Abstract
The gene encoding the miR-34a microRNA is a transcriptional target of the p53 tumor suppressor protein and subject to epigenetic inactivation in colorectal cancer and numerous other tumor types. Here, we combined pulsed SILAC (pSILAC) and microarray analyses to identify miR-34a-induced changes in protein and mRNA expression. pSILAC allowed to quantify the de novo protein synthesis of 1206 proteins after activation of a conditional miR-34a allele in a colorectal cancer cell line. ∼19% of the detected proteins were differentially regulated, with 113 proteins being down- and 115 up-regulated. The proteins with a miR-34a seed-matching-sequence in the 3′-untranslated region (UTR) of the corresponding mRNA showed a clear bias toward translational repression. Proteins involved in DNA replication, e.g. the MCM proteins, and cell proliferation, were over-represented among indirectly down-regulated proteins lacking a miR-34a seed-match. The decrease in de novo protein synthesis of direct miR-34a targets correlated with reduced levels of the corresponding mRNA in most cases, indicating an interdependence of both types of regulation. In addition, 43 mRNAs encoding proteins not detected by pSILAC were down-regulated after miR-34a expression and contained miR-34a seed-matches. The direct regulation of selected miR-34a target-mRNAs was confirmed using reporter assays. Via down-regulation of the proteins encoded by these mRNAs miR-34a presumably inhibits glycolysis (LDHA), WNT-signaling (LEF1), invasion/migration (AXL) and lipid metabolism (ACSL1, ACSL4). Furthermore, miR-34a may activate p53 by inhibiting its acetylation (MTA2, HDAC1) and degradation (YY1). In summary, miR-34a presumably participates in multiple tumor suppressive pathways by directly and indirectly suppressing the expression of numerous, critical proteins.
The transcription factor encoded by the p53 tumor suppressor gene is activated by numerous cellular insults such as γ-irradiation or deregulated oncogene expression, which have the commonality of inducing DNA damage. Activated p53 regulates numerous genes, which mediate tumor suppressive processes as inhibition of cell-cycle progression and induction of apoptosis (1). Loss of p53 function is commonly observed during tumor development. Apart from regulating protein-coding genes p53 also controls microRNA (miRNA)1 encoding genes (2–3). miRNAs represent an abundant class of small ∼21-nucleotide-long, noncoding RNAs involved in post-transcriptional control of gene expression. The influence of miRNAs on gene expression is predicted to be widespread, with more than 60% of human protein coding genes being subject to regulation by miRNAs (4). Among the p53-regulated miRNAs miR-34a seems to display the most pronounced induction by p53 (5–10). Ectopic expression of miR-34a induces apoptosis, senescence, cell cycle arrest and inhibits migration and invasion (2–3, 11). Therefore, miR-34a may be an important mediator of p53's tumor-suppressive activities. Interestingly, miR-34a is silenced by CpG methylation in numerous types of tumors, among them colorectal cancer, and may therefore itself represent a tumor suppressor gene (10, 12). Several mRNAs have been shown to be direct miR-34a targets (11), which encode factors required for G1/S transition (c-MYC, E2F, CDK4, CDK6), anti-apoptotic proteins (Bcl2, SIRT1), but also proteins involved in invasion (c-MET). However, it is likely that miR-34a regulates additional, as yet unconfirmed targets, because bioinformatic predictions suggest that several hundred mRNAs contain matches to the miR-34a seed sequence.
MiRNAs regulate their targets via association of a ∼7 nucleotide stretch, the so-called seed-sequence, located in their 5′-portion with a complementary sequence in the 3′-UTR of the target mRNA (13, 14). Additional base pairing may occur via nucleotides in the middle and 3′-portion of the miRNA. Along with binding of the miRNA to target mRNAs the RISC/Ago2 complex is recruited to the mRNA. This complex mediates inhibition of translation initiation through interfering with eIF4F-cap recognition and 40S small ribosomal subunit recruitment and/or by enhancing mRNA degradation through recruitment of the CCR4-NOT1 deadenylase complex. Because the relatively short seed-region is the primary determinant of target recognition, a single miRNA presumably regulates dozens or even hundreds of target mRNAs (15). However, because of the shortness of the seed-regions bioinformatic predictions of miRNA/mRNA interactions are not reliable for the identification of biologically relevant miRNA targets. Therefore, several attempts have been made to identify the messenger RNAs that are subject to regulation by a specific miRNA using unbiased genome- or proteome-wide experimental approaches (16).
Because targeting of mRNAs through miRNAs often leads to degradation of the respective mRNA, microarray analysis of mRNA levels after ectopic expression of a miRNA can be used to identify miRNA targets. However, this approach is limited as it cannot detect miRNA targets that are solely regulated at the level of translational repression. Assuming that miRNAs in most cases only cause modest decreases in protein translation, the miRNA-mediated regulation of proteins with long half-lives may not be detected by measuring steady-state protein levels using standard proteomic quantification as SILAC (stable isotope labeling by amino acids in cell culture) (17). This problem was solved by the introduction of pSILAC (pulsed SILAC), which facilitated the quantification of differences in protein translation rates caused by miRNAs (18).
Here we describe a global analysis of the effect of miR-34a expression on mRNA and protein expression using a combined microarray and pSILAC analysis. Our results indicate that miR-34a regulates numerous cellular pathways in addition to those described previously by modulating the expression of a large number of diverse proteins and mRNAs. Interestingly, almost all of the miR-34a-mediated regulations have the potential of contributing to tumor suppression.
EXPERIMENTAL PROCEDURES
Generation of Cell Pools with Conditional miR-34a Expression
The colorectal cancer cell line SW480 was kept in Dulbecco's modified Eagle's medium and 10% fetal calf serum (Invitrogen). SW480 cells were transfected with the episomal expression vector pRTS-miR-34a (9, 19) using Lipofectamine2000 (Invitrogen). Polyclonal cell pools were generated by selection with hygromycin (450 μg/ml) for 10 days. For characterization of miR-34a targets, SW480 cells were transfected with the episomal pRTR-miR-34a expression vector. Polyclonal cell pools were generated by selection with puromycin (2 μg/ml) for 10 days. The percentage of RFP/GFP-positive cells was determined 48 h after addition of doxycycline at a final concentration of 100 ng/ml.
Generation of Episomal Vectors for miR-34a Expression
The pRTR vector is an improved version of the pRTS vector (Georg W. Bornkamm and Christian Berens, unpublished results). The pRTR expression vector was generated by replacing the KRAB repressor domain containing Tet-trans-silencer of the pRTS (20) vector with an alternative trans-silencer containing a CtBP-recruiting PLDLS repression motif (21), and a relaxed effector specificity (22), together with an IRES-coupled puromycin-resistance gene. To generate the episomal pRTR-miR-34a vector, the pri-mir-34a cDNA sequence was excised with SfiI from pRTS-miR-34a and ligated into pRTR via the SfiI sites. The insert orientation and the miR-34a portion were verified by sequencing.
GFP-Expression and Cell-cycle Analysis by Flow Cytometry
Cells were seeded in six-well plates (2 × 105 cells/well) and cultured in the presence and absence of 100 ng/ml doxycycline, respectively. For flow cytometry, cells were trypsinized after 40 h for mRFP analysis and after 72 h for cell-cycle analysis. For the analysis of mRFP expression, cells were fixed in phosphate-buffered saline containing 3.5% (w/v) paraformaldehyde and 0.5% (v/v) Nonidet P-40 (Fluka) for 30 min on ice. Fixed cells were collected by centrifugation (200 × g, 5 min at 4 °C) and re-suspended in phosphate-buffered saline for FACS analysis.
For cell-cycle analysis cells were fixed with 70% (v/v) ethanol overnight at −20 °C, collected by centrifugation (200 × g, 5 min at 4 °C) followed by a washing step in phosphate-buffered saline, and stained with 0.6 mg/ml propidium iodide (MP Biochemicals, Solon, OH) in the presence of 0.1% (w/v) Triton X-100 (Sigma) and 0.5 mg/ml RNaseA (Sigma). RNA digestion was carried out before FACS analysis at room temperature for 1 h. For DNA content analysis 10,000 cells and for detection of mRFP expression 25,000 cells per sample were analyzed with a FACScalibur device (BD Bioscience).
Pulsed-SILAC Labeling
An aliquot of 5 × 105 SW480 cells, harboring the pRTS-miR-34a vector, were seeded onto 10-cm dishes and grown in light DMEM (PAN Biotech) supplemented with light l-arginine (84 mg/l) and l-lysine (40 mg/l) and containing 10% dialyzed FBS (Hyclone, Thermo Scientific), 100 units/ml penicillin and 0.1 mg/ml streptomycin. Sixteen hours after pri-miR-34a induction with 100 ng/ml doxycycline cells were labeled by shifting them to heavy SILAC medium (84 mg/l 13C615N4-l-arginine and 40 mg/l 13C615N2-l-lysine), whereas the noninduced control cells were shifted to medium-heavy media (84 mg/l 13C6-l-arginine and 40 mg/L 2H4-l-lysine). All amino acids were purchased from Cambridge Isotope Laboratories. After 24 h, cells were lysed with RIPA buffer and lysates were subjected to one-dimensional SDS-PAGE. In total, three independent pSILAC experiments were performed and subjected to further analysis.
Gel Electrophoresis and Tryptic Digestion of Proteins
Proteins were separated on a 4–12% NuPageTM Bis-Tris gradient gel (Invitrogen) according to the manufacturer's instructions and stained with colloidal Coomassie Brilliant Blue G-250. Gel lanes were cut into 20 slices, which were immediately destained, washed, subjected to tryptic digestion, and prepared for LC/MS analysis as described previously (23).
Nano-HPLC/ESI-MS/MS Analysis
Tryptic digests were analyzed by nano high performance liquid chromatography (HPLC)/electrospray ionization (ESI)-tandem MS (MS/MS) using the UltiMateTM 3000 HPLC system (Dionex LC Packings, Idstein, Germany) online coupled to an linear trap quadrupole (LTQ)-Orbitrap XL instrument (Thermo Fisher Scientific, Bremen, Germany). Separation of peptides by nanoflow reversed-phase capillary HPLC was carried out as follows: peptide mixtures were loaded onto one of two C18 μ-pre-columns (0.3 mm inner diameter × 5 mm, particle size 5 μm; PepMap, Dionex LC Packings) equilibrated with 0.1% (v/v) trifluoroacetic acid (TFA), washed and preconcentrated for 5 min at a flow rate of 30 μl/min. The precolumn was then switched in line with a C18 RP nano LC column (75 μm inner diameter × 150 mm, particle size 5 μm; PepMap, Dionex LC Packings) and peptides were eluted using a binary solvent system consisting of 0.1% (v/v) formic acid (solvent A) and 0.1% (v/v) formic acid in 84% (v/v) acetonitrile (solvent B) with the following linear gradient: 5–40% solvent B in 150 min and 40–95% solvent B in 2 min. The column was then washed for 3 min with 95% solvent B and equilibrated with 5% solvent B (20 min) at a flow rate of 300 nl/minute. Precolumns were washed as follows: 5 min with 0.1% (v/v) TFA, 20 min with 0.1% (v/v) TFA in 50% (v/v) acetonitrile, and 10 min with 0.1% (v/v) TFA in 84% (v/v) acetonitrile before the column was re-equilibrated with 0.1% TFA.
The LTQ-Orbitrap XL instrument was equipped with a nano-electrospray ion source (Thermo Fisher Scientific) and distal coated SilicaTips (FS360–20-10-D, New Objective, Woburn, MA). The instrument was externally calibrated using standard compounds. To provide high mass accuracy, lock masses were routinely used for internal calibration. The general mass spectrometric parameters were as follows: spray voltage, 1.5 kV; capillary voltage, 4 V; capillary temperature, 200 °C; tube lens voltage, 100 V. For data-dependent MS/MS analyses, the software XCalibur 2.0 SR 2 (Thermo Fisher Scientific) was used. Full scan MS spectra (m/z 300 to 2000; resolution of 60,000 at m/z 400) were acquired in the orbitrap. Automatic gain control was set to 5 × 105 ions and a maximum fill time of 750 ms. After a brief survey scan, the six most intense multiply charged ions were selected for fragmentation by low energy collision-induced dissociation in the linear ion trap simultaneous with the completion of the MS scan in the orbitrap. The automatic gain control of the LTQ was set to 10,000 ions and a maximum fill time of 150 milliseconds. Fragmentation was carried out at a normalized collision energy of 35% with an activation q = 0.25 and an activation time of 30 milliseconds. The ion selection threshold was set to 5000. Fragmentation of previously selected precursor ions was dynamically excluded for the following 45 s.
Mass Spectrometric Data Analysis
Mass spectrometric data were processed using the software MaxQuant (version 1.0.13.13) (24). For peptide and protein identification, generated peak lists of MS/MS spectra were filtered to contain at most six peaks per 100 Da interval and correlated with the International Protein Index (25) human protein database (version 3.52) containing 73,928 proteins, to which 175 commonly observed contaminants and the reversed sequences of all proteins had been added (26). Species restriction in database searches to human was justified by the fact that the cell line SW480 used in this work is a human colon adenocarcinoma cell line. Database searches using Mascot [version 2.2, Matrix Science (27)] were performed with tryptic specificity allowing one missed cleavage and two labeled amino acids as well as an initial mass tolerance of 7 ppm for precursor ions and 0.4 Da for fragment ions. Oxidation of methionine was considered as variable modification; no fixed modifications were included. 2H4-lysine and 13C6-arginine were set as medium labels and 13C6-15N4-arginine and 13C6-15N2-lysine as heavy labels. A false discovery rate of < 1% on the peptide and protein level, calculated as described in (24) was applied. Proteins were considered identified when supported by at least two peptides, at least one of them unique, with a minimum length of six amino acids. In case a set of identified peptides was assignable to several proteins or completely contained in the set of peptides of another protein (e.g. in case of multiple isoforms not distinguishable by unique peptides), these proteins were grouped together by MaxQuant and reported as single protein group.
Relative peptide and protein quantification by MaxQuant, based on two-dimensional centroid intensities of differentially labeled peptide species (see (24) for details), were performed automatically using the following settings: quantification was based on unique and “razor” peptides; “Re-quantify” and “Filter labeled amino acids” were enabled; low-scoring versions of identified peptides were excluded from quantification. Protein ratios reported by MaxQuant are the median of all peptide ratios assigned to a distinct protein or protein group. The variability (in %) is determined as standard deviation of the natural logarithms of all peptide ratios used to calculate the protein ratio multiplied by 100 (26). Systematic deviations such as mixing errors are corrected for by the MaxQuant algorithm by normalizing all protein ratios such that the median of all log-transformed ratios is zero (24). Raw mass spectrometric files, Mascot search results (dat/msm files), and MaxQuant output files are stored at the scientific file sharing network and data repository Tranche. Data associated with this manuscript may be downloaded from the Tranche website www.ProteomeCommons.org using the hash: drMpxhWY0iVgjjrlc LC8v8BI25dXgU2bX1t/V1PJBdmvrRI3fexqMYEpfOGonyiWgLMG60 DKG/n4eq7d8H1biWERyuwAAAAAAAA9lw== or will be provided on request.
Microarray Analysis
Expression of pri-miR-34a was induced in SW480 cells bearing the pRTS-miR-34a vector with 100 ng/μl doxycycline for 40 h. cDNA was generated from total RNA and amplified using the GeneChip WT cDNA Amplification Kit (Affymetrix, Santa Clara, CA). cDNA was labeled with the GeneChip WT terminal labeling kit and hybridized to Affymetrix Human Exon ST 1.0 arrays (Affymetrix) according to the manufacturer's instructions. Arrays were hybridized, washed, stained and scanned according to the manufacturer's recommendations. Expression values were derived from CEL files using the Expression Console implemented by Affymetrix. The normalization method of choice was the robust-multichip average (RMA) provided by the software.
Bioinformatic miR-34a Target Identification
For bioinformatic identification of mRNAs containing miR-34a seed matches, we used a combined target prediction set generated by the TargetScan and Pictar algorithms (30–31). The data sets were obtained from http://www.targetscan.org and http://pictar.mdc-berlin.de. When using TargetScan, only phylogenetically conserved predictions were included.
Cloning of 3′-UTRs
The 3′-UTRs of the indicated target mRNAs containing putative miR-34a binding sites were PCR-amplified from oligo-dT-primed cDNA from SW480 cells with the Verso cDNA kit (Thermo Scientific). The 3′-UTRs were cloned into pGL3-control-MCS (32) and verified by sequencing. Seed matching sequences were mutated with the Quick Change Mutagenesis kit according to manufacturer's instructions (Stratagene). Oligonucleotides used for cloning and mutagenesis are given in supplemental Tables S5 and S6, respectively.
Luciferase Assays
H1299 cells were seeded in 12-well format at 1 × 104 cells/well, and transfected after 48 h with 100 ng of the indicated firefly luciferase reporter plasmid, 20 ng of Renilla reporter plasmid as a normalization control, and 25 nm of miR-34a pre-miRNA (Ambion, Austin, TX; PM11030) or a negative control oligonucleotide (Ambion, neg. control #1). The miR-34as vector contains the exact complementary sequence of miR-34as and served as a positive control (32). Luciferase assays were carried out after 48 h with the Dual Luciferase Reporter assay system (Promega) according to manufacturer's instructions. Fluorescence intensities were measured with a luminometer (Berthold) in 96-well format and analyzed with the simplicity software package (DLR).
Western Blot Analysis
SW480 cells harboring the pRTR-miR-34a vector were treated with 100 ng/μl doxycycline. SDS-PAGE and Western blotting were performed according to standard protocols. Cells were lysed in RIPA buffer (50 mm Tris/HCl, pH 8.0, 250 mm NaCl, 1% Nonidet P-40, 0.5% (w/v) sodium deoxycholate, 0.1% sodium dodecylsulfate, complete mini protease inhibitors (Roche)). Lysates were sonicated and centrifuged at 16,060 × g for 15 min at 4 °C. Per lane 40 μg of whole cell lysate was separated using 7.5% or 10% SDS-acrylamide gels and transferred on polyvinylidene difluoride membranes (Immobilon, Millipore). Antibodies used to detect the indicated proteins were: LDHA (Epitomics, 2091-1), LEF1 (Cell Signaling, C12A5), HDAC1 (Epitomics, 3426-1), MTA2 (Cayman, 13778), and AXL (Cell Signaling, #4977).
Quantitative Real-Time PCR
RNA from SW480 cells was prepared with the RNeasy Kit (Qiagen) according to manufacturer′s instructions. Oligo-dT-primed cDNA was prepared with the Verso cDNA kit (Thermo Scientific). Real-Time PCR was performed with a Light Cycler (Roche). A list of qPCR-primers is provided in supplemental Table S7.
RESULTS
Conditional miR-34a Expression
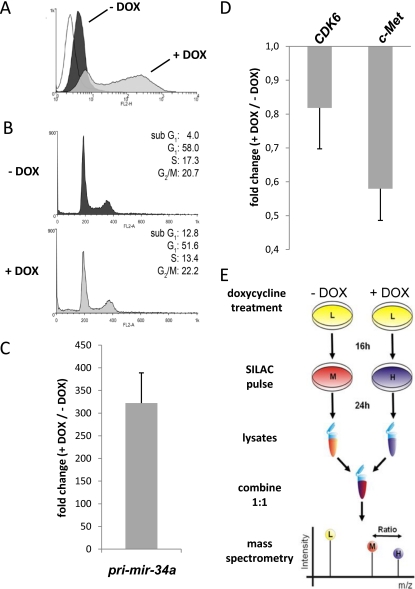
The colorectal cancer cell line SW480 was transfected with an episomal vector driving the expression of miR-34a pri-miRNA and mRFP from a bidirectional doxycycline (DOX)-inducible promoter. After selection of a hygromycin-resistant pool of cells, mRFP expression was determined by flow-cytometry (Fig. 1A). In the absence of DOX the vector containing cells displayed a minor increase in fluorescence. After treatment with DOX the fraction of positive cells increased to 73.8% with a more than 30 times higher mean fluorescence. Seventy-two hours after addition of DOX a threefold increase in the sub-G1 phase was detected, indicating the induction of apoptosis by ectopic miR-34a (Fig. 1B). This type of pro-apoptotic effect is known to result from ectopic expression of miR-34a (9). Ectopic expression of pri-miR-34a was verified by qPCR. We detected a ∼300-fold increase of pri-miR-34a levels in DOX-treated cells after 72 h compared with the untreated control cells (Fig. 1C). Also the processed miR-34a displayed a pronounced increase (supplemental Fig. S1). Moreover, expression levels of the previously identified miR-34a targets CDK6 and c-MET (5, 7, 10) decreased by ∼20% (CDK6) and ∼40% (c-MET) after induction pri-miR-34a expression in these cells (Fig. 1D). As the ectopic expression of miR-34a resulted in the expected effects in SW480 cells, we employed these cells for pSILAC and microarray analyses.
Fig. 1.
Characterization and pSILAC analysis of SW480 cells ectopically expressing miR-34a. A, Characterization of SW480 cells harboring an episomal pRTS-miR-34a plasmid by FACS analysis. RFP fluorescence before (dark gray) and after induction (light gray) of miR-34a with DOX for 72 h. The fluorescent signal of the parental SW480 cell line (white plot) was set to a false positive rate of 5%, which resulted in a mean fluorescence of 3.8 a.u. B, Cell cycle analysis by PI staining before (dark gray plot) and after (light gray plot) addition of DOX for 72 h. C, qPCR confirmation of pri-miR-34a expression 72 h after addition of DOX. D, Down-regulation of known miR-34a targets CDK6 and c-MET 72 h after induction of pri-miR-34a by addition of DOX. Expression levels were normalized to β-actin and calculated as fold change (+ DOX/- DOX). E, Schematic outline of the pSILAC analysis. SW480 cells were treated with DOX for 16 h in normal, unlabeled medium (light, L; in yellow). Then the medium of untreated cells was replaced with medium containing amino-acids labeled with medium-heavy (M) isotopes (13C6-l-arginine and 2H4-l-lysine; in red) and the medium of DOX-treated cells was replaced with medium containing heavy (H) isotope-coded amino acids (13C615N4-l-arginine and 13C615N2-l-lysine; in dark blue) and DOX for 24 h as depicted. Then cell lysates of both conditions were prepared, combined 1:1 and subjected to mass spectrometric analysis. Only the newly synthesized proteins, either labeled with medium-heavy (M) or heavy (H) amino acids, were included in the analysis, whereas pre-existing proteins labeled with light (L) amino acids were excluded.
Pulsed SILAC Analysis of miR-34a-Induced Changes in Protein Synthesis
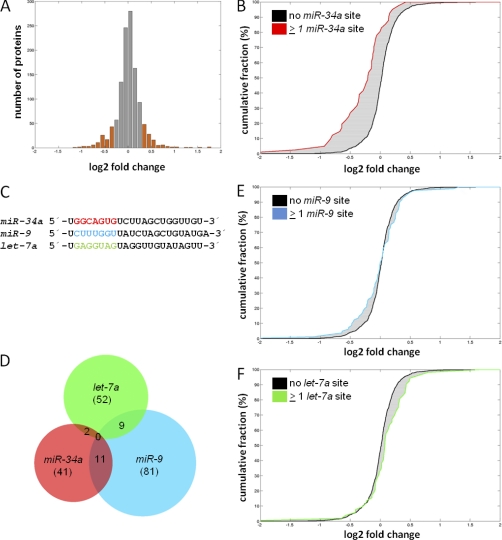
To identify miR-34a targets, we performed pulsed SILAC (pSILAC) as outlined in Fig. 1E and described before (18, 33). In short, we induced ectopic pri-miR-34a expression for 40 h by addition of DOX. During the last 24 h the miR-34a expressing cells were pulsed with heavy (H) medium and the untreated control cells with medium-heavy (M) medium. Subsequently, protein samples were prepared and subjected to mass spectrometric analysis. These analyses were performed with biological triplicates. In total, 1818 proteins were identified, when detection of at least two matching peptides per protein was set as a requirement for unambiguous identification. Of these, 1206 proteins could be reliably quantified in at least two experiments and were therefore considered for further analysis (for identification and quantification of proteins and peptides see supplemental Table S1A and S1B). Remarkably, ectopic expression of miR-34a caused only moderate changes in overall protein synthesis, with the majority of proteins having log2 (H/M) ratios between −0.3 and 0.3 (Fig. 2A). Similar, overall moderate effects on global protein synthesis after ectopic miRNA expression have been reported previously (18). Among the 1206 proteins quantified in at least two experiments, 228 (∼19%) were differentially regulated with log2 fold changes ≤–0.3 or ≥ 0.3 (+DOX versus –DOX). Among these the protein synthesis was down-regulated for 113 and up-regulated for 115 proteins (supplemental Table S1A).
Fig. 2.
miR-34a expression results in specific changes in protein expression. A, Histogram of changes in protein expression after ectopic miR-34a expression for all 1206 proteins quantified in at least two out of three biological replicates. Translationally regulated genes with a log2 fold change ≤ –0.3/≥ 0.3 are highlighted as orange bars. Fold change denotes the ratio of peptide intensities of doxycycline-treated, heavy-labeled versus untreated, medium-heavy labeled cells. B, Cumulative distribution of miR-34a targets with ≥1 seed-matching sequence in the 3′-UTR (red line) among proteins detected by pSILAC without the respective seed-matching sequence in the 3′-UTR of their mRNAs (black line). C, Sequences of the mature miR-34a, miR-9 and let-7a microRNAs. The seed sequences are high-lighted in red, blue and green, respectively. D, Venn diagram of bioinformatically predicted targets of miR-34a, miR-9 and let-7a with ≥1 seed-matching sequence in the 3′-UTR of the mRNAs corresponding to proteins identified by pSILAC. The total number of detected targets for each miRNA is given in brackets. E, F, Cumulative distributions of miR-9 and let-7a targets with ≥ 1 seed-matching sequence in the 3′-UTR (blue or green lines) among proteins detected by pSILAC without the respective seed-matching sequence in the 3′-UTR of their mRNAs (black lines).
Highly abundant ribosomal proteins, RNA binding proteins and metabolic enzymes were overrepresented among detected proteins, whereas proteins with generally low abundance, as transcription factors, were rarely detected (Fig. S2). Nonetheless, the fraction of bioinformatically predicted miR-34a targets among the proteins detected by pSILAC (41/1206 or approx. 3.4%) was in the range of miR-34a targets predicted based on the frequency of miR-34a seed-matching sequences present in mRNAs (623/17627 or approx. 3.5%, based on the number of annotated genes represented by the Affymetrix GeneChip Exon 1.0 ST array employed here), indicating that the quantitative proteomics approach does not per se cause a decrease in the fraction of detectable miRNA targets.
A cumulative distribution analysis of proteins with miR-34a seed-matching sequences in their respective 3′-UTRs revealed a reduced de novo protein synthesis when compared with proteins without miR-34a seed-matches (Fig. 2B). Moreover, the only miRNA binding site that was overrepresented in the 3′-UTRs of the down-regulated candidate proteins was that of miR-34a (supplemental Fig. S3). In order to determine whether the shift in protein synthesis as detected by pSILAC was dependent on the presence of predicted miR-34a binding sites in the 3′-UTRs, we performed the same analysis for proteins with predicted binding sites for the unrelated oncogenic miRNA miR-9 (34, 35) or the tumor suppressive miRNA let-7a (36) in their mRNAs. Both, miR-9 and let-7a have seed sequences that differ from the miR-34a sequence (Fig. 2C) and were not enriched in 3′-UTR of mRNAs targeted by miR-34a (Fig. 2D). Proteins that contain either miR-9 or let-7a binding sites in their 3′-UTRs did not show reduced protein synthesis after activation of miR-34a (Fig. 2E, F), nor was there a bias for the presence of either miR-9 or let-7a binding sites among the differentially translated proteins toward the down-regulated proteins, as detected for miR-34a (Fig. S4). These results indicate that even though the overall effects of miR-34a on global protein translation are moderate, miR-34a causes the specific translational repression of several proteins that is dependent on the presence of miR-34a binding sites in 3′-UTRs of their corresponding mRNAs.
Microarray Analysis of miR-34a Targets
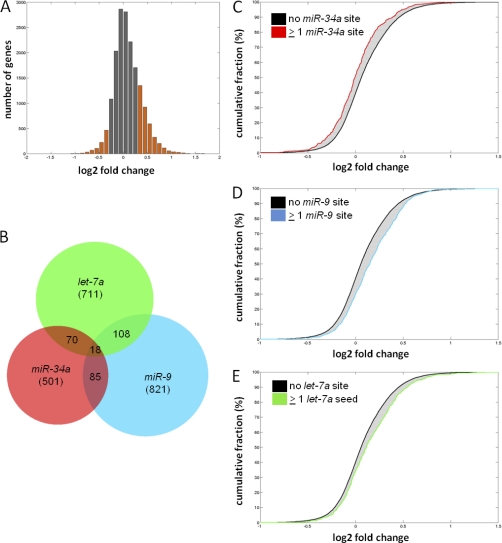
Next, genome-wide mRNA expression was determined by microarray analysis 40 h after ectopic expression of pri-miR-34a in SW480 cells. Thereby, transcriptional down-regulation of 930 genes was detected, when mRNAs with a log2 fold change ≤–0.3 (+DOX versus –DOX) were considered (Fig. 3A, supplemental Table S2).
Fig. 3.
Identification of transcriptionally regulated miR-34a targets by microarray analysis. A, Histogram showing changes in mRNA levels after induction of ectopic pri-miR-34a expression for 40 h in SW480 cells by addition of DOX. Transcriptionally regulated genes with a log2 fold change ≤ –0.3/≥ 0.3 are highlighted as orange bars. Fold change denotes the ratio of gene expression values of DOX-treated versus untreated cells. B, Venn diagram of the predicted target mRNAs of miR-34a, miR-9 and let-7a with ≥1 seed-matching binding site in the 3′-UTR among all detected 17,326 mRNAs illustrate minimal overlap between the targets of these miRNAs. The total number of detected targets for each microRNA is given in brackets. C, D, E, Cumulative distributions of miR-34a, miR-9 and let-7a targets with ≥1 binding site in the 3′-UTR compared with mRNAs with no binding site.
We analyzed whether the transcriptional changes could be ascribed specifically to expression of miR-34a. Compared with bioinformatically predicted targets of the unrelated miR-9 and let-7a miRNAs, which only showed minimal overlap with predicted miR-34a targets (Fig. 3B), 501 putative miR-34a target mRNAs with miR-34a seed-matching sequences in their 3′-UTRs were clearly biased toward lower expression levels when compared with mRNAs without miR-34a seed-matches (Figs. 3C, 3D, 3E). Therefore, the modest transcriptional down-regulation of a subset of mRNAs is specifically caused by expression of miR-34a and dependent on the presence of miR-34a seed-matching sequences in their 3′-UTR. Similar to the results obtained by pSILAC for proteins, the only seed-matching site that was over-represented among the down-regulated mRNAs was that corresponding to miR-34a (supplemental Fig. S5).
miR-34a Simultaneously Affects Protein Synthesis and mRNA Abundance
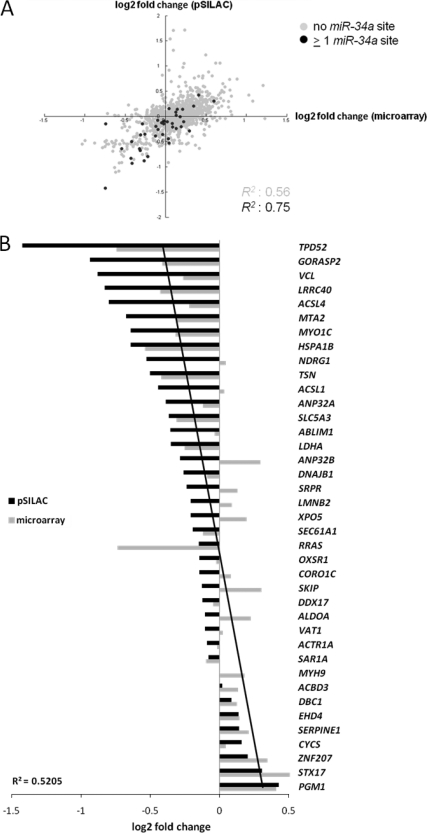
MiRNAs mediate the reduction of protein synthesis through inhibition of translation initiation and/or mRNA degradation. However, the relative contributions of each of these mechanisms to the down-regulation of the proteins encoded by the affected mRNAs and the order of events leading to reduced protein synthesis have not been fully resolved yet (37). To determine how miR-34a regulates expression of its target mRNAs, we compared changes in de novo protein synthesis with changes in mRNA abundance. The correlation between the changes in abundance of mRNAs and the encoded proteins was enhanced for mRNAs with miR-34a seed-matches, when compared with mRNAs lacking miR-34a seed-matches (Fig. 4A). Therefore, translational repression and mRNA abundance are presumably co-regulated for most miR-34a targets.
Fig. 4.
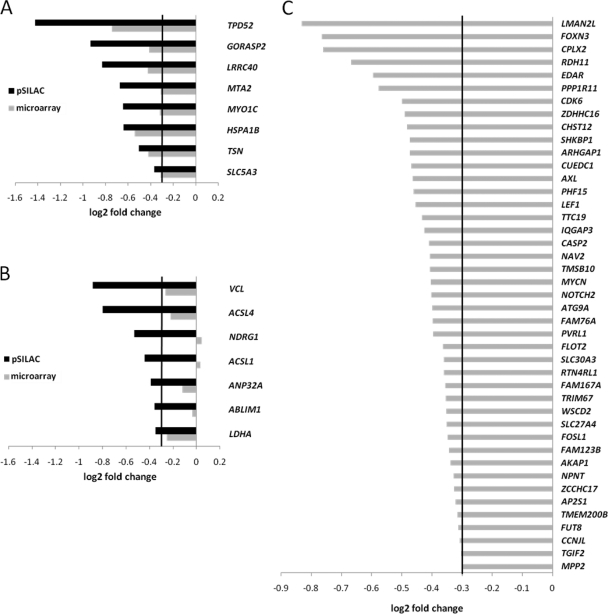
miR-34a-mediated changes in protein synthesis correlate with changes in mRNA abundance. A, Scatter plot correlating changes in protein synthesis as measured by pSILAC with changes in mRNA abundance as measured by microarray analysis. The Pearson′s correlation coefficient R2 is shown for genes with predicted miR-34a seed-matching sequences compared with genes without miR-34a seed matches. B, Representation of 39 miR-34a targets detected both by pSILAC and microarray analysis containing miR-34a seed-matching sequences. The miR-34a targets are sorted on the basis of log2 fold changes in protein synthesis as measured by pSILAC and are represented as horizontal bars. In addition, the corresponding log2 fold changes in mRNA abundance for each miR-34a target as determined by microarray analysis are shown. The linear regression for changes in mRNA abundance is depicted as a black line. The regression coefficient R2 is indicated.
By combining the results generated by pSILAC and microarray analyses, we obtained a set of 39 predicted miR-34a targets that were amenable to quantification in both types of analysis (Fig. 4B). We found that the extent of translational regulation correlated with changes in abundance of the corresponding mRNA in most cases. For less pronounced regulations on the protein level exceptions to this pattern were detected, as for RRAS and SKIP.
Identification of miR-34a Targets
Next we determined which of the predicted miR-34a targets showed differential regulation either at the level of protein synthesis and/or at the level of mRNA expression as detected by pSILAC and microarray analysis, respectively. When considering proteins with a predicted miR-34a seed-matching site among the proteins detected by pSILAC, we found that 15 predicted targets were significantly down-regulated (log2 fold change ≤–0.3), whereas only two predicted targets (STX17, PGM1) were translationally up-regulated with a log2 fold change ≥0.3 (Figs. 5A, 5B, supplemental Fig. S4A). Out of these 15 translationally down-regulated targets, eight were regulated both on the protein and mRNA level (Fig. 5A), whereas seven were mainly regulated at the level of de novo protein synthesis (with a log2 fold change ≤–0.3 being the cutoff for the mRNA expression; Fig. 5B). These results indicate that the extent of regulation on the level of mRNA abundance of the miR-34a targets is high, but does not account for the effects on all target genes. At least some of the miR-34a targets, as NDRG1, ACSL1, and ABLIM1, seem to be only regulated at the level of de novo protein synthesis. In addition, we identified 43 mRNAs with miR-34a seed-matches that displayed decreased abundance, of which the corresponding proteins were not detected by pSILAC (Fig. 5C). We detected additional miR-34a targets by using the Miranda search algorithm (38), which requires less stringent seed-pairing for target prediction compared with TargetScan and Pictar (15) and thus resulted in an increased number of predicted miR-34a targets. Although the Miranda-generated set of predicted miR-34a targets contained a lower fraction of experimentally verified down-regulated genes both in the pSILAC and microarray analysis compared with a composite Target Scan/Pictar set (supplemental Fig. S6), we nevertheless could identify 132 additional, putative miR-34a targets that were down-regulated at the level of protein synthesis and/or mRNA abundance (supplemental Fig. S7, supplemental Table S4).
Fig. 5.
Identification of miR-34a targets by pSILAC and microarray analysis. A, Predicted miR-34a targets down-regulated translationally and on the mRNA level with log2 fold changes ≤–0.3. B, Predicted miR-34a targets down-regulated translationally with log2 fold changes ≤–0.3, but showing no or minor changes at the mRNA level (log2 fold changes >–0.3). C, miR-34a targets that were not detected by pSILAC, but showed down-regulation at the mRNA level with log2 fold changes ≤–0.3.
Pathways Affected by miR-34a Activation
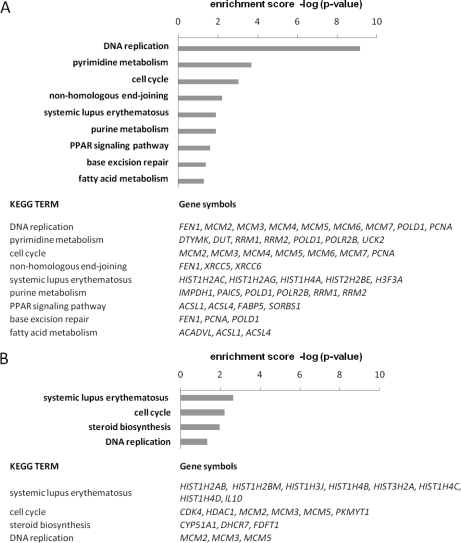
A KEGG pathway analysis of the 113 proteins that were translationally down-regulated according to pSILAC revealed a striking enrichment for proteins that are involved in DNA replication, as well as a moderate enrichment for proteins involved in pyrimidine metabolism, and cell cycle regulation (Fig. 6A). Remarkably, we found almost all members of the DNA replication initiation complex (MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7) to be translationally repressed. For the 115 translationally up-regulated proteins, we only detected minor enrichments of proteins involved in endocytosis as well as in several metabolic pathways (supplemental Table S3). Remarkably, some of the down-regulated proteins are common markers of cell proliferation (PCNA) or potential therapeutic targets in cancer therapy, such as RRM2 (39). The data indicate that one of the effects of miR-34a induction is an inhibition of DNA replication, either by regulation of the DNA replication machinery itself or by interference with the metabolic requirements of de novo DNA synthesis. A KEGG pathway analysis of the differentially regulated mRNAs showed that genes involved in cell cycle progression (p = 0.006) and DNA replication (p = 0.045) were slightly over-represented among the down-regulated mRNAs (log2 fold change ≤–0.3, p < 0.05), further substantiating the influence of miR-34a on DNA replication initiation and cell cycle arrest (Fig. 6B). Among the up-regulated mRNAs, we found a significant increase in genes involved in the p53 signaling pathway (p = 0.011) and in apoptosis (p = 0.016; supplemental Table S3).
Fig. 6.
KEGG pathway associations of down-regulated proteins and mRNAs. The enrichment scores of biological processes are shown as –log(p-value). For each KEGG pathway, the gene symbols of differentially down-regulated genes are shown in the table given below. Translationally down-regulated candidate proteins (A) and mRNAs (B) down-regulated with log2 fold change ≤–0.3, p < 0.05, were subjected to KEGG pathway enrichment analysis (http://david.abcc.ncifcrf.gov/) irrespective of the presence of miR-34a binding sites.
Confirmation of Direct Regulations by miR-34a
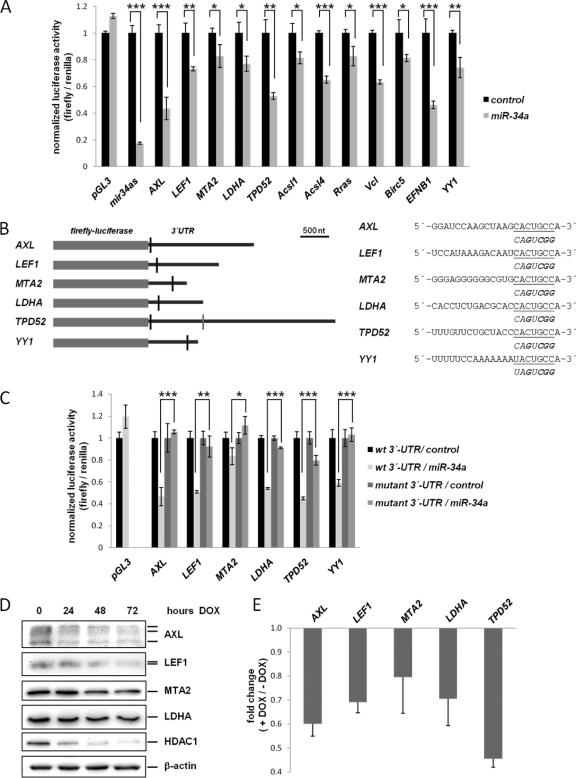
To validate whether the mRNAs and/or proteins down-regulated by miR-34a are direct targets of miR-34a, we performed dual reporter assays using reporter constructs containing the 3′-UTRs of representative mRNAs (Fig. 7A). Remarkably, all 12 tested reporter constructs were significantly repressed by ∼20 to ∼60% after cotransfection with a miR-34a mimic, whereas a control miRNA had no effect. To further validate the directness of the regulation by miR-34a, we mutated the miR-34a seed-matching sequences in six selected 3′-UTRs (Fig. 7B). Thereby, the repressive effect of miR-34a on the luciferase reporters of AXL, LEF1, MTA2, LDHA, and YY1 was reversed (Fig. 7C). In the case of TPD52 mutation of the indicated seed-matching sequence only caused a partial resistance towards inhibition by miR-34a. This may be because of an additional weakly conserved miR-34a binding site in the 3′-UTR of TPD52, which was not disrupted by mutation (Fig. 7B). YY1, which was not identified in our experimental screen, was analyzed on the basis of a bioinformatically predicted miR-34a binding site.
Fig. 7.
Experimental validation of miR-34a target regulation. A, Dual reporter assay in H1299 cells transfected with miR-34a mimics (pre-miR-34a) or control oligos and the indicated 3′-UTR-reporter constructs. Birc5/Survivin and EFNB1 have been identified as miR-34a targets based on the Miranda algorithm and their repression at the mRNA level (see supplemental Table S4). YY1 was analyzed on the basis of a bioinformatically predicted miR-34a binding site. Data are represented as mean ± S.D. (n = 3). B, Left, Schematic depiction of miR-34a seed-matching sequences and their targeted mutation in the 3′-UTRs of selected mRNAs. Black vertical bars: miR-34a seed-matching sequences subjected to mutagenesis. The TPD52 3′-UTR contains an additional weakly conserved binding site (gray vertical bar), which was not modified here. Right, 3′-UTR sequences of the miR-34a binding site of each target gene are given in the 5′- to 3′-orientation. The seed-matching sequence is underlined. Below each miR-34a binding site the mutant seed-matching sequence is shown in italics with exchanged nucleotides high-lighted in bold letters. C, Luciferase assay as in A, with selected 3′-UTR reporter constructs and variants with mutant seed-matching sequences (Asterices indicate *: p < 0.05, **: p < 0.01, ***: p < 0.001). D, Western blot analysis of selected miR-34a targets after activation of ectopic miR-34a expression for the indicated periods in SW480 cells. β-actin served as a loading control. E, Validation of mRNA regulation of the indicated miR-34a targets by qPCR 72 h after induction of pri-miR-34a (see also Fig. 1). Expression levels were normalized to β-actin and calculated as fold change (+ DOX/- DOX).
In addition, the expression of endogenous proteins encoded by miR-34a target genes was determined after ectopic pri-miR-34a expression (Fig. 7D). For all selected proteins a down-regulation was observed after ectopic miR-34a expression. We detected a reduction in protein levels between 24 and 72 h of miR-34a induction, albeit with different kinetics for every protein, which may be because of divergent protein half-lives or target mRNA abundance. AXL and LEF1 displayed an early reduction (i.e. after 24 h) of protein levels, whereas LDHA showed a detectable decrease only after 72 h. The histone deacetylase HDAC1 is a predicted miR-34a target that we found to be down-regulated by microarray analysis (supplemental Tables S2, S4). Although we could not unambiguously prove that HDAC1 is directly regulated by miR-34a using reporter assays, HDAC1 also displayed a robust decrease in protein levels after induction of miR-34a (Fig. 7D). At the mRNA level a decrease of all miR-34a target genes selected for analysis was detected 72 h after induction of ectopic pri-miR-34a expression (Fig. 7E).
DISCUSSION
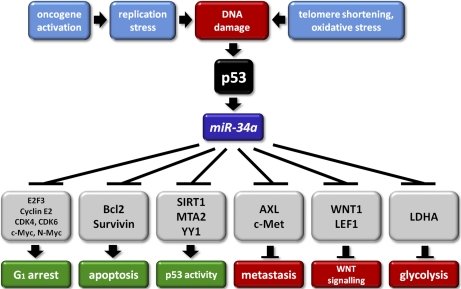
The analyses presented here revealed numerous new direct miR-34a targets by proteomic and/or microarray analysis, which presumably represent mediators of the effects of miR-34a (summarized in Fig. 8). The directness of the regulations by miR-34a was confirmed by reporter gene assays for selected miR-34a target mRNAs. These exemplary analyses showed that the miR-34a-mediated repression of these proteins requires the presence of seed-matching sequences in the respective 3′-UTRs. On a proteome-wide scale ectopic expression of miR-34a caused moderate changes in protein translation. This is in accordance with a previous pSILAC study of miRNA-mediated changes in protein expression (18). However, the rather subtle changes in protein translation detected in this study were specific for miR-34a and in representative cases dependent on the presence on miR-34a seed-matching sequences in the 3′-UTR of the respective mRNAs. Similarly, we detected only minor, yet miR-34a-specific changes in mRNA abundances by microarray analysis.
Fig. 8.
The p53-miR-34 pathway and its effects on multiple biological pathways. Schematic model of the regulation of and by the miR-34a miRNA. The indicated pathways are deduced from the known functions of the miR-34a targets identified in this and previous studies.
Among the down-regulated proteins and mRNAs, we noted an overrepresentation of proteins involved in chromatin assembly (e.g. histones), DNA-replication initiation and cell-cycle regulation. Even though the down-regulation of these proteins, which do not contain predicted miR-34a binding sites in their 3′-UTRs, presumably is a secondary consequence of the direct miR-34a-mediated repression of cell cycle regulators (e.g. CDK4, CDK6, E2F, c-MYC), it reflects the inhibition of DNA replication and a cell cycle arrest as a major consequence of miR-34a activation.
Furthermore, we found that miR-34a targets exhibiting the most pronounced decrease in protein translation also tended to have the strongest decreases in mRNA levels. Even though the mechanistic connections of inhibition of protein translation and mRNA decay have not been fully elucidated, both act in concert on a large proportion of miR-34a target genes. In the following sections the putative biological consequences of the down-regulation of new, direct miR-34a targets are discussed.
The metabolic switch from oxidative phosphorylation to aerobic glycolysis, commonly referred to as the Warburg effect, is a hallmark of cancer cells (40). p53 inhibits glycolysis by at least two mechanisms (41), namely induction of TIGAR or SCO2 (42, 43) and thereby mediates tumor suppression. Here, we provide evidence that p53 may also inhibit glycolysis through activation of miR-34a, which directly represses lactate dehydrogenase (LDHA), a key enzyme required for aerobic glycolysis. LDHA converts pyruvate, the end product of glycolysis, to lactate and pharmacological or siRNA-mediated inhibition of LDHA prevents cancer cell proliferation (44). Therefore, the miR-34a-mediated down-regulation of LDHA may contribute to the tumor suppressive action of p53. Interestingly, LDHA is activated by both the c-MYC protein and by HIF1-α, which senses hypoxic environments (45).
The newly identified miR-34a target LEF1/TCF1-α is a transcription factor that associates with nuclear β-catenin upon activation of Wnt signaling to induce the transcription of downstream target genes such as c-MYC. Wnt1 has previously been reported to be a direct miR-34a target (46), corroborating that miR-34a presumably suppresses oncogenic Wnt signaling. Constitutive activation of Wnt signaling through loss of APC or activating mutations of β-catenin has been observed in ca. 90% of all colorectal cancers (25). We observed inactivation of miR-34a by CpG methylation in 84 (74%) of 114 colorectal cancers analyzed (12). Therefore, it is likely that loss of miR-34a function cooperates with mutational activation of the APC/β-catenin pathway. In line with this hypothesis, inactivation of the miR-34a-related miR-34b/c gene by CpG methylation was found to occur in nearly all colorectal cancers analyzed (12, 47).
The NAD-dependent deacetylase SIRT1, which also deacetylates p53, has been described as a miR-34a target (48). It has been proposed that p53, miR-34a and SIRT1 form a positive feed-back-loop, in which repression of SIRT1 leads to further activation of p53. Here we identified additional negative regulators of p53 as miR-34a targets: MTA2, HDAC1 and YY1. Therefore, it is tempting to speculate that miR-34a enhances p53 activity and tumor suppression by the combined down-regulation of these proteins. MTA2 and HDAC1 are components of the NURD complex, which mediates deacetylation and destabilization of the p53 protein (49). YY1, which modulates MDM2-mediated ubiquitination of p53 (50, 51), was directly repressed by miR-34a. While this manuscript was in preparation YY1 was also identified as a miR-34a target by others (52). HDAC1 is a histone deacetylase, which is required for cell cycle progression in normal and transformed cells (53, 54). Interestingly, the p53 target gene p21 is repressed by HDAC1 (55, 56). Although HDAC1 may not to be a direct miR-34a target, we detected a robust decrease in both mRNA and protein levels of HDAC1 after expression of miR-34a. Thus, the inhibition of HDAC1 after induction of miR-34a may be one of several mechanisms by which miR-34a exerts its growth inhibitory effects.
AXL is a member of the TAM (Tyro-AXL-Mer) receptor tyrosine kinase (RTK) family. It was originally identified as a transforming gene in chronic myeloid leukemia (57, 58). AXL is involved in diverse aspects of tumor formation such as cell migration, proliferation and invasion (59, 60). Elevated expression of AXL has been associated with increased metastatic potential of breast cancer cells (61). Interestingly, another RTK, which confers metastatic potential, c-MET, is a direct target of miR-34a (62). Therefore, miR-34a might mediate suppression of metastasis by the combined down-regulation of several RTKs involved in invasion and metastasis.
Birc5/Survivin is an anti-apoptotic protein that is overexpressed in most human cancers because of activation by several oncogenic transcription factors, such as NFκB, STAT3, Notch and TCF4/β-catenin (63). The concomitant down-regulation of several anti-apoptotic proteins such as Survivin and Bcl2 (5) may therefore represent a mechanism that accounts for the tumor-suppressive function of miR-34a. Ephrin-B1 (EFNB1), which was identified as a miR-34a target in this study, is one of several ligands for the Ephrin-B1 receptor tyrosine kinase, which has been implicated in tissue architecture and organogenesis, as well as cancer progression (64).
Among the genes down-regulated either at the protein and/or mRNA level identified in this study that had a putative miR-34a binding site in their 3′-UTR, only CDK6, MYCN and Notch2 have previously been reported as direct miR-34a targets (7, 10, 32, 65). ACSL1 and AXL have been reported as direct miR-34a targets (66, 67) by others while this manuscript was in preparation.
Recently, the systemic treatment with miR-34a mimetics was shown to inhibit the growth of xenograft tumors in mice and the proliferation of prostate cancer stem cells (68–70). Therefore, ectopic miR-34a expression may serve as a tumor therapeutic means for human tumors in the future. A comprehensive knowledge of the targets and effects of miR-34a is of clinical relevance in order to evaluate the potential side effects of such therapies. The study presented here is an important step in the direction of obtaining a complete catalogue of miR-34a targets and will have to be extended to additional cellular systems and conditions. In the future, improvements in the sensitivity and dynamic range of proteomic and transcriptomic analysis may facilitate a more complete coverage of miRNA targets using this type of combined approach.
Acknowledgments
We thank Christian Berens (University of Nuremberg-Erlangen, Germany) and Georg W. Bornkamm (Helmholtz Center, Munich, Germany) for providing the pRTR vector, and Raymond L. Stallings (Royal College of Surgeons, Dublin, Ireland) for miR-34as and pGL3-control-MCS reporter plasmids.
Footnotes
* This study was in part supported by the Friedrich-Baur-Stiftung, the Deutsche Forschungsgemeinschaft, the Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS) and the Ministerium für Innovation, Wissenschaft, Forschung und Technologie des Landes Nordrhein-Westfalen.
 This article contains supplemental Figures S1 to S7 and Tables S1 to S7.
This article contains supplemental Figures S1 to S7 and Tables S1 to S7.
1 The abbreviations used are:
- miRNA
- microRNA
- SILAC
- stable isotope labeling with amino acids in cell culture
- RIPA
- radioimmunoprecipitation assay
- HPLC
- high performance liquid chromatography
- ESI
- electrospray ionization
- MS/MS
- tandem MS
- TFA
- trifluoroacetic acid
- UTR
- untranslated regions.
REFERENCES
- 1. Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 2. Hermeking H. (2007) p53 enters the microRNA world. Cancer Cell 12, 414–418 [DOI] [PubMed] [Google Scholar]
- 3. He X., He L., Hannon G. J. (2007) The guardian's little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 67, 11099–11101 [DOI] [PubMed] [Google Scholar]
- 4. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bommer G. T., Gerin I., Feng Y., Kaczorowski A. J., Kuick R., Love R. E., Zhai Y., Giordano T. J., Qin Z. S., Moore B. B., MacDougald O. A., Cho K. R., Fearon E. R. (2007) p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 17, 1298–1307 [DOI] [PubMed] [Google Scholar]
- 6. Chang T. C., Wentzel E. A., Kent O. A., Ramachandran K., Mullendore M., Lee K. H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C. J., Arking D. E., Beer M. A., Maitra A., Mendell J. T. (2007) Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 26, 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He L., He X., Lim L. P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., Hannon G. J. (2007) A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. (2007) Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 26, 731–743 [DOI] [PubMed] [Google Scholar]
- 9. Tarasov V., Jung P., Verdoodt B., Lodygin D., Epanchintsev A., Menssen A., Meister G., Hermeking H. (2007) Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6, 1586–1593 [DOI] [PubMed] [Google Scholar]
- 10. Lodygin D., Tarasov V., Epanchintsev A., Berking C., Knyazeva T., Körner H., Knyazev P., Diebold J., Hermeking H. (2008) Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7, 2591–2600 [DOI] [PubMed] [Google Scholar]
- 11. Hermeking H. (2010) The miR-34 family in cancer and apoptosis. Cell Death Differ. 17, 193–199 [DOI] [PubMed] [Google Scholar]
- 12. Vogt M., Munding J., Grüner M., Liffers S. T., Verdoodt B., Hauk J., Steinstraesser L., Tannapfel A., Hermeking H. (2011) Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 458, 313–322 [DOI] [PubMed] [Google Scholar]
- 13. Fabian M. R., Sonenberg N., Filipowicz W. (2010) Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379 [DOI] [PubMed] [Google Scholar]
- 14. Huntzinger E., Izaurralde E. (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12, 99–110 [DOI] [PubMed] [Google Scholar]
- 15. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomas M., Lieberman J., Lal A. (2010) Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 17, 1169–1174 [DOI] [PubMed] [Google Scholar]
- 17. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 18. Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 19. Epanchintsev A., Jung P., Menssen A., Hermeking H. (2006) Inducible microRNA expression by an all-in-one episomal vector system. Nucleic Acids Res. 34, e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bornkamm G. W., Berens C., Kuklik-Roos C., Bechet J. M., Laux G., Bachl J., Korndoerfer M., Schlee M., Hölzel M., Malamoussi A., Chapman R. D., Nimmerjahn F., Mautner J., Hillen W., Bujard H., Feuillard J. (2005) Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 33, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Molloy D. P., Barral P. M., Bremner K. H., Gallimore P. H., Grand R. J. (2001) Structural determinants outside the PXDLS sequence affect the interaction of adenovirus E1A, C-terminal interacting protein and Drosophila repressors with C-terminal binding protein. Biochim. Biophys. Acta. 1546, 55–70 [DOI] [PubMed] [Google Scholar]
- 22. Krueger C., Danke C., Pfleiderer K., Schuh W., Jäck H. M., Lochner S., Gmeiner P., Hillen W., Berens C. (2006) A gene regulation system with four distinct expression levels. J. Gene Med. 8, 1037–1047 [DOI] [PubMed] [Google Scholar]
- 23. Wiese S., Gronemeyer T., Ofman R., Kunze M., Grou C. P., Almeida J. A., Eisenacher M., Stephan C., Hayen H., Schollenberger L., Korosec T., Waterham H. R., Schliebs W., Erdmann R., Berger J., Meyer H. E., Just W., Azevedo J. E., Wanders R. J., Warscheid B. (2007) Proteomics characterization of mouse kidney peroxisomes by tandem mass spectrometry and protein correlation profiling. Mol. Cell Proteomics 6, 2045–2057 [DOI] [PubMed] [Google Scholar]
- 24. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 25. Wood L. D., Parsons D. W., Jones S., Lin J., Sjöblom T., Leary R. J., Shen D., Boca S. M., Barber T., Ptak J., Silliman N., Szabo S., Dezso Z., Ustyanksky V., Nikolskaya T., Nikolsky Y., Karchin R., Wilson P. A., Kaminker J. S., Zhang Z., Croshaw R., Willis J., Dawson D., Shipitsin M., Willson J. K., Sukumar S., Polyak K., Park B. H., Pethiyagoda C. L., Pant P. V., Ballinger D. G., Sparks A. B., Hartigan J., Smith D. R., Suh E., Papadopoulos N., Buckhaults P., Markowitz S. D., Parmigiani G., Kinzler K. W., Velculescu V. E., Vogelstein B. (2007) The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 [DOI] [PubMed] [Google Scholar]
- 26. Pan C., Olsen J. V., Daub H., Mann M. (2009) Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol. Cell Proteomics 8, 2796–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 28. Motulsky H. (1995) Intuitive Biostatistics. Oxford University Press, New York [Google Scholar]
- 29. Tukey J. W. (1990) Data-based graphics: visual display in the decades to come. Stat. Sci. 5, 327–339 [Google Scholar]
- 30. Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da, Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 31. Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Welch C., Chen Y., Stallings R. L. (2007) MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 26, 5017–5022 [DOI] [PubMed] [Google Scholar]
- 33. Schwanhäusser B., Gossen M., Dittmar G., Selbach M. (2009) Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 9, 205–209 [DOI] [PubMed] [Google Scholar]
- 34. Lujambio A., Calin G. A., Villanueva A., Ropero S., Sánchez-Céspedes M., Blanco D., Montuenga L. M., Rossi S., Nicoloso M. S., Faller W. J., Gallagher W. M., Eccles S. A., Croce C. M., Esteller M. (2008) A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. U.S.A. 105, 13556–13561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma L., Young J., Prabhala H., Pan E., Mestdagh P., Muth D., Teruya-Feldstein J., Reinhardt F., Onder T. T., Valastyan S., Westermann F., Speleman F., Vandesompele J., Weinberg R. A. (2010) miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 12, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. (2005) RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 [DOI] [PubMed] [Google Scholar]
- 37. Djuranovic S., Nahvi A., Green R. (2011) A parsimonious model for gene regulation by miRNAs. Science 331, 550–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004) Human MicroRNA targets. PLoS Biol. 2, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davis M. E., Zuckerman J. E., Choi C. H., Seligson D., Tolcher A., Alabi C. A., Yen Y., Heidel J. D., Ribas A. (2010) Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464, 1067–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Warburg O. (1956) On the origin of cancer cells. Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 41. Levine A. J., Puzio-Kuter A. M. (2010) The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330, 1340–1344 [DOI] [PubMed] [Google Scholar]
- 42. Bensaad K., Tsuruta A., Selak M. A., Vidal M. N., Nakano K., Bartrons R., Gottlieb E., Vousden K. H. (2006) TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120 [DOI] [PubMed] [Google Scholar]
- 43. Matoba S., Kang J. G., Patino W. D., Wragg A., Boehm M., Gavrilova O., Hurley P. J., Bunz F., Hwang P. M. (2006) p53 regulates mitochondrial respiration. Science 312, 1650–1653 [DOI] [PubMed] [Google Scholar]
- 44. Fantin V. R., St-Pierre J., Leder P. (2006) Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9, 425–434 [DOI] [PubMed] [Google Scholar]
- 45. Dang C. V., Kim J. W., Gao P., Yustein J. (2008) The interplay between MYC and HIF in cancer. Nat. Rev. Cancer 8, 51–56 [DOI] [PubMed] [Google Scholar]
- 46. Hashimi S. T., Fulcher J. A., Chang M. H., Gov L., Wang S., Lee B. (2009) MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood 114, 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Toyota M., Suzuki H., Sasaki Y., Maruyama R., Imai K., Shinomura Y., Tokino T. (2008) Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 68, 4123–4132 [DOI] [PubMed] [Google Scholar]
- 48. Yamakuchi M., Ferlito M., Lowenstein C. J. (2008) miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Luo J., Su F., Chen D., Shiloh A., Gu W. (2000) Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408, 377–381 [DOI] [PubMed] [Google Scholar]
- 50. Grönroos E., Terentiev A. A., Punga T., Ericsson J. (2004) YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. U.S.A. 101, 12165–12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sui G., Affar el B., Shi Y., Brignone C., Wall N. R., Yin P., Donohoe M., Luke M. P., Calvo D., Grossman S. R., Shi Y. (2004) Yin Yang 1 is a negative regulator of p53. Cell 117, 859–872 [DOI] [PubMed] [Google Scholar]
- 52. Chen Q. R., Yu L. R., Tsang P., Wei J. S., Song Y. K., Cheuk A., Chung J. Y., Hewitt S. M., Veenstra T. D., Khan J. (2011) Systematic Proteome Analysis Identifies Transcription Factor YY1 as a Direct Target of miR-34a. J. Proteome Res. 10, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Senese S., Zaragoza K., Minardi S., Muradore I., Ronzoni S., Passafaro A., Bernard L., Draetta G. F., Alcalay M., Seiser C., Chiocca S. (2007) Role for histone deacetylase 1 in human tumor cell proliferation. Mol. Cell. Biol. 27, 4784–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamaguchi T., Cubizolles F., Zhang Y., Reichert N., Kohler H., Seiser C., Matthias P. (2010) Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 24, 455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lagger G., Doetzlhofer A., Schuettengruber B., Haidweger E., Simboeck E., Tischler J., Chiocca S., Suske G., Rotheneder H., Wintersberger E., Seiser C. (2003) The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol. Cell. Biol. 23, 2669–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zupkovitz G., Grausenburger R., Brunmeir R., Senese S., Tischler J., Jurkin J., Rembold M., Meunier D., Egger G., Lagger S., Chiocca S., Propst F., Weitzer G., Seiser C. (2010) The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol. Cell. Biol. 30, 1171–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Janssen J. W., Schulz A. S., Steenvoorden A. C., Schmidberger M., Strehl S., Ambros P. F., Bartram C. R. (1991) A novel putative tyrosine kinase receptor with oncogenic potential. Oncogene 6, 2113–2120 [PubMed] [Google Scholar]
- 58. O'Bryan J. P., Frye R. A., Cogswell P. C., Neubauer A., Kitch B., Prokop C., Espinosa R., 3rd, Le Beau M. M., Earp H. S., Liu E. T. (1991) axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol. Cell. Biol. 11, 5016–5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Holland S. J., Powell M. J., Franci C., Chan E. W., Friera A. M., Atchison R. E., McLaughlin J., Swift S. E., Pali E. S., Yam G., Wong S., Lasaga J., Shen M. R., Yu S., Xu W., Hitoshi Y., Bogenberger J., Nör J. E., Payan D. G., Lorens J. B. (2005) Multiple roles for the receptor tyrosine kinase axl in tumor formation. Cancer Res. 65, 9294–9303 [DOI] [PubMed] [Google Scholar]
- 60. Vajkoczy P., Knyazev P., Kunkel A., Capelle H. H., Behrndt S., von Tengg-Kobligk H., Kiessling F., Eichelsbacher U., Essig M., Read T. A., Erber R., Ullrich A. (2006) Dominant-negative inhibition of the Axl receptor tyrosine kinase suppresses brain tumor cell growth and invasion and prolongs survival. Proc. Natl. Acad. Sci. U.S.A. 103, 5799–5804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang Y. X., Knyazev P. G., Cheburkin Y. V., Sharma K., Knyazev Y. P., Orfi L., Szabadkai I., Daub H., Kéri G., Ullrich A. (2008) AXL is a potential target for therapeutic intervention in breast cancer progression. Cancer Res. 68, 1905–1915 [DOI] [PubMed] [Google Scholar]
- 62. Corney D. C., Hwang C. I., Matoso A., Vogt M., Flesken-Nikitin A., Godwin A. K., Kamat A. A., Sood A. K., Ellenson L. H., Hermeking H., Nikitin A. Y. (2010) Frequent downregulation of miR-34 family in human ovarian cancers. Clin. Cancer Res. 16, 1119–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guha M., Altieri D. C. (2009) Survivin as a global target of intrinsic tumor suppression networks. Cell Cycle 8, 2708–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Merlos-Suárez A., Batlle E. (2008) Eph-ephrin signalling in adult tissues and cancer. Curr. Opin. Cell Biol. 20, 194–200 [DOI] [PubMed] [Google Scholar]
- 65. Bouhallier F., Allioli N., Lavial F., Chalmel F., Perrard M. H., Durand P., Samarut J., Pain B., Rouault J. P. (2010) Role of miR-34c microRNA in the late steps of spermatogenesis. Rna. 16, 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mudduluru G., Ceppi P., Kumarswamy R., Scagliotti G. V., Papotti M., Allgayer H. (2011) Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene 30, 2888–2889 [DOI] [PubMed] [Google Scholar]
- 67. Li W. Q., Chen C., Xu M. D., Guo J., Li Y. M., Xia Q. M., Liu H. M., He J., Yu H. Y., Zhu L. (2011) The rno-miR-34 family is upregulated and targets ACSL1 in dimethylnitrosamine-induced hepatic fibrosis in rats. Febs J. 278, 1522–1532 [DOI] [PubMed] [Google Scholar]
- 68. Liu C., Kelnar K., Liu B., Chen X., Calhoun-Davis T., Li H., Patrawala L., Yan H., Jeter C., Honorio S., Wiggins J. F., Bader A. G., Fagin R., Brown D., Tang D. G. (2011) The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 17, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Trang P., Wiggins J. F., Daige C. L., Cho C., Omotola M., Brown D., Weidhaas J. B., Bader A. G., Slack F. J. (2011) Systemic delivery of tumor suppressor microrna mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 19, 1116–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wiggins J. F., Ruffino L., Kelnar K., Omotola M., Patrawala L., Brown D., Bader A. G. (2010) Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 70, 5923–5930 [DOI] [PMC free article] [PubMed] [Google Scholar]