Abstract
The application of small interfering RNA (siRNA) for cancer treatment is a promising strategy currently being explored in early phase clinical trials. However, efficient systemic delivery limits clinical implementation. We developed and tested a novel delivery system comprised of (i) an internalizing streptavidin-conjugated monoclonal antibody (mAb-SA) directed against CD22 and (ii) a biotinylated diblock copolymer containing both a positively charged siRNA condensing block and a pH-responsive block to facilitate endosome release. The modular design of the carrier facilitates the exchange of different targeting moieties and siRNAs to permit its usage in a variety of tumor types. The polymer was synthesized using the reversible addition fragmentation chain transfer (RAFT) technique and formed micelles capable of binding siRNA and mAb-SA. A hemolysis assay confirmed the predicted membrane destabilizing activity of the polymer under acidic conditions typical of the endosomal compartment. Enhanced siRNA uptake was demonstrated in DoHH2 lymphoma and transduced HeLa-R cells expressing CD22 but not in CD22 negative HeLa-R cells. Gene knockdown was significantly improved with CD22-targeted vs. nontargeted polymeric micelles. Treatment of DoHH2 cells with CD22-targeted polymeric micelles containing 15 nmol/l siRNA produced 70% reduction of gene expression. This CD22-targeted polymer carrier may be useful for siRNA delivery to lymphoma cells.
Introduction
Over 65,000 new cases of non-Hodgkin lymphoma will be diagnosed in the United States alone in 2010.1 Despite advances in available treatments, >20,000 people will die from non-Hodgkin lymphoma, making this hematologic malignancy one of the top 10 causes of cancer-related deaths. Recently developed chemotherapeutic regimens and biologics such as rituximab have improved overall survival, however, most patients still relapse and innovative treatments are urgently needed.
Oligonucleotide-based drugs represent one promising strategy. The discovery of RNA interference has stimulated considerable research directed toward utilizing this endogenous pathway for therapeutic purposes including treatment of cancer.2,3 Synthetic double-stranded small interfering RNA (siRNA) activates the RNA interference pathway and directs the cleavage of target mRNA in the cytoplasm by the RNA-induced silencing complex culminating in the reduction of the encoded protein. Silencing of oncogene expression in tumors may promote apoptosis or enhance sensitivity to chemotherapy, thereby improving clinical outcome.3
A major obstacle to the use of therapeutic siRNA is the lack of an effective delivery system. A safe and reliable mode of systemic siRNA delivery in humans has yet to be established although early clinical trials are in progress.2,3,4 An ideal carrier protects siRNA from exogenous nucleases, prolongs its systemic half-life, and promotes specific uptake into diseased tissues. Additionally, the appropriate intracellular trafficking of siRNA from the endosome to the cytoplasmic RNA-induced silencing complex is necessary for gene silencing. Escape from the endosomal compartment is believed to be a major rate-limiting step for many delivery approaches.5 Furthermore, activation of toll-like receptors located within the endosome may result in cytokine release and potential clinical toxicity which may be a limitation to this intracellular delivery mechanism.2
Targeting delivery of siRNA via internalizing cell surface receptors is an appealing strategy to enhance tumor-specific uptake.6 We explored the use of a monoclonal antibody directed against CD22, a transmembrane protein preferentially expressed on mature B-lymphocytes and detected in 60–80% of B-cell malignancies.7,8,9 CD22 constitutively internalizes and binding of anti-CD22 antibodies induces rapid receptor-mediated endocytosis, making CD22 an attractive gateway for intracellular delivery of drugs.10,11,12,13 Monoclonal antibodies and antibody-drug conjugates directed against CD22 for non-Hodgkin lymphoma have been investigated.14,15,16,17,18,19 However, antibodies bound to CD22 are destined for lysosomal degradation unless endosomal escape occurs.10,11
Our group has developed a new class of pH-responsive diblock copolymers using reversible addition fragmentation chain transfer (RAFT) polymerization.20,21 The polymers form micelles that bind siRNA and undergo a functional transition to a membrane-destabilizing state in response to the acidic conditions found within the endosomal compartment. A biotin incorporated at a specified polymer chain-end enables the binding of a CD22 streptavidin-conjugated monoclonal antibody (mAb-SA) for specific cellular targeting. We demonstrate that this polymeric micelle system enhances siRNA uptake and mRNA knockdown in CD22-expressing cells.
Results
Synthesis and characterization of the biotinylated diblock copolymer
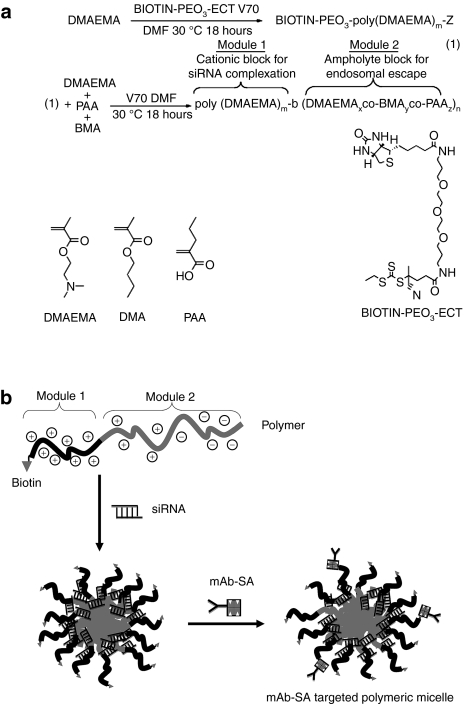
The biotinylated diblock copolymer was synthesized via controlled RAFT polymerization employing a biotin functionalized RAFT agent.20,21 This produced a linear polymer consisting of a single biotin molecule covalently attached to a cationic siRNA binding poly(DMAEMA) block followed by a second pH-responsive block containing propylacrylic acid (PAA), butyl methacrylate (BMA), and additional DMAEMA units (Figure 1a). The polymer chains spontaneously self-assemble under aqueous conditions to form micelles with a poly(DMAEMA) corona stabilizing the pH-responsive core. The addition of hydrophobic butyl methacrylate residues in the second block increases the hydrophobicity and membrane destabilizing activity of the copolymer and tunes the pKa of the propylacrylic acid carboxylate residues upward to endosomal values. The optimal incorporation of butyl methacrylate residues to achieve protonation within the desired pH range was between 40 and 50 mol% of the second block. As the micelles encounter an acidic pH gradient, the neutralization of the propylacrylic acid carboxylates together with protonation of the DMAEMA amino groups induces micelle and endosomal membrane destabilization to enhance siRNA delivery into the cytoplasm.
Figure 1.
Illustration of polymer synthesis and antibody-targeted polymeric micelle formation. (a) Polymerization utilizes a biotin functionalized reversible addition fragmentation chain transfer (RAFT) agent to produce a biotinylated poly(DMAEMA) block that condenses small interfering RNA (siRNA). Additional DMAEMA, PAA, and BMA subunits polymerize to form the second pH-responsive block for endosomal escape. Chemical structures of DMAEMA, BMA, and PAA monomers are shown. Details of chemical synthesis are described in the Supplementary Materials and Methods. (b) Polymeric micelle formation begins with incubation of siRNA with polymer and binding via electrostatic interaction to the poly(DMAEMA) cationic block. Subsequent addition of mAb-SA results in streptavidin binding to available surface biotin and generation of an antibody-targeted polymeric micelle. BMA, butylmethacrylate; DMAEMA, dimethylaminoethyl methacrylate; ECT (ethylsulfanylthiocarbonyl)sulfanyl pentanoic acid; mAb-SA, monoclonal antibody-streptavidin conjugate; PAA, polyacrylic acid; siRNA, small interfering RNA.
The formulation of targeted micelle siRNA carriers was initiated with the binding of siRNA to the polymer cationic block followed by addition of streptavidin-conjugated monoclonal antibody (mAb-SA) which binds to free biotin on the surface of the polymeric micelle (Figure 1b). Measurement of free biotin residues in the polymeric micelle using the 4-hydroxyazobenzene-2'carboxylic acid (HABA) assay indicated ~1 mol of available biotin sites per 8.6 mol of polymer chains (Supplementary Figure S1). This indicated that only one in eight biotin residues was available for mAb-SA binding likely due to steric-blocking effects. Electrostatic complexation of the cationic block of the carrier and negatively charged siRNA was confirmed by agarose gel electrophoresis (Supplementary Figure S2a). A minimum polymer to siRNA molar ratio of 2:1 was required for complete complexation (Supplementary Figure S2b). Addition of the mAb-SA conjugate did not interfere with siRNA complexation. Polyacrylamide gel protein electrophoresis under nondenaturing conditions showed specific binding between mAb-SA conjugate and polymer which was unaffected by siRNA complexation (Supplementary Figure S2c). Dynamic light scattering analysis demonstrated that polymers formed micelles measuring 35 nm in size. Complexation of siRNA or mAb-SA conjugates did not significantly change the particle size (34 and 38 nm, respectively). Each particle consists of ~35 polymer molecules, contains between 7 to 12 siRNA molecules, and bears 2–4 mAb-SA conjugates on its surface.
Evaluation of pH-dependent endosomolytic activity
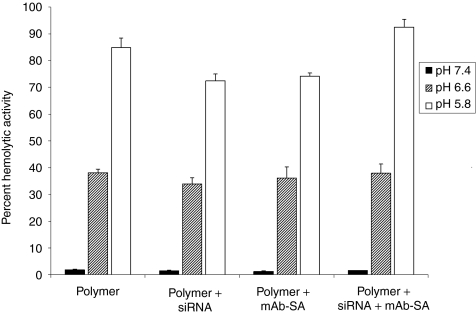
The red blood cell hemolysis assay provides a surrogate measurement of pH-dependent endosomolytic polymer activity that has previously been shown to correlate with delivery.20,29 Red blood cells were incubated in the presence of polymer alone or polymer complexed with siRNA, with or without mAb-SA conjugates, in buffered solutions at pH 7.4, 6.6, or 5.8, mimicking conditions in physiologic, early endosomal and late endosomal compartments, respectively. The polymer destabilized lipid membranes in a pH-dependent fashion that was unaffected by either complexation with siRNA or mAb-SA conjugate (Figure 2). Polymer hemolytic activity increased with rising buffer acidity whereas no significant hemolysis was observed at the physiological pH of 7.4. Hemolytic activity markedly rose at pH 6.6 which corresponds to the early endosomal compartment (range 33.9–38.1%). Maximal hemolysis by mAb-SA bearing polymeric micelles occurred at pH 5.8 (92.5 ± 3% red blood cell lysis). A similar profile of pH-dependent hemolytic activity was seen over a range of polymer concentrations (5–40 µg/ml) and mAb-SA conjugate ratios (data not shown). Notably, no significant hemolytic activity was observed at physiologic pH even at the highest polymer concentration.
Figure 2.
Evaluation of pH-dependent membrane destabilizing activity of polymeric micelles with and without mAb-SA using the red blood cell hemolysis assay. Comparative hemolytic activity was assessed in the presence of varying pH buffers. Values are relative to the positive control of 1% Triton X-100 (corresponding to 100% hemolytic activity). Disruption of the red blood cell membrane causes the release of free hemoglobin that is quantified by absorbance at 541 nm wavelength. Error bars represent the mean + s.d. from a single experiment conducted in triplicate. Similar results were obtained in three independent experiments. mAb-SA, monoclonal antibody-streptavidin conjugate; siRNA, small interfering RNA.
Antibody-mediated polymeric micelle internalization in CD22+ HeLa-R cells
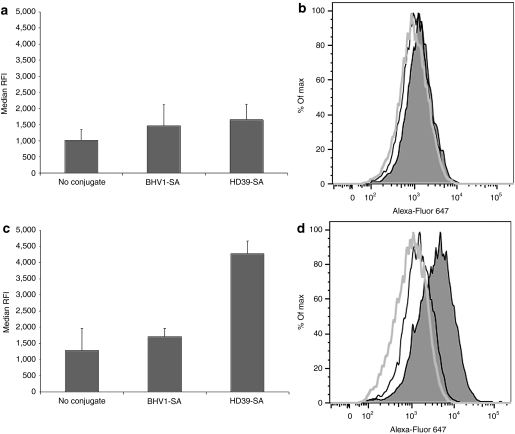
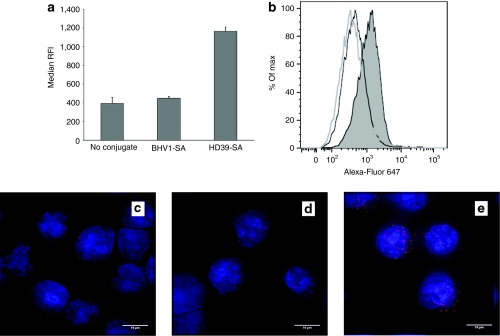
Paired cervical carcinoma cell lines consisting of HeLa-R cells and HeLa-R cells transduced with a vector encoding the human CD22 antigen (HeLa-R CD22+) were used to assess the extent of targeting conferred by the anti-CD22 mAb-SA HD39-SA. Cells were incubated with polymeric micelles containing fluorescently labeled siRNA and bearing HD39-SA, BHV1-SA or no conjugate and then analyzed for fluorescence by flow cytometry. BHV1-SA recognizes a bovine herpes virus antigen not expressed in human cells and served as a nontargeting control. No difference in median relative fluorescence intensity was observed between HeLa-R cells treated with HD39-SA, BHV1-SA, or naked polymeric micelles (Figure 3a,b). In contrast, the median relative fluorescence intensity of HeLa-R CD22+ cells treated with HD39-SA targeted polymeric micelle markedly exceeded that of naked polymeric micelle alone or BHV1-SA-targeted polymeric micelle indicating specificity of HD39-SA targeting and siRNA uptake to CD22-expressing cells (Figure 3c,d). Endosomal release of siRNA was demonstrated in HeLa-R cells treated with the related nonbiotinylated polymer. Dispersion of fluorescence in a cytosolic pattern was observed in cells after 24 hours of incubation with polymeric micelles (Supplementary Figure S3).
Figure 3.
Internalization of polymeric micelles containing Alexa-Fluor 647-labeled small interfering RNA (siRNA) with or without monoclonal antibody-streptavidin conjugate (mAb-SA) targeting. (a,b) HeLa-R or (c,d) HeLa-R CD22+ cells were incubated at 37 °C in the presence of polymeric micelles containing 20 nmol/l of Alexa-Fluor 647-labeled siRNA and bearing anti-CD22 HD39-SA conjugate, nontargeting BHV1-SA conjugate or no conjugate. After 1.5 hours of treatment, cells were trypsinized and rinsed extensively with phosphate-buffered saline (PBS) followed by acid wash to strip surface bound polymeric micelles then analyzed for fluorescence by flow cytometry. (a,c) Median relative fluorescence intensity (RFI) + s.d. of triplicate samples is shown. Data were concordant in three independent experiments. Representative histograms depict fluorescence intensity (horizontal axis) of (b) HeLa-R cells or (d) HeLa-R CD22+ cells treated with polymeric micelles bearing HD39-SA (shaded peak), BHV1-SA (black line, unshaded peak) or no conjugate (gray line, unshaded peak). Vertical axis is percentage of maximum intensity (% of Max).
Functional activity of siRNA delivered by antibody-targeted polymeric micelle
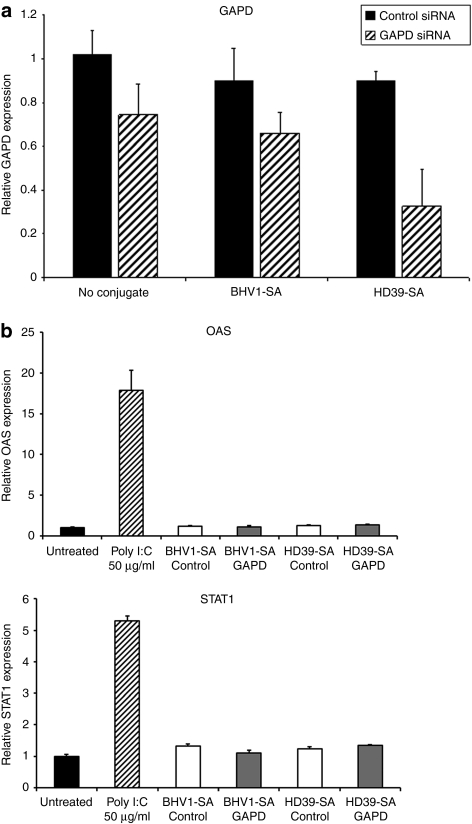
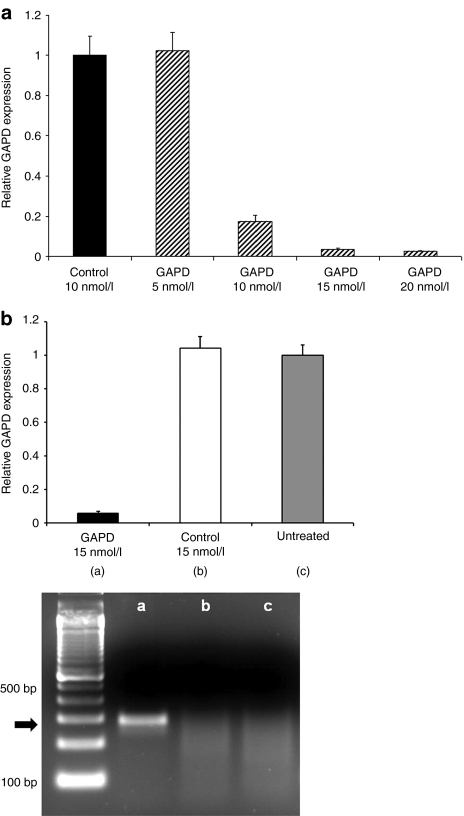
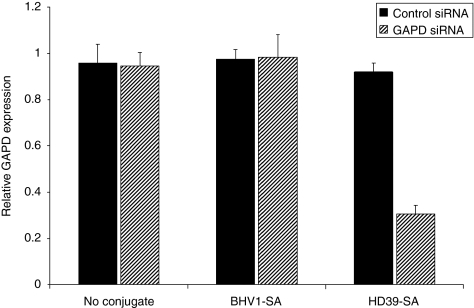
Polymer-mediated intracellular delivery should release internalized siRNA into the cytoplasm after receptor-mediated endocytosis, resulting in target gene transcript reduction. HeLaR-CD22+ cells were incubated with either naked or mAb-SA conjugate-bearing polymeric micelles containing 10 nmol/l siRNA directed against the housekeeping gene GAPD (glyceraldehyde-3-dehydrogenase) or a negative control siRNA with no sequence homology to any known human gene. Targeting polymeric micelles with HD39-SA improved knockdown of GAPD mRNA levels compared to BHV1-SA and no conjugate (67% vs. 26–35%) after 48 hours of treatment (Figure 4a). Treatment with polymeric micelles at a 15 nmol/l siRNA dose did not upregulate expression of the interferon response genes OAS (2′–5′ oligoadenylate synthetase) or STAT1 (signal transducer and activator of transcription-1) when compared to treatment with polyinosinic–polycytidylic acid (Poly I:C), a synthetic analog of double-stranded RNA that activates TLR-3 (toll-like receptor 3) (Figure 4b). Evaluation of interferon-β similarly did not show increased transcript levels after treatment with polymeric micelles (data not shown). Additionally, no excessive toxicity was detected after 24-hour treatment with nontargeted or CD22-targeted polymeric micelles at a 15 nmol/l siRNA dose. Cell viability ranged between 96 to 100% of untreated cells (Supplementary Figure S4). Dose titration of siRNA containing HD39-SA bearing polymeric micelles showed that a significant reduction of target gene expression was achievable with a siRNA dose of 10 nmol/l and almost complete silencing at 20 nmol/l (Figure 5a). Conversely, targeting of polymeric micelles with HD39-SA conjugate did not improve GAPD gene knockdown in HeLa-R cells not expressing CD22 (data not shown). Remaining GAPD protein levels were assessed by measuring GAPD protein activity. Cells were treated with polymeric micelles bearing either HD39-SA or BHV1-SA and containing 6.6 nmol/l of GAPD siRNA for 72 hours. Treatment with HD39-SA bearing polymeric micelles resulted in 24% greater reduction in GAPD enzyme activity compared to BHV1-SA bearing polymeric micelles (data not shown). Confirmation of the site specific cleavage of GAPD mRNA by siRNA was performed using the 5′-RNA-linker-mediated Rapid Amplification of cDNA Ends (5′-RLM-RACE) assay. The expected size PCR product (281 base pairs) was detected in cells treated with HD39-SA polymeric micelles containing 15 nmol/l GAPD siRNA after 24 hours but not in cells treated with HD39-SA polymeric micelles containing negative control siRNA or untreated cells (Figure 5b). Sequencing of the DNA extracted from the band verified its identity as the expected 3′ GAPD mRNA transcript cleavage product ligated to the RNA oligonucleotide linker. We have furthermore demonstrated specific reduction in mRNA levels of additional genes by quantitative reverse transcription-PCR using siRNA sequences directed against NAC1 and Mcl-1 transcripts (Supplementary Figure S5).
Figure 4.
Glyceraldehyde-3-phosphate dehydrogenase (GAPD) suppression and evaluation of inflammatory response mediated by monoclonal antibody-streptavidin conjugate (mAb-SA) targeted polymeric micelles. (a) GAPD mRNA levels were assayed by quantitative reverse transcription (qRT)-PCR 48 hours after transfection of HeLa-R CD22+ cells with polymeric micelles bearing HD39-SA (CD22-targeted conjugate), BHV1-SA (nontargeted conjugate) or no conjugate containing 10 nmol/l small interfering RNA (siRNA) directed against GAPD or a negative control siRNA with no sequence homology to known human genes. Values are normalized to the housekeeping gene PPIA (Cyclophilin A) and relative to GAPD expression in untreated cells. Error bars represent the mean GAPD expression + s.d. of triplicate samples. Results are representative of three independent experiments. (b) Evaluation of expression of interferon response genes OAS and STAT1 by qRT-PCR after treatment of HeLa-R CD22+ cells with HD39-SA or BHV1-SA polymeric micelles containing 15 nmol/l siRNA or Poly I:C (0.5, 5, and 50 µg/ml) for 24 hours. Values are normalized to the housekeeping gene PPIA (Cyclophilin A). Error bars represent the mean + s.d. of triplicate samples. Results are representative of three independent experiments. OAS, 2′–5′ oligoadenylate synthetase; Poly I:C, polyinosinic–polycytidylic acid; STAT1, signal transducer and activator of transcription-1.
Figure 5.
Dose response and 5′-RLM-RACE analysis of HeLa-R CD22+ cells treated with polymeric micelles bearing HD39-SA conjugate containing small interfering RNA (siRNA) directed against glyceraldehyde-3-phosphate dehydrogenase (GAPD) or a negative control siRNA with no sequence homology to known human genes. (a) Cells were harvested after 48 hours of treatment and analyzed using quantitative reverse transcription (RT)-PCR. Values are normalized to the housekeeping gene PPIA (Cyclophilin A) and relative to GAPD expression in untreated cells. Error bars represent the mean GAPD expression + s.d. of triplicate samples. (b) RNA was harvested from untreated HeLa-R CD22+ cells and HeLa-R CD22+ cells treated for 24 hours with polymeric micelles bearing HD39-SA conjugate at a dose of 15 nmol/l of GAPD siRNA or negative control siRNA. Relative GAPD expression normalized to the housekeeping gene PPIA was evaluated using quantitative RT-PCR. Error bars represent s.d. of triplicate samples. Specific detection of GAPD mRNA cleavage products using 5′-RLM-RACE assay was performed using RNA from each treatment group: GAPD siRNA (lane a), negative control siRNA (lane b), and untreated cells (lane c). PCR products were analyzed by agarose gel electrophoresis and run alongside a 100 base pair DNA ladder. The correct cleavage product indicated by the arrow has a size of 281 base pairs.
Antibody-mediated polymeric micelle internalization in CD22+ lymphoma cells
The ability of HD39-SA conjugate to enhance siRNA delivery in cells that naturally express CD22 was evaluated using the transformed follicular lymphoma cell line DoHH2. DoHH2 cells were incubated with polymeric micelles containing fluorescently labeled siRNA with and without HD39-SA or BHV1-SA, then analyzed by flow cytometric analysis (Figure 6a,b). Median relative fluorescence intensity in cells treated with polymeric micelles bearing HD39-SA conjugate was threefold higher compared to polymeric micelles without conjugate or bearing BHV1-SA, consistent with improved uptake with CD22 targeting. Microscopic examination of treated cells (Figure 6c–e) revealed a punctuate fluorescent pattern in the cytoplasm of the majority of cells after 30 minutes of incubation with HD39-SA-bearing polymeric micelles indicating rapid internalization of siRNA. Conversely, DoHH2 cells treated with polymeric micelles containing nontargeting BHV1-SA or no conjugate exhibited minimal fluorescence.
Figure 6.
Internalization of polymeric micelles containing Alexa-Fluor 647-labeled small interfering RNA (siRNA) with and without monoclonal antibody-streptavidin conjugate (mAb-SA) conjugate. DoHH2 lymphoma cells were incubated at 37 °C for 30 minutes with polymeric micelles bearing HD39-SA, BHV1-SA, or no conjugate then washed extensively with phosphate-buffered saline (PBS) then acid wash to remove surface based polymeric micelles prior to measuring fluorescence intensity by flow cytometric analysis. (a) The median relative fluorescence intensity (RFI) + s.d. of triplicate samples is shown. (b) A representative histogram depicting the relative fluorescence of DoHH2 cells treated with polymeric micelles bearing HD39-SA (shaded peak), BHV1-SA (black line, unshaded peak) or no conjugate (gray line, unshaded peak) is shown. Treated cells were cytospun and stained with DAPI nuclear stain (blue fluorescence). Representative random images captured from cells treated with polymeric micelles bearing no conjugate (c), nontargeting BHV1-SA (d), or CD22-targeted HD39-SA (e) are shown. Bar = 10 µm.
Gene expression knockdown in CD22+ lymphoma cells
Reduction of gene expression mediated by polymeric micelles targeted with HD39-SA in DoHH2 cells was evaluated using quantitative reverse transcription-PCR (Figure 7). The previous flow cytometry experiments with DoHH2 indicated that shorter incubation periods with polymeric micelles may be as efficacious as longer treatment times and may more closely simulate transient exposure to polymeric micelles in vivo. To test this, DoHH2 cells were treated with siRNA-containing polymeric micelles for 2 hours, rinsed, and then returned to culture in fresh media without polymeric micelles for 48 hours before assessment of gene expression. Treatment of DoHH2 cells with HD39-SA bearing polymeric micelles containing 15 nmol/l of GAPD siRNA yielded 70% reduction of GAPD expression relative to untreated cells. In contrast, treatment with negative control siRNA caused no change in GAPD expression. Polymeric micelles bearing BHV1-SA or no conjugate yielded no significant reduction in GAPD expression. GAPD protein levels were assessed using a GAPD enzyme activity assay. Treatment with HD39-SA bearing polymeric micelles containing 15 nmol/l of GAPD siRNA resulted in reduction of enzyme activity by 66% relative to untreated cells whereas treatment with control BHV1-SA polymeric micelles resulted in only 20% reduction in activity (data not shown).
Figure 7.
Anti-CD22 antibody-mediated reduction of gene expression in lymphoma cells. DoHH2 cells were treated with polymeric micelles bearing HD39-SA, BHV1-SA, or no conjugate and containing glyceraldehyde-3-phosphate dehydrogenase (GAPD) or negative control small interfering RNA (siRNA) for 48 hours. GAPD expression relative to untreated cells was measured by quantitative reverse transcription (RT)-PCR. Results are normalized to the housekeeping gene PPIA (Cyclophilin A) and relative to GAPD expression in untreated cells. Error bars represent the mean GAPD expression + s.d. of triplicate samples. GAPD, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
A major hurdle in the development of siRNA as a cancer therapeutic is the lack of a reliable mode of delivery in disseminated disease. Localized administration of siRNA via intravitreal or intradermal routes is effective in early phase clinical trials but systemic administration remains to be optimized.2,3 Recently, reduction of mRNA and protein expression in melanoma tumors was demonstrated after intravenous delivery of siRNA using nanoparticles bearing a transferrin ligand, providing a proof of principle that RNA interference may be utilized in human tumors.31 Previously, we described a new class of pH-responsive diblock copolymers that effectively delivers siRNA.20,21 We have now synthesized and tested a related biotinylated pH-responsive diblock copolymer that binds both siRNA and an anti-CD22 streptavidin-conjugated antibody that could be used for systemic treatment of non-Hodgkin lymphoma.
Our strategy involves a combinatorial approach utilizing both an internalizing antibody and an endosomolytic polymer to address some of the major obstacles that must be overcome for optimal administration of siRNA. Inefficient penetration of siRNA through the outer cell and endosomal membranes limits effective gene silencing. The enhanced permeability and retention effect permits accumulation of macromolecules in tumors based on abnormal vasculature and is the basis of tumor selectivity of liposomal formulations of drugs.32,33 However, intracellular uptake can be greatly enhanced by addition of internalizing ligands or antibodies to drug carriers.6 We hypothesized that polymeric micelles bearing an antibody targeting CD22 would selectively enhance siRNA uptake in cells expressing this antigen. In order to attach antibody to polymeric micelles, we utilized streptavidin as an adaptor molecule. Other groups have used avidin-conjugated antibodies or streptavidin-conjugated aptamers to directly bind biotinylated siRNA for delivery, however, the limited amount of siRNA conjugated per antibody are drawbacks to this approach.34,35,36 Furthermore, functional activity of siRNA is possible only if siRNA is able to escape into the cytoplasm after receptor-mediated endocytosis. We rationalized that this rate-limiting step would be overcome by attaching an endosomolytic polymer.
As expected, we observed improved internalization and gene silencing with CD22-targeted polymeric micelles in CD22-expressing cells. Reduction of gene expression through the RNA interference mechanism was confirmed by detection of the expected mRNA cleavage products only in cells treated with GAPD siRNA using the 5′-RLM-RACE assay. No significant toxicity or induction of immune response genes (STAT1, OAS, and interferon-β) was detected after treatment with polymeric micelles. Experiments using HeLa-R cell lines differing only in CD22 expression showed that enhanced uptake of polymeric micelles occurred only in cells expressing CD22. Some gene knockdown was noted in HeLa-R CD22+ cells with nontargeted polymeric micelles but this was anticipated under the in vitro transfection conditions employed. Polymeric micelles have a net positive charge and associate with the negatively charged cell membrane. This enhances nonreceptor mediated uptake with prolonged continuous incubation.37 Many adherent cell lines including HeLa-R are extremely permissive to transfection perhaps due to a higher basal rate of fluid phase pinocytosis.38 Consistent with these assumptions, a short 2-hour treatment period yielded reduction of both GAPD mRNA and protein activity levels in the DoHH2 cell line after treatment with CD22-targeted polymeric micelles whereas nontargeted polymeric micelles produced no effect. A 2-hour incubation time was similarly effective for uptake of CD22-targeted liposomal doxorubicin in studies by another group.16 The rapid kinetics of CD22 uptake may be advantageous for siRNA delivery in vivo because this minimizes loss through urinary excretion or nuclease degradation.12
Reduction of siRNA dose is important to reduce systemic off-target effects. Encouragingly, we observed >60% reduction of target GAPD expression using a relatively low dose of 15 nmol/l siRNA (7.5 pmol) in DoHH2 lymphoma cells. Nonadherent hematopoietic cells are notoriously difficult to transfect in vitro and typically require high doses of siRNA or physical methods such as nucleofection.38,39,40,41 Other investigators have also reported the use of antibodies to facilitate siRNA delivery to leukocytes but have required higher amounts of siRNA than used in our experiments to achieve the desired effect.42,43,44,45,46
Some limitations must be noted for our study. First, in vitro transfection does not precisely simulate conditions in vivo and evaluating siRNA delivery in a tumor xenograft model is the next essential step in the development of this technology. The potential immunogenicity of streptavidin-antibody conjugates is another concern. Although many cancer patients undergoing chemotherapy are immunocompromised, studies will be needed to evaluate the level of neutralizing antibody in cancer patients. Unpublished data by our group following human anti-mouse antibody formation in lymphoma patients treated with radiolabeled antibodies show that 18.6% of individuals (49 of 264) formed human anti-mouse antibody, suggesting neutralizing antibody formation may be low in this patient population. Alternatively, substitution of streptavidin with a mutant form engineered for reduced immunogenicity may be possible.47,48
In conclusion, we show that a streptavidin-conjugated anti-CD22 antibody enhances the cellular uptake of a biotinylated pH-responsive polymer resulting in efficient siRNA-mediated gene knockdown. Further studies will investigate the use of this carrier with clinically relevant siRNA gene targets in lymphoma and its efficacy in mouse tumor models. The modular design of the polymeric micelle system facilitates the exchange of different targeting moieties and siRNAs to permit its direct application to a variety of tumor types. Studies testing other streptavidin-conjugated antibodies are ongoing. This approach may be useful for the systemic delivery of siRNA for cancer treatment.
Materials and Methods
Polymer synthesis. Synthesis of biotin functional poly(dimethylaminoethyl methacrylate) [poly(DMAEMA)] macro chain transfer agent and corresponding poly[(DMAEMA)-b-(BMA)-co-(DMAEMA)-co-(PAA)] diblock copolymer was conducted as described previously with the addition of a biotinylated RAFT chain transfer agent and is described in detail in the Supplementary Figures S1–S5 and Supplementary Materials and Methods.20,21,22,23 Briefly, DMAEMA (2.5 g, 15.9 mmol), BIOTIN-PEO3-ECT (73 mg, 0.106 mmol), and V70 (3.27 mg, 0.01 mmol) were dissolved in N,N-dimethylformamide (DMF) (5 g). The solution was purged with nitrogen for 30 minutes and then allowed to react at 30 °C for 18 hours. The resultant polymer was isolated by repeated precipitation from ether into a 50-fold excess of pentane:ether (3:1 vol/vol). Following precipitation, the polymer was dissolved in deionized water and further purified by passing it through PD10 desalting columns. The final dry polymer was obtained by lyophilization.
Monoclonal antibody streptavidin conjugates. The hybridoma expressing HD39 (a murine IgG1 antihuman CD22 antibody) was a gift from Dr Edward Clark (University of Washington, Seattle, WA). HD39 was purified as previously described.24,25 BHV1 (a nontargeting isotype-matched IgG1 human anti-bovine herpes virus-1 antibody) was produced from a hybridoma obtained from American Type Culture Collection (Manassas, VA) and purified from ascites fluid over a HiTrap Protein G HP column (GE Healthcare, Piscataway, NJ). Streptavidin conjugation to HD39 and BHV1 antibodies was performed as previously described.26,27
Cell lines. The DoHH2 cell line was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany) and used within 6 months of the receipt or resuscitation of frozen aliquots. Authentication of the cell line was assured by the provider. The human HeLa-R cervical carcinoma cell line was a kind gift from Dr David Gius (National Institutes of Health). All cell culture reagents were obtained from Invitrogen (Carlsbad, CA). DoHH2 cells were grown in RPMI containing 10% fetal bovine serum, 100 IU/ml penicillin, 100 µg/ml streptomycin, and 2 mmol/l -glutamine. HeLa-R cells were grown in Dulbecco's modified Eagle's medium containing 5% fetal bovine serum, 100 IU/ml penicillin, 100 µg/ml streptomycin, and 2 mmol/l -glutamine. Cells were grown at 37 °C in 95% air/5% CO2 incubator. The HeLa-R CD22+ cell line was maintained in the same growth media as its parental line and was generated by transduction of HeLa-R cells with the retroviral LZRS pBMN vector-containing human CD22 cDNA as described previously.28 Transduced cells were selected for CD22 expression by labeling with antihuman CD22-PE/Cy5 antibody (BD Biosciences; San Diego, CA) then sorted using a BD FACS Vantage SE cell sorter.
Polymeric micelle formation and transfection. Cells were plated in 12-well tissue culture plates at a cell density of 50,000/well on the day before treatment (HeLa-R and HeLa-R CD22+) or 150,000/well on the day of treatment (DoHH2). Lyophilized polymer was dissolved in 100% ethanol at 20 mg/ml then diluted to a stock concentration of 1 mg/ml in phosphate-buffered saline (PBS) to form micelles. Polymer was then added to GAPD siRNA (sense strand 5′-GGUCGGAGUCAACGGAUUUTT-3′ Integrated DNA Technologies; Coralville, IA) or Silencer Negative Control #1 siRNA (Catalog AM4611; Ambion, Austin, TX) diluted in PBS at a polymer:siRNA molar ratio of 5:1 (for HeLa-R and HeLa-R CD22+ cells) or 3:1 (for DoHH2 cells) and incubated for 30 minutes at room temperature. The mAb-SA conjugates were then added to polymeric micelles at either 1:4 (for HeLa-R and HeLa-R CD22+ cells) or 1:1 (for DoHH2 cells) conjugate to available biotin molar ratios as determined by HABA assay (see Supplementary Figures S1–S5 and Supplementary Materials and Methods) and incubated for an additional 45 minutes at room temperature before treating cells in triplicate in 500 µl/well of Optimem or growth media containing 5% fetal bovine serum. Cells were exposed to the polymeric micelles continuously (HeLa-R and HeLa-R CD22+) or for a 2 hour pulse treatment followed by a media change (DoHH2) at 37 °C in a humidified incubator in 95% air/5% CO2. Cells were harvested at 24 or 48 hours for RNA extraction and at 72 hours for protein extraction.
Dynamic light scattering. Polymeric micelles were formed as described above with or without GAPD siRNA at a 3:1 polymer to siRNA ratio and with and without the addition of mAb-SA at a 1:1 mAb-SA to available biotin ratio. Polymeric micelles were characterized for size using Zetasizer NanoZS size analyzer (Malvern, Worcestershire, UK). Particle sizes expressed as effective diameters were measured at 25 °C using ZetaPals zeta potential analysis software (Brookhaven Instruments, Holtsville, NY) and calculated using the viscosity and refractive index of water at 25 °C.
Red blood cell hemolysis assay. A red blood cell hemolysis assay was used to assess pH-dependent disruption of lipid membranes as previously described.20,29 Polymeric micelles bearing mAb-SA were prepared as above. Samples containing 5–40 µg/ml of polymer were added to red blood cell suspensions and incubated at 37 °C for 1 hour. A portion of red blood cells was treated with 1% Triton X-100 as a positive control. Intact red blood cells were pelleted and the supernatant transferred to 96-well flat bottom assay plates. Disruption of the red blood cell membrane causes the release of free hemoglobin that is quantified by absorbance at 541 nm wavelength. Hemoglobin content in the supernatant was measured at absorbance of 541 nm with reference measured at absorbance 600 nm. Percent hemolysis was calculated relative to Triton X-100 treated samples.
Polymeric micelle internalization and microscopy. HeLaR-CD22 cells were plated at 200,000 per well in 12-well plates and allowed to adhere overnight. DoHH2 cells were plated on the day of transfection at 250,000 per well. Polymeric micelles were formed as above using AlexaFluor 647-labeled AllStars Negative Control siRNA (#1027287; Invitrogen) and added to cells in media containing 5% fetal bovine serum at a final concentration of 20 nmol/l of siRNA. After incubation at 37 °C to allow internalization for 30 to 90 minutes, cells were rinsed twice in cold PBS, trypsinized, rinsed again in PBS and then washed twice with 2 minute incubations in ice-cold RPMI (pH 2). After a final rinse in PBS, cells were resuspended in PBS with 0.1% fetal bovine serum and intracellular fluorescence was measured on a BD FACS Canto flow cytometer. Untreated cells were used to set the background reference. For microscopy, cells were treated as above but instead cytospun on slides, dried, then fixed with 10% neutral buffered formalin and rinsed in PBS prior to being coverslipped with Prolong Gold antifade reagent with DAPI (Invitrogen). Random fields were imaged with a DeltaVision RT wide-field deconvolution microscope (Applied Precision, Issaquah, WA) fitted with a Photometrics HQ scientific grade cooled CCD camera and an Olympus 100×/1.4 Plan Apochromatic objective. Three-dimensional data sets consisting of optical sections at 0.2 microns spacing were collected with the manufacturer's SoftWoRx software, and deconvolved using a constrained iterative algorithm. Scale bars were added with public domain software ImageJ.
Quantitative reverse transcription-PCR. RNA was isolated using the RNeasy mini kit (Qiagen, Valencia, CA) and reverse transcribed using TaqMan Reverse Transcription reagents (Applied Biosystems, Carlsbad, CA). PCR were run in duplicate using an ABI Prism 7900HT real-time PCR instrument. Primers with FAM and VIC-labeled compatible probes for multiplex PCR were obtained from Applied Biosystems: target gene GAPD (Catalog#4352934E), STAT1 (Catalog#Hs01013996_m1), OAS1 (Hs00973637_m1), IFNB1 (Catalog#Hs01077958_s1), and cyclophilin A (PPIA) internal control (Catalog#4326316E). Relative quantification of GAPD expression was based on the reference value from the untreated control.30
5′-RLM-RACE and sequencing. 5′ RLM-RACE was performed using the GeneRacer Kit (Invitrogen) with some modification as previously described.49 Briefly, 100 ng total RNA was directly ligated to 250 ng GeneRacer RNAOligo with T4 ligase. After phenol/chloroform extraction and ethanol precipitation, cDNA was synthesized using random primers. From this reaction, 1 µl was used for first round 5′RACE reaction using the GeneRacer 5′ primer and GAPD-specific reverse primer (5′-CCTGCAAATGAGCCCCAGCCTTCTC-3′) with the following cycling conditions: 1 cycle of 94 °C for 2 minutes, then 5 cycles of 94 °C for 30 seconds and 72 °C for 1 minute, then 5 cycles of 94 °C for 30 seconds and 70 °C for 1 minute, then 20 cycles of 94 °C for 30 seconds, and 68 °C for 1 minute. Second-round 5′ RACE reaction was then performed using 1 µl of the first-round reaction and internal GeneRacer 5′ nested and GAPD-specific nested (5′-CGCCAGCATCGCCCCACTTGATTTT-3′) primers using the above cycling conditions except for an extension time of 15 seconds and 25 cycles. PCR were performed using an Eppendorf Mastercycler thermocycler. PCR products were run on a 3% agarose gel containing ethidium bromide then excised and extracted using a QIAquick Gel Extraction kit (Qiagen). Sequencing was performed using the ABI BigDye Terminator v3.1 Cycle Sequencing kit and subsequently analyzed on an ABI-3730xl DNA Analyzer (Applied Biosystems) per manufacturer′s protocol.
GAPD protein assay. GAPD activity was measured using the KDAlert GAPDH assay kit (Ambion). Cells were treated in quadruplicate then lysed 72 hours later. Lysate was transferred to a 96-well plate. KDAlert master mix was added and the fluorescence output was immediately read over a 4 minute period with an excitation wavelength of 560 nm and emission wavelength of 590 nm.
SUPPLEMENTARY MATERIAL Figure S1. HABA (4-hydroxyazobenzene-2'carboxylic acid) assay to determine the amount of free biotin present on polymeric micelles. Figure S2. Gel shift assays demonstrating polymer binding to siRNA and mAb-SA conjugate. Figure S3. Fluorescence microscopy of HeLa-R cells after treatment with nonbiotinylated polymeric micelle carrier containing 100 nmol/l of Cy3-labeled siRNA (red fluorescence). Figure S4. HeLa-R CD22+ cell viability after 24 hours of treatment with BHV1-SA or HD39-SA targeted micelles at a dose of 15 nmol/l siRNA. Figure S5. Suppression of Mcl-1 and NAC1 gene expression by mAb-SA targeted polymeric micelles. Materials and Methods.
Acknowledgments
We thank Aimee Kenoyer and Scott James at Fred Hutchinson Cancer Research Center for providing BHV-1 antibody and retroviral supernatant, respectively, Don Hamlin (University of Washington) for producing antibody conjugates, and Wilo Dietrich for graphics support. We acknowledge helpful discussions with Bilal Ghosn, John Wilson, Craig Duvall, and Danielle Benoit (University of Washington). This research was supported by funding from the following grants: NIH 5R01EB002991-07 (P.S.S, O.W.P); Washington State Life Sciences Discovery Fund #2496490 (P.S.S., O.W.P.); NCI 2K12CA076930-11 (M.C.P-W); Lymphoma Research Foundation Fellowship Award (M.C.P-W). O.W.P. and P.S.S. are cofounders and consultants for PhaseRx, a company that has licensed some of the drug delivery technology represented in this research. However, this work was conducted independently of PhaseRx under NIH and WSLSDF support.
Supplementary Material
HABA (4-hydroxyazobenzene-2'carboxylic acid) assay to determine the amount of free biotin present on polymeric micelles.
Gel shift assays demonstrating polymer binding to siRNA and mAb-SA conjugate.
Fluorescence microscopy of HeLa-R cells after treatment with nonbiotinylated polymeric micelle carrier containing 100 nmol/l of Cy3-labeled siRNA (red fluorescence).
HeLa-R CD22+ cell viability after 24 hours of treatment with BHV1-SA or HD39-SA targeted micelles at a dose of 15 nmol/l siRNA.
Suppression of Mcl-1 and NAC1 gene expression by mAb-SA targeted polymeric micelles.
REFERENCES
- Jemal A, Siegel R, Xu J., and, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Tiemann K., and, Rossi JJ. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol Med. 2009;1:142–151. doi: 10.1002/emmm.200900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalon C, Rao DD., and, Nemunaitis J. Potential use of RNA interference in cancer therapy. Expert Rev Mol Med. 2010;12:e26. doi: 10.1017/S1462399410001584. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R., and, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominska M., and, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123 Pt 8:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- Ikeda Y., and, Taira K. Ligand-targeted delivery of therapeutic siRNA. Pharm Res. 2006;23:1631–1640. doi: 10.1007/s11095-006-9001-x. [DOI] [PubMed] [Google Scholar]
- Haas KM, Sen S, Sanford IG, Miller AS, Poe JC., and, Tedder TF. CD22 ligand binding regulates normal and malignant B lymphocyte survival in vivo. J Immunol. 2006;177:3063–3073. doi: 10.4049/jimmunol.177.5.3063. [DOI] [PubMed] [Google Scholar]
- Vitetta ES, Stone M, Amlot P, Fay J, May R, Till M.et al. (1991Phase I immunotoxin trial in patients with B-cell lymphoma Cancer Res 514052–4058. [PubMed] [Google Scholar]
- Coleman M, Goldenberg DM, Siegel AB, Ketas JC, Ashe M, Fiore JM.et al. (2003Epratuzumab: targeting B-cell malignancies through CD22 Clin Cancer Res 910 Pt 23991S–3994S. [PubMed] [Google Scholar]
- Shan D., and, Press OW. Constitutive endocytosis and degradation of CD22 by human B cells. J Immunol. 1995;154:4466–4475. [PubMed] [Google Scholar]
- Press OW, Farr AG, Borroz KI, Anderson SK., and, Martin PJ. Endocytosis and degradation of monoclonal antibodies targeting human B-cell malignancies. Cancer Res. 1989;49:4906–4912. [PubMed] [Google Scholar]
- Carnahan J, Wang P, Kendall R, Chen C, Hu S, Boone T.et al. (2003Epratuzumab, a humanized monoclonal antibody targeting CD22: characterization of in vitro properties Clin Cancer Res 910 Pt 23982S–3990S. [PubMed] [Google Scholar]
- Du X, Beers R, Fitzgerald DJ., and, Pastan I. Differential cellular internalization of anti-CD19 and -CD22 immunotoxins results in different cytotoxic activity. Cancer Res. 2008;68:6300–6305. doi: 10.1158/0008-5472.CAN-08-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JP, Schuster SJ, Emmanouilides C, Couture F, Teoh N, Wegener WA.et al. (2008Durable complete responses from therapy with combined epratuzumab and rituximab: final results from an international multicenter, phase 2 study in recurrent, indolent, non-Hodgkin lymphoma Cancer 1132714–2723. [DOI] [PubMed] [Google Scholar]
- Leonard JP, Coleman M, Ketas JC, Chadburn A, Furman R, Schuster MW.et al. (2004Epratuzumab, a humanized anti-CD22 antibody, in aggressive non-Hodgkin's lymphoma: phase I/II clinical trial results Clin Cancer Res 105327–5334. [DOI] [PubMed] [Google Scholar]
- O'Donnell RT, Martin SM, Ma Y, Zamboni WC., and, Tuscano JM. Development and characterization of CD22-targeted pegylated-liposomal doxorubicin (IL-PLD) Invest New Drugs. 2010;28:260–267. doi: 10.1007/s10637-009-9243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield E, Amlot P, Pastan I., and, FitzGerald DJ. Recombinant RFB4 immunotoxins exhibit potent cytotoxic activity for CD22-bearing cells and tumors. Blood. 1997;90:2020–2026. [PubMed] [Google Scholar]
- Bogner C, Dechow T, Ringshausen I, Wagner M, Oelsner M, Lutzny G.et al. (2010Immunotoxin BL22 induces apoptosis in mantle cell lymphoma (MCL) cells dependent on Bcl-2 expression Br J Haematol 14899–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetie MA, May RD, Till M, Uhr JW, Ghetie V, Knowles PP.et al. (1988Evaluation of ricin A chain-containing immunotoxins directed against CD19 and CD22 antigens on normal and malignant human B-cells as potential reagents for in vivo therapy Cancer Res 482610–2617. [PubMed] [Google Scholar]
- Convertine AJ, Benoit DS, Duvall CL, Hoffman AS., and, Stayton PS. Development of a novel endosomolytic diblock copolymer for siRNA delivery. J Control Release. 2009;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Convertine AJ, Diab C, Prieve M, Paschal A, Hoffman AS, Johnson PH.et al. (2010pH-Responsive Polymeric Micelle Carriers for siRNA Drugs Biomacromoleculesepub ahead of print) [DOI] [PMC free article] [PubMed]
- Bathfield M, D'Agosto F, Spitz R, Charreyre MT., and, Delair T. Versatile precursors of functional RAFT agents. Application to the synthesis of bio-related end-functionalized polymers. J Am Chem Soc. 2006;128:2546–2547. doi: 10.1021/ja057481c. [DOI] [PubMed] [Google Scholar]
- Hong C, Pan C. Direct synthesis of biotinylated stimuli-responsive polymer and diblock copolymer by RAFT polymerization using biotinylated trithiocarbonate as RAFT agent. Macromolecules. 2006;39:3517–3524. [Google Scholar]
- Pagel JM, Pantelias A, Hedin N, Wilbur S, Saganic L, Lin Y.et al. (2007Evaluation of CD20, CD22, and HLA-DR targeting for radioimmunotherapy of B-cell lymphomas Cancer Res 675921–5928. [DOI] [PubMed] [Google Scholar]
- Dörken B, Moldenhauer G, Pezzutto A, Schwartz R, Feller A, Kiesel S.et al. (1986HD39 (B3), a B lineage-restricted antigen whose cell surface expression is limited to resting and activated human B lymphocytes J Immunol 1364470–4479. [PubMed] [Google Scholar]
- Hylarides MD, Mallett RW., and, Meyer DL. A robust method for the preparation and purification of antibody/streptavidin conjugates. Bioconjug Chem. 2001;12:421–427. doi: 10.1021/bc0001286. [DOI] [PubMed] [Google Scholar]
- Pagel JM, Hedin N, Subbiah K, Meyer D, Mallet R, Axworthy D.et al. (2003Comparison of anti-CD20 and anti-CD45 antibodies for conventional and pretargeted radioimmunotherapy of B-cell lymphomas Blood 1012340–2348. [DOI] [PubMed] [Google Scholar]
- James SE, Greenberg PD, Jensen MC, Lin Y, Wang J, Till BG.et al. (2008Antigen sensitivity of CD22-specific chimeric TCR is modulated by target epitope distance from the cell membrane J Immunol 1807028–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank C, Oberhauser B, Mechtler K, Koch C., and, Wagner E. The influence of endosome-disruptive peptides on gene transfer using synthetic virus-like gene transfer systems. J Biol Chem. 1994;269:12918–12924. [PubMed] [Google Scholar]
- Livak KJ., and, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA.et al. (2010Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles Nature 4641067–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Nakamura H, Maeda H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv Drug Deliv Rev. 2010. pp. 136–151. [DOI] [PubMed]
- Matsumura Y., and, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46 12 Pt 1:6387–6392. [PubMed] [Google Scholar]
- Xia CF, Boado RJ., and, Pardridge WM. Antibody-mediated targeting of siRNA via the human insulin receptor using avidin-biotin technology. Mol Pharm. 2009;6:747–751. doi: 10.1021/mp800194y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia CF, Zhang Y, Zhang Y, Boado RJ., and, Pardridge WM. Intravenous siRNA of brain cancer with receptor targeting and avidin-biotin technology. Pharm Res. 2007;24:2309–2316. doi: 10.1007/s11095-007-9460-8. [DOI] [PubMed] [Google Scholar]
- Chu TC, Twu KY, Ellington AD., and, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner SD., and, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- Goldmacher VS, Tinnel NL., and, Nelson BC. Evidence that pinocytosis in lymphoid cells has a low capacity. J Cell Biol. 1986;102:1312–1319. doi: 10.1083/jcb.102.4.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearling JL, Park EJ, Dunning P, Baker A, Fahey F, Treves ST.et al. (2010Detection of intestinal inflammation by MicroPET imaging using a (64)Cu-labeled anti-beta(7) integrin antibody Inflamm Bowel Dis 161458–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkerova M, Klamova H, Brdicka R., and, Bruchova H. Targeting of gene expression by siRNA in CML primary cells. Mol Biol Rep. 2007;34:27–33. doi: 10.1007/s11033-006-9006-x. [DOI] [PubMed] [Google Scholar]
- Seiffert M, Stilgenbauer S, Döhner H., and, Lichter P. Efficient nucleofection of primary human B cells and B-CLL cells induces apoptosis, which depends on the microenvironment and on the structure of transfected nucleic acids. Leukemia. 2007;21:1977–1983. doi: 10.1038/sj.leu.2404863. [DOI] [PubMed] [Google Scholar]
- Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL.et al. (2008T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice Cell 134577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM.et al. (2005Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors Nat Biotechnol 23709–717. [DOI] [PubMed] [Google Scholar]
- Peer D, Zhu P, Carman CV, Lieberman J., and, Shimaoka M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc Natl Acad Sci USA. 2007;104:4095–4100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer D, Park EJ, Morishita Y, Carman CV., and, Shimaoka M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Peer D, Kumar P, Subramanya S, Wu H, Asthana D.et al. (2010RNAi-mediated CCR5 silencing by LFA-1-targeted nanoparticles prevents HIV infection in BLT mice Mol Ther 18370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DL, Schultz J, Lin Y, Henry A, Sanderson J, Jackson JM.et al. (2001Reduced antibody response to streptavidin through site-directed mutagenesis Protein Sci 10491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinol M, Casalini P, Maggiolo M, Canevari S, Omodeo ES, Caliceti P.et al. (1998Biochemical modifications of avidin improve pharmacokinetics and biodistribution, and reduce immunogenicity Br J Cancer 78189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasham A, Herbert M, Coppieters 't Wallant N, Patel R, Feng S, Eszes M.et al. (2010A rapid and sensitive method to detect siRNA-mediated mRNA cleavage in vivo using 5' RACE and a molecular beacon probe Nucleic Acids Res 38e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HABA (4-hydroxyazobenzene-2'carboxylic acid) assay to determine the amount of free biotin present on polymeric micelles.
Gel shift assays demonstrating polymer binding to siRNA and mAb-SA conjugate.
Fluorescence microscopy of HeLa-R cells after treatment with nonbiotinylated polymeric micelle carrier containing 100 nmol/l of Cy3-labeled siRNA (red fluorescence).
HeLa-R CD22+ cell viability after 24 hours of treatment with BHV1-SA or HD39-SA targeted micelles at a dose of 15 nmol/l siRNA.
Suppression of Mcl-1 and NAC1 gene expression by mAb-SA targeted polymeric micelles.