Abstract
Langerhans cell histiocytosis (LCH) is a rare disease characterized by heterogeneous lesions including CD207+/CD1a+ dendritic cells that can result in significant morbidity and mortality. The etiology of LCH remains speculative, and neoplastic and inflammatory origins have been debated for decades. A recent study identified abundant interleukin-17 (IL-17A) protein in dendritic cells in LCH lesions as well as in plasma from patients with active disease. Furthermore, it identified dendritic cells as a novel source of IL-17A expression. However, subsequent studies from our research group failed to identify any IL-17A gene expression from CD207+ dendritic cells or CD3+ T cells in LCH lesions. In this study, further investigation once again fails to identify any cells in LCH lesions with IL-17A gene expression. Furthermore, IL-17A antigen is undetectable in LCH lesion lysates with western blotting, immunoprecipitation, spectral analysis, and enzyme-linked immunosorbent assay (ELISA). Western blots, immunoprecipitation, and ELISA experiments also demonstrate that antibodies used in original studies that established the IL-17A hypothesis for pathogenesis of LCH recognize nonspecific proteins. We conclude that evidence for IL-17A as a significant factor in LCH remains inadequate and clinical trials targeting IL-17A remain unjustified.
Introduction
Langerhans cell histiocytosis (LCH) is a rare disease characterized by heterogeneous lesions including characteristic histiocytes. LCH is estimated to arise in 5–9 children per million and 1–2 adults per million.1,2,3,4 As the name indicates, the origin of the cells has been thought to be epidermal Langerhans cells of the skin due to the common findings of Birbeck granules with electron microscopy and co-expression of CD1a and CD207 cell surface proteins.5,6,7 More recent studies show that CD207 is not restricted to Langerhans cells, and gene expression profiles of LCH lesion CD207+ cells have features of immature myeloid dendritic cells, suggesting that the pathogenic cells in LCH may arise from circulating dendritic cells.8,9 The etiology of LCH remains speculative with debates spanning decades regarding inflammatory versus neoplastic origins.10,11,12,13 The clinical presentation of LCH ranges from single lesions that can be treated with local therapy to systemic disease that requires intensive chemotherapy or bone marrow transplant. Strategies for treating LCH are based on a lymphoma model of general immune suppression and cytotoxicity to rapidly proliferating cells. While outcomes have improved with coordinated efforts of international clinical trials, rational therapies are required for further advances. Patients with recurrent and refractory disease remain a particular challenge. Therapy for patients with LCH-associated neurodegenerative disease is also lacking.
A study in Nature Medicine by Coury et al.14 described high levels of interleukin-17A (IL-17A) protein in serum from patients with LCH and identified dendritic cells in LCH lesions as the source of IL-17A. This was a landmark report for several reasons, including evidence for an immune etiology of LCH and the first observation of IL-17A synthesized in vivo by dendritic cells. IL-17A is a proinflammatory cytokine produced primarily by a subset of T cells, Th17 cells, that activate an inflammatory response important in clearing bacterial, fungal, viral, and protozoal infections. IL-17A is especially important for maintaining host immune defense at mucosal surfaces (reviewed in refs. 15,16,17). IL-17A is also associated with autoimmune diseases including rheumatoid arthritis, psoriasis, inflammatory bowel disease, and multiple sclerosis (reviewed in refs. 18,19,20). Many features of LCH make a central functional role for IL-17A in LCH pathogenesis plausible: histology of LCH resembles a granuloma with recruitment of presumably normal leukocytes; bony destruction of LCH lesions resembles bony destruction by stimulated osteoclasts in rheumatoid arthritis; and possible autoimmune etiology of neurodegeneration in LCH.21 Pathologic IL-17A expression is now regularly cited as a likely factor for disease manifestations of LCH.22,23,24 Targeted therapies against IL-17A are in phase 1 and phase 2 trials for autoimmune diseases,25,26 and are being considered for treatment of patients with LCH.
Despite the arguments and evidence presented for a central role for IL-17A in pathogenesis of LCH, our group has been unable to substantiate this hypothesis. We previously reported our inability to identify evidence of IL-17A expression in LCH lesions.27 Due to the significant scientific and clinical implications, we further examined cell-specific IL-17A gene expression and IL-17A protein in LCH lesions in order to reconcile the conflicting data. We remain unable to identify evidence supporting IL-17A as an important factor in LCH pathogenesis.
Results
IL-17A RNA is not detectable in LCH lesions
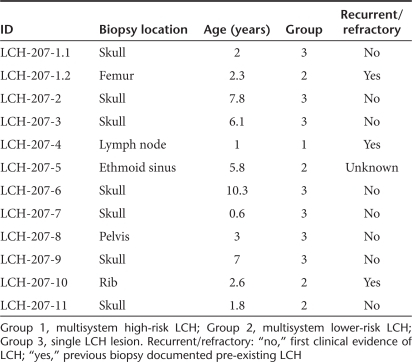
In our earlier study, we tested IL-17A RNA expression in two unsorted LCH lesions and in purified CD3+ and CD207+ cells from 14 LCH lesions.27 Delprat et al. suggested that our failure to detect IL-17A in these populations may be due to (i) tissue-specific rates of IL-17A RNA degradation, (ii) the IL-17A primer sequence used in the reverse transcription-PCR may correspond to isoforms with differential expression from canonical IL-17A, and (iii) restricted IL-17A expression by a CD1A+/CD207− population of dendritic cells (response to ref. 27). To address these issues, we tested quantitative IL-17A expression in a new series of LCH lesions as well as control tonsils that were harvested and viably frozen within 2 hours of surgical biopsy with identical processing protocols. Real-time PCR using primer sets that together include sequence from all known IL-17A exons failed to detect any IL-17A message in complementary DNA (cDNA) samples generated from purified CD207+ cells from 12 different LCH lesions (Figure 1). Quality and identity of the cDNA samples was verified with CD207 and SPP1 assays. SPP1 is highly expressed in LCH lesion CD207+ cells.8 Subjects in this series represent a range of clinical groups from single lesion to high-risk multisystem LCH. Clinical details of the LCH lesions used in this study are described in Table 1.
Figure 1.
Langerhans cell histiocytosis (LCH) lesion CD207+ cell complementary DNA (cDNA) real-time PCR. Interleukin-17A (IL-17A) gene expression in purified CD207+ cells from LCH lesions is quantified using real-time PCR. cDNA was amplified from RNA isolated from purified cell populations: CD207+ from LCH lesions and CD3+ from control tonsils. The primer sets cover all three known exons of IL-17A: IL-17A-1 (exon 2–3), IL-17A-2 (exon 2), and IL-17A-3 (exon 1–2). Expression of CD207 and SPP1 (encoding osteopontin) are included as positive controls for quality and identity of the LCH lesion CD207+ cells. Gene expression is described by the Ct, internally corrected for GAPDH expression with the following ΔCt equation: {20-[Ct(Probe)-Ct(GAPDH)]}. The result is quantitative gene expression where absent gene expression equals zero and greater abundance of cDNA, or higher levels of expression, is reflected by a larger number (log2 scale). These results demonstrate that none of the IL-17A probes amplify detectable message in any of the 12 LCH lesion cDNA samples, where IL-17A is detected by all three probes in control CD3+ cells isolated from tonsils processed in an identical fashion. As expected, CD207 and OPN expression are detected at high levels in all LCH CD207+ samples, but absent from the tonsil CD3+ samples.
Table 1. Langerhans cell histiocytosis (LCH) biopsy samples.
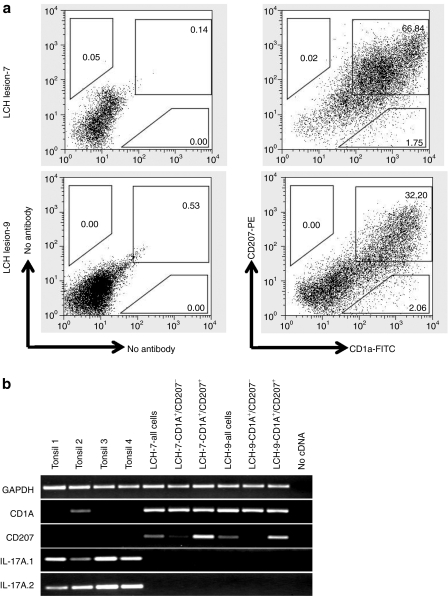
We further evaluated the possibility that IL-17A could be expressed in a dendritic cell or lymphocyte population other than CD207+ cells. Specifically, we aimed to test the suggestion from Delprat et al. that our previous failure to detect IL-17A message in LCH lesions may be due to specific expression by a CD1a+/CD207− population. Two representative LCH lesions were sorted into several populations: all viable cells, CD1a+/CD207− and CD1a+/CD207+. Finally, we used the IL-17A primer set from our first study as well as an additional IL-17A primer set based on coding sequence from a previously published study to evaluate for the presence of IL-17A mRNA.28 The scatterplots of LCH lesions are representative of all LCH lesions we have evaluated, where the vast majority of dendritic cells co-express CD1a+ and CD207+ (Figure 2a). The origin of the rare CD1a+/CD207− population has not yet been well-characterized in LCH lesions, though CD1a is less specific than CD207 and may be expressed on some lymphocytes as well as dendritic cells.29 The standard clinical histopathologic description of LCH lesions is generally defined by histiocytes that co-express CD1a and CD207.5,6 We were once again unable to amplify IL-17A cDNA from any LCH lesions, including samples enriched for CD1a+/CD207+ and CD1a+/CD207− cells, but did amplify IL-17A cDNA from control tonsil tissues processed in parallel. As noted in Figure 2b, cDNA for GAPDH is amplified in all specimens, and CD207 and CD1a are amplified as expected. Furthermore, IL-17A mRNA was undetectable in a separate study analyzing gene expression in CD207+ cells isolated from 13 LCH lesions by the IL-17A–specific probes on Affymetrix U133 Plus 2.0 chips.8 Taken together, these data are consistent with our previous report that failed to find evidence of any known forms of IL-17A expression in any cells in multiple LCH lesions.27
Figure 2.
Cell-specific interleukin-17A (IL-17A) expression in Langerhans cell histiocytosis (LCH) lesions. (a) Representative scatterplots from two typical LCH lesions show that CD207+ cells that define LCH lesions co-express CD1a. CD207+/CD1a− cells are not identified, and CD1+/CD207− cells are rare. (b) Semi-quantitative PCR of complementary DNA (cDNA) isolated from all viable cells from LCH lesions, CD1a+/CD207− cells from LCH lesions, CD1a+/CD207+ cells from LCH lesions, and control tonsil cells show that IL-17A expression is not detectable in any LCH lesion cell population using two different primer sets. IL-17A expression is detected in multiple control tonsil samples. CD1A and CD207 as well as control GAPDH cDNA are amplified in specific cell populations as expected.
IL-17A protein is not detectable in LCH lesions
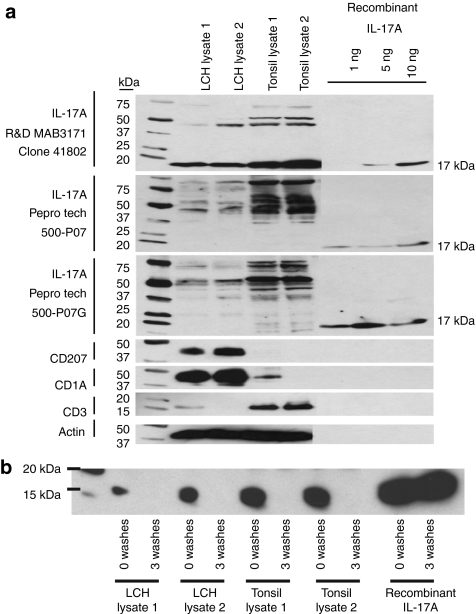
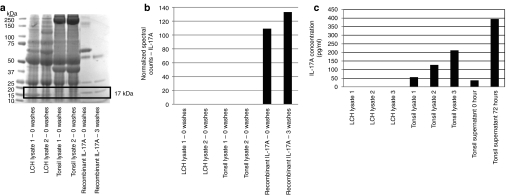
In the absence of RNA expression, it is difficult to explain the presence of IL-17A protein. However, in order to further evaluate the possibility of IL-17A protein expression in LCH lesions, we tested three of the antibodies from the study by Coury et al.: R&D Systems (Minneapolis, MN)—MAB3171 used for immunohistochemistry; PeproTech (Rocky Hill, NJ)—#500-P07 and PeproTech—#500-P07G used for enzyme-linked immunosorbent assay (ELISA) studies.14 We found that all three antibodies detected recombinant IL-17A with detection starting at 1 ng for the PeproTech antibodies and 5 ng for the R&D Systems antibody. However, all three antibodies also bound other nonspecific proteins (Figure 3a). The R&D Systems antibody strongly detected a nonspecific 17 kDa protein in LCH lesion lysate as well as in control tonsil lysate. While IL-17A protein typically runs at 17 kDa in sodium dodecyl sulfate gels, this protein was not detected by either of the PeproTech antibodies (Figure 3a). Further evidence that the 17 kDa protein bound by the R&D antibody is not IL-17A is demonstrated by differential affinity in immunoprecipitation studies (Figure 3b). The R&D antibody can pull down recombinant IL-17A, and recombinant IL-17A binding was not disrupted by three washes with NP40 buffer. However, the 17 kDa protein pulled down from LCH lysates and tonsil lysates that was detected before washing beads with NP40 wash buffer was disrupted by three washes with NP40 buffer. In order to further characterize the proteins bound by IL-17A antibody, we performed immunoprecipitation, cut Coomassie blue-stained protein bands ranging from 15 to 18 kDa from sodium dodecyl sulfate polyacrylamide gel electrophoresis (Figure 4a), and analyzed the immunoprecipitation products with mass spectroscopy. While IL-17A was detected by mass spectroscopy from the immunoprecipitation experiments including recombinant IL-17A protein, no IL-17A peptide sequence was detected from the immunoprecipitates from LCH lysates or tonsil lysates (Figure 4a,b). If the 17 kDa band recognized by the R&D antibody with similar intensity as the purified IL-17A by western blot (Figure 3a) was endogenous IL-17A, it should have been recognized in tonsils and LCH lesions with this method if it were present in these tissues at the concentration suggested by the intensity of the western blot 17 kDa bands.
Figure 3.
Interleukin-17A (IL-17A) protein detection in Langerhans cell histiocytosis (LCH) Lesions. (a) Western blot analysis of LCH lesion lysate, tonsil lysate, and recombinant IL-17A using antibodies to IL-17A, CD207, CD1a, CD3, and actin. All of the tested antibodies to IL-17A bind recombinant IL-17A as well as nonspecific proteins in LCH lesions and tonsils. Antibodies to CD207 and CD1a are positive controls for the LCH lesions, and antibody to CD3 is a positive control for tonsil lysate. The CD3 antigen detected in LCH lesion 1 reflects T-cell infiltrate, which is typical in LCH lesions. The CD1a detected in Tonsil lysate 1 likely reflects T-cell expression of that protein. Antibody to actin is used as a loading control. (b) Immunoprecipitation with IL-17A R&D MAB 3171 antibody demonstrates differential affinity for 17 kDa band in LCH lesion and tonsil lysates compared to recombinant IL-17A. The protein species bound by the antibody in the LCH and tonsil lysates is displaced after three washes with buffer, where recombinant IL-17A binding remains stable.
Figure 4.
Quantitative analysis of interleukin-17A (IL-17A) protein in Langerhans cell histiocytosis (LCH) lesions. (a) Rectangular box indicates the 15–18 kDa gel area excised from Coomassie-blue staining of protein bands immunoprecipitated with IL-17A R&D MAB 3171 antibody. (b) Gel slices were quantitatively analyzed for IL-17A sequence with mass spectroscopy. The y-axis normalized count of the spectra matching any of the peptides in IL-17A protein. Endogenous IL-17A was not identified in the samples from LCH lysate or tonsil lysate, but was identified in the recombinant IL-17A immunoprecipitate. (c) IL-17A concentration in LCH lysates, tonsil lysates, unstimulated and stimulated tonsil supernatants by enzyme-linked immunosorbent assay. Endogenous IL-17A was detected in tonsil lysate and supernatant from stimulated tonsil lymphocytes as predicted levels, but no IL-17A was detected in protein lysates from three different LCH lesions.
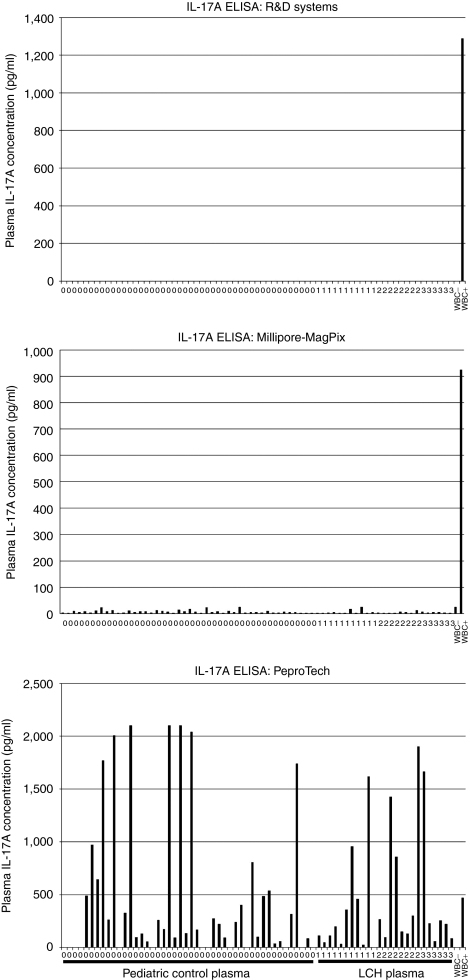
Because the limit of detection for IL-17A by western blot analysis is 1 ng, ELISAs were performed to determine if lower levels of IL-17A were present in LCH lysates. No IL-17A was detected in LCH lysates but IL-17A protein could be detected in lysates from tonsils processed with the same protocol that expressed IL-17A in concentrations ranging from 56 to 212 pg/ml (Figure 4c). The IL-17A protein concentration in the control tonsils correlates with the relative abundance of RNA from expression studies described above. As a positive control for the IL-17A ELISA, endogenous IL-17A levels increased in media supernatant after tonsil lymphocytes were stimulated with PMA and ionomycin (Figure 4c). Finally, IL-17A ELISA kits from three different manufacturers were used to determine abundance of IL-17A in plasma from patients with LCH (Figure 5). While the R&D Systems and Millipore-Luminex kits (Millipore, Billerica, MA) failed to identify increased concentrations of IL-17A in plasma from patients with active LCH, the PeproTech IL-17A Development Kit demonstrated levels up to 2 ng/ml in both pediatric control and LCH plasma samples. The PeproTech kit is the system used by Coury et al. in their original LCH study.14 Due to the very high levels of IL-17A in control and LCH plasma in the PeproTech experiment as well as the component IL-17A antibody analysis described above, we conclude that these results are likely due to nonspecific binding. As a positive control, increased IL-17A concentration was observed in supernatant from stimulated peripheral white blood cells in all of these ELISA assays. Overall, these analyses demonstrate that the antibodies to IL-17A from the study that proposed a relationship between IL-17A and LCH pathogenesis are able to detect true IL-17A peptide, but they also detect a much larger mass of nonspecific proteins.
Figure 5.
Comparison of plasma interleukin-17A (IL-17A) enzyme-linked immunosorbent assay (ELISA) studies. ELISA was performed on plasma samples for 46 pediatric control subjects and 26 patients with active Langerhans cell histiocytosis (LCH), including 11 with multisystem high-risk disease, 8 with multisystem lower-risk disease, and 6 with single lesion disease. While the R&D Systems and Millipore assays fail to identify increased IL-17A plasma protein in LCH subjects, the PeproTech Development Kit demonstrates increased signal for both control and LCH subjects. As a positive control, IL-17A is detected in supernatant from peripheral white blood cells (WBCs) stimulated with PMA/ionomycin in all of the assays. On the figure, 0 = control, 1 = multisystem high-risk LCH, 2 = multisystem lower-risk LCH, 3 = single LCH lesion.
Discussion
LCH is an extremely difficult disease to study. In addition to being a rare condition, there are no validated in vitro or animal models. Therefore, research has relied primarily on immunohistochemical analysis of frozen or paraffin-embedded biopsies. While this approach may be useful, interpretation of data is challenged by lack of appropriate control tissues, difficulty quantitatively analyzing results and variability in antibody specificity. Our recent study analyzing cell-specific gene expression in LCH found that many of the genes previously reported as “upregulated” or “downregulated” in LCH lesions had no difference in gene expression between LCH lesion CD207+ cells and control skin CD207+ cells.8 Quantitative differences in results from gene expression and protein analysis may occur due to differential turnover, small interfering RNA-mediated RNA degradation or technical differences in assays, but gene expression is a prerequisite for protein translation. In a biological sample one would expect to be able to validate identification of an abundant protein with some evidence of gene expression.
In this case of IL-17A in LCH, our data suggest that nonspecific antibodies are likely responsible for the incongruent RNA expression and protein data from our studies and those of Coury et al.14 Multiple primers and probes failed to detect IL-17A message in any cells from LCH lesions, and the antibodies used to establish a role for IL-17A in LCH demonstrate nonspecific binding to many proteins in the lesions. It is possible that in other clinical contexts, IL-17A may play an important role in dendritic cell function and granuloma formation.14 However, we conclude that evidence for a role for IL-17A in LCH pathogenesis remains inadequate and therapeutic strategies targeting IL-17A or the IL-17A pathway in LCH are not warranted.
Materials and Methods
Study enrollment. All studies were conducted under Baylor College of Medicine institutional review board–approved protocols. Classifications of single-system disease (single lesion excluding lungs, liver, spleen or bone marrow), multisystem low-risk (multiple lesions excluding lungs, liver, spleen or bone marrow), and multisystem high-risk (multiple lesions including at least one lesion in lungs, liver spleen or bone marrow) are based on clinical risk categories described by the Histiocyte Society.30 Control plasma samples were collected from discarded clinical samples from the hematology lab, and specimens with diagnosis consistent with inflammation, infection or malignancy were excluded.
Isolation of specific cell populations from LCH lesions and control tonsils. LCH lesions and control tonsils were collected within 2 hours of surgical removal, viably frozen as single-cell suspension in Recovery Medium (Invitrogen, Carlsbad, CA) and stored in liquid nitrogen. Samples were thawed, then incubated with conjugated antibodies including CD207-PE (Beckman-Coulter, Miami, FL), CD1A (BD Biosciences, Franklin Lakes, NJ), and CD3-FITC (BD Biosciences). Viable (propidium-iodide negative) cells were isolated with a MoFlo flow cytometry sorter as described previously.8 Cells from all samples were sorted directly into PicoPure RNA Extraction Buffer (Molecular Devices, Sunnyvale, CA). RNA was then processed according to manufacturer's protocol. cDNA amplification was performed with the WT-Ovation Pico System (NuGen, San Carlos, CA) according to manufacturer's protocol.
Quantitative real-time PCR. Real-time PCR reactions were performed with TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA) including FAM dye-labeled TaqMan MGB Probe (250 nmol/l final concentration). Single stranded cDNA was generated from CD207+ cells from LCH lesions or from CD3+ cells from control tonsils as described above. Each reaction included 20 ng cDNA. TaqMan Fast Universal PCR Master Mix (Applied Biosystems) was used for 25-µl reaction in 96-well plates on a iQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Assays were performed in duplicate. Thermal cycling conditions were 3 min at 95 °C, then 40 cycles of 95 °C for 10 seconds and 55 °C for 30 seconds. Quantitative RNA expression was estimated by the following ΔCt equation: {20−[Ct(Probe)−Ct(GAPDH)]}. This method standardizes each sample according to GAPDH concentration with typical Ct = 20 cycles. TaqMan probe sets included IL17A-1 (Hs00174383), IL17A-2 (Hs99999082_m1), IL17A-3 (Hs00936345), GAPDH (Hs03929097_g1), CD207 (Hs00210453-m1), and SPP1 (Hs00960942_m1). (Relative gene expression of IL-17A of in LCH CD207 cells have been evaluated in cDNA from pooled LCH lesion CD207 cells in previous studies using primer (Hs00174383).8 None of the LCH CD207 cDNAs in this series have been used in previous publications.)
Reverse transcription-PCR. RNA was isolated from tonsil lymphocytes, whole LCH lesions, LCH CD1a+/CD207+ and CD1a+/CD207− cell populations using PicoPure RNA Isolation Kit (Applied Biosystems). RNA was then reverse-transcribed into cDNA with the WT-Ovation Pico Amplification System (NuGen) and 20 ng of cDNA was PCR-amplified using template-specific primers:
Interleukin 17A.1-F 5′-CCCCTAGACTCAGGCTTCCT-3′
Interleukin 17A.1-R 5′-TCAGCTCCTTTCTGGGTTGT-3′
Interleukin 17A.2-F 5′-GAAGGCAGGAATCACAATC-3′
Interleukin 17A.2-R 5′-GCCTCCCAGATCACAGA-3′
CD207-F 5′-CAACAATGCTGGGAACAATG-3′
CD207-R 5′-GGGGAAGAAAGAGGCATTTC-3′
CD1A-F 5′-GCTCCAGACACACCTGAACA-3′
CD1A-R 5′-GGAAGGCCCTCTGGAGTAAG-3′
GAPDH-F 5′-GGCCTCCAAGGAGTAAGACC-3′
GAPDH-R 5′-AGGGGTCTACATGGCAACTG-3′
Amplified cDNA was run on a 2% agarose gel with ethidium bromide, then visualized and photographed under UV light with a Gel Doc imager (Bio-Rad). (IL17A.1-F/IL17A.1-R, CD207-F/CD207-R, and GAPDH-F/GAPDH-R primers were used in a previous publication.8 None of the cDNA samples in this experiment have been used in previously published studies.)
Protein lysate preparation. LCH lesions and tonsils were collected within 2 hours of surgical removal, the tissue was snap-frozen then stored at −80 °C. Tissue was then lysed in T-PER Tissue Protein Extraction Reagent (Thermo Scientific, Rockford, IL) containing protease inhibitors (Protease Inhibitor Cocktail Kit; Thermo Scientific). Lysates were collected and stored at −80 °C. Final protein concentrations ranged from 1 to 5 mg·ml−1. Lymphocytes isolated from fresh tonsils or peripheral blood were incubated in RPMI (Invitrogen) containing 10% fetal bovine serum and penicillin/streptomycin. IL-17A was induced by incubating 5 × 106 cells in 2 ml RPMI with 1 µg·ml−1 ionomycin (Sigma-Aldrich, St Louis, MO) and 20 ng·ml−1 PMA for 0 or 72 hours.
Western blotting. Proteins in lysates and supernatants were separated on 4–12% Bis-Tris gels (Invitrogen) using MOPS buffer (Invitrogen) and transferred onto polyvinylidine fluoride membranes. Membranes were blocked for half an hour using StartingBlock T20 Blocking Buffer (Thermo Scientific). Membranes were then incubated in primary antibody diluted in blocking buffer for 1 hour at room temperature or overnight at 4 °C. After washing three times for 5 minutes each in Tris buffered Saline- Tween 20 (Thermo Scientific), membranes were incubated in horseradish peroxidase-linked secondary antibody diluted in blocking buffer for 1 hour at room temperature. After washing three times for 5 minutes each in Tris buffered Saline- Tween 20, protein of interested was detected using SuperSignal West Pico or SuperSignal West Dura Chemiluminescent Substrate (Thermo Scientific).
Recombinant human IL-17A was purchased from eBioscience (San Diego, CA). Primary antibodies used in western blot analysis were IL-17A (1. MAB3171, Clone41802; R&D Systems, 2. 500-P07; PeproTech, 3. 500-P07G; PeproTech), CD3 (ab699; AbCam, Cambridge, MA), CD207 (sc22620; SantaCruz Biotechnology, Santa Cruz, CA), CD1a (ab26531; AbCam), Actin (ab3280; AbCam). Secondary antibodies were anti-mouse horseradish peroxidase (P0447; Dako, Carpinteria, CA), anti-rabbit horseradish peroxidase (P0448; Dako) and anti-goat horseradish peroxidase (P0449; Dako).
Immunoprecipitation. Cell lysate (1,000 µg), cell supernatant (1 ml) or recombinant IL-17A (2 µg) was incubated with 8 µg IL-17A antibody (MAB3171, Clone41802; R&D Systems) in NP40 buffer overnight at 4 °C with gentle rotation. Rec-Protein G-Sepharose 4B beads (Invitrogen) were then added for 1–2 hours. Beads were collected and underwent either 0 washes or 3 washes with NP40 buffer. Beads were then heated with sample buffer to 100 °C for 10 minutes, supernatant was collected, and proteins were analyzed by western blotting and mass spectrometry.
Spectral analysis. Immunoprecipitation was run on a 4–12% Bis-Tris gel (Invitrogen), stained with Coomassie blue, then gel slices corresponding to 15–18 kDa were removed, then analyzed by NextGen Sciences (Ann Arbor, MI) by mass spectroscopy. Solution digests were analyzed using liquid chromatography-tandem mass spectrometry with a 30-minute gradient on a LTQ Orbitrap XL mass spectrometer. Product ion data were searched against the concatenated forward and reverse IPI Human v3.67 protein database, and Mascot output files were parsed into the Scaffold program for collation into nonredundant lists per sample. Spectral counts per protein are a semi-quantitative measure of abundance of peptides across samples that reflects the number of matched peptides and the number of times those peptides were observed.
ELISA. L17A levels were measured in tissue lysate and cell culture supernatants using the Human IL-17A Quantikine ELISA Kit (D1700; R&D Systems). IL-17A levels were also tested in plasma samples from pediatric controls and pediatric subjects with active LCH with Human IL-17A Quantikine ELSA Kit (D1700; R&D Systems), MilliplexMag Human Cytokine/Chemokine Magnetic Bead Panel Immunoassay (HCYTMAG-60K-PX39; Millipore), and the Peprotech IL-17A ELISA Development Kit (900-K84; PeproTech). (Some of the LCH plasma samples in this study were also used in IL-17A ELISA studies with the R&D Systems kit in a previous publication.27 They are included in this manuscript to demonstrate discrepant results between different assays.)
Acknowledgments
This work in this report was supported by grants from the Histiocytosis Association of America, the Histiocytosis Association of Canada, the Hoag Foundation (C.E.A.), the Thrasher Research Fund (C.E.A.), and the American Society of Hematology (C.E.A.). We would also like to acknowledge the continued contributions of the Kontoyannis family in supporting scientific discussions of histiocytic diseases through the Nikolas Symposium.
REFERENCES
- Aricò M, Girschikofsky M, Généreau T, Klersy C, McClain K, Grois N.et al. (2003Langerhans cell histiocytosis in adults. Report from the International Registry of the Histiocyte Society Eur J Cancer 392341–2348. [DOI] [PubMed] [Google Scholar]
- Baumgartner I, von Hochstetter A, Baumert B, Luetolf U., and, Follath F. Langerhans'-cell histiocytosis in adults. Med Pediatr Oncol. 1997;28:9–14. doi: 10.1002/(sici)1096-911x(199701)28:1<9::aid-mpo3>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Carstensen H., and, Ornvold K. [Langerhans-cell histiocytosis (histiocytosis X) in children] Ugeskr Laeg. 1993;155:1779–1783. [PubMed] [Google Scholar]
- Ornvold K, Ralfkiaer E., and, Carstensen H. Immunohistochemical study of the abnormal cells in Langerhans cell histiocytosis (histiocytosis x) Virchows Arch A Pathol Anat Histopathol. 1990;416:403–410. doi: 10.1007/BF01605145. [DOI] [PubMed] [Google Scholar]
- Chikwava K., and, Jaffe R. Langerin (CD207) staining in normal pediatric tissues, reactive lymph nodes, and childhood histiocytic disorders. Pediatr Dev Pathol. 2004;7:607–614. doi: 10.1007/s10024-004-3027-z. [DOI] [PubMed] [Google Scholar]
- Lau SK, Chu PG., and, Weiss LM. Immunohistochemical expression of Langerin in Langerhans cell histiocytosis and non-Langerhans cell histiocytic disorders. Am J Surg Pathol. 2008;32:615–619. doi: 10.1097/PAS.0b013e31815b212b. [DOI] [PubMed] [Google Scholar]
- Nezelof C, Basset F., and, Rousseau MF. Histiocytosis X histogenetic arguments for a Langerhans cell origin. Biomedicine. 1973;18:365–371. [PubMed] [Google Scholar]
- Allen CE, Li L, Peters TL, Leung HC, Yu A, Man TK.et al. (2010Cell-specific gene expression in Langerhans cell histiocytosis lesions reveals a distinct profile compared with epidermal Langerhans cells J Immunol 1844557–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Ginhoux F., and, Collin M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat Rev Immunol. 2008;8:935–947. doi: 10.1038/nri2455. [DOI] [PubMed] [Google Scholar]
- Arceci RJ. The histiocytoses: the fall of the Tower of Babel. Eur J Cancer. 1999;35:747–67; discussion 767. doi: 10.1016/s0959-8049(99)00039-8. [DOI] [PubMed] [Google Scholar]
- Egeler RM, Annels NE., and, Hogendoorn PC. Langerhans cell histiocytosis: a pathologic combination of oncogenesis and immune dysregulation. Pediatr Blood Cancer. 2004;42:401–403. doi: 10.1002/pbc.10464. [DOI] [PubMed] [Google Scholar]
- Fadeel B., and, Henter JI. Langerhans-cell histiocytosis: neoplasia or unbridled inflammation. Trends Immunol. 2003;24:409–10; author reply 410. doi: 10.1016/s1471-4906(03)00171-6. [DOI] [PubMed] [Google Scholar]
- Laman JD, Leenen PJ, Annels NE, Hogendoorn PC., and, Egeler RM. Langerhans-cell histiocytosis ‘insight into DC biology'. Trends Immunol. 2003;24:190–196. doi: 10.1016/s1471-4906(03)00063-2. [DOI] [PubMed] [Google Scholar]
- Coury F, Annels N, Rivollier A, Olsson S, Santoro A, Speziani C.et al. (2008Langerhans cell histiocytosis reveals a new IL-17A-dependent pathway of dendritic cell fusion Nat Med 1481–87. [DOI] [PubMed] [Google Scholar]
- Miossec P, Korn T., and, Kuchroo VK. Interleukin-17 and type 17 helper T cells. N Engl J Med. 2009;361:888–898. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- Pappu R, Ramirez-Carrozzi V, Ota N, Ouyang W., and, Hu Y. The IL-17 family cytokines in immunity and disease. J Clin Immunol. 2010;30:185–195. doi: 10.1007/s10875-010-9369-6. [DOI] [PubMed] [Google Scholar]
- Zou W., and, Restifo NP. T(H)17 cells in tumour immunity and immunotherapy. Nat Rev Immunol. 2010;10:248–256. doi: 10.1038/nri2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S., and, Lindén A. Interleukin-17 as a drug target in human disease. Trends Pharmacol Sci. 2009;30:95–103. doi: 10.1016/j.tips.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Sarkar S., and, Fox DA. Targeting IL-17 and Th17 cells in rheumatoid arthritis. Rheum Dis Clin North Am. 2010;36:345–366. doi: 10.1016/j.rdc.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Segal BM. Th17 cells in autoimmune demyelinating disease. Semin Immunopathol. 2010;32:71–77. doi: 10.1007/s00281-009-0186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grois N, Prayer D, Prosch H., and, Lassmann H, CNS LCH Co-operative Group Neuropathology of CNS disease in Langerhans cell histiocytosis. Brain. 2005;128 Pt 4:829–838. doi: 10.1093/brain/awh403. [DOI] [PubMed] [Google Scholar]
- Egeler RM, van Halteren AG, Hogendoorn PC, Laman JD., and, Leenen PJ. Langerhans cell histiocytosis: fascinating dynamics of the dendritic cell-macrophage lineage. Immunol Rev. 2010;234:213–232. doi: 10.1111/j.0105-2896.2009.00883.x. [DOI] [PubMed] [Google Scholar]
- Gavhed D, Akefeldt SO, Osterlundh G, Laurencikas E, Hjorth L, Blennow K.et al. (2009Biomarkers in the cerebrospinal fluid and neurodegeneration in Langerhans cell histiocytosis Pediatr Blood Cancer 531264–1270. [DOI] [PubMed] [Google Scholar]
- Nichols KE., and, Arceci RJ. BRAF, a piece of the LCH puzzle. Blood. 2010;116:1825–1827. doi: 10.1182/blood-2010-06-289934. [DOI] [PubMed] [Google Scholar]
- Genovese MC, Van den Bosch F, Roberson SA, Bojin S, Biagini IM, Ryan P.et al. (2010LY2439821, a humanized anti-interleukin-17 monoclonal antibody, in the treatment of patients with rheumatoid arthritis: A phase I randomized, double-blind, placebo-controlled, proof-of-concept study Arthritis Rheum 62929–939. [DOI] [PubMed] [Google Scholar]
- Hueber W, Patel DD, Dryja T, Wright AM, Koroleva I, Bruin G, Psoriasis Study Group; Rheumatoid Arthritis Study Group; Uveitis Study Group et al. Effects of AIN457, a fully human antibody to interleukin-17A, on psoriasis, rheumatoid arthritis, and uveitis. Sci Transl Med. 2010;2:52ra72. doi: 10.1126/scitranslmed.3001107. [DOI] [PubMed] [Google Scholar]
- Allen CE., and, McClain KL. Interleukin-17A is not expressed by CD207(+) cells in Langerhans cell histiocytosis lesions. Nat Med. 2009;15:483–4; author reply 484. doi: 10.1038/nm0509-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupedo T, Crellin NK, Papazian N, Rombouts EJ, Weijer K, Grogan JL.et al. (2009Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells Nat Immunol 1066–74. [DOI] [PubMed] [Google Scholar]
- Blumberg RS, Gerdes D, Chott A, Porcelli SA., and, Balk SP. Structure and function of the CD1 family of MHC-like cell surface proteins. Immunol Rev. 1995;147:5–29. doi: 10.1111/j.1600-065x.1995.tb00085.x. [DOI] [PubMed] [Google Scholar]
- Gadner H, Grois N, Pötschger U, Minkov M, Aricò M, Braier J, Histiocyte Society et al. Improved outcome in multisystem Langerhans cell histiocytosis is associated with therapy intensification. Blood. 2008;111:2556–2562. doi: 10.1182/blood-2007-08-106211. [DOI] [PubMed] [Google Scholar]