Abstract
Like many effective therapeutics, interleukin-12 (IL-12) therapy often causes side effects. Tumor targeted delivery may improve the efficacy and decrease the toxicity of systemic IL-12 treatments. In this study, a novel targeting approach was investigated. A secreted alkaline phosphatase (SEAP) reporter gene-based screening process was used to identify a mini-peptide which can be produced in vivo to target gene products to tumors. The coding region for the best peptide was inserted into an IL-12 gene to determine the antitumor efficacy. Affinity chromatography, mass spectrometry analysis, and binding studies were used to identify a receptor for this peptide. We discovered that the linear peptide VNTANST increased the tumor accumulation of the reporter gene products in five independent tumor models including one human xenogeneic model. The product from VNTANST-IL-12 fusion gene therapy increased accumulation of IL-12 in the tumor environment, and in three tumor models, VNTANST-IL-12 gene therapy inhibited distal tumor growth. In a spontaneous lung metastasis model, inhibition of metastatic tumor growth was improved compared to wild-type IL-12 gene therapy, and in a squamous cell carcinoma model, toxic liver lesions were reduced. The receptor for VNTANST was identified as vimentin. These results show the promise of using VNTANST to improve IL-12 treatments.
Introduction
Interleukin-12 (IL-12), discovered by Giorgio Trinchieri in 1989,1 bridges the innate and adaptive immune responses by inducing interferon-γ (IFN-γ) production primarily from natural killer and T cells. Cancer therapy with IL-12 exploits its natural immune functions to polarize T cells to the Th1 phenotype, boost effector T cells, downregulate angiogenesis, remodel the extracellular matrix, and alter the levels of immune suppressive cytokines.2 Due to these activities, IL-12 is one of the most promising cytokines for immunomodulatory cancer therapy.
The initial clinical trials with IL-12 resulted in grave toxicities including deaths, which severely downgraded the reputation and potential application of this effective cytokine. In reality, most anticancer drugs or biological modalities are associated with systemic toxicity. It is imperative to solve this problem to effectively and safely treat the extremely high number of cancer patients.2
A popular strategy for sequestering the effects of cytokine therapies in the tumor environment is targeting cellular markers that are upregulated exclusively in the tumor cells or the tumor microenvironment. Indeed, conjugating IL-12 to tumor-specific antibodies, such as L193 and HER2,4 and tumor vasculature–specific peptides, such as RGD5 and CNGRC,6 improves the efficacy of treatments; however, the necessarily high frequency of administrations of recombinant cytokines increases the immunogenicity, toxicity, and cost of treatments. A gene therapy approach would reduce these limitations.
Intratumoral IL-12 gene therapy is able to eradicate 40% of tumors in an SCCVII model while systemic delivery via intramuscular administration fails to eradicate any tumors;7 however, direct injection into tumor sites is rarely available noninvasively or postsurgically. Several methods have been developed to target the IL-12 effect to the tumor after systemic delivery. For example, modifying viral vectors with tissue-specific gene promoters such as the CALC-I promoter,8 capsid-expressed tumor-specific peptides,9 and polyethylene glycol or other nanoparticles10,11 increases tumor-specific expression and decreases systemic expression; however, the fenestrated vasculature of the tumor environment allows for the gene products to leak out of the tumor environment leading to systemic toxicities.12 Therefore, a gene product that can interact with and remain in the tumor environment will increase the level of therapeutic efficacy and decrease systemic toxicity.
To this end, we used an in vivo reporter gene-mediated screening strategy13 to identify a new tumor-targeting peptide, VNTANST.14 A DNA fragment encoding VNTANST was inserted directly before the stop codon of the IL-12 encoding sequence in plasmid DNA. Transfection of this plasmid DNA via intramuscular (i.m.) electroporation into muscle tissue distal from the tumor site inhibited tumor growth and extended survival in multiple tumor models and two mouse strains and reduced lung metastasis in a spontaneous metastatic model. Due to this broad targeting nature and to simplify the description, the peptide VNTANST was renamed the Comprehensive Carcinoma Homing Peptide (CHP). Also, we discovered the receptor for CHP to be vimentin, which is upregulated in several tumor types. Vimentin experessin in tumors is associated with the epithelial-to-mesenchymal transition and increased malignancy and metastasis in tumors.15,16,17,18,19 Lastly, this gene product-targeted approach minimized the risk of IL-12–induced toxicity.
Results
CHP increases accumulation of the fusion reporter gene product and biotin-CHP conjugate into tumors
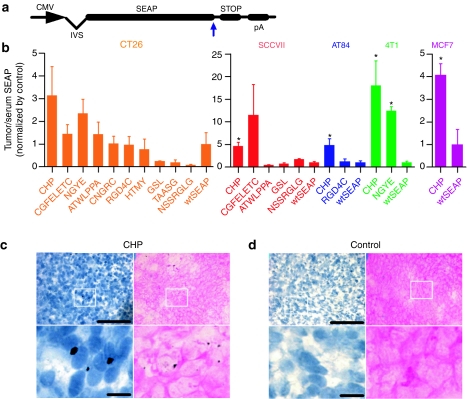
Several fusion gene constructs were cloned by inserting peptide-encoding DNA sequences directly prior to the stop codon in a secreted alkaline phosphatase (SEAP) reporter plasmid DNA (Figure 1a).13 These peptide-SEAP fusion gene constructs were delivered via i.m. electroporation of the anterior tibialis muscles in mice bearing tumors located 1 cm craniodorsal of the tail. After 72 hours, tumors and serum were collected and analyzed for SEAP distribution. It has been shown that inserting peptides into the SEAP plasmid can alter SEAP activity but not protein production.20 To compensate for the altered SEAP activity, we used the ratio of the SEAP activity between tumors and serum (T/S SEAP). The uncorrected values for the tumor and serum SEAP activities are shown in Supplementary Figure S1.
Figure 1.
Accumulation of peptide-SEAP reporter gene products and CHP-biotin in tumors. (a) The peptide-SEAP constructs with insertion of the peptide-coding sequence directly before the stop codon (blue arrow). (b) T/S SEAP levels 72 hours after intramuscular EP of peptide-SEAP plasmid DNA in syngeneic CT26 (orange, n = 3), SCCVII (red, n = 4), AT84 (blue, n = 4), and 4T1 (green, n = 4) tumor-bearing mice as well as xenogeneic MCF7 tumor-bearing mice (purple, n = 4). Columns represent the ratio of the control-normalized SEAP/protein (pg/mg) in tumor to SEAP (pg/ml) in the serum and error bars represent SEM (*P < 0.05 compared to wtSEAP). (c,d) DAB staining of tumor tissues from CHP-biotin and control-peptide-biotin treated mice counterstained with either hematoxylin (blue) or eosin (pink). The bottom images are larger versions of the areas within the white squares. Bar = 100 µm in the top panels and bar = 200 µm in the bottom panels. CHP, carcinoma homing peptide; CMV, cytomegalovirus promoter; EP, electroporation; IVS, intron; pA, bovine growth hormone polyadenylation signal; SEAP, secreted alkaline phosphatase-coding sequence; STOP, stop codon.
CHP, a linear peptide, repeatedly increased the T/S SEAP levels in several tumor models compared to wtSEAP. In Balb/c mice bearing colon carcinomas (CT26), CHP showed the greatest increase in T/S SEAP (orange; Figure 1b). To identify the peptides with potential for targeting multiple tumor models, some of these peptides were also tested in other models. In two squamous cell carcinoma models (SCCVII and AT84) in C3H mice, five and sevenfold increases in T/S SEAP were seen, respectively, for CHP-SEAP (red and blue, respectively; Figure 1b). In a breast adenocarcinoma model (4T1) in Balb/c mice, T/S SEAP was increased 15-fold for CHP-SEAP compared to wtSEAP (green; Figure 1b). Importantly, the gene product targeting property of CHP was also confirmed in a xenogeneic human breast cancer model (MCF7) with a fourfold increase in T/S SEAP (purple; Figure 1b), which suggests that this peptide has potential application for human tumors.
In addition to the quantitative T/S SEAP data above, we were interested in visualizing the CHP distribution in the tumors and throughout the body. To easily detect the localization of this targeted peptide, synthetic CHP-biotin conjugate or a control-peptide-biotin conjugate (control-biotin) were injected into the tail vein of SCCVII tumor–bearing C3H mice. CHP preferentially accumulated deep into the tumor environment, and, as seen in slides counterstained with either hematoxylin (Figure 1c, top and bottom left) or eosin (Figure 1c, top and bottom right), the CHP-biotin localized in the tumor tissue (19.6 ± 1.3 positive per field, n = 5 fields). In contrast, control-biotin was unable to penetrate deep into the tumor tissues (1.8 ± 0.37 positive per field, n = 5 fields, P < 0.0001 compared to CHP-biotin) (Figure 1d). Negligible levels of biotin were accumulated in the hearts, lungs, livers, and kidneys of mice treated with either CHP- or control-biotin; however, similar levels of CHP-biotin and control-biotin were detected in the spleens (Supplementary Figure S2), most likely due to nonspecific uptake by the efficient mononuclear phagocytes bounding splenic red pulp sinuses.
CHP-IL-12 gene product maintains targeting and biological functions
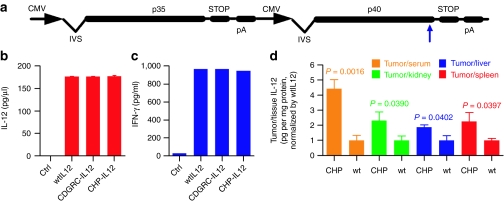
Peptide-IL-12 fusion gene constructs were generated by inserting the peptide-coding sequences directly before the stop codon of the p40 subunit in an IL-12 plasmid DNA (Figure 2a).20 CHP-IL-12, CDGRC-IL-12, wild-type IL-12 (wtIL-12), or empty vector plasmid DNA were transfected into 4T1 cells. After 24 hours, equivalent levels, ~175 pg/µl, of the IL-12p70 heterodimer were detected in the medium of all three IL-12 gene plasmid DNA-transfected cells, and negligible IL-12p70 was detected in the control wells (Figure 2b). Transferring the IL-12 containing medium to splenocytes induced similar levels of IFN-γ, a hallmark of IL-12 function (Figure 2c) indicating that these fusion IL-12 proteins possess the same biological function as wtIL-12.
Figure 2.
Biological activity and targeting ability of peptide-IL-12 fusion gene products. (a) The peptide-IL-12 constructs with insertion of the peptide-coding sequence directly before the stop codon in the p40 coding region (blue arrow). (b) Expression of interleukin-12 (IL-12) after in vitro transfection of 4T1 cells with control, wtIL-12, and peptide-IL-12 (n = 3) and (b) induction of interferon-γ (IFN-γ) from splenocytes after transfer of condition medium containing control, wild-type IL-12 (wtIL-12), or peptide-IL-12 gene products. (d) IL-12 accumulation in tumor-bearing IL-12−/− mice treated with carcinoma homing peptide (CHP)-IL-12 or wtIL-12 determined via an IL-12p70 enzyme-linked immunosorbent assay. Columns represent the wtIL-12-normalized level of IL-12/protein (pg/mg) in tumor per IL-12/protein (pg/mg) in kidneys (green), livers (blue), and spleens (red) and IL-12 pg/ml serum (orange, n = 4). Error bars represent SEM (*P < 0.05 compared to all groups). CMV, cytomegalovirus promoter; IVS, intron; pA, bovine growth hormone polyadenylation signal; SEAP, secreted alkaline phosphatase-coding sequence; STOP, stop codon.
The distribution of CHP-IL-12 in the tumor, kidney, liver, and serum was determined via IL-12p70 enzyme-linked immunosorbent assay 72 hours after treating CT26 tumor-bearing IL-12 knockout Balb/c (IL-12−/−) mice with the CHP-IL-12 and wtIL-12 plasmid DNA. The CHP-IL-12 protein localized in the tumor environment as seen by the fourfold increase in T/S IL-12 ratio compared to wtIL-12 (orange, Figure 2d). Likewise, CHP-IL-12 increased the tumor/kidney, tumor/liver, and tumor/spleen IL-12 ratios compared to wtIL-12 (green, blue, and red, respectively; Figure 2d). Therefore, a single copy of the CHP peptide is capable of targeting each IL-12 molecule to the tumor microenvironment.
CHP-IL-12 fusion gene therapy increases inhibition of primary and metastatic tumor growth and extends survival
Balb/c mice bearing 4T1 or CT26 tumors and C3H mice bearing SCCVII tumors were treated via i.m. electroporation with empty (control), wtIL-12, and CHP-IL-12 (CHP-IL-12) fusion gene plasmid DNA. The treatments were repeated 10 days later. In the highly aggressive syngeneic 4T1 model, CHP-IL-12 gene therapy, compared to wtIL-12 gene therapy, significantly inhibited tumor growth (P < 0.05 at day 30 and P < 0.001 from day 33 until day 42), while both wtIL-12 and CHP-IL-12 treated tumors were less voluminous than control DNA treated mice (Figure 3a). Likewise, CHP-IL-12 treatments extend survival further than wtIL-12 and control (P < 0.05 compared to wtIL-12 plasmid DNA and P < 0.001 compared to control plasmid DNA; Figure 3c). In the same tumor model and treatment regimen, CHP-IL-12 gene therapy reduced by half the number of spontaneous metastatic nodules in the lungs compared to wtIL-12 (P < 0.05 compared to wtIL-12 plasmid DNA; Figure 3b). Similarly, in the SCCVII model CHP-IL-12 improves tumor growth inhibition compared to wtIL-12 (P < 0.05 on days 17 and 20; Figure 3d) and extends survival of mice compared to both control and wtIL-12 (P < 0.05; Figure 3e).
Figure 3.
Inhibition or regression of primary tumor growth, decrease in metastatic tumor incidence, and increase in survival time by systemic delivery of CHP-IL-12 plasmid DNA. (a) Tumor growth following treatments with CHP-IL-12, wild-type IL-12 (wtIL-12), and control plasmid DNA in 4T1 tumor-bearing balb/c mice (n = 5; *P < 0.05 at day 30 and P < 0.001 from day 33 until day 42 compared to wtIL-12 plasmid DNA and P < 0.01 at day 21 and P < 0.001 from day 24 to day 33 compared to control plasmid DNA). (b) Metastatic nodules in the lungs of mice (n = 5) treated as in (a) and killed 17 days after the second treatment (*P < 0.05 compared to wtIL-12 plasmid DNA; #P < 0.001 compared to control plasmid DNA). (c) Kaplan–Meier survival analysis of the same mice treated in (a) (*P < 0.05 compared to wtIL-12 plasmid DNA; #P < 0.001 compared to control plasmid DNA). (d) Tumor growth following treatments as in a in SCCVII tumor-bearing C3H mice (n = 5; *P < 0.05 on days 17 and 20 compared to wtIL-12 plasmid DNA and control plasmid DNA). (e) Kaplan–Meier survival analysis of the same mice treated in (d) (*P < 0.05 compared to wtIL-12 and control plasmid DNA). (f) Treatments as in a in CT26 tumor-bearing balb/c mice (n = 5; *P < 0.05 compared to wtIL-12 plasmid DNA, n = 4, on day 25, and control plasmid DNA, n = 3, on days 19 through 25). Black arrows represent treatments, and error bars represent SEM. CHP, carcinoma homing peptide; IL-12, interleukin-12.
In a third syngeneic model, CT26, CHP-IL-12 treatments inhibit tumor volumes starting only a few days after one treatment (P < 0.05 compared to wtIL-12 plasmid DNA on day 25 and control plasmid DNA on days 19 through 25), and tumors begin to regress after the second treatment (Figure 3f). After day 25, tumors in both wtIL-12 and CHP-IL-12 treated mice began to be eradicated. By day 55, 100% of mice treated with CHP-IL-12 were tumor-free while only 75% of wtIL-12 treated mice were tumor-free.
The CHP-IL-12 treatments increased the immune response to the tumor cells. To understand the mechanism by which CHP-IL-12 boosts inhibition of tumor growth as compared to wtIL-12, both cytotoxic T lymphocytes (CTL) activity and tumor microenvironment immune cell profiling were analyzed. The rationale is that intratumoral injection, associated with a high level of IL-12 in tumors, boosted antitumor immune responses as compared to i.m. injection of IL-12 plasmid DNA, which is associated with a very low level of IL-12 in tumors.7 Treatment with CHP-IL-12 increased the number of the tumor-infiltrating mature dendritic cells in the tumor environment as determined by fluorescence-activated cell sorting for CD11c+/CD80hi expression (Figure 4a). Tumors from wtIL-12 treated mice contained 76.9% mature dendritic cells, an increase from 73.7% in control treated mice. This population in CHP-IL-12 treated mice was even higher at 82.3% (Figure 4a). In agreement with this increase in mature dendritic cells in tumors, tumor-specific CTL activity was increased with CHP-IL-12 treated mice compared to wtIL-12 treated mice (Figure 4b). Furthermore, CHP-IL-12 treatments did not cause any further increase in serum IFN-γ levels, so these immune responses are not the result of widespread IL-12 activity (Figure 4c). These results suggest that CHP-IL-12 improves the antitumor immune response of effector cells in the tumor microenvironment.
Figure 4.
Increased antitumor immune response by CHP-IL-12 fusion gene treatments. (a) Fluorescence-activated cell sorting analysis of tumor-infiltrating cells isolated from SCCVII tumors from C3H mice treated as described in Figure 5 collected 7 days after the second treatment. The top right quadrant of the colored dot plot representation of cells gated for CD11c+ represents activated dendritic cells (CD80hi). (b) Tumor-specific cytotoxic T lymphocytes (CTL) activity from wtIL-12 and CHP-IL-12 fusion gene plasmid DNA treated mice bearing orthotopic EMT6 tumors collected (*P <0.05). (c) Serum interferon-γ (IFN-γ) levels from 4T1-tumor bearing Balb/c 3 days after treatments with CHP-IL-12, wtIL-12, and control plasmid. Error bars represent SEM. CHP, carcinoma homing peptide; IL-12, interleukin-12; wtIL-12, wild-type IL-12.
As with any treatment which includes the introduction of a foreign substance, CHP has the potential of eliciting the production of CHP-specific antibodies. To determine whether these antibodies against the CHP-IL-12 gene product were being produced, a sandwich enzyme-linked immunosorbent assay was developed using wells coated with buffer only, a control peptide, or CHP (capture “antibody”) and biotin conjugated goat anti-mouse immunoglobulin G (detection antibody) with avidin–horseradish peroxidase as the signal. There were no significant differences in the levels of immunoglobulin G retained among serum samples from EMT6 tumor-bearing mice treated with control, wtIL-12 or CHP-IL-12 plasmid DNAs showing that CHP-IL-12 is not immunogenic (Supplementary Figure S3, Supplementary Materials and Methods).
CHP homes to vimentin expressed in the tumor environment
CHP-biotin was used to isolate the CHP receptor from a pool of cell-surface receptor proteins isolated from SCCVII cells. Mass spectrometry of the isolated protein (Figure 5a, white arrow) identified this protein as vimentin. To validate this receptor, CHP-biotin was added to wells of a polystyrene plate that were coated with coating buffer only, GST, or recombinant vimentin-GST. Indeed, CHP interacts with vimentin as the vimentin-coated wells retained a significantly higher level of horseradish peroxidase activity (Figure 5b). To confirm that the interaction of CHP and vimentin did not inhibit the IL-12 activity, CHP-IL-12 and wtIL-12 were transferred to separate wells coated with either vimentin or bovine serum albumin (1:1 molar ratio of IL-12 and vimentin), and then an hour later murine splenocytes were added to the wells. Interestingly, the CHP-IL-12 bound to vimentin-coated wells induced a small but significantly higher level of IFN-γ than both CHP-IL-12 and wtIL-12 in the bovine serum albumin–coated wells (n = 3, P < 0.05, Supplementary Figure S4, Supplementary Materials and Methods).
Figure 5.
Identification of carcinoma homing peptide (CHP) interacting protein: vimentin. (a) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of potential receptors for CHP isolated via affinity chromatography of a pool of cell-surface proteins isolated from SCCVII cells. The only distinct band (white arrow) was located in the second fraction, and mass spectrometry identified this band as vimentin. (b) Interaction of CHP-biotin with recombinant vimentin-GST (Vimentin), GST, and coating buffer only (control) coated wells of a polystyrene plate (n = 6; *P < 0.001 compared to both GST and control, errors bars represent SEM). (c) Western blot analysis of vimentin expression in an (1) SCCVII tumor and (2) heart, (3) lung, (4) liver, (5) kidney, (6) spleen, and (7) serum from SCCVII-tumor bearing C3H mice. (d) Western blot analysis of vimentin expression in in vitro and ex vivo tumor samples. (e) Accumulation of peptide-biotin in syngeneic SCCVII tumor-bearing C3H mice following intravenous (i.v.) injection of either control-biotin (top left and right) or CHP-biotin (bottom left and right) without (top and bottom left) or with (top and bottom right) depletion of vimentin with a coinjection of purified polyclonal goat antivimentin (100 µg) in the same i.v. injection as the peptide-biotin. BSA, bovine serum albumin; GAPDH (glyceraldehyde 3-phosphate dehydrogenase).
To determine the levels of vimentin expression in normal tissues versus tumors, tissue lysates from an SCCVII tumor, heart, lung, liver, kidney, spleen, and serum from a C3H mouse were probed for vimentin expression via western blot analysis. Very low levels of vimentin were detected in the heart, liver, kidney, spleen, and serum (Figure 5c, lanes 2 and 4 through 7) while high levels of vimentin were detected in the tumor and lung (Figure 5c, lanes 1 and 3). Similarly, analysis of vimentin expression in SCCVII, CT26, 4T1, and B16F10 (melanoma cell line derived from C57Bl/6 mice) tumor cell lines and ex vivo tumor tissues shows that all these tumor cell lines and their respective tumor models express vimentin (Figure 5d), which explains universal tumor-homing property as illustrated in Figure 1.
To determine that vimentin is the receptor protein interacting with CHP which is responsible for tumor homing, we performed tail vein injections of control-biotin and CHP-biotin with or without blocking vimentin using purified polyclonal goat antivimentin in SCCVII tumor-bearing C3H mice. As expected, injection of control-biotin with or without antivimentin did not result in any accumulation of peptide in the tumor (Figure 5e, top left and top right, 1.8 ± 0.37 with antivimentin and 2.6 ± 0.51 without antivimentin, n = 5). However, CHP-biotin did accrue in the tumor environment (Figure 5e, bottom left, 19.6 ± 1.3 positive per field, n = 5, P < 0.0001 compared to all other groups), but coadministration with antivimentin almost completely inhibited the tumor-targeting ability of CHP (Figure 5e, bottom right, 1.2 ± 0.58 positive per field, n = 5).
CHP-IL-12 reduces the level of toxic lesions in the liver
IL-12 induces liver toxicity (data not shown) and it was our expectation that the tumor-homing CHP-IL-12 may reduce the toxicity. To test this hypothesis, SCCVII tumor-bearing C3H mice were treated with two treatments of 1 µg (2 × 1 µg), 2 µg (2 × 2 µg), or 10 µg (2 × 10 µg) or three treatments of 2 µg (3 × 2 µg) of wtIL-12 or CHP-IL-12, and mice were killed on days 1, 3, and 30 after the final treatment. At low levels of plasmid DNA administration, 2 × 1 µg and 2 × 2 µg, there were no differences between wtIL-12 and CHP-IL-12 treatments; however, at the therapeutic level, 2 × 10 µg, and the triple treatment, 3 × 2 µg, CHP-IL-12 treatments caused toxic lesions in only one mouse while wtIL-12 treatments had significantly higher numbers (Figure 6a). Serum chemistry profiles of these mice revealed that there were no differences between any treatment, regimen, or time points, and all levels for the hallmarks of toxicity, such as alanine transaminase, were within the normal range (Figure 6d). So, CHP-IL-12 reduces the level of toxicity in the liver and does not cause any other detectable systemic toxicities.
Figure 6.
Decreased liver toxicity of interleukin-12 (IL-12) treatments with carcinoma homing peptide (CHP)-IL-12. (a) Number of SCCVII tumor-bearing C3H mice with toxic lesions on the liver following two treatments of 1 µg (2 × 1 µg), 2 µg (2 × 2 µg), or 10 µg (2 × 10 µg) or three treatments of 2 µg (3 × 2 µg) of wild-type IL-12 (wtIL-12) or CHP-IL-12 (n = 12). (b) Representative image of a normal liver area. (c) Representative image of a toxic lesion. Bar = 50 µm in (b) and (c). (d) Levels of alanine transaminase (ALT), a key indicator of liver function, for both plasmid DNA treatments at all DNA levels and time points.
Discussion
Tumor targeting can be achieved via the screening of various libraries to select tumor-targeted peptides, DNA/RNA aptamers, antibodies, etc; however, the only mechanism that can be used for homing gene products from systemically injected genes will be tumor-targeted mini-peptide encoding DNA. The small size of these peptides eliminates the concern of immunogenicity, as seen in a CHP-specific antibody assay (Supplementary Figure S3) and reduces the effect on the biological function of the gene product, though some minipeptides may boost or inhibit gene function.20 The tiny peptide-encoding DNA sequences can be easily fused with any therapeutic gene. Finally, these peptides can complement existing tumor-targeting approaches such as transcriptional targeting,8 translational targeting,21 and targeted delivery.3,4,5,6
In this study, we have identified a 7 amino acid targeted peptide, CHP. In this gene therapy method, CHP was capable of increasing the T/S SEAP levels much more than the known cyclic tumor-homing peptides such as CNGRC and RGD4C (Figure 1), which rely on disulfide bonds to maintain the structure of the targeting peptides.22,23 This linear structure may decrease the effect of the peptide on the activity of the fusion protein and vice versa. These differences in tumor-targeting show that not all tumor-targeting peptides will work in this novel system.
Other tumor-targeting peptides can deliver small molecules with only one copy for each small-molecule payload but require multiple copies of the peptide to target larger molecules such as a full length cytokine.24 In this study, we demonstrate that fusion of a single copy of CHP encoding DNA can boost the accumulation of IL-12 in tumors, suggesting one copy of CHP is sufficient to carry one copy of IL-12 to the tumor site. Of course, increasing the number of CHP peptides per molecule may also increase the targeting efficiency.
Currently, most tumor-targeting strategies are based on extremely specific interactions, and the ability to target the tumor environment is constrained to a single cell type or specific type of tumor. We have shown that CHP can increase the efficacy of IL-12 gene therapy to inhibit tumor growth in the three tumor cell lines, breast adenocarcinoma, squamous cell carcinoma, and colon carcinoma, and in two different mouse strains (Figure 3). Subsequently, CHP-IL-12 extends survival more than wtIL-12 treatments in both the breast adenocarcinoma and squamous cell carcinoma (Figure 3c,e). Similarly, CHP-IL-12 treatments inhibit the development of spontaneous lung metastasis, which is the primary killer of cancer patients (Figure 3b). This increase in antitumor response is associated with increases in both tumor-specific CTL activity (Figure 4) and IL-12 accumulation in tumors (Figure 2d). This result is in agreement with the result that intratumoral delivery of IL-12 yields better antitumor efficacy than systemic delivery.7 The discovery of CHP permits us to use systemic delivery to target IL-12 to tumors without the need of intratumoral delivery, which is not realistic for treating internal tumors, metastatic tumors, and residual tumor cells after standard therapy. In this regard, this peptide will be extremely valuable, and further study is warranted.
We were able to identify vimentin as a receptor for CHP (Figure 5). Vimentin is an intermediate filament protein conventionally regarded as an intracellular structural protein in cells of mesenchymal origin such as fibroblasts, chondrocytes, and macrophages.15 Vimentin expression has been reported to be increased in several tumor models, including human prostate, colon,17 hepatocellular,16 and gemcitabine-resistant pancreatic cancers,19 and the tumor stromal cells in human colorectal tumors.18 The upregulation of vimentin is associated with the epithelial-to-mesenchymal transition, which is important for motility as well as metastasis in several tumors.
Though vimentin is upregulated in tumors, two remaining questions involve determining how the CHP targets an intracellular receptor and how the peptide avoids targeting lungs where vimentin is also expressed. These questions were perfectly addressed recently by the discovery that vimentin is expressed on the cell surface of tumor cells25 and epithelial cells during angiogenesis.26 Additionally, some human tumor-initiating cells remaining after treatment overexpress vimentin on the tumor cell surface.27 Another important aspect of vimentin is the conserved sequences among mouse, rat, dog, and humans.28 This information along with the T/S SEAP accumulation in the xenogeneic human tumor model (Figure 1b) strongly suggests that CHP targeting will crossover to human treatments.
We also confirmed that vimentin is expressed at very low levels in the heart, liver, kidney, spleen, and serum of C3H mice, yet it is highly expressed in lung tissue. However, since most general expression of vimentin is intracellular,15,29,30 this expression should not be a target of CHP. As seen in Supplementary Figure S1, there was no accumulation of CHP-biotin in the lung sections which supports this notion. Conversely, vimentin is highly expressed in SCCVII tumors in C3H mice, and CHP-biotin did accumulate in the SCCVII tumors (Figure 2). Likewise, the tumor cells and corresponding syngeneic tumors both express detectable levels of vimentin (Figure 5). The differences seen between expression in tumor cell lines and the respective tumor tissues is due to the heterogeneous nature and multiple cell types in the tumor microenvironment.
A potential problem with using CHP for targeting immunomodulatory agents is the cellular fate of the surface-bound peptide and its payload. Internalization of peptide-conjugated proteins has been shown for several tumor-targeted peptides, including RGD peptides.23,31 Additionally, RGD4C-IL-12 and RGD-TNF recombinant proteins have been shown to elicit antitumor immune responses.23 After being synthesized by and excreted from the muscle tissue, the CHP-IL-12 circulates in the blood stream, where it may be activating natural killer and T cells or perhaps even guiding these cells to the tumor tissue. Additionally, Ise et al. have recently described the internalization of vimentin-bound ligands.32 According to their studies, the vimentin-bound ligands are still located on the cell surface after 2 hours. The half-life of circulating IL-12 is between 2.5 and 3.3 hours;33 therefore internalization would not affect the IL-12 activity. Further, if CHP is eventually internalized by the tumor cells, then CHP would also be a potent targeting modality for using intracellular antitumor agents, which is an angle that requires further investigation.
We have developed a fully functional tumor-targeting IL-12 gene construct that can be delivered systemically for treating distally located neoplastic diseases. Inserting peptide-encoding sequences directly prior to the stop codon in the p40 gene of an IL-12 plasmid did not interfere with transcription, translation, post-translational modifications, or therapeutic functionality of the IL-12 gene product (Figure 2). Also, CHP maintains its tumor-targeting ability as seen in IL-12−/− mice (Figure 2) and can increase the therapeutic efficacy of systemic IL-12 gene therapy treatments (Figure 3) while decreasing liver toxicity (Figure 6). Future studies will include investigating the fate of vimentin-bound CHP, methods to increase the targeting efficiency, the potential for CHP to target other modalities, and the efficacy of other CHP-antitumor fusion gene constructs.
Materials and Methods
Plasmid DNA preparation. All SEAP gene constructs were generated via direct PCR as previously described.13 The wild-type IL-12 gene construct (wtIL-12) was obtained from Valentis (San Francisco, CA),34 and gene sequences encoding the peptide sequences were inserted directly prior to the stop codon of the IL-12 p40 subunit encoding region using the primer sequences listed in Supplementary Table S1. The IL-12 plasmid includes both the p35 and p40 subunits. The control plasmid DNA (control) consisted of a deletion of the IL-12 gene from the IL-12 construct. All plasmid DNAs were manufactured with the Qiagen (Alameda, CA) EndoFree plasmid preparation kit.
Cell lines, in vitro gene transfer, and IFN-γ induction. CT26, SCCVII, 4T1, EMT6, and B16F10 cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA), the AT84 cell line was a generous gift from Dr Edward Shillitoe (State University of New York Upstate Medical School, Syracuse, NY), and MCF7 cells were provided by Dr Bolin Liu (University of Colorado Denver School of Medicine, Aurora, CO). All cell lines were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (Life Technologies, Carlsbad, CA) at 37 °C and 5% CO2.
For in vitro transfections, 4T1 cells were suspended at a concentration of 1 × 107 cells/ml Opti-mem medium (Life Technologies), and 100 µl of this suspension were transferred to individual electroporation cuvettes and 2 µg of control, wtIL-12, CDGRC-IL-12, or CHP-IL-12 plasmid DNA was added (n = 3). Each cuvette was pulsed with one 75-ms pulse of 150 V, and the suspensions were transferred to individual wells of a 6-well plate containing 900 µl Dulbecco's modified Eagle's medium. The next day, 900 µl of medium was collected, placed on ice, and analyzed for the presence of IL-12p70 using an IL-12p70 ELISA (eBiosciences, San Diego, CA) as per the manufacturer's instructions. The spleen from a naive Balb/c mouse was placed in serum-free RPMI-1640 containing Pen/Strep/Glu (RPMI), splenocytes were filtered through a 70 µm cell strainer, and suspended in 10 ml RPMI. After the cell suspension was centrifuged for 10 minutes at 1,000 rpm, the supernatant was removed, cells resuspended in 10 ml red blood cell lysis solution, centrifuged again, and then resuspended in RPMI at a concentration of 2 × 106 cells per 100 µl. 2 × 106 cells were placed into wells of a 6-well plate. Condition medium from the plasmid DNA-transfected cells containing 150 pg/ml IL-12 was transferred to these wells and the volume was adjusted to 1 ml with Dulbecco's modified Eagle's medium. The next day, the mediums were collected and assayed for the presence of IFN-γ using an IFN-γ ELISA (eBiosciences) as per manufacturer's instructions.
Animal models, tumor inoculations, in vivo gene transfer, protein extraction, and therapeutic analyses. All animals used in this study were maintained under and animal protocols were performed following National Institutes of Health guidelines, approved by the Institutional Animal Care and Use Committee of Louisiana State University (Baton Rouge, LA). Balb/c mice were obtained from the in-house breeding colony, and C3H, Nude, and wtIL-12−/− mice were obtained from Charles River Laboratories (Wilmington, MA). All mice were six- to eight-weeks-old upon initiation of experiments. Tumor models were initiated via subcutaneous inoculations of 30 µl cell suspension containing 1 × 105 4T1 cells or 2 × 105 cells for all other cell lines in 1 × phosphate-buffered saline (PBS). Orthotopic EMT6 tumors were initiated by inoculating 1 × 105 cells in the mammary fat pads of female Balb/c mice.
For in vivo i.m. gene transfections, plasmid DNA was diluted in 0.45% NaCl to a concentration of 5 µg/30 µl was injected into each rear tibialis muscle, and the muscles were immediately subjected to electroporation as previously described.20 When 4T1 tumors were 3–4 mm in diameter or all other tumor models were 4.0–4.5 mm in diameter, the first treatment was performed, and a second identical treatment was performed 10 days later. Tumor volumes were determined as previously described.13 To determine the distribution of the fusion gene products peptide-SEAP and peptide-IL-12, the treatments were performed when tumors reached 6–7 mm in diameter; 72 hours after treatment, mice were killed via CO2 asphyxiation, and then tissues were collected, wrapped in foil, and flash-frozen in liquid nitrogen. To extract proteins, the frozen tissues were smashed with a hammer, placed in 1 × lysis buffer (Promega, Madison, WI), beaten for 1 minute with a mini-beadbeater 8 (Biospec, Bartlesville, OK), and spun at 16,000g for 5 minutes. The supernatant was transferred to a new tube. Serum was collected by extracting blood from the left ventricle, transferring it to Serum Separator Tubes (BD, Franklin Lakes, NJ), and spinning at 5,000g for 5 minutes. The serum was then transferred to a new 1.5 ml tube.
India ink inflation was performed to determine the level of lung metastasis. After CO2 asphyxiation, the thoracic cavity was opened, the trachea exposed, and the trachea clipped with a hemostat. 1.5 ml 15% India ink was injected into the lung which was then transferred into 20 ml Fekete's solution and incubated overnight. The next day, white metastatic nodules were counted using a dissecting microscope.
For fluorescence-activated cell sorting analyses, tumor-infiltrating lymphocytes were isolated by extracting the tumors, cutting them into pieces, and resuspending the mixture in sterile PBS (without Ca2+ and Mg2+) containing a mixture of collagenase IV, hyaluronidase V (Sigma-Aldrich, St Louis, MO), and DNase II (Fisher, Pittsburgh, PA). The tissue suspension was placed in a shaker at 37 °C for 1–2 hours, and then poured through a 70 µm cell strainer, followed by washing twice in PBS with Ca2+ and Mg2+. The isolated cells were stained with the fluorescein isothiocyanate conjugated anti-CD11c (AbD Serotec, Raleigh, NC) and goat anti-mouse CD80 (R&D, Minneapolis, MN) for 30 minutes at 4 °C, washed with PBS, and then stained with phycoerythrin conjugated anti-goat immunoglobulin G (Cedarlane Laboratories, Burlington, NC). The expression of the proteins was analyzed on FACSCalibur (BD Biosciences, San Jose, CA) and analyzed with FCS Express 3 (De Novo Software, Los Angeles, CA). Splenocytes were also isolated from Balb/c mice bearing orthotopic EMT6 tumors, and a CTL assay was performed as described previously.13 Serum was collected from 4T1-tumor bearing mice 3 days after treatments with control, wtIL-12, and CHP-IL-12 plasmid DNA as described above. The serum was analyzed for the presence of IFN-γ as described above.
Peptide-biotin distribution, vimentin depletion, and tissue staining. CHP-biotin was synthesized by United Biochemical Research (Seattle, WA) at >95% purity, resuspended in H2O with 5% glycerol, and stored at −80 °C. The peptide sequence is NH2-VNTANSTGG-biotin. Control-biotin was created by conjugating a nonspecific peptide (CTSTSPLPPPSHSTSKKG, Alpha Diagnostics, San Antonio, TX) to EZ-Link Amine-PEG2-Biotin via 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide cross-linking (Pierce, Rockford, IL) following the manufacturer's instructions. The peptide-biotin conjugates (10 µg/100 µl normal saline) were injected into the tail vein of C3H mice bearing SCCVII tumors with 6–7 mm diameters. For vimentin depletion studies, goat polyclonal antivimentin (Millipore, Billerica, MA) was purified via protein-G antibody purification (Pierce), and 150 µg was added to the peptide-biotin conjugate solutions for administration. One hour after intravenous administration, mice were perfused via injection of 10 ml 1 × PBS into the left ventricle after cutting the right atrium. The tissues were immediately removed, wrapped in foil, and flash frozen in liquid nitrogen. Four- to five-micron sections were placed on poly-L-lysine coated slides.
The sections were fixed in ice-cold acetone, nonspecific interactions were blocked with 1% bovine serum albumin in PBS, and endogenous peroxidase activity was suppressed with stable peroxidase suppressor (Pierce). Since the peptides already contained biotin, the peptide were incubated with the avidin-HRP reagent Vectastain ABC (Vector Biolabs, Philadelphia, PA) for 45 minutes, washed in PBS, and then incubated with 1 × Metal Enhanced DAB (Pierce). The sections were then counterstained with Meyer's hematoxylin (blue) for 2 minutes or eosin (pink) for 20 seconds.
Isolation of cell-surface proteins, identification of CHP receptor, and western blot. Cell-surface proteins were isolated from SCCVII cells by using the Cell Surface Protein Isolation Kit (Pierce) and following the manufacturer's instructions. A streptavidin agarose column (Peirce) was loaded with CHP-biotin (3 mg/ml in PBS for 10 minutes), and the cell-surface protein suspension was incubated on the column overnight at room temperature. Next, five fractions were eluted with 8 mol/l Guanidine-HCl, pH 1.5, and then 1 mol/l Na2HPO4 was added to the fractions at a 1:10 ratio. From these fractions, volumes containing 40 µg of protein were mixed with 2 × sodium dodecyl sulfate loading buffer and added to wells of a 12% polyacrylamide gel and an electric field was applied. The gel was then incubated with Coomassie Brilliant Blue R250 followed by destaining solution (10% acetic acid and 20% MeOH). Images were captured with a VersDoc Model 1000 and Quantity One Version 4.4.1 software (Bio-Rad, Hercules, CA).
To identify the protein from fraction two, the protein in the gel was extracted using the Trypsin Profile IGD Kit (Sigma, St Louis, MO) with the ProteoPrep Reduction and Alkylation Kit (Sigma) following the manufacturer's instructions. Liquid chromatography electrospray tandem mass spectrometry was used to analyze peptide mixture extracted from gel spots. Tryptic digests of gel spots (~6 µl) were diluted with 0.1% formic acid (10 µl) and 10 µl injected by microplate autosampler (Famos; Dionex Corporation, Sunnyvale, CA) onto a 0.3 × 1 mm trapping column (PepMap C18; Dionex Corporation) using a nano LC system equipped with Switchos and Ultimate 2000 pumps (Dionex Corporation), at a flow rate of 10 µl/minute. The switchos valve was set on loading position prior to sample loading. After sample loading, the trapping column was washed with 0.1% formic acid at flow rate of 5 µl/minute for additional 5 minutes and then switchos valve was switched to inject position. Peptides were then eluted at 200 nl/minute and chromatographed on a 75 µm × 15 cm Biobasic C18 column (Vydac HPLC Columns, Grace Davison, IL), with a gradient of 5–40% acetonitrile over 60 minutes followed by 80% acetonitrile for 5 minutes. The eluent was directed into a quadrupole time-of-flight mass spectrometer (Q-Star; Applied Biosystems MDS Sciex, Foster City, CA) and ionized immediately using electrospray source (Nano spray II; Applied Biosystems MDS Sciex) at high voltage of 2.5 kv with nebulizer gas at level 2. The mass spectrometer was operated in information dependent acquisition mode with the three most intense ions in each survey scan subjected to tandem mass spectrometry analysis using collision energies ranging from 20 eV to 50 eV. Tandem mass spectrometry data obtained from Q-Star was processed for database search using Mascot search engine (Matrix science, London, UK). A Mascot search was performed using the following parameters: type of search, tandem mass spectrometry ion search; database, nrNCBI; taxonomy, all; enzyme, trypsin; fixed modification, carbamidomethyl (C); mass values, monoisotopic; protein mass, unrestricted; peptide mass tolerance, +0.2 Da; fragment mass tolerance, +0.2 Da; and maximum miss cleavage, 1.
A cell-free assay was developed to confirm that vimentin interacts with CHP. Wells of a microtiter plate were coated with 50 µl of 100 mmol/l NaHCO3 (coating buffer) or 5 µg/ml of either vimentin-GST or GST (ProSpec, East Brunswick, NJ) in coating buffer and incubated at 4 °C overnight. After two washes with PBS, nonspecific binding was blocked by incubating the wells with 100 µl 1 × bovine serum albumin for 2 hours at room temperature. After another wash (twice), 100 µl of PBS containing 10 ng CHP-biotin was added to each well (n = 6 for each coat), incubated for 2 hours at room temperature, and then washed four times with PBS. Avidin-HRP (100 µl, eBiosciences) was added to each well, incubated for 30 minutes, and the wells were washed seven times with PBS. Lastly, 100 µl TMB substrate (eBiosciences) was added to each well for 15 minutes followed by 50 µl Stop solution (eBiosciences), and the absorbance at 450 nm was read using a SpectraCount and PlateReader Version 3.0 software (PerkinElmer, Waltham, MA).
To prepare cells for western blot analysis of cellular expression of vimentin, when SCCVII, CT26, 4T1, and B16F10 cells were 95% confluent in individual wells of 6-well plates, the cells were directly lysed with 60 µl Laemmli sample buffer. For preparation of ex vivo samples, tissues and tumors were processed as described above. Volumes of the tissue lysates containing 40 µg of protein were mixed with 2 × sodium dodecyl sulfate loading buffer. Twenty microliter volumes of the cell lysates or tissue lysates were added to a 12% polyacrylamide gel and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a TransBlot Transfer Medium nitrocellulose membrane (Bio-Rad Laboratories). Immunoblotting of the membrane was performed with a 1:100 dilution of polyclonal Goat antivimentin (Millipore, Billerica, MA) and a 1:5,000 dilution of the secondary horseradish peroxidase conjugated rabbit anti-goat immunoglobulin G. The peroxidase signal was generated with the Western Lightning ECL (PerkinElmer) and visualized with a Kodak Image Station 440CF using the 1D Image Analysis Software v3.6 (PerkinElmer).
Analysis of toxicity induced by gene therapy treatments. SCCVII tumors were induced in C3H mice as described above, and allowed to grow to a volume of 300 mm3. Groups of four mice for each treatment at each time point were treated with either wild-type IL-12 or CHP-IL-12 as described above at a dose of 1 µg, 2 µg, and 10 µg plasmid DNA; a fourth set of mice received three treatments of 2 µg. Mice were killed on days 1, 3, and 30 after the second treatment, blood was collected in serum separator tubes, and livers were fixed in 10% neutral-buffered formalin.
Serum chemistry profiles were analyzed by a private GLP-certified diagnostic laboratory (Antech Diagnostics, Memphis, TN). Formalin-fixed tissue was cut-in, embedded in paraffin, and sectioned into 5 µm sections. Sections were mounted on glass slides and stained with hematoxylin and eosin prior to microscopic examination by a pathologist. A liver toxicity scoring system based on the number of characteristic liver lesions (foci of hepatocellular necrosis with Kupffer cell hyperplasia) per ×200 field was used. The sections were scored blindly and recorded for analysis.
Statistical analyses. All statistical analyses were performed with GraphPad Prism version 5.00 for Windows, (GraphPad Software, San Diego, CA). One-way analysis of variance with Bonferroni's post hoc test was used to analyze the following data: ratios of tissue/serum (T/S) SEAP levels, production of fusion gene products from in vitro transfected 4T1 cells, inhibition of metastasis, IFN-γ serum levels, and CHP/vimentin interaction. Tumor versus normal tissue distributions of exogenous IL-12 or CHP-IL-12 gene products in IL-12−/− mice and CTL data were analyzed via one-tailed unpaired T tests. All tumor growth experiments were analyzed via two-way analysis of variance plus Bonferroni's post hoc test. Mantel–Cox tests were used to analyze differences in survival of mice. Liver toxicity was first analyzed using blind pathological scores of the liver tissues, but no differences were seen among time points, so the data was pooled to create a larger sample size and then analyzed with one-sided Fisher's exact tests comparing the number of mice having lesions from CHP-IL-12 and wtIL-12 fusion plasmid DNA treated mice.
SUPPLEMENTARY MATERIAL Figure S1. Uncorrected peptide-SEAP tumor and serum data. Figure S2. Lack of CHP homing to toxicity-sensitive organs. Figure S3. Lack of immunogenicity against VNTANST. Figure S4. Activity of CHP-SEAP when bound to vimentin. Table S1. Primers for PCR. Materials and Methods.
Acknowledgments
This work was supported by National Institutes of Health (NIH) RO1CA120895. This work was performed in Baton Rouge, LA and Houston, TX. The authors declared no conflict of interests.
Supplementary Material
Uncorrected peptide-SEAP tumor and serum data.
Lack of CHP homing to toxicity-sensitive organs.
Lack of immunogenicity against VNTANST.
Activity of CHP-SEAP when bound to vimentin.
Primers for PCR.
REFERENCES
- Kobayashi M, Fitz L, Ryan M, Hewick RM, Clark SC, Chan S.et al. (1989Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes J Exp Med 170827–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Vecchio M, Bajetta E, Canova S, Lotze MT, Wesa A, Parmiani G.et al. (2007Interleukin-12: biological properties and clinical application Clin Cancer Res 134677–4685. [DOI] [PubMed] [Google Scholar]
- Halin C, Rondini S, Nilsson F, Berndt A, Kosmehl H, Zardi L.et al. (2002Enhancement of the antitumor activity of interleukin-12 by targeted delivery to neovasculature Nat Biotechnol 20264–269. [DOI] [PubMed] [Google Scholar]
- Dela Cruz JS, Trinh KR, Morrison SL., and, Penichet ML. Recombinant anti-human HER2/neu IgG3-(GM-CSF) fusion protein retains antigen specificity and cytokine function and demonstrates antitumor activity. J Immunol. 2000;165:5112–5121. doi: 10.4049/jimmunol.165.9.5112. [DOI] [PubMed] [Google Scholar]
- Dickerson EB, Akhtar N, Steinberg H, Wang ZY, Lindstrom MJ, Padilla ML.et al. (2004Enhancement of the antiangiogenic activity of interleukin-12 by peptide targeted delivery of the cytokine to alphavbeta3 integrin Mol Cancer Res 2663–673. [PubMed] [Google Scholar]
- Colombo G, Curnis F, De Mori GM, Gasparri A, Longoni C, Sacchi A.et al. (2002Structure-activity relationships of linear and cyclic peptides containing the NGR tumor-homing motif J Biol Chem 27747891–47897. [DOI] [PubMed] [Google Scholar]
- Li S, Zhang L, Torrero M, Cannon M., and, Barret R. Administration route- and immune cell activation-dependent tumor eradication by IL12 electrotransfer. Mol Ther. 2005;12:942–949. doi: 10.1016/j.ymthe.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Zhang R, Straus FH, Messina M, Robinson BG, Hashizume K.et al. (2002Effective gene therapy for medullary thyroid carcinoma using recombinant adenovirus inducing tumor-specific expression of interleukin-12 Gene Ther 964–74. [DOI] [PubMed] [Google Scholar]
- Okada Y, Okada N, Mizuguchi H, Takahashi K, Hayakawa T, Mayumi T.et al. (2004Optimization of antitumor efficacy and safety of in vivo cytokine gene therapy using RGD fiber-mutant adenovirus vector for preexisting murine melanoma Biochim Biophys Acta 1670172–180. [DOI] [PubMed] [Google Scholar]
- Gao JQ, Eto Y, Yoshioka Y, Sekiguchi F, Kurachi S, Morishige T.et al. (2007Effective tumor targeted gene transfer using PEGylated adenovirus vector via systemic administration J Control Release 122102–110. [DOI] [PubMed] [Google Scholar]
- Wang H, Chen K, Cai W, Li Z, He L, Kashefi A.et al. (2008Integrin-targeted imaging and therapy with RGD4C-TNF fusion protein Mol Cancer Ther 71044–1053. [DOI] [PubMed] [Google Scholar]
- Maeda H, Fang J, Inutsuka T., and, Kitamoto Y. Vascular permeability enhancement in solid tumor: various factors, mechanisms involved and its implications. Int Immunopharmacol. 2003;3:319–328. doi: 10.1016/S1567-5769(02)00271-0. [DOI] [PubMed] [Google Scholar]
- Craig R, Cutrera J, Zhu S, Xia X, Lee YH., and, Li S. Administering plasmid DNA encoding tumor vessel-anchored IFN-alpha for localizing gene product within or into tumors. Mol Ther. 2008;16:901–906. doi: 10.1038/mt.2008.40. [DOI] [PubMed] [Google Scholar]
- Work LM, Büning H, Hunt E, Nicklin SA, Denby L, Britton N.et al. (2006Vascular bed-targeted in vivo gene delivery using tropism-modified adeno-associated viruses Mol Ther 13683–693. [DOI] [PubMed] [Google Scholar]
- Dandachi N, Hauser-Kronberger C, Moré E, Wiesener B, Hacker GW, Dietze O.et al. (2001Co-expression of tenascin-C and vimentin in human breast cancer cells indicates phenotypic transdifferentiation during tumour progression: correlation with histopathological parameters, hormone receptors, and oncoproteins J Pathol 193181–189. [DOI] [PubMed] [Google Scholar]
- Matos JM, Witzmann FA, Cummings OW., and, Schmidt CM. A pilot study of proteomic profiles of human hepatocellular carcinoma in the United States. J Surg Res. 2009;155:237–243. doi: 10.1016/j.jss.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisan E., and, Girard D. Cell surface expression of intermediate filament proteins vimentin and lamin B1 in human neutrophil spontaneous apoptosis. J Leukoc Biol. 2006;79:489–498. doi: 10.1189/jlb.0405190. [DOI] [PubMed] [Google Scholar]
- Ngan CY, Yamamoto H, Seshimo I, Tsujino T, Man-i M, Ikeda JI.et al. (2007Quantitative evaluation of vimentin expression in tumour stroma of colorectal cancer Br J Cancer 96986–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS.et al. (2009Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway Cancer Res 692400–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutrera J, Dibra D, Xia X., and, Li S. Enhancement of reporter gene detection sensitivity by insertion of specific mini-peptide-coding sequences. Cancer Gene Ther. 2010;17:131–140. doi: 10.1038/cgt.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoff-Khalili MA, Rivera AA, Nedeljkovic-Kurepa A, DeBenedetti A, Li XL, Odaka Y.et al. (2008Cancer-specific targeting of a conditionally replicative adenovirus using mRNA translational control Breast Cancer Res Treat 10843–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti A, Curnis F, Arap W., and, Pasqualini R. The neovasculature homing motif NGR: more than meets the eye. Blood. 2008;112:2628–2635. doi: 10.1182/blood-2008-04-150862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temming K, Schiffelers RM, Molema G., and, Kok RJ. RGD-based strategies for selective delivery of therapeutics and imaging agents to the tumour vasculature. Drug Resist Updat. 2005;8:381–402. doi: 10.1016/j.drup.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Garanger E, Boturyn D, Jin Z, Dumy P, Favrot MC., and, Coll JL. New multifunctional molecular conjugate vector for targeting, imaging, and therapy of tumors. Mol Ther. 2005;12:1168–1175. doi: 10.1016/j.ymthe.2005.06.095. [DOI] [PubMed] [Google Scholar]
- Huet D, Bagot M, Loyaux D, Capdevielle J, Conraux L, Ferrara P.et al. (2006SC5 mAb represents a unique tool for the detection of extracellular vimentin as a specific marker of Sezary cells J Immunol 176652–659. [DOI] [PubMed] [Google Scholar]
- Bhattacharya R, Gonzalez AM, Debiase PJ, Trejo HE, Goldman RD, Flitney FW.et al. (2009Recruitment of vimentin to the cell surface by beta3 integrin and plectin mediates adhesion strength J Cell Sci 122Pt 91390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A.et al. (2009Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features Proc Natl Acad Sci USA 10613820–13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Gilles C, Polette M, Zahm JM, Tournier JM, Volders L, Foidart JM.et al. (1999Vimentin contributes to human mammary epithelial cell migration J Cell Sci 112 (Pt 24)4615–4625. [DOI] [PubMed] [Google Scholar]
- Nieminen M, Henttinen T, Merinen M, Marttila-Ichihara F, Eriksson JE., and, Jalkanen S. Vimentin function in lymphocyte adhesion and transcellular migration. Nat Cell Biol. 2006;8:156–162. doi: 10.1038/ncb1355. [DOI] [PubMed] [Google Scholar]
- Sancey L, Lucie S, Garanger E, Elisabeth G, Foillard S, Stéphanie F.et al. (2009Clustering and internalization of integrin alphavbeta3 with a tetrameric RGD-synthetic peptide Mol Ther 17837–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafner V, Trachsel E., and, Neri D. An engineered antibody-interleukin-12 fusion protein with enhanced tumor vascular targeting properties. Int J Cancer. 2006;119:2205–2212. doi: 10.1002/ijc.22101. [DOI] [PubMed] [Google Scholar]
- Gafner S, Dietz BM, McPhail KL, Scott IM, Glinski JA, Russell FE.et al. (2006Alkaloids from Eschscholzia californica and their capacity to inhibit binding of [3H]8-Hydroxy-2-(di-N-propylamino)tetralin to 5-HT1A receptors in Vitro J Nat Prod 69432–435. [DOI] [PubMed] [Google Scholar]
- Zhu S, Lee DA., and, Li S. IL-12 and IL-27 sequential gene therapy via intramuscular electroporation delivery for eliminating distal aggressive tumors. J Immunol. 2010;184:2348–2354. doi: 10.4049/jimmunol.0902371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Uncorrected peptide-SEAP tumor and serum data.
Lack of CHP homing to toxicity-sensitive organs.
Lack of immunogenicity against VNTANST.
Activity of CHP-SEAP when bound to vimentin.
Primers for PCR.