Abstract
Angiogenesis and osteogenesis are tightly coupled during bone development. We studied the effect of inhibition of Hif1α and Runt-related protein 2 (Runx2) on the formation of heterotopic ossification (HO). We constructed lentivirus vectors expressing Hif1α small interfering RNA (siRNA) and Runx2 siRNA. The inhibition of Hif1α function impaired osteoblast proliferation while osteoblasts differentiated normally. Osteoblasts lacking Runx2 proliferated normally while the differentiation was impaired. The osteoblast differentiation was significantly inhibited by co-Runx2 and Hif1α siRNA treatment. The formation of HO by inhibiting Runx2 and Hif1α in an animal model induced by Achilles tenotomy was investigated. The results showed that lacking of Runx2 and Hif1α could inhibit HO formation. Inhibition of Hif1α prevented HO formation only at the initial step and inhibition of Runx2 worked both at the initial step and after chondrogenesis. Angiogenesis and the expressions of osteogenic genes were downregulated in the Hif1α siRNA group. We found synergistic inhibition of endochondral bone formation by silencing Hif1α and Runx2. Our study provided new insight into the roles of Hif1α and Runx2 during the processes of endochondral bone formation, and had important implications for the new therapeutic methods to inhibit HO or to enhance bone formation.
Introduction
Heterotopic ossification (HO) is characterized by pathologic endochondral bone formation at nonskeletal sites and can lead to devastating consequences. The clinical spectrum of HO is wide, and it often results from traumatic injury. Additionally there is the rare hereditary form known as fibrodysplasia ossificans progressive.1 The histological features of developing HO are inflammation followed by woven bone formation, which later remodels to form lamellar bone. The exact etiology for acquired HO remains unclear.
During endochondral bone development chondrocytes undergo proliferation, hypertrophic differentiation, mineralization of the surrounding matrix, death, blood vessel invasion, and finally replacement of cartilage with bone. Angiogenesis and osteogenesis are tightly coupled during bone development.2 The hypoxia-inducible factor 1α (Hif1α) pathway has been identified as a key component in this process.3 Mesenchymal cells in the developing stroma elicit angiogenic signals to recruit new blood vessels into bone. Hif1α is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis by regulating Sox9.4 Dilling et al. found that vessel formation is induced before the appearance of cartilage in bone morphogenetic protein 2 (BMP2)-mediated HO, and suggested that vascular remodeling and growth may be essential to modify the microenvironment and enable engraftment of the necessary progenitors to form endochondral bone.5 Hypoxia is able to upregulate BMP2 in osteoblasts via the activation of Hif1α signaling pathway.6
In the process of osteoblasts differentiation a set of bone-specific genes (e.g., alkaline phosphatase (ALP) and osteocalcin genes) are activated. Runt-related protein 2 (Runx2) is a key regulator of osteocalcin and ALP gene promoters and cooperates with BMP-specific R-Smads.7 Runx2 is shown to be a master regulator of osteoblastic differentiation and is considered as a molecular switch in osteoblast biology.8,9 Furthermore, disruption of Runx2 by antisense oligonucleotides in osteoblast cultures inhibited expression of osteoblastic differentiation markers and formation of mineralized nodules.10 Our previous study showed that Runx2-specific small interfering RNA (siRNA) partially inhibits the formation of HO by inhibition of osteogenesis.11,12
In this study, RNA interference technology was used to silence the expression of Hif1α and Runx2 in vitro and in vivo. We studied the effects of lacking of Hif1α and Runx2 on the formation of HO at different phases by inhibition of angiogenesis and osteogenesis in animal model. And we attempted to outline the related cellular and molecular processes.
Results
Efficient inhibition of Runx2 and Hif1α expression by lentivirus-mediated transduction of siRNA against Runx2 and Hif1α in primary osteoblasts
Real-time reverse-transcription (RT)-PCR and western blot analysis revealed Runx2 siRNA and Hif1α siRNA led to remarkable suppression of Runx2 and Hif1α expression, whereas no change was observed after lentivirus containing scrambled siRNA transfection (Figure 1a,b). The inhibition effects persisted for at least 4 weeks after transduction. These results indicated that lentivirus-mediated transduction of siRNA into primary osteoblastic cells was feasible.
Figure 1.
The effects of small interfering RNAs (siRNAs) expressing against runt-related protein 2 (Runx2) and Hif1α on Runx2 and Hif1α expression in primary osteoblasts. (a) The results of real-time reverse-transcription (RT)-PCR analysis of Runx2 and Hif1α expression. (b) Corresponding western blot analysis for Runx2 and Hif1α expression. The inhibition effects persisted for at least 4 weeks after transduction.
Inhibition of Runx2 and Hif1α affects osteoblast differentiation in vitro
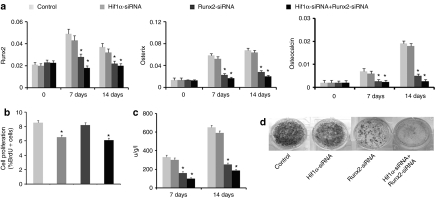
To evaluate the effects of inhibiting Runx2 and Hif1α expression on osteoblast differentiation, we examined the expression of osteoblast phenotypic genes. The expression levels of Runx2, osterix, and osteocalcin were clearly downregulated in the cells transduced with Runx2 siRNA, and were similar in scramble siRNA and Hif1α siRNA transduced cells (Figure 2a). We also examined the osteoblast proliferation to evaluate the effect of inhibition of Runx2 and Hif1α expression on the cells. The inhibition of Hif1α expression was associated with impaired osteoblast proliferation, as BrdU incorporation was significantly decreased in Hif1α siRNA transfected osteoblasts compared with controls. However, osteoblasts lacking Runx2 proliferated normally (Figure 2b).We measured the ALP activity and mineralized bone nodule formation in primary osteoblasts. The results showed that ALP activity increased as cells progressed through differentiation in control cells on days 7 and 14. However, this increase was blocked by Runx2 siRNA treatment, and no effects were found in scramble siRNA and Hif1α siRNA treatment (Figure 2c). Osteoblast terminal differentiation was determined by von Kossa staining and the results showed that mineralization was significantly suppressed in the presence of Runx2 siRNA, and no effects were found in scramble siRNA and Hif1α siRNA treatment (Figure 2d). The inhibition of Hif1α function impaired osteoblasts proliferation while osteoblasts differentiated normally. Osteoblasts lacking Runx2 proliferated normally while the differentiation was impaired. The osteoblast differentiation was significantly inhibited by co-Runx2 and Hif1α siRNA treatment.
Figure 2.
The effects of small interfering RNAs (siRNAs) against runt-related protein 2 (Runx2) and Hif1α on osteoblast differentiation in primary osteoblasts. (a) The results of real-time reverse-transcription (RT)-PCR of Runx2, osterix, and osteocalcin expression on day 0, 7, and 14. (b) The effects of lacking of Runx2 and Hif1α expression on the osteoblast proliferation. (c) Alkaline phosphatase (ALP) activity in the cell lysate was measured on days 7 and 14. ALP activity was significantly decreased by Runx2 siRNA treatment. (d) The terminal differentiation was determined by von Kossa staining. The results showed that mineralization was significantly suppressed in the presence of Runx2 siRNA on day 21. The inhibition of Hif1α function impaired osteoblast proliferation whereas osteoblasts differentiated normally. Osteoblasts lacking Runx2 proliferated normally whereas the differentiation was impaired. The osteoblast differentiation was significantly inhibited by co-Runx2 and Hif1α siRNA treatment.
The affects on HO after Achilles tenotomy by inhibition of Runx2 and Hif1α
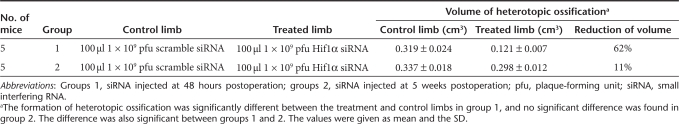
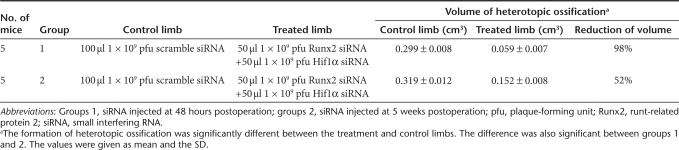
To evaluate the roles of Runx2 and Hif1α in the development of endochondral bone formation, we investigated the formation of HO by inhibiting Runx2 and Hif1α in an animal model induced by Achilles tenotomy. This model was used for the study of HO and the expressions of bone- and cartilage-related genes were found during the formation of new bone.13 At different phases of HO development, 48 hours (at the initial step) and 5 weeks (after chondrogenesis) after Achilles tenotomy, each Achilles site was injected with 100-µl lentivirus vectors.
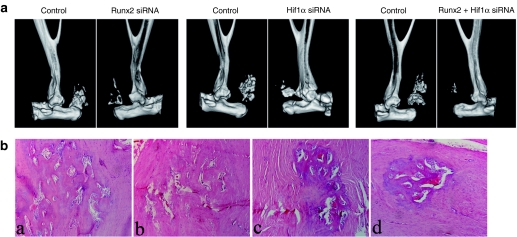
In the Runx2 siRNA-treated limbs less HO was observed with 86% reduction in group 1 and 37% reduction in group 2. The results were summarized in Table 1. In the Hif1α siRNA-treated limbs less HO was observed with 62% reduction in group 1 and 11% reduction in group 2. The results were summarized in Table 2. We further evaluated the ability of cotransduction of siRNA against Runx2 and Hif1α to inhibit HO after Achilles tenotomy. Significantly less HO was observed with 98 and 52% reduction, respectively. The results were summarized in Table 3. The representative three-dimensional computerized tomography scans and histological examinations were shown in Figure 3. When treated siRNAs were administered at 5 weeks postoperation in group 2 (after chondrogenesis) after Achilles tenotomy the volumes of HO were not decreased as much as those treated at 48 hours postoperation (at the initial step) in group 1. Inhibition of Hif1α prevented HO formation only at the initial step and inhibition of Runx2 worked both at the initial step and after chondrogenesis.
Table 1. Animals treated with Runx2 siRNA and the results.
Table 2. Animals treated with Hif1α siRNA and the results.
Table 3. Animals treated with Runx2 and Hif1α siRNA and the results.
Figure 3.
Micro-computerized tomography (CT) scans and histological examination demonstrated the effects of small interfering RNA (siRNAs) against Runt-related protein 2 (Runx2) and Hif1α on heterotopic ossification formation in animal models by 10 weeks. (a) Representative three-dimensional CT scans showed the results of siRNA injection at 48 hours postoperation. When treated siRNAs were administered at 5 weeks (after chondrogenesis) after Achilles tenotomy the volumes of HO were not decreased as much as those treated at 48 hours (data not shown). (b) Hematoxylin and eosin (HE) staining for the heterotopic ossification formation (magnification ×20). (a) Control limb. (b) Hif1α siRNA-treated limb. (c) Runx2 siRNA-treated limb. (d) co-Runx2 and Hif1α siRNA-treated limb.
Changes of bone- and cartilage-related gene expressions and angiogenesis by inhibition of Runx2 and Hif1α
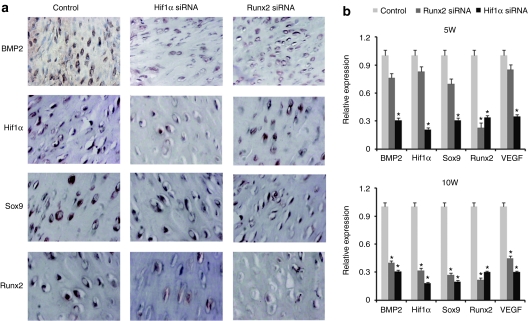
Our previous study showed that the expressions of BMP2, Hif1α, Sox9, and Runx2 were presented within the HO tissues.13 These cytokines expressions were also examined by immunohistochemical staining in the present study. All of the cytokines expressions were presented in the Hif1α siRNA-treated tissues, and stained weakly compared with the control tissues at 5 weeks postoperation (Figure 4a). The mRNA expressions of BMP2, vascular endothelial growth factor (VEGF), Hif1α, Sox9, and Runx2 were downregulated confirmed by real-time RT-PCR in the Hif1α siRNA-treated tissues (Figure 4b). The expressions of BMP2, Hif1α, Sox9 in Runx2 siRNA-treated groups were not significantly different compared with the control group (Figure 4b).
Figure 4.
The changes of bone- and cartilage-related genes expressions by inhibition of runt-related protein 2 (Runx2) and Hif1α. (a) The expressions of BMP2, Sox9, Runx2, and Hif1α were presented in the Hif1α siRNA-treated tissues, and stained weakly compared with the control tissues at 5 weeks postoperation. (b) The mRNA expressions of BMP2, vascular endothelial growth factor (VEGF), Hif1α, Sox9, and Runx2 were downregulated confirmed by real-time reverse-transcription (RT)-PCR in the Hif1α siRNA-treated tissues.
Angiogenesis in the ectopic bone formation was investigated using immunohistochemistry for CD31, a specific tissue marker of endothelial cells.14 Angiogenesis was less active in the tissues of the Hif1α siRNA group than in those of the control and Runx2 siRNA group at 7 days postoperation. Representative photos were shown in Figure 5. By 14 days postoperation, more extensive capillary network was formed in the control and Runx2 siRNA group compared with those in the Hif1α siRNA group. Representative photos were shown in Figure 5. The angiogenesis between Runx2 and control group was not significantly different. At 10 weeks postoperation, angiogenesis in all groups subsided (data not shown).
Figure 5.
Angiogenesis in the ectopic bone formation. (a,b) Angiogenesis was less active in the tissues of the Hif1α small interfering RNA (siRNA) group than in those of the control group at 7 days postoperation. (c,d) By 14 days postoperation, more extensive capillary network was formed in the control group compared with those in the Hif1α siRNA group. The angiogenesis between Runx2 and control group was not significant difference. Histomorphometry demonstrated that the relative capillary density was significantly lower in Hif1α siRNA group than control group at 14 days. At 10 weeks postoperation, angiogenesis in all groups subsided (data not shown).
Discussion
The physiological steps of bone formation in HO are similar with that during endochondral bone formation. In both cases, bone formation is a two-stage mechanism: first chondrocytes shape a template, on which osteoblasts then differentiate to form bone.5 Our findings demonstrated that inhibition of Hif1α prevented the HO formation induced by Achilles tenotomy at the initial step. Lacking of Runx2 inhibited the HO formation at initial steps and after chondrogenesis. More importantly, our results showed that inhibition of Hif1α and Runx2 acted synergistically to prevent HO formation. The mechanisms of inhibition HO by lacking of Hif1α could lie in the antiangiogenic (vessel formation) and antiosteogenic (bone-and cartilage-related genes, BMP2 and Runx2) pathways.
Hif1α is believed to play an important roles in endochondral ossification which coincides both spatially and temporally with capillary in-growth and angiogenesis. Evidences have shown that invoke a critical role for angiogenesis in the increased bone volume observed when the Hif1a/VEGF pathway is upregulated.15 Several studies showed that VEGF improved BMPs induce bone formation and bone healing through modulation of angiogenesis.16,17 Over expression of Hif1 in mature osteoblasts through disruption of the von Hippelb–Lindau (Vhl) protein profoundly increases angiogenesis and osteogenesis; these processes appear to be coupled by cell nonautonomous mechanisms involving the action of VEGF on the endothelial cells. Mice lacking Hif1a in osteoblasts had the reverse skeletal phenotype: long bones were significantly thinner and less vascularized than those of controls.2,18 Our study showed that lacking of Hif1a impaired the proliferation of osteoblast while osteoblasts differentiated normally in vitro. The VEGF expression was downregulated and angiogenesis was less active in the tissues of HO animal model treated with Hif1α siRNA group. We believe that antiangiogenesis may be a primary reason for inhibition of HO formation.
The chondrogenesis evoked by chondrogenic growth factors is further enhanced by hypoxia. And hypoxia also increased Akt phosphorylation and this was associated with a downstream increase in nuclear translocation and transactivation of Hif1α. The hypoxia-induced enhancement of chondrogenesis was abolished by siRNA-mediated knockdown of Hif1a.19 Hypoxia exposure increased BMP2 expression in cultured osteoblastic cells in a Hif-dependent manner involving activation of the ILK/Akt/mTOR pathway.6 Our study confirmed that the expressions of BMP2, Hif1α, Sox9, and Runx2 were downregulated in the Hif1α siRNA-treated tissues compared with the control tissues. So less expression of bone and cartilage-related genes could be one of factors for inhibition of HO.
Runx2, a runt-related transcription factor, is essential for osteoblast differentiation and regulates the expression of several osteoblastic genes including type I collagen, osteopontin, bone sialoprotein, and the skeletal-specific osteocalcin gene.7,8,9,10 Runx2–/– calvarial cells can not differentiate into osteoblasts both in vitro and in vivo, even in the presence of BMP2.20 Runx2–/– chondrocytes also spontaneously differentiated into adipocytes.21 Inhibition of Runx2 expression did not affect the angiogenesis in the HO formation. Our results demonstrated that Runx2 siRNA is capable of inhibiting the formation of HO induced by Achilles tenotomy, further confirmed that Runx2 is a master regulator of osteoblastic differentiation and a molecular switch in osteoblast biology.
We further evaluated of the ability of cotransduction of siRNA against Runx2 and Hif1α to inhibit HO after Achilles tenotomy. Significantly less HO was observed. TNP-470 is an antiangiogenic agent that strongly inhibits neovascular formation in vivo. Mori et al. showed that TNP-40 inhibits the biological activity of BMP2 in early stage of bone induction.22 BMP signals have pivotal roles in cartilage and bone development. BMP antagonists have been used to interrupt BMP signals to explore new methods for HO treatment. Small molecule inhibition of BMP type I receptor activity has been found to reduce HO.23 In the present study, combination inhibition of angiogenesis and osteoblast differentiation by Hif1α siRNA and Runx2 siRNA leads to dramatically prevent the formation of endochondral bone formation.
HO after tenotomy is usually occurred in rats and mice. It is not possible to decide the origin of the cartilage cells in this study. Damage of the capillary and haematoma may cause low oxygen tension in the impaired Achilles tendon. These factors combined most likely contribute to reduce oxygen tension in this model system and then upregulate Hif1α. Vascular invasion of the growth plate has been well documented to precede the recruitment of osteoblast progenitors to form the new bone.24,25,26,27 However, recruitment from the surrounding tissue is equally likely. Kaplan et al. showed local stem and progenitor contribution to heterotopic bone formation in a murine model of stem cell transplantation, and this process may require new vessel formation for establishment of these cells.28 Inhibition of Hif1α leads to reduce vessel formation, which may fail migration of host mesenchymal stem cells to the bone regeneration site. In this study, inhibiting Hif1α after chondrogenesis did not show significant reduction of HO. There are two possible reasons: (i) partial inhibition the expression of Hif1α by siRNA, (ii) the existing of the formed vessels in the initial step of endochondral ossification.
Lentiviral vectors can infect both dividing and nondividing cells, be produced in high titers and efficiently integrate into target cells, thus allowing the therapeutic genes to be expressed permanently.29 The formation of HO in animal model used in this study is a long period of 10 weeks. So, we chose lentiviral vectors for siRNA production in animal study. In clinical applications, lentiviral vectors must meet a higher safety standard. It is feasible to use lentivirus vectors in animal models, but it is a long way to go for clinical applications.
In summary, angiogenesis and osteogenesis are tightly coupled in endochondral bone formation. Angiogenic factor Hif1α and osteogenic factor Runx2 are essential for vessel and bone development, respectively. In this study, we found synergistic inhibition of endochondral bone formation by silencing Hif1α and Runx2 in trauma-induced HO. These results allow us to design more targeted therapies to reduce HO or enhance the bone formation.
Materials and Methods
Construction of lentiviral vectors expressing Hif1αsiRNA and Runx2 siRNA. The siRNA sequences specifically targeting rat Hif1α (accession no. NM024359) and Runx2 (accession no. NM053470) were designed through siRNA Target Finder (Ambion, Austin, TX) and the target site was at 407–427 for Hif1α and at 1057–1077 for Runx2.12,30 Scrambled siRNA was used as the negative control. Packaging, purification, and titer determination of the lentivirus were performed as described previously.31,32 All recombinant lentiviruses were produced by transient transfection of HEK293T cells according to standard protocols. Briefly, HEK293T cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (HyClone, Logan, UT) and penicillin/streptomycin (100 U/ml; North Pharmaceutical, Beijing, China). The subconfluent cells in a 10-cm culture dish were co-transfected with lentiviral vector (10 µg), and the lentiviral packaging vectors pRSV-REV (2 µg), pMDLg/pRRE (5 µg), and the vesicular stomatitis virus G glycoprotein expression vector pMD2G (3 µg). The viruses were collected from the culture supernatants on days 2 and 3 post-transfection, concentrated by ultracentrifugation for 1.5 hours at 25,000 r.p.m., and resuspended in phosphate-buffered saline (PBS). Titers were determined by infecting HEK293T cells with serial dilutions of concentrated lentivirus and counting enhanced green fluorescent protein-expressing cells after 48 hours under fluorescent microscopy. For a typical preparation, the titer was ~10 × 108 infectious units/ml.
Cell culture and transduction. Primary osteoblasts were isolated from the calvariae of 21-day fetal rats as previously described.12 In brief, cells were obtained from the calvariae (the dura and periosteum removed) by five sequential digestions of 10 min at 37 °C in PBS containing 0.05% collagenase. Cells from the forth and fifth digests were used in the present study. Cells were suspended in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone), penicillin/streptomycin (100 units/ml) at 37 °C in a humidified atmosphere of 5% carbon dioxide and 95% air. Osteogenic differentiation was initiated by adding 50 µg/ml ascorbic acid and 10 mmol/l β-glycerol phosphate. For lentiviral transduction, the cells were transduced at different multiplicity of infection in the presence of polybrene (8 mg/ml; Sigma, Carlsbad, CA). The target cell transduction efficiency was monitored and analyzed under fluorescent microscopy as described previously.32
Osteoblast proliferation. Osteoblast proliferation was assessed by flow cytometry as described previously.18 Briefly, osteoblasts were plated in 6-well plates at 5,000 cells/cm2 and cultured in DMEM containing 1% fetal bovine serum for 48 hours. BrdU (10 mmol/l) was added to the medium for the last 24 hours before harvesting the cells. The cells were stained with anti-BrdU-APC and 7-amino-actinomycin D and analyzed by FACSCalibur (BD Biosciences, San Jose, CA). Twenty thousand events were collected for each sample, and the results were analyzed.
Quantitative real-time RT-PCR analysis. Total RNA of the specimens was extracted with Trizol reagent (Invitrogen, Carlsbad, CA) and quantified by UV spectroscopy at assigned time points postinduction. Complementary DNA synthesis was performed using total RNA (1 µg) as a template by oligo(dT) priming using the Superscript First Strand Synthesis System for RT-PCR (Invitrogen). Real-time RT-PCR was performed with an optional continuous fluorescence detection system (MJ Research, Waltham, MA). One micro liter of reverse transcribed product and 1× SYBR green (Molecular Probes, Eugene, OR) were included in 25-µl reaction mixture (10 mmol/l Tris–HCl, pH 8.3, 50 mmol/l KCL, 1.5 mmol/l MgCl2, 200 µmol/l of dNTP mix, 0.2 µmol/l of each primer and 1 unit of Taq DNA polymerase). Oligonucleotide primers (Supplementary Table S1) were designed using Oligo 6 primer analysis software. mRNA levels were normalized to GAPDH using the comparative cycle threshold (CT) method.13
Western blotting analysis. The harvested medium was collected on the indicated days. Protein concentration was measured by BCA protein assay kit (Pierce Biotechnology, Rockford, IL) using bovine serum albumin as the standard. Proteins were run on SDS-PAGE gels and electrotransferred to nitrocellulose membrane at 4 °C for 2 hours. The blots were probed with anti-Runx2 (Santa Cruz, CA) and Hif1α (Abcam Limited, Cambridge, UK) at 1:500 dilutions overnight at 4 °C. The proteins were detected by chemiluminescence according to manufacture's recommendations (ECL, Amersham Arlington Heights, IL). GAPDH was used as an internal control.
Osteoblast differentiation. For ALP activity assay, on days 7 and 14, cell lysates were harvested and ALP activity was measured using an ALP assay kit (Zhongsheng Biochemical, Beijing, China). The enzyme activity was normalized against the protein concentration and expressed as U/g/l. Protein concentration was measured by BAC protein assay kit (Pierce Biotechnology) using bovine serum albumin as the standard.
In order to determine extracellular matrix mineralization, on day 21 the cells were fixed for von Kossa silver staining. Briefly, fixed cells were stained with 5% silver nitrate for 30 min under sunlight, fixed with sodium thiosulfate for 5 minutes, rinsed with distilled water and mounted for inspection.
Animal experiments. All animal experimental protocols were approved by the Animal Care and Use Committee of Peking University Third Hospital and conformed to National Institutes of Health Guidelines.
Sprague–Dawley rats (male, 4 weeks) were randomly divided into designated groups and underwent bilateral midpoint Achilles tenotomy through a posterior approach. Incision was routinely closed with an interrupted 4-0 silk suture. At designated time point (at 48 hours and 5 weeks postoperation, respectively), each Achilles site was injected with 100 µl lentivirus vectors. The details of each group were shown in Tables 1–3. The surgeon was blinded with regard to which limb received treatment during the surgery. Another four rats in the groups (siRNA injected at 48 hours postoperation) were used for the analysis of cartilage and bone-related genes expression at 5 weeks postoperation.
At 10 weeks, the animals were scanned by Siemens Inveon Micro-computerized tomography (German). Images were obtained using a 30-µm isotropic voxel size imaging system with the standard algorithm with 80 kV, 80 µA, and 200 ms integration time. Three-dimensional reconstruction and bone volumes were performed with Inveon software workstation. The gastrocnemius muscles were removed and fixed in 10% neutral buffered formalin, and then the ossified tissues were decalcified with decalcifying solution composed 10% HCL and 0.1% EDTA. The samples were embedded in paraffin and sectioned. The slides were stained with hematoxylin and eosin.
Immunohistochemical staining analysis for bone and cartilage-related genes and blood vessels. For immunohistochemical staining, paraffin-embedded sections were deparaffinated and hydrated through graded alcohols. Primary antibodies were diluted in PBS as follows: BMP2 (Abcam) 1:100; BMP4 (Abcam) 1:100; sox9 (Abcam) 1:100; Runx2 (Santa Cruz, CA) 1:100, HIF1α (Abcam) 1:400; CD31 (Dako, Glostrup, Denmark) 1:250. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide in PBS. After blocking with goat serum (1:100) and incubation with the primary antibody at 4 °C overnight, sections were washed with PBS. After three times of washing, secondary antimouse immunoglobulin G was added and incubated for 1 hour at 37 °C. The colorization developed in DAB solution and counterstained by hematoxylin. Control sections were incubated in PBS without the primary antibody.
Microvessel numbers were counted in six images captured from Achilles areas using MicroImage v4.5 imaging software (Olympus, America, Long Island, NY). The number of microvessels was calculated as the mean of the numbers from the six pictures.33
Statistical analysis. All comparisons between treated and control limbs were made in the same animals so that animal-to-animal variations in the ability to form HO would not affect the results. The significance between the treated and control limbs was determined with use of a paired two-tailed test. The significance of multiple comparisons was determined using the Student–Newman–Keuls test. Data were considered statistically significant at P ≤ 0.05.
SUPPLEMENTARY MATERIAL Table S1. Specific primers used for real-time RT-PCR.
Acknowledgments
This research was supported by Beijing NOVA Program (2008B04) sponsored by Beijing Municipal Science and Technology Commission and Doctoral Program Foundation of Institutions of Higher Education of China. This work was performed in Institute of Sports Medicine, Peking University Third Hospital, Beijing, China. The authors declared no conflict of interest.
Supplementary Material
Specific primers used for real-time RT-PCR.
REFERENCES
- Pape HC, Marsh S, Morley JR, Krettek C., and, Giannoudis PV. Current concepts in the development of heterotopic ossification. J Bone Joint Surg Br. 2004;86:783–787. doi: 10.1302/0301-620x.86b6.15356. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wan C, Deng L, Liu X, Cao X, Gilbert SR.et al. (2007The hypoxia-inducible factor alpha pathway couples angiogenesis to osteogenesis during skeletal development J Clin Invest 1171616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araldi E., and, Schipani E. Hypoxia, HIFs and bone development. Bone. 2010;47:190–196. doi: 10.1016/j.bone.2010.04.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarilio R, Viukov SV, Sharir A, Eshkar-Oren I, Johnson RS., and, Zelzer E. HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development. 2007;134:3917–3928. doi: 10.1242/dev.008441. [DOI] [PubMed] [Google Scholar]
- Dilling CF, Wada AM, Lazard ZW, Salisbury EA, Gannon FH, Vadakkan TJ.et al. (2010Vessel formation is induced prior to the appearance of cartilage in BMP-2-mediated heterotopic ossification J Bone Miner Res 251147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng WP, Yang SN, Lai CH., and, Tang CH. Hypoxia induces BMP-2 expression via ILK, Akt, mTOR, and HIF-1 pathways in osteoblasts. J Cell Physiol. 2010;223:810–818. doi: 10.1002/jcp.22104. [DOI] [PubMed] [Google Scholar]
- Alliston T, Choy L, Ducy P, Karsenty G., and, Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K.et al. (1997Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts Cell 89755–764. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR.et al. (1997Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development Cell 89765–771. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL., and, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Lin L, Chen L, Wang H, Wei X, Fu X, Zhang J.et al. (2006Adenovirus-mediated transfer of siRNA against Runx2/Cbfa1 inhibits the formation of heterotopic ossification in animal model Biochem Biophys Res Commun 349564–572. [DOI] [PubMed] [Google Scholar]
- Xue T, Mao Z, Lin L, Hou Y, Wei X, Fu X.et al. (2010Non-virus-mediated transfer of siRNAs against Runx2 and Smad4 inhibit heterotopic ossification in rats Gene Ther 17370–379. [DOI] [PubMed] [Google Scholar]
- Lin L, Shen Q, Xue T., and, Yu C. Heterotopic ossification induced by Achilles tenotomy via endochondral bone formation: expression of bone and cartilage related genes. Bone. 2010;46:425–431. doi: 10.1016/j.bone.2009.08.057. [DOI] [PubMed] [Google Scholar]
- Muller WA, Ratti CM, McDonnell SL., and, Cohn ZA. A human endothelial cell-restricted, externally disposed plasmalemmal protein enriched in intercellular junctions. J Exp Med. 1989;170:399–414. doi: 10.1084/jem.170.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Shao J, Gilbert SR, Riddle RC, Long F, Johnson RS.et al. (2010Role of HIF-1alpha in skeletal development Ann N Y Acad Sci 1192322–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Usas A, Olshanski A, Ho AM, Gearhart B, Cooper GM.et al. (2005VEGF improves, whereas sFlt1 inhibits, BMP2-induced bone formation and bone healing through modulation of angiogenesis J Bone Miner Res 202017–2027. [DOI] [PubMed] [Google Scholar]
- Peng H, Wright V, Usas A, Gearhart B, Shen HC, Cummins J.et al. (2002Synergistic enhancement of bone formation and healing by stem cell-expressed VEGF and bone morphogenetic protein-4 J Clin Invest 110751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomento SH, Wan C, Cao X, Faugere MC, Bouxsein ML, Clemens TL.et al. (2010Hypoxia-inducible factors 1alpha and 2alpha exert both distinct and overlapping functions in long bone development J Cell Biochem 109196–204. [DOI] [PubMed] [Google Scholar]
- Kanichai M, Ferguson D, Prendergast PJ., and, Campbell VA. Hypoxia promotes chondrogenesis in rat mesenchymal stem cells: a role for AKT and hypoxia-inducible factor (HIF)-1alpha. J Cell Physiol. 2008;216:708–715. doi: 10.1002/jcp.21446. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Gao Y, Ueta C, Yamaguchi A., and, Komori T. Multilineage differentiation of Cbfa1-deficient calvarial cells in vitro. Biochem Biophys Res Commun. 2000;273:630–636. doi: 10.1006/bbrc.2000.2981. [DOI] [PubMed] [Google Scholar]
- Enomoto H, Furuichi T, Zanma A, Yamana K, Yoshida C, Sumitani S.et al. (2004Runx2 deficiency in chondrocytes causes adipogenic changes in vitro J Cell Sci 117Pt 3417–425. [DOI] [PubMed] [Google Scholar]
- Mori S, Yoshikawa H, Hashimoto J, Ueda T, Funai H, Kato M.et al. (1998Antiangiogenic agent (TNP-470) inhibition of ectopic bone formation induced by bone morphogenetic protein-2 Bone 2299–105. [DOI] [PubMed] [Google Scholar]
- Yu PB, Deng DY, Lai CS, Hong CC, Cuny GD, Bouxsein ML.et al. (2008BMP type I receptor inhibition reduces heterotopic [corrected] ossification Nat Med 141363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuru S, Tamai K, Yamazaki T, Yoshikawa H., and, Kaneda Y. Bone marrow-derived osteoblast progenitor cells in circulating blood contribute to ectopic bone formation in mice. Biochem Biophys Res Commun. 2007;354:453–458. doi: 10.1016/j.bbrc.2006.12.226. [DOI] [PubMed] [Google Scholar]
- Rüster B, Göttig S, Ludwig RJ, Bistrian R, Müller S, Seifried E.et al. (2006Mesenchymal stem cells display coordinated rolling and adhesion behavior on endothelial cells Blood 1083938–3944. [DOI] [PubMed] [Google Scholar]
- Reddi AH. Bone and cartilage differentiation. Curr Opin Genet Dev. 1994;4:737–744. doi: 10.1016/0959-437x(94)90141-o. [DOI] [PubMed] [Google Scholar]
- Suda RK, Billings PC, Egan KP, Kim JH, McCarrick-Walmsley R, Glaser DL.et al. (2009Circulating osteogenic precursor cells in heterotopic bone formation Stem Cells 272209–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan FS, Glaser DL, Shore EM, Pignolo RJ, Xu M, Zhang Y.et al. (2007Hematopoietic stem-cell contribution to ectopic skeletogenesis J Bone Joint Surg Am 89347–357. [DOI] [PubMed] [Google Scholar]
- Chang LJ., and, Gay EE. The molecular genetics of lentiviral vectors–current and future perspectives. Curr Gene Ther. 2001;1:237–251. doi: 10.2174/1566523013348634. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Kojima I, Ohse T, Inagi R, Miyata T, Ingelfinger JR.et al. (2005Hypoxia-inducible factor modulates tubular cell survival in cisplatin nephrotoxicity Am J Physiol Renal Physiol 289F1123–F1133. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J.et al. (2003A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference Nat Genet 33401–406. [DOI] [PubMed] [Google Scholar]
- Hu W, Huang J, Mahavadi S, Li F., and, Murthy KS. Lentiviral siRNA silencing of sphingosine-1-phosphate receptors S1P1 and S1P2 in smooth muscle. Biochem Biophys Res Commun. 2006;343:1038–1044. doi: 10.1016/j.bbrc.2006.03.079. [DOI] [PubMed] [Google Scholar]
- Hou Y, Mao Z, Wei X, Lin L, Chen L, Wang H.et al. (2009Effects of transforming growth factor-beta1 and vascular endothelial growth factor 165 gene transfer on Achilles tendon healing Matrix Biol 28324–335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specific primers used for real-time RT-PCR.