotein D, ceruloplasmin, myeloperoxidase, S100A8, and A9 were more abundant in samples 1 and 3, whereas the ECM proteins perlecan, aggrecan, versican, collagen XIV, and the matrix associated enzyme eSOD were higher in samples 2 and 4 (Figs. 4A and 4B). These differences reflect the cellular composition observed in the SDS extracts (Fig. 1D). AAA specimens 1 and 3 had a higher macrophage content explaining the abundance of inflammatory factors whereas the loss of smooth muscle cells is consistent with the reduction in ECM components.
Validation of Proteomics and ECM Remodelling
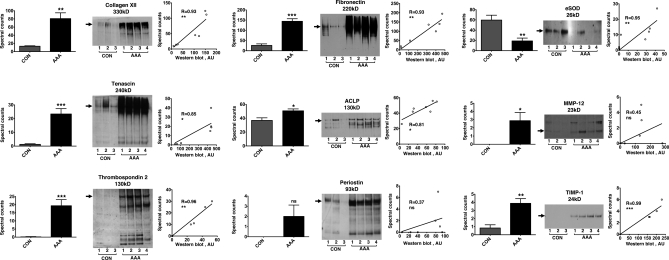
Six glycoproteins covering the detectable range of protein abundance in our samples were selected for further validation. According to spectral counting, collagen XII, tenascin, thrombospondin 2, fibronectin, ACLP, and periostin (Fig. 5, left panels) were increased in the 0.5 m NaCl extracts of the four AAA specimens. Immunoblotting confirmed higher levels in AAA (Fig. 5, middle panels). The loss of eSOD as well as the accumulation of MMP-12 and TIMP-1 were also validated by immunoblotting. Overall, there was a good correlation (Spearman correlation, r ≥ 0.8, p ≤ 0.05) between the protein abundance estimated by spectral counting and Western blotting (Fig. 5, right panels). For periostin and MMP-12 the correlation was not significant because spectral counting is less accurate when protein levels are low (23). Nonetheless, our methodology allowed the detection of matrix macromolecules as well as scarce secreted proteins in complex clinical samples.
Fig. 5.
Validation of ECM remodeling markers. The spectral counts (left panels) for collagen XII, tenascin, thrombospondin 2, fibronectin and ACLP (alternative name: adipocyte enchancer-binding protein 1; AEBP-1) were significantly higher in the 0.5 m NaCl extracts of AAA compared with controls. Values are mean ± S.E. *** p ≤ 0.001, ** p ≤ 0.01, ** p ≤ 0.05, ns not significant. Statistical analysis was performed using two-tailed, unpaired t test. Immunoblotting (middle panels) confirmed increased levels as well as fragmentation of these ECM proteins in AAA. The loss of eSOD and the accumulation of MMP-12 and TIMP-1 in AAA were also validated by immunoblotting. Arrows indicate the expected molecular mass of full-length proteins. With the exception of periostin and MMP-12, there was a high correlation (Spearman correlation r ≥ 0.8, p ≤ 0.05) in protein abundance determined by spectral counting and Western blotting (right panels).
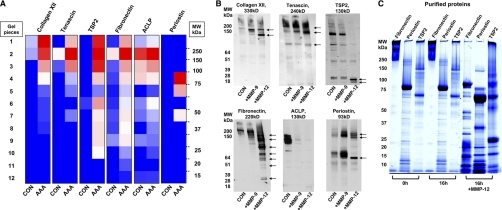
ECM Remodelling and the Role of MMP-12
The Western blots presented in Fig. 5 highlight the fragmentation of collagen XII, tenascin, thrombospondin 2, fibronectin, ACLP, and periostin in the AAA samples. Indeed, detailed examination of the gel-LC-MS/MS data revealed that a high number of MS/MS spectra were identified in gel segments below the expected molecular weight of these proteins (Fig. 6A) confirming the presence of proteolytic products in the aneurysmal tissue. Although various MMPs, including MMP-12, have been linked to matrix proteolysis during aneurysm formation (24–26), MMP-12 was the only metalloproteinase that was significantly increased in our proteomics analysis (Fig. 4). To investigate the possible relationship between the observed fragmentation of these glycoproteins and the presence of MMP-12, we incubated macroscopically normal, human aortic specimens with recombinant MMP-12 in vitro (Fig. 6B). MMP-9, another metalloproteinase implicated in AAA pathology (27), served as an additional control. MMP-12 induced extensive fragmentation of fibronectin and cleaved collagen XII, tenascin, and periostin. Thrombospondin 2 was degraded producing two small cleavage products (≈20 kDa). In comparison to MMP-9, MMP-12 had more pronounced effects on the degradation of fibronectin, tenascin, collagen XII, and thrombospondin 2, whereas MMP-9 induced the release of full-length periostin from the aortic matrix, presumably by limited proteolysis. Intriguingly, treatment with both proteinases resulted in an almost complete loss of ACLP immunostaining. With the exception of fibronectin (28), all other glycoproteins are novel substrates for MMP-12. The proteinase had a dose (0–50 pm, supplemental Fig. S4) and time-dependent (2–16h, supplemental Fig. S5) effect on these glycoproteins in the aortic explants. Importantly, its proteolytic effect on the tissue was almost completely abolished by 10 mm EDTA (supplemental Fig. S6). In addition, MMP-12 was able to cleave human recombinant periostin and thrombospondin 2 in vitro (Fig. 6C). Human fibronectin, a known MMP-12 substrate, was used as a positive control.
Fig. 6.
Novel MMP-12 targets. A, The color-coded heat map demonstrates the number of identified spectra for the 6 glycoproteins in the 0. 5 m NaCl extracts in control and AAA samples and their molecular weight distribution after SDS-PAGE. B, A normal aortic specimen was dissected into three equal blocks and treated with either 50 pm MMP-9, 50 pm MMP-12, or left untreated (CON) in MMP reaction buffer for 24 h at 37 °C. The resulting supernatants were analyzed by immunoblotting for collagen XII, tenascin, thrombospondin 2 (TSP2), fibronectin, ACLP, and periostin. Arrows indicate proteolytic fragments generated by MMP-12 treatment. C, 10 μg human purified fibronectin and 10 μg human recombinant periostin and TSP2 were incubated for 16h at 37 °C either in plain MMP reaction buffer or supplemented with 50 pm MMP-12 (+MMP-12). The protein mixtures were separated by one-dimensional SDS-PAGE and stained in a 0.05% w/v Coomasie G-250, 5% v/v glacial acetic acid solution to monitor substrate degradation.
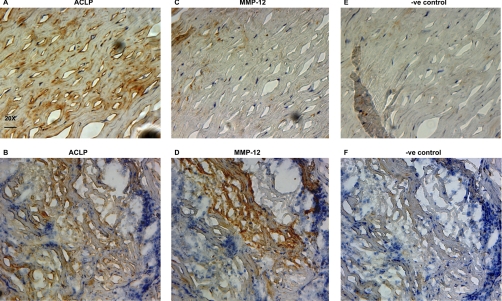
ACLP and MMP-12
The proteolytic effect of MMP-12 on ACLP was very pronounced and considering the lack of knowledge on this collagen-associated glycoprotein (29) in aneurysms, we sought to examine its localization by immunohistochemistry. In AAA, extracellular ACLP staining was present both in the hypocellular regions of the media (Fig. 7A), where tissue architecture is more preserved with intact elastic laminae and absence of inflammatory infiltrates, and in the amorphous, inflammatory areas, where connective tissue loss is evident (Fig. 7B). In consecutive serial sections, MMP-12 staining was predominantly found in the inflammatory areas (Figs. 7C and 7D) and there was an inverse correlation in the staining intensity of the two proteins, supporting the proteolytic effect of MMP-12 on ACLP in the tissue. Negative controls are shown in Figs. 7E and 7F.
Fig. 7.
Immunohistochemical staining for ACLP and MMP-12. Consecutive serial sections of AAA tissue were stained with antibodies to ACLP (A and B) or MMP-12 (C and D). ACLP was found in both the hypocellular areas with the remaining elastic laminae (A) and in the amorphous, inflammatory regions of the media with evidence of connective tissue destruction (B). MMP-12 was predominantly found in the inflammatory regions (D). Positive immunostaining is indicated by brown color obtained with diaminobenzidine (DAB) development of horseradish peroxidase-conjugated secondary antibodies. Negative controls were incubated with isotype control antibodies (E and F). Nuclei were counterstained with Mayer's hematoxylin. 20X magnification. Scale bar: 25 μm.
DISCUSSION
The destruction of the aortic connective tissue in aneurysms is driven by a severe inflammatory reaction and involves the excessive degradation of the aortic ECM. However, the abundance of cellular proteins in the aorta and the biochemical properties of matrix components have so far limited the characterization of the ECM in vascular pathologies (30). In this study we employed a solubility-based, protein subfractionation methodology that enabled the detailed characterization of the ECM and its associated proteins in AAA. Importantly, this approach offered novel insights into the pathological remodeling process and allowed us to relate changes in the turnover of certain ECM proteins to the increased levels of MMP-12 in AAA.
Changes in the Extracellular Proteome of AAA
Proteomics revealed marked differences in the ECM composition between control and AAA specimens (Figs. 4 and 5). As a consequence of the pathological remodeling, the salt-soluble protein fraction of aneurysms was enriched in ECM fragmentation products. Many of the identified protein changes have not been previously reported in AAA, including collagen XII, thrombospondin 2, periostin, and ACLP. Collagen XII is a fibril-associated collagen and accumulates in tissues exposed to high tensile forces (31) whereas periostin and thrombospondin 2 are glycoproteins involved in cellular adhesion and spreading (32, 33). ACLP is an extracellular glycoprotein, which is known to accumulate in the ECM associated with collagen fibers (29), but its function is not well understood. Recently, Schissel et al., showed that ACLP is a positive regulator of fibrosis in the injured lung (34). Fibronectin is the primary component of granulation tissue after wound healing (35) and this may explain its accumulation in the injured aortic wall. Increased tenascin mRNA expression has been previously reported in AAA and was associated with the accumulation of macrophages (36). Our proteomics data also revealed the fragmentation of these ECM proteins (Fig. 5). Degradation products reflect the pathological remodeling process in AAA.
In the 4 m guanidine extracts of AAA there was a reduction in the three major proteoglycans versican, perlecan, and aggrecan, as well as link protein, the glycoprotein which links aggregating proteoglycans to hyaluronic acid. Although a previous study demonstrated that versican was reduced in aneurysms (37), not much is known about the contribution of perlecan and aggrecan to this pathology (38, 39). The loss of these major proteoglycans is likely to have major impact on SMC attachment, proliferation, and migration but it will also affect the binding and storage of various regulatory molecules in the extracellular space including extracellular enzymes and growth factors (40). For example, extracellular superoxide dismutase (eSOD) was reduced in AAA (Fig. 5). This potent antioxidant enzyme is located in the vascular extracellular space bound to subendothelial heparan sulfate proteoglycans such as perlecan (21). A loss of these proteoglycans could result in reduced retention of eSOD and other extracellular proteins, thereby affecting the overall integrity of the vascular wall.
MMP-12 in AAA
Studies investigating the proteolytic activity in human tissues rely on mRNA expression, immunostaining or zymography and tend to focus on individual proteinases and their inhibitors. MMPs -1, -2, -3, -7, -9, and -12 have previously been detected in macrophage-rich areas of AAA and their proteolytic activity has been linked to aneurysm formation (24–26). Interestingly, little is known about their relative abundance in AAA. A limitation of antibody-based detection is that the staining intensity is not only dependent on the protein amount but also on the binding affinity and specificity of the antibody. Hence, staining with different antibodies cannot be directly compared. Instead, a proteomics approach provides an unbiased assessment of proteinases and potential degradation products in diseased tissue. Whereas previous proteomics studies primarily focused on cellular proteins and failed to identify MMPs in AAA (8, 9), our subfractionation method enabled the detection of MMP-12. This does not exclude the presence of other MMPs, but suggests that MMP-12 is probably more abundant and underscores its role in the pathology of AAA (41, 42). MMP-2, cathepsins D, G, S, and Z, and the mast cell proteinases chymase and tryptase β-1 were also identified (Table I and Table II), but only cathepsin G was significantly increased in AAA (Fig. 4B). Interestingly, cathepsin G was recently shown to degrade intraluminal thrombi in aneurysms (43).
Novel Targets of MMP-12 in the Aortic Tissue
Apart from its well-established elastolytic and collagenolytic properties (24), little is known about the ability of MMP-12 to degrade other substrates in the aorta. Thus far, the presence of proteolytic products from known MMP targets has been mainly studied by immunostaining using neoepitope antibodies against the cleaved substrates (44). Our subfractionation methodology allowed a more comprehensive analysis of the ECM remodelling in AAA. For instance, in the salt-soluble protein fraction, we observed not only an increase but also a partial degradation of collagen XII, fibronectin, tenascin, thrombospondin 2, ACLP, and periostin. When healthy aortic tissue was incubated with recombinant MMP-12, the proteinase was able to cleave these substrates. Its effect, at least in vitro, was more potent than MMP-9, suggesting that MMP-12 might be responsible for the degradation of these proteins in AAA. Of course, the fragmentation pattern after MMP-12 treatment was not identical to the laddering observed in the 0.5 m NaCl extracts. This is expected because the proteolytic activity in tissues is a highly dynamic process regulated by the interplay of MMPs with TIMPs and other inhibitors. In agreement with previous studies (45), we found that TIMP-1 and -3 were significantly increased in AAAs (Figs. 4A and 4B) indicating a compensatory anticatabolic response. One of the fragmented proteins in AAA was ACLP (Fig. 5). MMP-12 was able to degrade it in vitro (Fig. 6B). Further analysis by immunohistochemistry confirmed an inverse correlation of the staining intensities for ACLP and MMP-12 in the inflammatory regions of AAA (Figs. 7B and 7D), providing additional evidence that the proteinase could target ACLP in vivo.
Strengths and Limitations
The gel-based separation employed in this study, preserves the molecular mass of proteins and allows the assessment of proteolytic degradation in diseased tissues. This key information on protein fragmentation is lost in conventional shotgun proteomics. The label-free quantitation based on spectral counting allows the relative estimation of protein abundance (46) and shows a good correlation with approaches based on isotopic labels (47) and precursor peak area intensity measurements (23). However, its accuracy is limited to large differences and proteins with reasonable abundance. Hence, caution should be taken in the interpretation of modest changes and the quantitation of scarce proteins (48). In this study we analyzed simplified subproteomes and nine extracellular proteins were validated by Western blotting to ensure the accuracy of our proteomics findings. The observed changes between healthy and aneurysmal tissues were pronounced allowing reliable conclusions to be made despite the low number of clinical specimens available. Because AAAs are currently treated by endovascular stenting rather than open repair interventions, surgical specimens are increasingly rare. Additionally, healthy control tissue could only be obtained from the thoracic part of the aorta.
Conclusion
This study is the first to employ a solubility-based extraction procedure to study the ECM composition in human AAA by proteomics. Our methodology led to the identification of novel candidate markers of pathological tissue remodeling and identified new proteolytic targets of MMP-12, one of the key proteinases in AAA.
Footnotes
* This work was funded by the British Heart Foundation and the Oak Foundation. M.M. is a Senior Fellow of the British Heart Foundation.
 This article contains supplemental Figs. S1 to S6 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S6 and Tables S1 to S3.
1 The abbreviations used are:
- AAA
- Abdominal aortic aneurysm
- ACLP
- Aortic carboxypeptidase-like protein
- ECM
- Extracellular matrix
- eSOD
- extracellular superoxide dismutase
- LTQ
- Linear trap quadrupole
- MMP
- Matrix metalloproteinase
- TIMP
- Tissue inhibitor of metalloproteinases
- LC
- liquid chromatography
- MS/MS
- tandem MS.
REFERENCES
- 1. Alcorn H. G., Wolfson S. K., Jr., Sutton-Tyrrell K., Kuller L. H., O'Leary D. (1996) Risk factors for abdominal aortic aneurysms in older adults enrolled in The Cardiovascular Health Study. Arterioscler. Thromb. Vasc. Biol. 16, 963–970 [DOI] [PubMed] [Google Scholar]
- 2. Baumgartner I., Hirsch A. T., Abola M. T., Cacoub P. P., Poldermans D., Steg P. G., Creager M. A., Bhatt D. L. (2008) Cardiovascular risk profile and outcome of patients with abdominal aortic aneurysm in out-patients with atherothrombosis: data from the Reduction of Atherothrombosis for Continued Health (REACH) Registry. J Vasc Surg 48, 808–814 [DOI] [PubMed] [Google Scholar]
- 3. Golledge J., Tsao P. S., Dalman R. L., Norman P. E. (2008) Circulating markers of abdominal aortic aneurysm presence and progression. Circulation 118, 2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hellenthal F. A., Buurman W. A., Wodzig W. K., Schurink G. W. (2009) Biomarkers of AAA progression. Part 1: extracellular matrix degeneration. Nat Rev Cardiol 6, 464–474 [DOI] [PubMed] [Google Scholar]
- 5. Shiraya S., Miyake T., Aoki M., Yoshikazu F., Ohgi S., Nishimura M., Ogihara T., Morishita R. (2009) Inhibition of development of experimental aortic abdominal aneurysm in rat model by atorvastatin through inhibition of macrophage migration. Atherosclerosis 202, 34–40 [DOI] [PubMed] [Google Scholar]
- 6. Shah P. K. (1997) Inflammation, metalloproteinases, and increased proteolysis: an emerging pathophysiological paradigm in aortic aneurysm. Circulation 96, 2115–2117 [DOI] [PubMed] [Google Scholar]
- 7. Shimizu K., Shichiri M., Libby P., Lee R. T., Mitchell R. N. (2004) Th2-predominant inflammation and blockade of IFN-gamma signaling induce aneurysms in allografted aortas. J. Clin. Invest. 114, 300–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liao M., Liu Z., Bao J., Zhao Z., Hu J., Feng X., Feng R., Lu Q., Mei Z., Liu Y., Wu Q., Jing Z. (2008) A proteomic study of the aortic media in human thoracic aortic dissection: implication for oxidative stress. J. Thorac. Cardiovasc. Surg. 136, 65–72, 72,, e61–63 [DOI] [PubMed] [Google Scholar]
- 9. Pilop C., Aregger F., Gorman R. C., Brunisholz R., Gerrits B., Schaffner T., Gorman J. H., 3rd, Matyas G., Carrel T., Frey B. M. (2009) Proteomic analysis in aortic media of patients with Marfan syndrome reveals increased activity of calpain 2 in aortic aneurysms. Circulation 120, 983–991 [DOI] [PubMed] [Google Scholar]
- 10. Didangelos A., Yin X., Mandal K., Baumert M., Jahangiri M., Mayr M. (2010) Proteomics characterization of extracellular space components in the human aorta. Mol. Cell Proteomics 9, 2048–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mayr M., Zhang J., Greene A. S., Gutterman D., Perloff J., Ping P. (2006) Proteomics-based development of biomarkers in cardiovascular disease: mechanistic, clinical, and therapeutic insights. Mol. Cell Proteomics 5, 1853–1864 [DOI] [PubMed] [Google Scholar]
- 12. Eisenstein R., Larsson S. E., Kuettner K. E., Sorgente N., Hascal V. C. (1975) The ground substance of the arterial wall. Part 1. Extractability of glycosaminoglycans and the isolation of a proteoglycan from bovine aorta. Atherosclerosis 22, 1–17 [DOI] [PubMed] [Google Scholar]
- 13. Groves W. E., Davis F. C., Jr., Sells B. H. (1968) Spectrophotometric determination of microgram quantities of protein without nucleic acid interference. Anal. Biochem. 22, 195–210 [DOI] [PubMed] [Google Scholar]
- 14. Yin X., Cuello F., Mayr U., Hao Z., Hornshaw M., Ehler E., Avkiran M., Mayr M. (2010) Proteomics analysis of the cardiac myofilament subproteome reveals dynamic alterations in phosphatase subunit distribution. Mol. Cell Proteomics 9, 497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Waanders L. F., Almeida R., Prosser S., Cox J., Eikel D., Allen M. H., Schultz G. A., Mann M. (2008) A novel chromatographic method allows on-line reanalysis of the proteome. Mol. Cell Proteomics 7, 1452–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 17. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 18. Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J. P. (2006) GenePattern 2.0. Nat. Genet. 38, 500–501 [DOI] [PubMed] [Google Scholar]
- 19. Mason R. M., Mayes R. W. (1973) Extraction of cartilage protein-polysaccharides with inorganic salt solutions. Biochem. J. 131, 535–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilson R., Diseberg A. F., Gordon L., Zivkovic S., Tatarczuch L., Mackie E. J., Gorman J. J., Bateman J. F. (2010) Comprehensive profiling of cartilage extracellular matrix formation and maturation using sequential extraction and label-free quantitative proteomics. Mol. Cell Proteomics 9, 1296–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karlsson K., Sandström J., Edlund A., Marklund S. L. (1994) Turnover of extracellular-superoxide dismutase in tissues. Lab. Invest. 70, 705–710 [PubMed] [Google Scholar]
- 22. Croce K., Gao H., Wang Y., Mooroka T., Sakuma M., Shi C., Sukhova G. K., Packard R. R., Hogg N., Libby P., Simon D. I. (2009) Myeloid-related protein-8/14 is critical for the biological response to vascular injury. Circulation 120, 427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 24. Curci J. A., Liao S., Huffman M. D., Shapiro S. D., Thompson R. W. (1998) Expression and localization of macrophage elastase (matrix metalloproteinase-12) in abdominal aortic aneurysms. J. Clin. Invest. 102, 1900–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knox J. B., Sukhova G. K., Whittemore A. D., Libby P. (1997) Evidence for altered balance between matrix metalloproteinases and their inhibitors in human aortic diseases. Circulation 95, 205–212 [DOI] [PubMed] [Google Scholar]
- 26. Pyo R., Lee J. K., Shipley J. M., Curci J. A., Mao D., Ziporin S. J., Ennis T. L., Shapiro S. D., Senior R. M., Thompson R. W. (2000) Targeted gene disruption of matrix metalloproteinase-9 (gelatinase B) suppresses development of experimental abdominal aortic aneurysms. J. Clin. Invest. 105, 1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Newman K. M., Ogata Y., Malon A. M., Irizarry E., Gandhi R. H., Nagase H., Tilson M. D. (1994) Identification of matrix metalloproteinases 3 (stromelysin-1) and 9 (gelatinase B) in abdominal aortic aneurysm. Arterioscler. Thromb. 14, 1315–1320 [DOI] [PubMed] [Google Scholar]
- 28. Fu J. Y., Lyga A., Shi H., Blue M. L., Dixon B., Chen D. (2001) Cloning, expression, purification, and characterization of rat MMP-12. Protein Expr. Purif. 21, 268–274 [DOI] [PubMed] [Google Scholar]
- 29. Layne M. D., Endege W. O., Jain M. K., Yet S. F., Hsieh C. M., Chin M. T., Perrella M. A., Blanar M. A., Haber E., Lee M. E. (1998) Aortic carboxypeptidase-like protein, a novel protein with discoidin and carboxypeptidase-like domains, is up-regulated during vascular smooth muscle cell differentiation. J. Biol. Chem. 273, 15654–15660 [DOI] [PubMed] [Google Scholar]
- 30. Mayr M., Chung Y. L., Mayr U., Yin X., Ly L., Troy H., Fredericks S., Hu Y., Griffiths J. R., Xu Q. (2005) Proteomic and metabolomic analyses of atherosclerotic vessels from apolipoprotein E-deficient mice reveal alterations in inflammation, oxidative stress, and energy metabolism. Arterioscler. Thromb. Vasc. Biol. 25, 2135–2142 [DOI] [PubMed] [Google Scholar]
- 31. Trächslin J., Koch M., Chiquet M. (1999) Rapid and reversible regulation of collagen XII expression by changes in tensile stress. Exp. Cell Res. 247, 320–328 [DOI] [PubMed] [Google Scholar]
- 32. Gillan L., Matei D., Fishman D. A., Gerbin C. S., Karlan B. Y., Chang D. D. (2002) Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 62, 5358–5364 [PubMed] [Google Scholar]
- 33. Lopes N., Gregg D., Vasudevan S., Hassanain H., Goldschmidt-Clermont P., Kovacic H. (2003) Thrombospondin 2 regulates cell proliferation induced by Rac1 redox-dependent signaling. Mol. Cell. Biol. 23, 5401–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schissel S. L., Dunsmore S. E., Liu X., Shine R. W., Perrella M. A., Layne M. D. (2009) Aortic carboxypeptidase-like protein is expressed in fibrotic human lung and its absence protects against bleomycin-induced lung fibrosis. Am. J. Pathol. 174, 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Valenick L. V., Hsia H. C., Schwarzbauer J. E. (2005) Fibronectin fragmentation promotes alpha4beta1 integrin-mediated contraction of a fibrin-fibronectin provisional matrix. Exp. Cell Res. 309, 48–55 [DOI] [PubMed] [Google Scholar]
- 36. Satta J., Soini Y., Pöllänen R., Pääkko P., Juvonen T. (1997) Tenascin expression is associated with a chronic inflammatory process in abdominal aortic aneurysms. J. Vasc. Surg. 26, 670–675 [DOI] [PubMed] [Google Scholar]
- 37. Theocharis A. D., Tsolakis I., Hjerpe A., Karamanos N. K. (2001) Human abdominal aortic aneurysm is characterized by decreased versican concentration and specific downregulation of versican isoform V(0). Atherosclerosis 154, 367–376 [DOI] [PubMed] [Google Scholar]
- 38. Melrose J., Whitelock J., Xu Q., Ghosh P. (1998) Pathogenesis of abdominal aortic aneurysms: possible role of differential production of proteoglycans by smooth muscle cells. J. Vasc. Surg. 28, 676–686 [DOI] [PubMed] [Google Scholar]
- 39. Baas A. F., Medic J., van't Slot R., de Vries J. P., van Sambeek M. R., Verhoeven E. L., Boll B. P., Grobbee D. E., Wijmenga C., Blankensteijn J. D., Ruigrok Y. M. (2010) The intracranial aneurysm susceptibility genes HSPG2 and CSPG2 are not associated with abdominal aortic aneurysm. Angiology 61, 238–242 [DOI] [PubMed] [Google Scholar]
- 40. Vincent T., Hermansson M., Bolton M., Wait R., Saklatvala J. (2002) Basic FGF mediates an immediate response of articular cartilage to mechanical injury. Proc. Natl. Acad. Sci. U.S.A. 99, 8259–8264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Longo G. M., Buda S. J., Fiotta N., Xiong W., Griener T., Shapiro S., Baxter B. T. (2005) MMP-12 has a role in abdominal aortic aneurysms in mice. Surgery 137, 457–462 [DOI] [PubMed] [Google Scholar]
- 42. Wang Y., Ait-Oufella H., Herbin O., Bonnin P., Ramkhelawon B., Taleb S., Huang J., Offenstadt G., Combadière C., Rénia L., Johnson J. L., Tharaux P. L., Tedgui A., Mallat Z. (2010) TGF-beta activity protects against inflammatory aortic aneurysm progression and complications in angiotensin II-infused mice. J. Clin. Invest. 120, 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dejouvencel T., Féron D., Rossignol P., Sapoval M., Kauffmann C., Piot J. M., Michel J. B., Fruitier-Arnaudin I., Meilhac O. (2010) Hemorphin 7 reflects hemoglobin proteolysis in abdominal aortic aneurysm. Arterioscler. Thromb. Vasc. Biol. 30, 269–275 [DOI] [PubMed] [Google Scholar]
- 44. Sukhova G. K., Schönbeck U., Rabkin E., Schoen F. J., Poole A. R., Billinghurst R. C., Libby P. (1999) Evidence for increased collagenolysis by interstitial collagenases-1 and -3 in vulnerable human atheromatous plaques. Circulation 99, 2503–2509 [DOI] [PubMed] [Google Scholar]
- 45. Mao D., VanVickle S. J., Curci J. A., Thompson R. W. (1999) Expression of matrix metalloproteinases and TIMPs in human abdominal aortic aneurysms. Ann. Vasc. Surg. 13, 236–237 [DOI] [PubMed] [Google Scholar]
- 46. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 47. Zybailov B., Coleman M. K., Florens L., Washburn M. P. (2005) Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 77, 6218–6224 [DOI] [PubMed] [Google Scholar]
- 48. Simper D., Mayr U., Urbich C., Zampetaki A., Prokopi M., Didangelos A., Saje A., Mueller M., Benbow U., Newby A. C., Apweiler R., Rahman S., Dimmeler S., Xu Q., Mayr M. (2010) Comparative proteomics profiling reveals role of smooth muscle progenitors in extracellular matrix production. Arterioscler. Thromb. Vasc. Biol. 30, 1325–1332 [DOI] [PubMed] [Google Scholar]