Abstract
Finding suitable nonviral delivery vehicles for nucleic acid–based therapeutics is a landmark goal in gene therapy. Cell-penetrating peptides (CPPs) are one class of delivery vectors that has been exploited for this purpose. However, since CPPs use endocytosis to enter cells, a large fraction of peptides remain trapped in endosomes. We have previously reported that stearylation of amphipathic CPPs, such as transportan 10 (TP10), dramatically increases transfection of oligonucleotides in vitro partially by promoting endosomal escape. Therefore, we aimed to evaluate whether stearyl-TP10 could be used for the delivery of plasmids as well. Our results demonstrate that stearyl-TP10 forms stable nanoparticles with plasmids that efficiently enter different cell-types in a ubiquitous manner, including primary cells, resulting in significantly higher gene expression levels than when using stearyl-Arg9 or unmodified CPPs. In fact, the transfection efficacy of stearyl-TP10 almost reached the levels of Lipofectamine 2000 (LF2000), however, without any of the observed lipofection-associated toxicities. Most importantly, stearyl-TP10/plasmid nanoparticles are nonimmunogenic, mediate efficient gene delivery in vivo, when administrated intramuscularly (i.m.) or intradermally (i.d.) without any associated toxicity in mice.
Introduction
The gene therapy field has experienced tremendous progress in the past decades and the application of gene expression modulation today seems more accessible than ever. Classically, the underlying principle of any such manipulation is to adjust the levels of otherwise deficient gene products to normal physiological levels. The simplest way to achieve this is to incorporate a gene of interest into bacterial plasmids and introduce these plasmids to specific cells or tissues. However, plasmids, similar to other nucleic acid–based molecules in general, share the inherent properties of having high molecular weight and negative net charge, making biological membranes essentially impermeable to them. In recent years, a myriad of different delivery vehicles have been developed and evaluated for plasmid delivery, of both viral and nonviral origin. Although viral vectors fulfil the criteria associated with efficient gene delivery vehicles, their applicability is severely hampered by possible side-effects deriving from their origin, such as immunogenicity, possible in vivo recombination, virus-induced chromosomal integrations, and tumorigenicity.1,2 These inherent drawbacks have paved the way for nonviral delivery vectors.3,4
Nonviral delivery of plasmid DNA (pDNA) is usually based on chemical vectors that form nanoparticles with pDNA. Most of these chemical vectors are based on natural and/or synthetic peptides, lipids or polymers.5,6 Cell-penetrating peptides (CPPs) are one such class of peptide-based vectors, which has demonstrated great potential for numerous delivery applications since being discovered in 1994.7 CPPs are relatively short, cationic, and/or amphipathic peptides that possess the ability to deliver a wide range of different macromolecules into cells both in vitro and in vivo (as recently reviewed in refs. 8,9,10,11,12). Plasmid delivery with CPPs has been reported numerous times, however, the relative transfection efficiencies in these reports have in general been poor.13,14,15
While the internalization mechanisms of CPPs are widely debated, it is generally accepted that most CPPs use some endocytic pathway to gain access to the interior of cells.16,17,18 Following endocytosis, CPP/cargo complexes/conjugates reside in endosomal vesicles and only a minor portion is able to escape these compartments, which severely reduces the bioavailability.19,20
To overcome this problem many strategies aiming at increasing endosomal escape have been investigated (as recently reviewed in refs. 8,21). One possibility to increase the activity of CPPs is to introduce a stearyl moiety. This approach has previously been reported to improve polyarginine-mediated delivery of small interfering RNAs22 and plasmids.23,24,25 However, the stearylation strategy does not seem to provide the same effects for all peptides. For example, we recently showed, in two separate reports, that the activity of stearyl-Arg9 is rather modest for the delivery of oligonucleotides.25,26 In contrast, stearylation of the more amphipathic peptides (RXR)4 and transportan 10 (TP10) had a dramatic impact on the delivery efficacy of oligonucleotides. In particular, stearyl-TP10 displayed superior delivery properties compared to other stearylated CPPs.
Based on these findings we therefore, in this paper, investigate the plasmid delivery efficacy of the stearyl-TP10 peptide, not only in vitro, but also in vivo. We show that serum-resistant nanoparticles are formed upon coincubation with pDNA and that these nanoparticles efficiently transfect cells ubiquitously in a nontoxic manner. We also take this technology one step further and show that stearyl-TP10/plasmid nanoparticles facilitate efficient dose-dependent gene delivery in vivo when being administered intramuscularly (i.m.) or intradermally (i.d.), without any associated toxicity or induction of immune response. These promising results show that stearyl-TP10 is an interesting nonviral, peptide-based vector for plasmid delivery, which is effective both in vitro and in vivo, and it opens an intriguing perspective for the future gene delivery applications.
Results
Noncovalent particle formation
pDNA can be vectorized with peptides by using a noncovalent complexation strategy. This strategy is based on the phenomenon that positively charged peptides can condense negatively charged oligonucleotides predominantly through electrostatic- and hydrophobic interactions and form complexes/nanoparticles.27,28
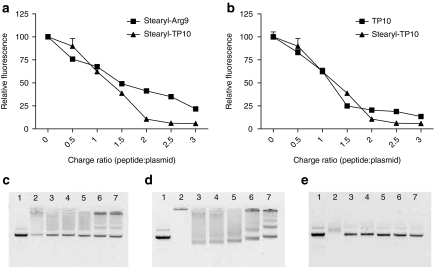
To assess the extent to which the CPPs are able to condense pDNA, we used an ethidium bromide (EtBr) exclusion assay, essentially as described in refs. 29,30. Both the stearylated CPPs, stearyl-Arg9 and stearyl-TP10, efficiently condensed pDNA (Figure 1a). However, the condensing effect of stearyl-TP10 was more pronounced at higher charge ratios (CRs). Unmodified TP10 also condensed pDNA, but to a lower extent than stearyl-TP10 (Figure 1b).
Figure 1.
Complex formation efficiency. (a) Ability of stearylated cell-penetrating peptides (CPPs) to form complexes with a pGL3 plasmid at different peptide:plasmid charge ratios (CRs) 0.5:1–3:1 (CR0.5–CR3), was analyzed using an ethidium bromide (EtBr) exclusion assay. (b) The EtBr exclusion assay carried out in the same manner, however, comparing unmodified transportan 10 (TP10) with stearyl-TP10. Effect of the addition of heparin sulphate to the CPP/plasmid complexes: stearyl-TP10/plasmid (c) at CR1, (d) at CR3 and (e) TP10/plasmid complexes at CR3. Lane numbers on the images on c–e represent: (1) naked plasmid, (2) CPP/plasmid complexes, (3–7) CPP/plasmid complexes treated with heparin sodium at the concentrations of 1.135, 2.27, 4.54, 9.08, 18.16 mg/ml, respectively.
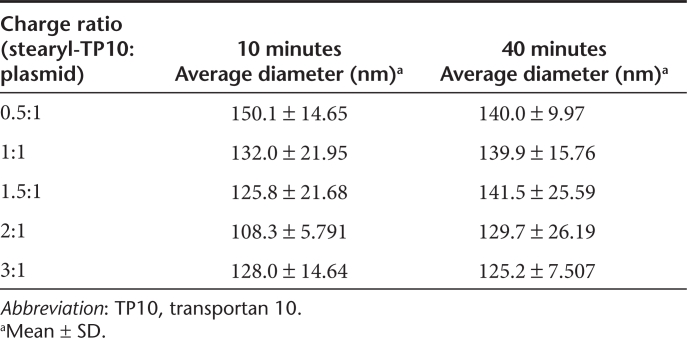
In order to study the physicochemical properties of stearyl-TP10/plasmid complexes, we carried out dynamic light scattering measurements. Particles were formed at different CRs and particle size was in line with the condensing effect seen in the EtBr exclusion assay. At higher CRs, particles were more condensed and concurrently smaller in size. In general, particles tended to grow over time; however, overall particle size remained in the range of 125–150 nm (Table 1). Additionally, we measured the ζ-potentials of the nanoparticles. ζ-Potential of stearyl-TP10/plasmid nanoparticles remained in the range of −5.0 to −11.4 mV in serum-supplemented media, which is in a suitable range for in vivo applicability31,32,33 (Supplementary Table S1).
Table 1. Size distribution and homogenicity of stearyl-TP10/plasmid nanoparticles.
To further evaluate the stability of the stearyl-TP10/plasmid nanoparticles, we analyzed the complexes using a heparin displacement assay, essentially as described in ref. 34. Stearyl-TP10 formed stable complexes with pDNA at all the studied CRs. At CR1, i.e., at a CR wherein all the negative charges of the pDNA had theoretically been neutralized with positive charges of the peptide, some of the DNAs were still detectable on the gel, implying that either a small portion of the plasmid remained noncomplexed or was easily dissociated from complexes by the electrophoresis process (Figure 1c). In contrast, at all other higher CRs, the plasmid was completely condensed into nanoparticles and hence completely shielded. At CR1, heparin sodium (heparin) easily displaced the peptide from the stearyl-TP10/plasmid complexes. At higher CRs, heparin either only displaced peptides at higher heparin concentrations or liberated only a small portion of plasmid (Figure 1d), that is except for CR1, stearyl-TP10/plasmid complexes were very stable. As mentioned above, TP10 also condensed DNA at higher peptide concentrations, however, addition of heparin completely dissociated TP10/plasmid complexes at all the heparin concentrations (Figure 1e). In contrast to the results from the EtBr exclusion assay, very little complexation was observed when analyzing the TP10 complexes on the gel (Figure 1e). These discrepancies might be due to the complexes that are very weakly associated and that during the course of electrophoresis, the complexes dissociate.
Next, the stability of stearyl-TP10/plasmid nanoparticles toward the degrading capacity of serum was assessed by incubating preformed nanoparticles in the presence of 10% of serum and analyzing degradation by agarose gel electrophoresis. At CR2, the peptide was able to completely shield the plasmid from degradation, for at least 24 hours. At CR1, plasmid was degraded to some extent, however, most of the complexes were still intact, whereas naked plasmid was completely degraded (Supplementary Figure S1).
Effect of N-terminal stearylation on plasmid transfection efficiency
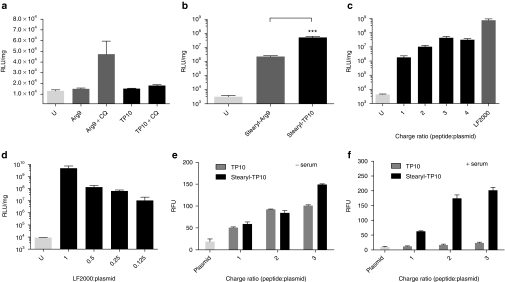
Plasmid delivery with unmodified CPPs has been described in a handful of papers, but transfection efficiencies have generally been relatively poor in most of these reports.13,14,15 To corroborate this, Chinese hamster ovary (CHO) cells were transfected with a luciferase-encoding plasmid complexed with either TP10 or Arg9. As expected, neither one of the peptides conferred transfections above the level of cells treated with only plasmid (Figure 2a). As aforementioned, despite being a field of intense investigation, it is widely accepted, that CPPs use endocytic pathways to enter cells and that subsequent entrapment of peptides in endosomes significantly reduces their activity.19,20 Cotreatment with chloroquine has been shown to enhance the release of endosomally entrapped material.35 Interestingly, only minor increases in gene expression were observed when cotreating with chloroquine, being slightly more pronounced in the case of Arg9 (Figure 2a). These results imply that although unmodified CPPs form complexes with plasmids to a certain extent, they are either not internalized in sufficient amounts (suggested by the lack of biological response even in presence of chloroquine) or complexes rapidly dissociate before or after being taken up by endocytic pathways and the plasmids are thus unable to reach the nucleus of cells.
Figure 2.
Effect of unmodified and stearylated cell-penetrating peptides (CPPs) on plasmid transfections compared to the Lipofectamine 2000 (LF2000). (a) To evaluate the ability of unmodified CPPs to mediate plasmid transfections, 4 × 104 Chinese hamster ovary cells were seeded 24 hours before experiment in 24-well plates. Cells were treated with complexes at different charge ratios (CRs), from CR1 to CR4 (in this case CR3 is shown), using 0.5 µg of plasmid per well, for 4 hours in serum-free media (alternatively with the addition of 100 µmol/l chloroquine (CQ)) followed by replacement to 10% serum containing medium and incubated additionally for 20 hours. Cells were washed with HEPES-buffered Krebs Ringer buffer and lysed in 0.1% Triton X-100, luciferase activity was measured and normalized against the protein content in each well. (b) Transfection comparison of stearyl-transportan 10 (TP10) and stearyl-Arg9 at CR3, carried out as described above. (c) Efficiency of stearyl-TP10 compared to LF2000. Stearyl-TP10 was formulated at different CRs as described above and LF2000 was used according to the manufacturer's protocol. (d) Decrease in LF2000-mediated luciferase plasmid transfections as a result of decreased LF2000 amounts compared to the standard protocol. Uptake of the fluorescenyl-labeled plasmid in complex with either (e) TP10 or stearyl-TP10 in U2OS cells in Opti-MEM or (f) serum containing media. Treatments were carried out as described above, however, cells were lysed for 1 hour and fluorescence was measured in black 96-well plate at 490/518 nm on a fluorometer. Fluorescence signal (RFU) from untreated cells was subtracted from the signals of treated cells. The values represent the mean of at least three independent experiments performed in duplicate (mean ± SEM). (b) ***P < 0.001, analysis of variance Dunnett's multiple comparison test.
After confirming that unmodified CPPs did not promote any significant plasmid delivery, we sought, based on our previous success with stearyl-TP10 for the delivery of splice-correcting oligonucleotides,26 to test it for plasmid transfection efficiency. Stearyl-TP10/plasmid nanoparticles enabled efficient transfection of pDNA and significantly increased luciferase gene expression levels in CHO cells, generating around four orders of magnitude higher levels over background (Figure 2b). The same modification on Arg9 also had an impact on plasmid delivery; however, transfection efficiency was ~100-times lower as compared to when using stearyl-TP10 (Figure 2b). Interestingly, stearyl-TP10 was able to significantly increase the luciferase activity at all tested CRs, showing that the transfection efficiency is not strictly dependent on peptide amounts (Figure 2c). In order to fully appreciate the potential of this peptide, we compared it with the widely used transfection reagent Lipofectamine 2000 (LF2000). Intriguingly, stearyl-TP10 almost reached the transfection levels of LF2000 in CHO cells (Figure 2c) Importantly, in contrast to stearyl-TP10, transfection levels were severely reduced when decreasing the amount of LF2000 (Figure 2d).
To delineate whether the increased activity imparted by the stearyl moiety was a result of increased cellular uptake or increased endosomal escape (or a combination of these factors), the uptake of fluoresceinyl-labeled plasmid complexed with TP10 or stearyl-TP10 was assessed. As seen in Figure 2e, both peptides mediated plasmid delivery to the same extent in serum-free media, displaying enhanced uptake with increased CRs. Interestingly, in the presence of serum, negligible uptake was observed for TP10/plasmid complexes while stearyl-TP10/plasmid complexes were efficiently internalized (Figure 2f). These results suggest that although the stearyl moiety slightly increases cellular uptake as compared to TP10 in serum-free media, the radical difference in transfection levels should, at least partially, be due to increased endosomal escape.
Transfection efficiency in different cell lines and impact of serum proteins
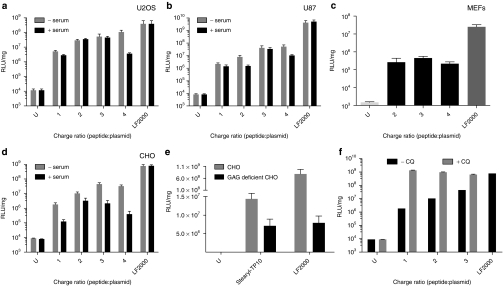
To validate the effectiveness of stearyl-TP10 in other cells as well, we conducted transfection experiments in different commonly used cell lines. As expected, and in accordance with the results in CHO cells (Figure 2c), stearyl-TP10 efficiently mediated plasmid delivery in U2OS, U87, and HEK293 cells (Figures 3a,b and 4b, respectively). In line with the condensation efficiency seen in the EtBr exclusion assay (Figure 1a,b), at higher CRs and, hence, at higher peptide concentrations, the increase in luciferase expression was more pronounced in all of these cell lines. Interestingly, CR3 seemed to be an optimal peptide-over-pDNA ratio, since CRs above that did not significantly increase the delivery efficiency further, or the effect was even decreased. The dynamic light scattering profile of the stearyl-TP10/plasmid nanoparticles (Table 1) supports this observation, as the size of the nanoparticles is more homogenous at higher CRs, especially at CR3.
Figure 3.
Delivery efficacy of stearyl-transportan 10 (TP10)/plasmid nanoparticles in different cell lines and importance of cell surface glucosaminoglycans (GAGs). (a) 5 × 104 U2OS, (b) U87, (c) 3 × 104 mouse embryonal fibroblast (MEF) and (d) 5 × 104 Chinese hamster ovary (CHO) cells were seeded 24 hours before experiment into 24-well plates. Cells were treated and analyzed as in Figure 2a,c, however, for the first 4 hours, transfections were also carried out in 10% serum containing medium, except in MEF cells. (e) 5 × 104 CHO and 5 × 104 GAG-deficient CHO cells were seeded 24 hours before experiment into 24-well plates. Cells were treated and analyzed as in Figure 2a,c. (f) Effect of the addition of chloroquine (CQ) to the transfection efficiency of stearyl-TP10/plasmid nanoparticles in CHO cells. Cells were treated and analyzed as in Figure 2a,c. The values represent the mean of at least three independent experiments performed in duplicate (mean ± SEM).
Figure 4.
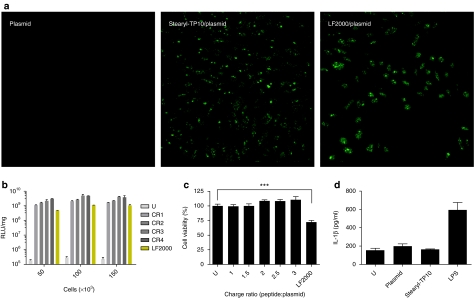
Evaluation of transfection efficiency, confluence dependency, toxicity profile, and induction of innate immunity by stearyl-transportan 10 (TP10)/plasmid nanoparticles compared to Lipofectamine 2000 (LF2000). (a) 7 × 104 Chinese hamster ovary (CHO) cells were seeded 24 hours before experiment into 24-well plates. Cells were treated with stearyl-TP10/plasmid nanoparticles expressing enhanced green fluorescent protein at charge ratio 3 (CR3) for 4 hours in serum-free media followed by replacement to 10% serum containing medium and incubated additionally for 20 hours (stearyl-TP10/plasmid). For negative control plasmid-treated cells were imaged (plasmid). LF2000 was used according to the manufacturer's protocol (LF2000/plasmid). Thereafter, cells were washed with phosphate-buffered saline and fixed by using 4% formaldehyde solution at room temperature for 10 minutes. Images were taken using confocal microscopy. (b) To assess the impact of increasing cell confluency on the transfection efficiency, 5 × 104, 1 × 105, 1.5 × 105 HEK293 cells were seeded 24 hours before experiment into 24-well plates. Cells were treated and analyzed as in Figure 2a,c. (c) Toxicity was assessed by WST-1 proliferation assay 24 hours after treatment of cells with stearyl-TP10/plasmid nanoparticles at different CRs (CR1–CR3) or lipofection. The values represent the mean of at least three independent experiments performed in duplicate (mean ± SEM). (d) Analysis of interleukin (IL)-1β induction in primed THP1 cells treated with stearyl-TP10/plasmid nanoparticles, mock plasmid or LF2000. Lipopolysaccharide (LPS) was used as a positive control. Supernatants were analyzed by enzyme-linked immunosorbent assay 24 hours after incubation.
Cargo-size is one factor that often limits the full potential of delivery vectors. To this end, we used a plasmid with a larger size. As seen in Supplementary Figure S2, a larger plasmid with a size of 6.4 kb was transfected into U87 cells with similar efficiency as the smaller vector (4.7 kb) used throughout most of the experiments in this study. Thus, at least in this size range, stearyl-TP10 is not restricted to certain plasmid size.
Next, we also wanted to investigate whether these nanoparticles would facilitate delivery of plasmids in more refractory primary cells. For this, primary mouse embryonal fibroblast cells were transfected with stearyl-TP10/plasmid complexes, which resulted in a 2.5 log increase in luciferase expression compared to untreated cells (Figure 3c). In line with the effects seen in regular cell lines, LF2000 was around a log more efficient. As expected, the transfection levels with both reagents were overall a log lower than that in the previously tested cell lines (Figures 3a,b,d and 4b).
In order to obtain information on the possible applicability in vivo, a setting that naturally necessitates maintained delivery activity also in the presence of serum proteins, we set out to mimic in vivo conditions. To address this, in vitro transfections were also carried out in full growth media. As seen in U2OS, U87, and CHO cells (Figure 3a–c, respectively), the presence of serum components only slightly affected the transfection efficiency. Even though overall modest decreases were observed in all of the cell lines, increased luciferase expression levels of around three logs over background were obtained.
It is widely accepted that most CPPs use endocytic pathways to enter cells and the first interaction with cellular membranes is commonly considered to involve glucosaminoglycans. To confirm the role of glucosaminoglycans, we assessed the transfection efficiency in a glucosaminoglycan-deficient CHO cell line. Transfection levels of stearyl-TP10/plasmid nanoparticles in this cell line were severely reduced as compared to the regular CHO cells, emphasizing the importance of glucosaminoglycans and that endocytic uptake is a prevailing mechanism for cellular uptake of stearyl-TP10/plasmid nanoparticles (Figure 3e), which was also evident in the case of LF2000. Furthermore, it was observed that coaddition of chloroquine substantially increased the biological activity of these nanoparticles, further underlining the involvement of endocytosis as the mechanism of internalization for stearyl-TP10/plasmid nanoparticles (Figure 3f).
Transfection of entire cell populations and cell confluency independency
One important aspect in the evaluation of new transfection agents is the ability to transfect entire cell populations. In order to address that, we used an enhanced green fluorescent protein (EGFP)-encoding plasmid and analyzed cells by confocal microscopy. Stearyl-TP10/plasmid complexes formulated at CR3 enabled transfection of almost 100% of the cell population, whereas naked plasmid was unable to induce gene expression in any cells (Figure 4a). In the case of lipofection, we confirmed the well-established observation that LF2000 complexes did not reach the entire cell population (Figure 4a).
Another important parameter is that the activity of the transfection reagent ideally should not be strictly dependent on cell confluences. To address this aspect, HEK293 cells were seeded at different densities (5 × 104, 1 × 105, and 1.5 × 105 cells per well) and transfections were carried out as described above. In general, the transfection efficiency was maintained at higher cell confluency both in the case of stearyl-TP10 and LF2000. In keeping with the earlier observations, higher peptide concentration (CRs) resulted in superior transfection levels at all tested cell densities (Figure 4b).
Comparison of toxicity profiles of stearyl-TP10 and LF2000
The most elementary in vitro cytotoxicity is usually evaluated using cell viability assays, which measure metabolic activity of mitochondria. It is well known that high transfection efficiency with lipofection correlates widely with toxicity. This has been described many times previously and we here confirm that LF2000 significantly reduces cell viability, as determined by the WST-1 assay (Figure 4c). After lipofection, we observed as much as a 30% decrease in cell viability over 24 hours. In contrast, stearyl-TP10/plasmid nanoparticles did not exert any toxic effect at any of the tested CRs (Figure 4c).
Inflammatory responses in vitro and in vivo
An important parameter for successful in vivo implementation of nucleic acids is to avoid triggering inflammatory responses. To address this, we evaluated the impact of stearyl-TP10/plasmid nanoparticles in transformed human monocytic THP1 cells by measuring the release of interleukin (IL)-1β, 24 hours after treatment. Stearyl-TP10/plasmid nanoparticles did not influence the release of IL-1β as compared to basal levels of untreated cells, whereas the positive control lipopolysaccharide induced a strong immunogenic response (Figure 4d). The lack of inflammatory responses was further corroborated in vivo, where only negligible levels of the inflammatory mediators IL-6 or tumor necrosis factor-α were observed 24 hours after i.d. or i.m. administration of either stearyl-TP10/plasmid complexes or naked plasmids (data not shown) and the level of C-reactive protein remained unchanged (data not shown).
Administration of stearyl-TP10/plasmid nanoparticles in vivo
After demonstrating efficient plasmid delivery in vitro and minimal influence on inflammatory pathways, we sought to go one step further and investigate the potential of the peptide to be used for plasmid delivery in vivo. To assess the delivery in vivo, we administered stearyl-TP10/plasmid nanoparticles via two different local administration routes; i.m. and i.d.
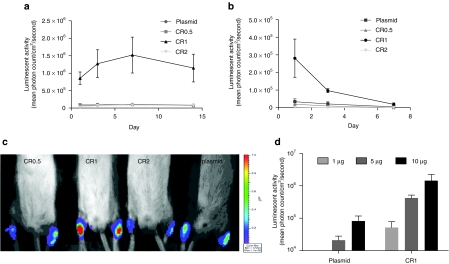
Mice were injected i.m. by bilateral injections into the Musculus tibialis anterior with stearyl-TP10/plasmid nanoparticles, formulated at different CRs, and luciferase activity was measured in anesthetized animals on days 1, 3, 7, and 14 postinjection using a bioluminescence imager. Control mice were injected with naked plasmid. Although animals treated with naked plasmid displayed significant luciferase expression, the expression levels were increased approximately by one log when using stearyl-TP10 nanoparticles formed at CR1, one day postinjection (Figure 5a,c). Particles formed at the other CRs were unable to increase plasmid delivery, as luciferase expression remained at the level of control group. Moreover, luciferase expression was increased on day 3 and reached its peak at day 7. Thereafter, expression started to decline at day 14. These results demonstrate that stearyl-TP10 improves gene delivery in vivo and that it allows relatively persistent expression over time. However, in contrast to in vitro transfections, using a defined CR1 is crucial in order to obtain increased gene expression levels.
Figure 5.
Delivery of stearyl-transportan 10 (TP10)/plasmid nanoparticles in vivo. (a) For the evaluation of delivery efficiency of stearyl-TP10/plasmid nanoparticles after intramuscular (i.m.) administration, 5 µg of pGL3 plasmid was complexed with stearyl-TP10 at charge ratio 0.5 (CR0.5), CR1 and CR2 in 5% glucose in 50 µl volume and injected locally in the Musculus tibialis anterior of Balb/c female mice. Control levels were obtained with only naked plasmid DNA. Luciferase expression was measured using bioluminescence imaging in a Xenogen IVIS100 imager. Error bars indicate SEM, n = 4 for each group. (b) Delivery efficiency of stearyl-TP10/plasmid nanoparticles locally to the skin was evaluated as described above, however, nanoparticles were injected in the dermis of Balb/c female mice. Error bars indicate SEM, n = 4 for each group. (c) In vivo bioluminescence imaging of luciferase expression at day 1 after i.m. administration of stearyl-TP10/plasmid nanoparticles at CR0.5, CR1, CR2 and mock plasmid as a control. (d) Dose-dependent luciferase expression of the stearyl-TP10/plasmid (pEGFPLuc) nanoparticles after intradermal administration at the different doses of plasmid (1, 5, and 10 µg). Luciferase expression was measured as described above.
Next we carried out injections through the i.d. route. Luciferase activity was measured in anesthetized animals using bioluminescence on days 1, 3, and 7 postinjection (Figure 5b). As keratinocytes undergo rapid cell division, extended follow-up was not possible. In accordance with the data obtained in the previous study on i.m. injections, one day postinjection gene expression was increased by nearly one log when using stearyl-TP10 at CR1 over plasmid as compared to animals treated with plasmid only (Figure 5b). Thereafter, expression started to decline and was diminished 7 days after administration of the nanoparticles, which is an expected result, considering the rapid cell division which dilutes episomally located plasmids. Again, peptides formulated at other CRs were unable to promote increased gene expression.
From a pharmacological point of view it is crucial that obtained effects are exerted in a dose-dependent manner. In order to verify this, mice were injected i.d. using different plasmid doses (1, 5, and 10 µg) complexed with stearyl-TP10 at CR1, where 5 µg represented the amount used in the decay kinetics experiments described above. Dose-dependent increases in luciferase expression were achieved with both naked plasmid and with plasmid complexed with stearyl-TP10. However, in line with earlier results, stearyl-TP10 promoted increases in gene expression of an additional order of magnitude as compared to the naked plasmid (Figure 5d). Interestingly, these dose-titration studies were carried out using a larger luciferase-EGFP fusion plasmid (6.4 kb, pEGFPLuc) as compared to the pGL3 plasmid (4.7 kb) used in the initial gene expression decay studies. These results are in accordance with the results from in vitro transfections where the size of plasmid did not affect the transfection efficiency (Supplementary Figure S2).
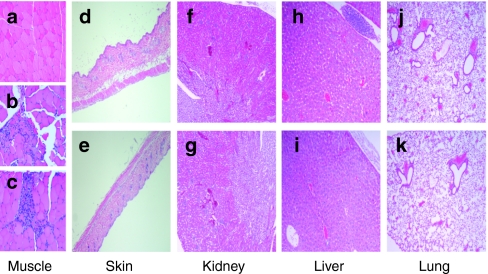
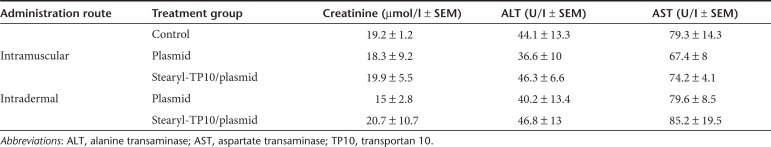
To rule out toxic effects associated with the treatment regimen of stearyl-TP10/plasmid nanoparticles we investigated clinical chemistry parameters, as well as carried out histopathological analysis of the treated tissues and several organs of the treated animals. Histopathological analysis of the treated tissues showed that as compared to normal tissue (Figure 6a) there was a local accumulation of inflammatory cells after i.m. injections in case of both plasmid and stearyl-TP10/plasmid nanoparticle treatments (Figure 6b,c), which probably emanates from local inflammation caused by the response to the injection trauma. However, i.d. treatments did not cause any adverse effects to the histology of the skin with neither treatment regimens (Figure 6d,e). To exclude systemic toxicity, we also conducted histopathological examinations of lung, kidney, and liver, where no signs of histological disturbances compared to normal tissues were detected (Figure 6f–k). This was also corroborated by clinical chemistry measurements. There were no deviations from the basal liver transaminase levels (alanine transaminase/aspartate transaminase) neither after after i.d. nor i.m. administration, indicating that treatments did not affect liver function. Furthermore, serum creatinine levels were also unaffected, thus confirming intact kidney function (Table 2). All clinical chemistry parameters can be found in Table 2.
Figure 6.
Histopathological examination of the treated animals. Animals were treated as described in Figure 5. Organs were dissected 24 hours after treatment and fixed in formalin, embedded in paraffin, and stained with eosin and hematoxylin. Images were taken on the Olympus BX45 microscope with a Sony DXC-S500 digital camera. Histopathological sections of: (a) muscle tissue of the negative control, (b) plasmid-treated and (c) stearyl-transportan 10 (TP10)/plasmid treated animals; (d) skin after treatment with plasmid or (e) stearyl-TP10/plasmid complexes; (f) kidney after treatment with plasmid or (g) stearyl-TP10/plasmid complexes; (h) liver after treatment with plasmid or (i) stearyl-TP10/plasmid complexes; (j) and lung after treatment with plasmid or (k) stearyl-TP10/plasmid complexes.
Table 2. Clinical chemistry parameters of the treated animals.
Discussion
Nucleic acids, e.g., pDNA, can be vectorized with natural and/or synthetic polycations, cationic peptides, lipids or polymers.4,5 The common denominator of all of these compounds is that they interact with pDNA via electrostatic and/or hydrophobic interactions and enable the condensation of pDNA into nanoparticles for cellular delivery.4,36
CPPs are one group of such nonviral peptide-based delivery vehicles, which have been shown to facilitate the delivery of pDNA. Even though a handful of papers have described the vectorization of pDNA by CPPs, overall efficiency has in general been poor.13,14,15 Moreover, with a few exceptions, these reports have only proven the effects in in vitro settings, and, to our knowledge, extrapolation to in vivo settings have only rarely been established.4 The explanation for poor efficiency is emanating from the fact that CPPs are internalized into cells via endocytic pathways and, unfortunately, CPPs and their cargo for the most part remain entrapped in nonavailable endocytic vesicles.16,17,18 Therefore, for successful peptide-based delivery, it is pivotal to find efficient measures to overcome this limitation. A wide variety of different strategies has been tested to find solutions to release complexes from endosomes. Although there are some simple methods for endosomal escape in vitro, e.g., coaddition of chloroquine,35 these are not applicable in vivo and, therefore, searches for CPPs with endosomolytic properties, or finding modifications of CPPs to facilitate endosomal escape are ongoing. For example, different membrane-disruptive peptides have been used to facilitate endosomal escape, most prominent being human influenza virus HA2 subunit or different modifications of that segment.37 Lundberg et al., for instance, designed an endosomolytic peptide, EB1, which by the introduction of histidines, formed helical structures in early/late endosomes and destabilized endosomal membranes.30
Addition of hydrophobic moieties to CPPs has been shown to be an efficient mean to increase the bioavailability of CPPs and associated cargo. Particularly, stearylation has proven to be a successful method to introduce hydrophobicity to CPPs, in the case of delivery of plasmids,23,24,25,38 splice-correcting oligonucleotides26 and small interfering RNAs.22,25 However, it is not a universal method, as we have previously modified dozens of different CPPs with stearic acid and only few have been active in noncovalent coincubation settings for the delivery of short nucleic acids (ref. 8 and data not shown). In our previous study, we showed that stearyl-TP10 was superior to other stearylated CPPs in terms of delivery of splice-correcting oligonucleotides, significantly exceeding the activity of arginine-rich CPPs such as Arg9.26 However, to what extent these findings could be extrapolated to the delivery of larger molecules such as plasmids remain unknown. Therefore, we assessed the delivery properties of stearyl-TP10 for its ability to convey pDNA into cells in vitro and in vivo.
Our results demonstrate that stearylation of the TP10 peptide has a significant impact on plasmid delivery in different cell lines. While TP10/plasmid complexes were completely inactive, stearylation of the peptide resulted in around four log increases in luciferase expression levels as compared to plasmid-treated cells (Figures 2b, 3a,b,d and 4b). However, it remains unclear exactly how the stearyl moiety mediates the increased biological effect. Clearly, stearic acid renders the peptide more hydrophobic and presumably hydrophobicity plays an important role in particle formation, as it enables more pronounced pDNA condensation and the formation of small, stable particles (Figure 1b and Table 1). This protects pDNA, making it more stable against heparin and the degrading capacity of serum enzymes (Figure 1c–e and Supplementary Figure S1). The drastic increase in activity of stearyl-TP10 compared to TP10 could also be a result of increased cellular uptake of particles. The lack of biological response with TP10/plasmid particles, even in presence of chloroquine, indicates that these complexes are not taken up by cells to a sufficient degree (Figure 2a). To corroborate this, the uptake levels of fluoresceinyl-labeled plasmid complexed with either of the peptides were assessed. Interestingly, only minor differences in uptake were observed between the two peptides, both promoting dose-dependent increases in plasmid uptake with increasing CRs in serum-free media (Figure 2e). Thus, the underlying reason for the absence of effect of TP10/plasmid could be rapid dissociation of peptides from plasmids in endosomes prohibiting further transport of the plasmid to the nucleus. The inability of TP10 to promote plasmid uptake in presence of serum further supports the heparin replacement data that these complexes are very labile (Figures 2f and 1e). These results suggest that stearyl-TP10 forms more stable particles with plasmids and promotes endosomal escape to a higher extent. It should however be emphasized that even though stearyl-TP10 triggers endosomal release to a greater extent than TP10, there is still plenty of room for improvement as seen in Figure 3f. Apparently, coaddition of chloroquine to stearyl-TP10/plasmid complexes further increases gene expression by another 1–2 logs.
It has been shown before that stearylation of polyarginines facilitates plasmid delivery.23 We observed that stearylation of Arg9 had an effect on plasmid transfection efficiency, however, compared to stearyl-TP10, stearyl-Arg9 was around 100-times less efficient (Figure 2b). Similar effects were seen in the case of other stearylated arginine-rich peptides, e.g., penetratin or M918 peptide, which were less effective than stearyl-Arg9 (data not shown). This is a further justification of the role of hydrophobicity, as TP10 is a known amphipathic CPP, i.e., partially hydrophobic, and stearylation of this peptide has more pronounced effects than on arginine-rich peptides such as Arg9.
Stearyl-TP10 fulfilled most of the critical criteria required for an efficient transfection agent in vitro, namely: transfection of the whole cell population, nontoxic nature, transfection of primary cells, insensitivity to serum proteins, and relative independence of cell confluence. Moreover, plasmids of different sizes were transfected with equal efficiency. Although being efficient, LF2000 fails to meet some of these criteria, as mentioned above. For being applicable in vivo, we also confirmed that stearyl-TP10/plasmid nanoparticles were in vitro essentially nontoxic and nonimmunogenic.
These stearyl-TP10/plasmid nanoparticles facilitated efficient gene delivery in muscle and skin, after i.m. or i.d. injections, respectively. In both cases, luciferase activity was increased around one log as compared to the background levels and these effects were shown to be dose-dependent (Figure 5d). We also confirmed that the gene delivery did not trigger any immune response in vivo and that these treatments were not associated with any systemic toxicity (Table 2, Figure 6 and data not shown). Interestingly, these effects were only seen with the nanoparticles formed at CR1, while at other CRs, no effect on luciferase activity was seen. The critical dependency on certain CR possibly emanates from the avidity of stearyl-TP10 toward pDNA and, therefore, the stability of these nanoparticles. Probably, at higher CRs release of pDNA from the complexes is perturbed and the affinity of stearyl-TP10 toward pDNA too great for in vivo applicability. Therefore, pDNA cannot escape from the nanoparticle complex and, consequently, does not reach to cell nuclei throughout the tissue with higher efficacy than naked pDNA. These differences between the avidity at different CRs were confirmed by heparin displacement assay, where at CR1 nanoparticles were easily accessible to heparin, whereas at higher CRs heparin only partially displaced peptides from the nanoparticles. These results underline that in vitro results do not inevitably correlate with in vivo results, as higher peptide concentrations in vitro facilitated increased gene expression, whereas in the more complex living organism, where conditions differ substantially, only CR1 was able to enhance the gene delivery. In comparison with other nanoparticle-based gene delivery vehicles, the relative efficiency of stearyl-TP10 is in similar range, as for i.m. delivery, gene expression increase between 10–100-fold has been reported with most efficient systems.39,40,41
Conclusively, taking into account that stearyl-TP10 facilitated efficient plasmid delivery in several cell lines with the efficiency being in line with the commercially used transfection agent, LF2000, and that stearyl-TP10/plasmid nanoparticles enabled efficient gene delivery in vivo, it makes stearyl-TP10 a highly interesting peptide-based plasmid delivery vector for the future utilization, both in vitro and in vivo.
Materials and Methods
Synthesis of peptides. All peptides (Supplementary Table S2) were synthesized in stepwise manner in a 0.1 mmol scale on an automated peptide synthesizer (ABI433A; Applied Biosystems, Carlsbad, CA) using fluorenylmethyloxycarbonyl solid-phase peptide synthesis strategy with Rink-amide methylbenzylhydrylamine resin (Fluka, Buchs, Switzerland) as a solid phase to obtain C-terminally amidated peptides. N-terminally stearylated peptides were prepared by treatment of peptidyl-resins with 4 eq. stearic acid (Sigma, St Louis, MO) and 4 eq. HOBt/HBTU (MultiSynTech, Witten, Germany) and 8 eq. DIEA (Fluka) in DMF for 60 min. The final cleavage was performed using standard protocol (95% trifluoroacetic acid/2.5% triisopropylsilane/2.5% water) for 2 hours at room temperature. Peptides were purified by reversed-phase high-performance liquid chromatography using C18 column and 5–80% acetonitrile (0.1% trifluoroacetic acid) gradient. The molecular weight of the peptides was analyzed by matrix-assisted laser desorption/ionization-time of flight mass-spectroscopy and purity was >90% as determined by analytical high-performance liquid chromatography.
Cell culture. CHO cells were grown at 37 °C, 5% CO2 in Dulbecco's modified Eagle's medium F12 with glutamax supplement with 0.1 mmol/l nonessential amino acids, 1.0 mmol/l sodium pyruvate, 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin (PAA Laboratories GmbH, Cölbe, Germany).
HEK293, U87, U2OS and mouse embryonal fibroblast cells were grown at 37 °C, 5% CO2 in Dulbecco's modified Eagle's medium with glutamax supplemented with 0.1 mmol/l nonessential amino acids, 1.0 mmol/l sodium pyruvate, 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin (PAA Laboratories GmbH).
Complex formation. 0.5 µg of pGL3 or pEGFP-C1 plasmid (4.7 kb), expressing luciferase or EGFP respectively, was mixed with CPPs at different peptide:plasmid CRs of 0.5:1–4:1 (CR0.5–CR4) in milli-Q water in 50 µl (1/10th of the final treatment volume). CRs were calculated theoretically, taking into account the positive charges of the peptide and negative charges of the plasmid. For instance, final concentration of stearyl-TP10 was 0.75 µmol/l at CR1. Complexes were formed for 1 hour at room temperature. Meanwhile, cell medium was replaced in 24-well tissue culture plates for fresh media (450 µl). In case of LF2000 (Invitrogen, Carlsbad, CA), the complexes were formed according to the manufacturer's protocol, using the recommended amounts for each cell line. Additional luciferase-expressing plasmids were used, namely pcDNA4/TO-Ubi-FFLuc (7kb) and pEGFPLuc (6.4 kb), both from Clontech (Mountain View, CA).
DNA condensation was analyzed using an EtBr (Sigma, Taufkirchen, Germany) exclusion assay. Briefly, complexes were formed as described above. After 1 hour incubation, 135 µl milli-Q water was added to each sample and transferred into a black 96-well plate (NUNC, Roskilde, Denmark). Thereafter, 15 µl of EtBr solution was added to give a final EtBr concentration of 400 nmol/l. After 10 minutes, fluorescence was measured on a Spectra Max Gemini XS fluorometer (Molecular Devices, Palo Alto, CA) at λex = 518 nm and λem = 605 nm. Results are given as relative fluorescence and a value of 100% is attributed to the fluorescence of naked DNA with EtBr.
Stability of stearyl-TP10/plasmid nanoparticles was evaluated in the presence of serum. Briefly, complexes were formed as described above. Thereafter, serum was added to the complexes at standard concentration (10%) and incubated over different periods of time. At 0, 1, 2, 4, and 24 hours samples were loaded on an agarose gel (2%) and imaged by staining the gel with EtBr (0.5 µg/ml).
Dynamic light scattering and zeta potential measurements. Hydrodynamic mean diameter of the DNA nanoparticles was determined by dynamic light scattering studies using a Zetasizer Nano ZS apparatus (Malvern Instruments, Worcester, UK). pDNA complexes resulting from the addition of stearyl-TP10 were formulated according to the protocol for in vitro transfection, as described above, and assessed in disposable low volume cuvettes. Briefly, pDNA complexes were formulated in deionized water, in 20 µl volume, at a final concentration of 0.01 µg/µl of pDNA. After 30 minutes incubation at room temperature, the DNA complexes were diluted in Opti-MEM into a final volume of 200 µl. All data was converted to “relative intensity” plots from where the mean hydrodynamic diameter was derived. ζ-Potential was measured in Opti-MEM supplemented with 10% fetal calf serum. Measurements were performed in ZS Malvern instrument, set to automode and a number of 5 runs.
Heparin displacement assay. For the analysis of their resistance to heparin, peptide formulations containing 100 ng of pDNA were incubated for 30 minutes at 37 °C in the presence of heparin sodium (Sigma-Aldrich, Taufkirchen, Germany) over a range of concentrations. After the incubation period, loading buffer was added and the reactions were analyzed on 0.8% agarose gels in 1× TAE buffer and visualized by staining with SYBR Gold (Invitrogen, Molecular Probes). Gels were documented using the Fluor-S system with a cooled CCD camera (Bio-Rad, Hercules, CA).
Plasmid delivery assay. 5 × 104 CHO, HEK293, U87, U2OS and 3 × 104 mouse embryonal fibroblast cells were seeded 24 hours before experiment into 24-well plates. Cells were treated with CPP/plasmid complexes at different CRs for 4 hours in serum-free or serum containing media followed by addition of 1 ml 10% serum containing medium and incubated for another 20 hours. Thereafter, cells were washed and lysed using 100 µl 0.1% Triton X-100 in HEPES-buffered Krebs Ringer buffer for 30 minutes at room temperature. Luciferase activity was measured using Promega's luciferase assay system on GLOMAX 96 microplate luminometer (Promega, Nacka, Sweden) and normalized to protein content (Lowry; Bio-Rad). LF2000 (Invitrogen) was used according to the manufacturer's protocol and results were taken as a positive control for measuring transfection efficiency.
In experiments with chloroquine, after complex formation and before treatment of cells, chloroquine (final concentration 100 µmol/l) was added to the complex solution. Four hours after addition of the complexes and chloroquine to cells, cell medium was replaced with fresh medium in order to avoid toxicity effects.
Spectrofluorometry analysis. 5 × 104 U2OS cells/well were seeded in 24-well plates 24 hours before cellular treatments with fluorescein-plasmid (Mirus, Madison, USA) complexed with TP10 or stearyl-TP10 as described in the complex formation section. Cells were treated for 24 hours either in Opti-MEM or in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Cells were washed twice with phosphate-buffered saline and once, briefly, with trypsin to remove membrane-bound complexes. Cells were thereafter lysed using 0.2% Triton in phosphate-buffered saline for 1 hour and lysates were transferred to a black 96-well plate. Fluorescence was measured on at 490/518 nm on a Spectra Max Gemini (Molecular Devices) fluorometer. Fluorescence signal (RFU) from untreated cells was subtracted from the signals of treated cells.
Confocal microscopy. 7 × 104 CHO cells were seeded 24 hours before experiment onto 13 mm tissue culture coverslips that were placed into a 24-well plate. Cells were treated with pEGFP-C1 plasmid and CPP complexes at different CRs (1–3) for 4 hours in serum-free media followed by addition of 1 ml of full growth media and incubated for another 20 hours at 37 °C. LF2000 (Invitrogen) was used according to the manufacturer's protocol. Thereafter, cells were washed with phosphate-buffered saline and fixed by using 4% formaldehyde solution at room temperature for 10 minutes. Images were captured using 60-fold objective on Nikon Eclipse TE2000-U inverted microscope and a digital camera DXM1200C, and processed with EZ-C1 software V.2.30 (Nikon, Tokyo, Japan).
WST-1 proliferation assay. Cell proliferation was studied with the Roche Wst-1 proliferation assay according to the manufacturer's instructions. Briefly, 104 cells were seeded one day before the experiment in a 96-well plate. Cells were treated with stearyl-TP10/plasmid nanoparticles at different CRs for 4 hours in serum-free medium followed by addition of 10% serum containing medium and incubated for another 20 hours. Transfection with LF2000 was carried out according to the manufacturer's protocol. WST-1 was added according to manufacturer's protocol (Roche Diagnostics Scandinavia AB, Bromma, Sweden). WST-1 measures the activity of mitochondrial dehydrogenases to convert tetrazolium salts to formazan, which absorbs light at 450 nm. Absorbance was measured on Digiscan absorbance reader (Labvision via AH Diagnostics AB, Varmdo, Sweden). Untreated cells were defined as 100% viable.
IL-1β, tumor necrosis factor-α, and IL-6 analysis. THP1 cells were differentiated using phorbol myristate acetate (10 ng/ml) for 48 hours and subsequently seeded into 24-well plates (2 × 105 cells/well). Cells were treated as previously. Lipopolysaccharide (15 µg/ml) was used as positive control. Culture supernatants were collected at 4 hours and 24 hours after treatment, and assayed for IL-1β and tumor necrosis factor-α by enzyme-linked immunosorbent assay according to manufacturer's protocol (R&D systems, Minneapolis, MN). IL-6 and tumor necrosis factor-α levels in blood were analyzed at 24 hours post i.d. or i.m. treatments of NMRI female mice. Blood was collected retro-orbitally and serum was purified using serum separation tubes (BD Bioscience, Stockholm, Sweden). An amount of 100 µl serum was assayed using enzyme-linked immunosorbent assay Max Deluxe Set (BioLegend, San Diego, CA) and absorbance measured on Spectra Max Gemini (Molecular Devices).
In vivo experiments. Female Balb/c mice aged 8–10 weeks were first anaesthetized with isoflurane gas (400 ml air flow and 4% isoflurane) and kept under anesthesia (220 ml air flow and 2.8% isoflurane) during the administration procedure. For the complex formation, 1, 5, or 10 µg of pGL3 (4.7 kb) or pEGFPLuc (6.4 kb) plasmid was mixed with stearyl-TP10 at CRs of 0, 0.5, 1, and 2 in 5% glucose using a total volume of 50 µl. In decay kinetics measurements 5 µg of pGL3 plasmid was used, while in dose-dependency experiments, the abovementioned doses of pEGFPLuc plasmid was used. Thereafter, stearyl-TP10/plasmid nanoparticles were injected i.d. or i.m. into M. tibialis anterior. Gene expression was assessed by imaging of the reporter gene (firefly luciferase) expression. Anesthetized mice were injected intraperitoneally with 150 mg/kg of D-Luciferin (Xenogen, Alameda, CA). Light signals (CCD) images were obtained using a IVIS 100 system (Xenogen). Luciferase expression was quantified by total flux using Living Image Software (Xenogen). The maximum photon/second of acquisition/cm2 pixel/steridian was determined within a region of interest to be the most consistent measure for comparative analysis. In general, acquisition times ranged from 10 seconds to 1 minute.
Clinical chemistry and histopathology. Clinical chemistry parameters (alanine transaminase/aspartate transaminase, C-reactive protein, and creatinine levels) in serum from Balb/c mice were analyzed after 24 hours post-treatments by the Clinical chemistry laboratory at Karolinska University Hospital using International Federation of Clinical Chemistry and Laboratory Medicine standardized techniques. Blood was collected retro-orbitally and serum was purified using serum separation tubes (BD Bioscience). Organs were dissected after 24 hours and fixed in formalin, embedded in paraffin, and stained with eosin and hematoxylin. Images were taken on the Olympus BX45 microscope with a Sony DXC-S500 digital camera. Histology sections were analyzed by the Department of Pathology at Karolinska University Hospital.
Ethical permission. The animal experiments were approved by The Swedish Local Board for Laboratory Animals. The experiments were performed in accordance with the ethical permission and were carried out in accordance to European Community directive (86/609/EEC). All animal experiments where designed to minimize the suffering and pain of the animals.
Statistics. Values in all experiments are represented as mean ± SEM of at least three independent experiments done in duplicate. Increase in delivery efficiency was considered significant at ***P < 0.001 using analysis of variance Dunnett's multiple comparison test or analysis of variance Bonferroni's multiple comparison test. In toxicity measurements, decrease in viability was considered significant at ***P < 0.001 using analysis of variance Dunnett's multiple comparison test.
SUPPLEMENTARY MATERIAL Figure S1. Serum stability of stearyl-TP10/plasmid nanoparticles. Figure S2. Ability of stearyl-TP10 to transfect plasmids with different size in U87 cells. Table S1. Sequences of cell-penetrating peptides. Table S2. ζ-Potential of stearyl-TP10/plasmid nanoparticles.
Acknowledgments
The work presented in this article was supported by Swedish Research Council (VR-NT); by Center for Biomembrane Research, Stockholm; by Knut and Alice Wallenberg's Foundation; by the EU through the European Regional Development Fund through the Center of Excellence in Chemical Biology, Estonia; by the targeted financing SF0180027s08 from the Estonian Government; by the DoRa Program of The European Social Fund; and by Archimedes Foundation; by KI faculty funds for funding of postgraduated students (to J.R.V.). S.E.L.A. was supported by the Swedish Society of Medical Research (SSMF), D.-M.C. was supported by a postdoctoral fellowship of the Estonian Science Foundation—Mobilitas—MJD64, E.M.Z is supported by a grant from the Egyptian Ministry of higher Education. We would like thank Kadi-Liis Veiman for assisting some of the experiments.
Supplementary Material
Serum stability of stearyl-TP10/plasmid nanoparticles.
Ability of stearyl-TP10 to transfect plasmids with different size in U87 cells.
Sequences of cell-penetrating peptides.
ζ-Potential of stearyl-TP10/plasmid nanoparticles.
REFERENCES
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P.et al. (2003LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1 Science 302415–419. [DOI] [PubMed] [Google Scholar]
- Thomas CE, Ehrhardt A., and, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- Glover DJ, Lipps HJ., and, Jans DA. Towards safe, non-viral therapeutic gene expression in humans. Nat Rev Genet. 2005;6:299–310. doi: 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- Viola JR, El-Andaloussi S, Oprea II., and, Smith CI. Non-viral nanovectors for gene delivery: factors that govern successful therapeutics. Expert Opin Drug Deliv. 2010;7:721–735. doi: 10.1517/17425241003716810. [DOI] [PubMed] [Google Scholar]
- Pack DW, Hoffman AS, Pun S., and, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- Petros RA., and, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- Derossi D, Joliot AH, Chassaing G., and, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- Mäe M, Andaloussi SE, Lehto T., and, Langel U. Chemically modified cell-penetrating peptides for the delivery of nucleic acids. Expert Opin Drug Deliv. 2009;6:1195–1205. doi: 10.1517/17425240903213688. [DOI] [PubMed] [Google Scholar]
- Fonseca SB, Pereira MP., and, Kelley SO. Recent advances in the use of cell-penetrating peptides for medical and biological applications. Adv Drug Deliv Rev. 2009;61:953–964. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Vivès E, Schmidt J., and, Pèlegrin A. Cell-penetrating and cell-targeting peptides in drug delivery. Biochim Biophys Acta. 2008;1786:126–138. doi: 10.1016/j.bbcan.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Eguchi A., and, Dowdy SF. siRNA delivery using peptide transduction domains. Trends Pharmacol Sci. 2009;30:341–345. doi: 10.1016/j.tips.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Said Hassane F, Saleh AF, Abes R, Gait MJ., and, Lebleu B. Cell penetrating peptides: overview and applications to the delivery of oligonucleotides. Cell Mol Life Sci. 2010;67:715–726. doi: 10.1007/s00018-009-0186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatovich IA, Dizhe EB, Pavlotskaya AV, Akifiev BN, Burov SV, Orlov SV.et al. (2003Complexes of plasmid DNA with basic domain 47-57 of the HIV-1 Tat protein are transferred to mammalian cells by endocytosis-mediated pathways J Biol Chem 27842625–42636. [DOI] [PubMed] [Google Scholar]
- Liu Z, Li M, Cui D., and, Fei J. Macro-branched cell-penetrating peptide design for gene delivery. J Control Release. 2005;102:699–710. doi: 10.1016/j.jconrel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Lo SL., and, Wang S. An endosomolytic Tat peptide produced by incorporation of histidine and cysteine residues as a nonviral vector for DNA transfection. Biomaterials. 2008;29:2408–2414. doi: 10.1016/j.biomaterials.2008.01.031. [DOI] [PubMed] [Google Scholar]
- Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R., and, Brock R. A comprehensive model for the cellular uptake of cationic cell-penetrating peptides. Traffic. 2007;8:848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- Zorko M., and, Langel U. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv Drug Deliv Rev. 2005;57:529–545. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Futaki S, Nakase I, Tadokoro A, Takeuchi T., and, Jones AT. Arginine-rich peptides and their internalization mechanisms. Biochem Soc Trans. 2007;35 Pt 4:784–787. doi: 10.1042/BST0350784. [DOI] [PubMed] [Google Scholar]
- El-Andaloussi S, Johansson HJ, Lundberg P., and, Langel U. Induction of splice correction by cell-penetrating peptide nucleic acids. J Gene Med. 2006;8:1262–1273. doi: 10.1002/jgm.950. [DOI] [PubMed] [Google Scholar]
- Lundin P, Johansson H, Guterstam P, Holm T, Hansen M, Langel U.et al. (2008Distinct uptake routes of cell-penetrating peptide conjugates Bioconjug Chem 192535–2542. [DOI] [PubMed] [Google Scholar]
- El-Sayed A, Futaki S., and, Harashima H. Delivery of macromolecules using arginine-rich cell-penetrating peptides: ways to overcome endosomal entrapment. AAPS J. 2009;11:13–22. doi: 10.1208/s12248-008-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Kogure K, Futaki S., and, Harashima H. Octaarginine-modified multifunctional envelope-type nano device for siRNA. J Control Release. 2007;119:360–367. doi: 10.1016/j.jconrel.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Futaki S, Ohashi W, Suzuki T, Niwa M, Tanaka S, Ueda K.et al. (2001Stearylated arginine-rich peptides: a new class of transfection systems Bioconjug Chem 121005–1011. [DOI] [PubMed] [Google Scholar]
- Khalil IA, Futaki S, Niwa M, Baba Y, Kaji N, Kamiya H.et al. (2004Mechanism of improved gene transfer by the N-terminal stearylation of octaarginine: enhanced cellular association by hydrophobic core formation Gene Ther 11636–644. [DOI] [PubMed] [Google Scholar]
- Lehto T, Abes R, Oskolkov N, Suhorutsenko J, Copolovici DM, Mäger I.et al. (2010Delivery of nucleic acids with a stearylated (RxR)4 peptide using a non-covalent co-incubation strategy J Control Release 14142–51. [DOI] [PubMed] [Google Scholar]
- Mäe M, El Andaloussi S, Lundin P, Oskolkov N, Johansson HJ, Guterstam P.et al. (2009A stearylated CPP for delivery of splice correcting oligonucleotides using a non-covalent co-incubation strategy J Control Release 134221–227. [DOI] [PubMed] [Google Scholar]
- Deshayes S, Morris M, Heitz F., and, Divita G. Delivery of proteins and nucleic acids using a non-covalent peptide-based strategy. Adv Drug Deliv Rev. 2008;60:537–547. doi: 10.1016/j.addr.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Meade BR., and, Dowdy SF. Enhancing the cellular uptake of siRNA duplexes following noncovalent packaging with protein transduction domain peptides. Adv Drug Deliv Rev. 2008;60:530–536. doi: 10.1016/j.addr.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen H, Kunath K, Martin AL, Stolnik S, Roberts CJ, Davies MC.et al. (2002Star-shaped poly(ethylene glycol)-block-polyethylenimine copolymers enhance DNA condensation of low molecular weight polyethylenimines Biomacromolecules 3926–936. [DOI] [PubMed] [Google Scholar]
- Lundberg P, El-Andaloussi S, Sütlü T, Johansson H., and, Langel U. Delivery of short interfering RNA using endosomolytic cell-penetrating peptides. FASEB J. 2007;21:2664–2671. doi: 10.1096/fj.06-6502com. [DOI] [PubMed] [Google Scholar]
- Tong AW, Jay CM, Senzer N, Maples PB., and, Nemunaitis J. Systemic therapeutic gene delivery for cancer: crafting Paris' arrow. Curr Gene Ther. 2009;9:45–60. doi: 10.2174/156652309787354630. [DOI] [PubMed] [Google Scholar]
- Bartsch M, Weeke-Klimp AH, Meijer DK, Scherphof GL., and, Kamps JA. Massive and selective delivery of lipid-coated cationic lipoplexes of oligonucleotides targeted in vivo to hepatic endothelial cells. Pharm Res. 2002;19:676–680. doi: 10.1023/a:1015318415705. [DOI] [PubMed] [Google Scholar]
- Furumoto K, Nagayama S, Ogawara K, Takakura Y, Hashida M, Higaki K.et al. (2004Hepatic uptake of negatively charged particles in rats: possible involvement of serum proteins in recognition by scavenger receptor J Control Release 97133–141. [DOI] [PubMed] [Google Scholar]
- Hao J, Sha X, Tang Y, Jiang Y, Zhang Z, Zhang W.et al. (2009Enhanced transfection of polyplexes based on pluronic-polypropylenimine dendrimer for gene transfer Arch Pharm Res 321045–1054. [DOI] [PubMed] [Google Scholar]
- Erbacher P, Roche AC, Monsigny M., and, Midoux P. Putative role of chloroquine in gene transfer into a human hepatoma cell line by DNA/lactosylated polylysine complexes. Exp Cell Res. 1996;225:186–194. doi: 10.1006/excr.1996.0169. [DOI] [PubMed] [Google Scholar]
- Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Wadia JS, Stan RV., and, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- Khalil IA, Kogure K, Futaki S, Hama S, Akita H, Ueno M.et al. (2007Octaarginine-modified multifunctional envelope-type nanoparticles for gene delivery Gene Ther 14682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello-Roufaï M, Lambert O., and, Pitard B. Relationships between the physicochemical properties of an amphiphilic triblock copolymers/DNA complexes and their intramuscular transfection efficiency. Nucleic Acids Res. 2007;35:728–739. doi: 10.1093/nar/gkl860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitard B, Pollard H, Agbulut O, Lambert O, Vilquin JT, Cherel Y.et al. (2002A nonionic amphiphile agent promotes gene delivery in vivo to skeletal and cardiac muscles Hum Gene Ther 131767–1775. [DOI] [PubMed] [Google Scholar]
- Namgung R, Nam S, Kim SK, Son S, Singha K, Kwon JS.et al. (2009An acid-labile temperature-responsive sol-gel reversible polymer for enhanced gene delivery to the myocardium and skeletal muscle cells Biomaterials 305225–5233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Serum stability of stearyl-TP10/plasmid nanoparticles.
Ability of stearyl-TP10 to transfect plasmids with different size in U87 cells.
Sequences of cell-penetrating peptides.
ζ-Potential of stearyl-TP10/plasmid nanoparticles.