Abstract
Mitochondrial dysfunction is associated with a variety of human diseases including inherited mitochondrial diseases, neurodegenerative disorders, diabetes mellitus, and cancer. Effective medical therapies for mitochondrial diseases will ultimately require an optimal drug delivery system, which will likely be achieved through innovations in the nanotechnology of intracellular trafficking. To achieve efficient mitochondrial drug delivery, two independent processes, i.e., “cytoplasmic delivery through the cell membrane” and “mitochondrial delivery through the mitochondrial membrane” are required. In previous studies, we developed an octaarginine (R8) modified nano carrier for efficient cytoplasmic delivery, showing that R8-modified liposomes were internalized into cells efficiently. On the other hand, we also constructed MITO-Porter for the mitochondrial delivery of macromolecules, a liposome-based carrier that delivers cargos to mitochondria via membrane fusion. Here, we report the development of a dual function MITO-Porter (DF-MITO-Porter), based on the concept of integrating both R8-modified liposomes and MITO-Porter. We show that the DF-MITO-Porter effectively delivers exogenous macro-biomolecules into the mitochondrial matrix, and provide a demonstration of its potential use in therapies aimed at mitochondrial DNA.
Introduction
Mitochondrial dysfunction is implicated in a variety of human diseases including mitochondrial inherited diseases, neurodegenerative disorders, diabetes mellitus, and cancer.1,2,3,4,5,6,7,8,9 Effective medical therapies for such diseases will ultimately require an optimal drug delivery system, which will likely be achieved through innovations in the nanotechnology of intracellular trafficking. In addition, the use of therapeutic agents will be needed to overcome the impregnable mitochondrial membrane to exert pharmacological effects inside mitochondria. Previous studies reported that the conjugation of mitochondrial targeting signal peptides to exogenous proteins and small linear DNAs enhanced their delivery to mitochondria,10,11 but this strategy failed, in cases of macromolecules and hydrophobic molecules, including mitochondrial DNA and proteins.12,13,14
To achieve efficient mitochondrial drug delivery, two independent processes i.e., “cytoplasmic delivery through the cell membrane” and “mitochondrial delivery through the mitochondrial membrane” are required.6 In previous studies, we reported on the development of an octaarginine (R8) modified nano carrier for efficient cytoplasmic delivery15,16 and a MITO-Porter for the mitochondrial delivery of macromolecules.17 The former, R8 (refs. 18,19) is a synthetic peptide that mimics the trans-activating transcriptional activator derived from the human immunodeficiency virus.20,21 We showed that high-density R8-modified liposomes were internalized primarily via macropinocytosis and are efficiently delivered to the cytosol, similar to an adenovirus vector.15,16 On the other hand, we proposed a MITO-Porter, which is different from mitochondrial targeting signal peptide system, for mitochondrial delivery.17 The MITO-Porter is a liposome-based nano carrier that delivers cargos to mitochondria via membrane fusion. Using the green fluorescence protein as a model macromolecule and analysis by confocal laser scanning microscopy (CLSM), we were able to confirm mitochondrial macromolecule delivery by the MITO-Porter. We were also able to verify that the MITO-Porter delivered cargoes to the mitochondrial matrix, which pools mitochondrial DNA (mtDNA), using this novel imaging method.22
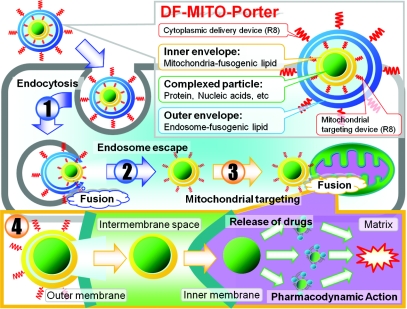
A nano-carrier system which achieves both efficient cytoplasmic delivery and mitochondrial macromolecule delivery could open new avenues for mitochondrial disease therapies. We report herein on the development of a dual function MITO-Porter (DF-MITO-Porter), which has the potential for serving as a nano-device for mitochondrial disease therapies, based on the concept integrating both high-density R8-modified liposomes and the conventional MITO-Porter (Figure 1). The critical structural elements of the DF-MITO-Porter include a complexed particle of cargos that are coated with two mitochondria-fusogenic inner membranes and two endosome-fusogenic outer membranes. Modification of the outer envelope-surface with a high density of R8 (refs. 18,19) greatly assists in the efficient internalization of the carriers into cells (1st step). Inside the cell, the carrier escapes from the endosome into the cytosol via membrane fusion, a process that is mediated by the outer endosome-fusogenic lipid membranes (2nd step). The carrier then binds to mitochondria via R8 (3rd step) and fuses with the mitochondrial membrane (4th step). Recent studies reported that transcriptional activator has the ability to deliver a certain cargo to mitochondria,23,24 and we also showed that modification of the liposome-surface with R8 significantly enhances binding to mitochondria, regardless of the lipid composition.6 Thus, we chose R8, not only as a cytoplasmic delivery device but also as a mitochondrial targeting device as well.
Figure 1.
Schematic diagram illustrating mitochondrial macromolecule delivery via a series of membrane fusions using dual function (DF)-MITO-Porter. Complexed particles of cargos are coated with mitochondria-fusogenic lipid envelope (inner) and endosome-fusogenic lipid envelope (outer). Octaarginine (R8) functions as a cell-uptake device in the outer envelope and as a mitochondrial targeting device in the inner envelope.
In this study, we attempted to prepare DF-MITO-Porter with mitochondria-fusogenic inner membranes and endosome-fusogenic outer membranes, and optimize the characteristics. Intracellular observation using CLSM permitted us to compare mitochondrial targeting activity between DF-MITO-Porter and conventional MITO-Porter. Moreover, we validated that DF-MITO-Porter delivered exogenous macro-biomolecules into mitochondria matrix. Finally, we screened the optimal lipid composition for the DF-MITO-Porter, for achieving a high mitochondrial membrane fusion activity and low cytotoxicity.
Results
Construction of DF-MITO-Porter
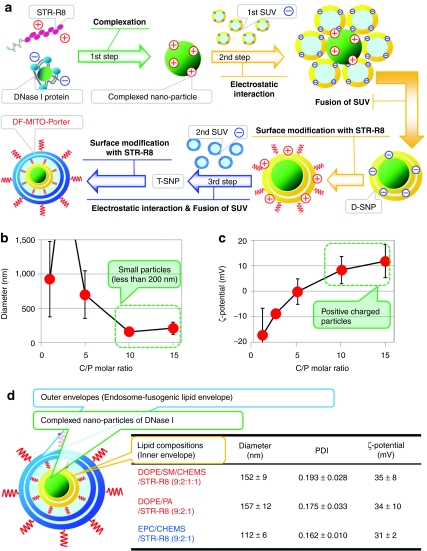
The findings reported herein, confirm that the DF-MITO-Porter has the ability to deliver macromolecules into the mitochondrial matrix, which pools mtDNA, over the mitochondrial double-membrane structure. DNase I protein was chosen as a model macromolecule, which permitted us to estimate mitochondrial matrix delivery of the carrier. It was expected that mtDNA would be digested, when the mitochondrial matrix delivery of DNase I progressed. The construction of the DF-MITO-Porter encapsulating DNase I requires the following three steps (Figure 2a): (i) the construction of nano particles containing DNase I; (ii) coating the nano particles with mitochondria-fusogenic envelope; (iii) further coating endosome-fusogenic envelope in a step-wise manner, based on our previous report regarding gene packaging with two-different types of lipid layers.25
Figure 2.
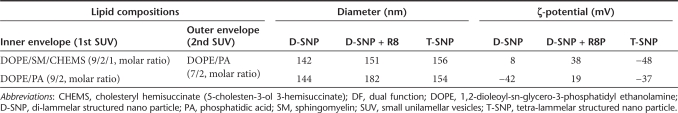
Construction of the dual function (DF)-MITO-Porter. (a) Schematic diagram of the preparation of the DF-MITO-Porter. D-SNP, di-lammelar structured nano particle; SUV, small unilamellar vesicles; T-SNP, tetra-lamellar structured nano particle. (b) Diameter and (c) ζ-potential of nano particles of DNase I complexed with stearyl octaarginine (STR-R8) at various molar ratios. Data are represented by the mean ± SD (n = 3–7). C/P, complex-inducer (STR-R8)/protein (DNase I). (d) Characteristics of the DF-MITO-Porter containing DNase I. Diameter, polydispersity index (PDI) and ζ-potential of carriers were measured. Data are represented by the mean ± SD (n = 3–4). CHEMS, cholesteryl hemisuccinate (5-cholesten-3-ol 3-hemisuccinate); DOPE, 1,2-dioleoyl-sn-glycero-3-phosphatidyl ethanolamine; PA, phosphatidic acid; SM, sphingomyelin.
We first determined the optimal conditions for complexation between DNase I protein (M.W. 36 kDa, weak negative charge) and stearyl R8 (STR-R8)26 that would be suitable for the complexation of proteins.27 We mixed DNase I with STR-R8 at several complex-inducer/protein (C/P) molar ratios to form nano particles and then determined their diameters and ζ-potentials (surface charges) (Figure 2b,c). Figure 2b shows data for the diameters of the complexes. Particles with diameters of less than 200 nm were formed at C/P molar ratios higher than 10. Figure 2c shows the ζ-potentials for the complexes. The charges of the particles were changed from minus to plus with increasing C/P ratios. In this study, we chose small positively charged particles formed at a C/P molar ratio of 10 for preparation of the DF-MITO-Porter. We also prepared small unilamellar vesicles (SUVs) with various lipid compositions (Supplementary Table S1). The positively charged particles were then coated with the first two-layered envelope by the assembly of negatively charged SUVs. The resulting structure was modified with STR-R8 to reverse the surface charge, and then coated with the second envelope via the fusion of negatively charged SUVs. Finally, the carrier-surface was modified with STR-R8 to give the final DF-MITO-Porter, which is a tetra-lamellar structured nano particle. The physiochemical properties of the intermediate particles are summarized in Table 1.
Table 1. Physicochemical properties of intermediated particles of the DF-MITO-Porter.
The size, polydispersity index as an indicator of the particle-size distribution, and the ζ-potential of the DF-MITO-Porter and control carrier are summarized in Figure 2d. The outer envelope of all carriers had a endosome-fusogenic composition [DOPE/PA/STR-R8 (7:2:1, molar ratio)].28 For the DF-MITO-Porter, inner envelope had a mitochondria-fusogenic composition [DOPE/SM/CHEMS/STR-R8 (9:2:1:1, molar ratio) or DOPE/PA/STR-R8 (9:2:1, molar ratio)],17 while a non-mitochondrial fusogenic composition [EPC/CHEMS/STR-R8 (9:2:1, molar ratio)]17 was used in the control inner envelope. As shown in Figure 2d, the prepared carriers were positively charged nano particles with a homogeneous structure. We also prepared a conventional MITO-Porter for purposes of checking the multi-layered structure value (Supplementary Figure S1).
Intracellular observation of DF-MITO-Porter using CLSM and evaluation for mitochondrial targeting activity
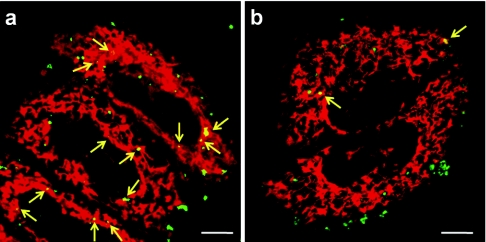
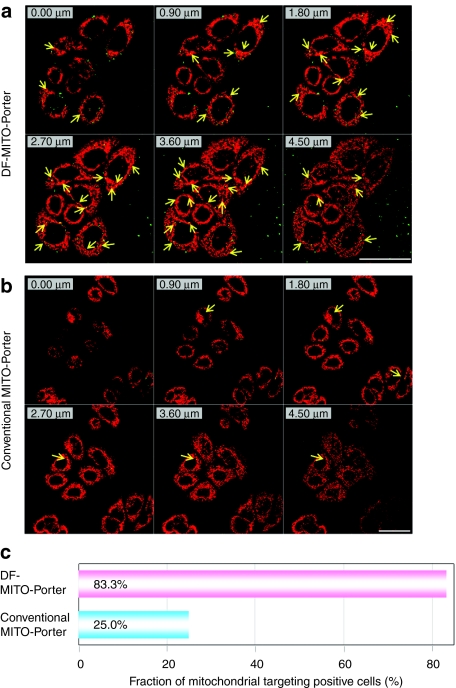
We observed the intracellular trafficking of the DF-MITO-Porter and the conventional MITO-Porter using CLSM to validate that the carrier was able to regulate its intracellular trafficking for efficient mitochondrial delivery. In the case of the DF-MITO-Porter, many yellow clusters were observed (Figure 3a), indicating that the NBD-DOPE in the carriers (green) are colocalized with mitochondria (stained red with MitoFluor Red 589). Moreover, it was confirmed that the DF-MITO-Porter accumulated in mitochondria more efficiently compared to the conventional MITO-Porter (Figure 3). We next evaluated the fraction of mitochondrial targeted positive cells based on images observed by CLSM (see Materials and Methods for details). The fraction of mitochondrial targeted positive cells of the total number of observed cells was calculated as follows;
Figure 3.
Intracellular distribution of carriers was analyzed by confocal laser scanning microscopy. (a) DF-MITO-Porter (SM) and (b) conventional MITO-Porter (SM) labeled with NBD-DOPE (green) were incubated with HeLa cells. Mitochondria were stained with MitoFluor Red 589 (red) prior to observation. The clusters present in the mitochondria (yellow) are indicated. Bars = 10 µm. DF, dual function; DOPE, 1,2-dioleoyl-sn-glycero-3-phosphatidyl ethanolamine; NBD, 7-nitrobenz-2-oxa-1, 3-diazole; SM, sphingomyelin.
where Nmt, Ntot represent the number of mitochondrial targeted positive cells and the total number of cells observed, respectively.
Figure 4a,b show Z-series images of the intracellular observation of the DF-MITO-Porter and the conventional MITO-Porter, respectively. The yellow pixel areas where carriers (green) are colocalized with mitochondria (red) were marked in each xy plane. We defined cells where carriers were colocalized with mitochondria in at least one Z-series image as mitochondrial targeted positive cells. Consequently, the fraction of mitochondrial targeted positive cells of the DF-MITO-Porter (more than 80%) was considerably higher than the corresponding value for the conventional MITO-Porter (Figure 4c). This suggests that a multi-layered structure can be very useful for mitochondrial delivery in living cells.
Figure 4.
Z-series images of intracellular observation and the fraction of mitochondrial positive targeted cells. (a) DF-MITO-Porter (SM) and (b) the conventional MITO-Porter (SM) labeled with NBD-DOPE were incubated with HeLa cells at 37 °C for 1 hour. After replacing the medium with fresh medium, the cells were incubated for a further 1 hour. Mitochondria were stained with MitoFluor Red 589 prior to observation. Fluorescent and bright-field images were captured using confocal laser scanning microscopy (LSM510). Images obtained from the bottom of the coverslip to the top of the cells were recorded by the LSM510 on a PC. The clusters present in the mitochondria are indicated as yellow arrows. Bars = 50 µm. (c) Mitochondrial targeting activity was calculated from the images shown in Figure 4a,b, (see Materials and Methods for details).
Relationship between mitochondrial membrane fusion activity and cytotoxicity
The possibility that the DF-MITO-Porter might induce cytotoxicity cannot be excluded, since this system fuses with the mitochondria, which play a role in the homeostasis of vital physiological functions. Therefore, we investigated the relationship between mitochondrial membrane fusion activity and cytotoxicity. In this experiment, we prepared an empty DF-MITO-Porter with various inner mitochondria-fusogenic envelopes as previously reported,17 and then evaluated their cell viabilities. The graph in Figure 5 shows the relationship between mitochondrial membrane fusion activity (x-axis) and cell viability (y-axis). We also calculated Pearson's correlation coefficient (r = 0.21), and determined if the correlation coefficient was significantly different from zero using a t-test (P > 0.05). We did not find a significant correlation between mitochondrial membrane fusogenic activity and cell viability. These results suggest that a high fusogenic activity with the mitochondrial membrane induces cytotoxicity is not universally true. The finding that a carrier with an inner envelope composed of sphingomyelin (SM) was optimal for the DF-MITO-Porter is particularly noteworthy, since it has a high mitochondrial membrane fusogenic activity but low cytotoxicity.
Figure 5.
Relationship between mitochondrial membrane fusion activity (x-axis) and cell viability (y-axis). For x-axis, we used data reported in our previous study17 (Supplementary Figure S2). For the y-axis, we calculated cell viability after the addition of carriers without DNase I to HeLa cells. Data are represented by the mean ± SE (n = 4–5). CHEMS, cholesteryl hemisuccinate (5-cholesten-3-ol 3-hemisuccinate); Chol, cholesterol; CL, cardiolipin; PA, phosphatidic acid; PG, phosphatidyl glycerol; PI, phosphatidyl inositol; PS, phosphatidyl serine; SM, sphingomyelin.
Evaluation of mtDNA-levels after the mitochondrial delivery of DNase I using the DF-MITO-Porter
To validate the conclusion that the DF-MITO-Porter is useful as a carrier of therapeutic cargos, we examined the delivery of DNase I protein to the mitochondrial matrix, since the latter is an active bio-molecule that functions inside mitochondria. In a previous study, we reported on the successful mitochondrial matrix delivery of the MITO-Porter, as evidenced by the use of our novel imaging method.22 In the paper, we used propidium iodide, a membrane-impermeable red-fluorescent dye for staining nucleic acids as an aqueous phase marker for the MITO-Porter. This dye permits mitochondrial matrix delivery to be confirmed, by the detection of red-fluorescent light, because the light is emitted as the result of the conjugation of the dye with mtDNA in the mitochondrial matrix. We first confirmed mitochondrial matrix delivery in isolated rat liver mitochondria using the MITO-Porter. CLSM analyses showed that this system can be used to efficiently visualize mtDNA, not only in isolated mitochondria, but in living cells as well. We also verified that no red-fluorescent light was produced when propidium iodide only was incubated with isolated mitochondria or living cells, indicating that propidium iodide did not gain access to mtDNA inside mitochondrial matrix. Finally, the fact that the red fluorescence could be detected in living cells in spectral imaging fluorescent microscopy experiments clearly demonstrated that the MITO-Porter successfully delivered its cargo to the mitochondrial matrix. Based on our previous report, we conclude that the DF-MITO-Porter has the ability to deliver cargoes to the mitochondrial matrix.
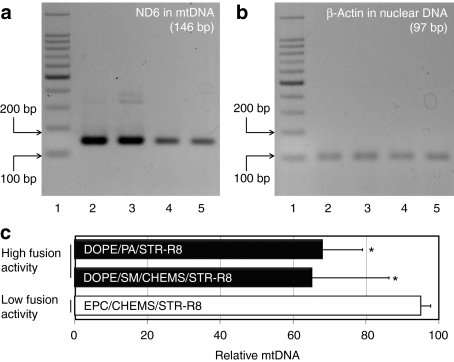
To verify that mtDNA-levels were decreased after the mitochondrial delivery of DNase I using the DF-MITO-Porter, we evaluated mtDNA-levels within cells using PCR. DNase I protein encapsulated in carriers were incubated with HeLa cells. The cellular DNA was then purified and subjected to the PCR. PCR assays to detect the ND6 and β-actin genes were performed in order to detect both mtDNA and nuclear DNA. The primers used in these experiments are summarized in Supplementary Table S2. Figure 6 shows agarose gel electrophoresis data for the PCR products derived from mtDNA (Figure 6a) and nuclear DNA (Figure 6b). In the case of the DF-MITO-Porter, decreases in mtDNA-levels were observed (lanes 4, 5 in Figure 6a), whereas carriers with a low mitochondrial fusion activity had a negligible effect on mtDNA-levels (lane 3 in Figure 6a). This result suggests that DNase I is delivered to mitochondria by the DF-MITO-Porter, as indicated by the digestion of mtDNA by DNase I to some extent. On the other hand, a decrease in nuclear DNA-levels was not detected in any of the carriers (Figure 6b). Moreover, we quantified mtDNA-levels within the cells using real-time PCR (Figure 6c). The results showed that the DF-MITO-Porter decreased mtDNA-levels compared with carriers with a low mitochondrial fusion activity. We also confirmed that the MITO-Porter has a high fusogenic activity for mitochondria, but that is failed to effectively fuse with the nucleus (data not shown). Based on these results, it is presumed that the mitochondrial specific fusion activity of the MITO-Porter might be involved in a pathway related to the selective digestion of mtDNA.
Figure 6.
Evaluation of mtDNA-levels after the mitochondrial delivery of DNase I using PCR. DNase I encapsulated in the DF-MITO-Porter (SM), the DF-MITO-Porter (PA) or the control carrier with low mitochondrial fusion activity were incubated with HeLa cells. Cellular DNA was then purified and subjected to the PCR as described in the Materials and Methods section. PCR assays for ND6 and β-actin genes detection were performed in order to detect (a) mitochondrial DNA (mtDNA) and (b) nuclear DNA, respectively. Lane 1, 100 bp DNA ladder; lane 2, non-treatment; lane 3, control carrier with low mitochondrial fusion activity (EPC/CHEMS/STR-R8); lane 4, DF-MITO-Porter (SM); lane 5, DF-MITO-Porter (PA). (c) We also quantified mtDNA-levels within cells using real-time PCR. Data are represented as the mean ± SD (n = 3–6). *Significant difference between control carrier (EPC/CHEMS/STR-R8) and other carriers (P < 0.05 by one-way analysis of variance, followed by Bonferroni correction). CHEMS, cholesteryl hemisuccinate (5-cholesten-3-ol 3-hemisuccinate); DF, dual function; EPC, egg yolk phosphatidyl choline; ND6, mitochondrial NADH dehydrogenase 6; PA, phosphatidic acid; SM, sphingomyelin; STR-R8, stearyl octaarginine.
Discussion
In this study, we attempted to achieve macromolecule packaging using a multifunctional envelope-type nano-device preparation technique. The multifunctional envelope-type nano-device consists of a condensed pDNA core and a lipid envelope equipped with various functional devices that mimic envelope-type viruses.29,30 The condensation of pDNA into a compact core prior to its inclusion in the lipid envelope has several advantages as follows: protection of pDNA from DNase; size control; and, improved packaging efficiency resulting from electrostatic interactions between the condensed core and the lipid envelope. To date, we have been successful in efficiently packaging not only pDNA, but also oligo nucleic acids, proteins, and other substances into a multifunctional envelope-type nano-device.27,31,32,33 On the other hand, the MITO-Porter system delivers cargos into mitochondria via a membrane fusion mechanism. Therefore, large cargoes could be delivered to mitochondria, provided that the cargoes can be encapsulated in the MITO-Porter. Accordingly, a combination of a MITO-Porter and a macromolecule packaging technique in multifunctional envelope-type nano-device preparations would likely achieve the mitochondrial delivery of macromolecules such as pDNA, mtDNA, oligo nucleic acids, and proteins.
We constructed the DF-MITO-Porter according to Figure 2a, using the multi-layering method.24 First, a di-lamellar structured nano particle (D-SNP) was prepared through membrane fusion of neighboring SUVs, triggered by the assembly of negatively charged SUVs around the positively charged complexed nano particle of DNase I protein. As previously proposed, it is possible that core particles can form a di-lamellar structure during liposome incubation.34 The surface of the resulting particles is then modified with STR-R8 to reverse the surface charge. Next, R8-modified D-SNP was coated with the second envelope, again through the fusion of negatively charged SUV to produce tetra-lamellar structured nano particle (T-SNP). If the 2nd SUVs (∼60 nm) only bind to the D-SNP (∼140 nm), the mean diameter would be expected to be ∼260 nm. However, the resulting particle had a diameter of ∼150 nm, suggesting that the 2nd SUVs fuse with each other, forming a T-SNP. The conversion of ζ-potential in each coating process strongly suggests that the complexed nano particles of DNase I protein underwent a step-wise encapsulation. Thus, the resulting DF-MITO-Porter consists of two types of lipid layers. Table 1 shows physicochemical properties of intermediate of DF-MITO-Porter.
We applied the confocal image-assisted three-dimensionally integrated quantification method to evaluate the fraction of mitochondrial targeted positive cells35,36 and counted number of mitochondrial targeted positive cells (see Materials and Methods for details). Z-series images of the cells after transduction were observed by the LSM510 as shown in Figure 4a,b, and the total brightness of each region of interest was then quantified by the confocal image-assisted three-dimensionally integrated quantification method.35,36 The yellow pixel areas where carriers (green) colocalized with mitochondria (red) were marked in each xy plane. Mitochondrial targeted positive cells were defined as cells where carriers were colocalized with mitochondria in at least one Z-series image, and have a significant effect on the fraction of mitochondrial targeted positive cells (%). As shown in Figure 4c, the fraction of mitochondrial targeted positive cells produced using the DF-MITO-Porter (more than 80%) was considerably higher than that for the conventional MITO-Porter (about 25%). This result indicates that the DF-MITO-Porter was threefold more efficient than a conventional MITO-Porter in numbers of mitochondrial targeted positive cells, suggesting that a multi-layered structure can be very useful for improving mitochondrial delivery.
Mitochondrial delivery requires the regulation of many intracellular trafficking steps. In this study, we attempted to improve endosomal escape using a multi-layered structure with endosomal fusogenic activity. As a result, the DF-MITO-Porter could deliver molecules to mitochondria more efficiently than a conventional carrier. However, it would be difficult to achieve selective mitochondrial delivery using the current DF-MITO-Porter. On the other hand, the conjugation of mitochondrial targeting signal peptides can achieve selective mitochondrial delivery,10,11 but this strategy fails, in cases of macromolecules and hydrophobic molecules.12,13,14 In the future, we plan to integrate mitochondrial targeting signal peptides and a MITO-Porter system to achieve the selective delivery of macromolecules to mitochondria. Studies concerning this are currently in progress.
The DF-MITO-Porter developed in this study, is an innovative nano-device for mitochondrial delivery, which has the ability to pass through endosomal and mitochondrial membranes via step-wise membrane fusion. The findings show that the DF-MITO-Porter delivers exogenous macro-biomolecules into the mitochondrial matrix. Moreover, we identified the optimal lipid composition for the DF-MITO-Porter, for achieving a high mitochondrial membrane fusion activity and low cytotoxicity. Our ultimate goal is to develop a mitochondrial drug delivery system, not only effective cargo delivery, but also a system that will be functional inside a mitochondrion for therapy. Future studies will involve attempts to improve the DF-MITO-Porter in terms of mitochondrial gene function with experts in the field of mitochondrial molecular biology. Studies directed toward this goal are currently in progress.
Materials and Methods
Materials. Cholesterol, 1,2-dioleoyl-sn-glycero-3-phosphatidyl ethanolamine (DOPE), 7-nitrobenz-2-oxa-1, 3-diazole labeled DOPE (NBD-DOPE), and rhodamine-DOPE were purchased from Avanti Polar lipids (Alabaster, AL). Egg yolk phosphatidyl choline (EPC) was obtained from Nippon Oil and Fats (Tokyo, Japan). Cholesteryl hemisuccinate (5-cholesten-3-ol 3-hemisuccinate; CHEMS), cardiolipin, phosphatidic acid (PA), phosphatidyl glycerol, phosphatidyl inositol, phosphatidyl serine, and SM were purchased from Sigma (St Louis, MO). STR-R8 (ref. 26) was obtained from KURABO Industries (Osaka, Japan). DNase I protein (from bovine pancreas, Grade II) was purchased from Roche Diagnostics (Mannheim, Germany). HeLa human cervix carcinoma cells were obtained from RIKEN Cell Bank (Tsukuba, Japan). Dulbecco's modified Eagle medium (DMEM) and fetal bovine serum were purchased from Invitrogen (Carlsbad, CA). All other chemicals used were commercially available reagent-grade products.
Preparation of complexed protein particles with STR-R8. DNase I protein was gently mixed with the STR-R8, as complex-inducer, in 100 µl of 10 mmol/l HEPES buffer (pH 7.4), followed by incubation for 15 minutes at 25 °C to form complexed protein particles. Particles of the DNase I/STR-R8 were formed using 50 µg of DNase I and STR-R8 in a series of molar ratios of complex-inducer/protein (C/P).
Construction of DF-MITO-Porter. The DNase I-encapsulated DF-MITO-Porter was constructed by the multi-layering method (Figure 2a), as previously reported.25 A solution of DNase I protein in HEPES buffer (1 mg/ml) was mixed with an STR-R8 solution to form complexed protein particles by gently pipetting at a C/P molar ratio of 10, followed by incubation for 15 minutes at 25 °C. For inner and outer envelopes, various SUVs were prepared. The suspended negatively charged SUVs and complexed protein particles were mixed at a ratio of 3: 1 (v/v) to coat the complexed protein particles with a double-lipid envelope to produce a D-SNP. These SUVs composed of mitochondria-fusogenic lipid [DOPE/PA = 9:2, DOPE/SM/CHEMS = 9:2:1 (molar ratio)] or non-fusogenic lipid [EPC/CHEMS = 9:2 (molar ratio)]. An STR-R8 solution (10 mol% of total lipid) was added to the suspension of D-SNPs to reverse the surface charge. This suspension was then mixed with endosome-fusogenic SUV [DOPE/PA = 7: 2 (molar ratio)] at a ratio of 1:2 (v/v) to generate particles with a double endosome-fusogenic envelope, hereafter referred to a T-SNP. The STR-R8 solution (10 mol% of endosome-fusogenic lipid) was added to the suspension of T-SNP to modify the outer envelope with R8. We refer to the R8 modified T-SNP with mitochondria-fusogenic inner envelope that contain PA and SM as the DF-MITO-Porter (PA) and the DF-MITO-Porter (SM), respectively.
Measurement of diameter and ζ-potential. Particle-diameter was measured by a quasi-elastic light scattering method and ζ-potentials were determined electrophoretically by means of an electrophoretic light scattering spectrophotometer (Zetasizer Nano ZS; Malvern Instruments, Herrenberg, Germany).
Intracellular trafficking observation of DF-MITO-Porter using CLSM. HeLa cells (4 × 104 cells/ml) were cultured in 35 mm dishes (BD Falcon, Franklin Lakes, NJ) with DMEM, which contained 10% fetal bovine serum, under an atmosphere of 5% CO2/air at 37 °C for 24 hours. The cells were washed with serum-free medium before incubation with carriers. The DF-MITO-Porter or conventional MITO-Porter, labeled with 0.5 mol% NBD-DOPE lipids, were incubated with HeLa cells (final lipid concentration, 13.5 µmol/l) to observe the intracellular trafficking of the carriers themselves. After a 1-hour incubation under an atmosphere of 5% CO2/air at 37 °C, the cells were washed with serum-free medium, and further incubated in medium with 10% serum for 100 minutes in the absence of carriers. After incubation, MitoFluor Red 589 (Molecular Probes, Eugene, OR) was applied to the medium at a final concentration of 100 nmol/l to stain the mitochondria. After incubation for 20 minutes, the cells were washed with serum-free medium and then observed by CLSM (LSM510; Carl Zeiss, Jena, Germany). The cells were excited with 488 nm and 568 nm light from an Ar/Kr laser. A series of images were obtained using an LSM510 microscope with a water immersion objective lens (Achroplan 63x/NA = 0.95) and a dichroic mirror (HFT488/568). The two fluorescence detection channels (Ch) were set to the following filters: Ch1: LP585 (red); Ch 2: BP 505–550 (green).
Fluorescence image processing for calculation of fraction of mitochondrial targeted positive cells. Fluorescent and bright-field images were captured using an LSM510 microscope. Images obtained from the bottom of the coverslip to the top of the cells were recorded by the LSM510 on a PC as shown in Figure 4a,b. Each 8-bit TIFF image was analyzed using the Image-Pro plus v.4.5.1.26 software (Media Cybernetics, Bethesda, MD) to quantify the total brightness of each region of interest based on confocal image-assisted three-dimensionally integrated quantification method.35,36 The fraction of mitochondrial targeted positive cells was determined as described below. First, the yellow pixel areas where carrier (green) colocalized with mitochondria (red) were marked in each xy plane. We define cells where carriers colocalized with mitochondria in at least one Z-series image as mitochondrial targeted positive cells. In some cases, it was difficult to distinguish the localization of the carrier between on and outside the mitochondria. Therefore, criteria were introduced to classify the carrier cluster as colocalized in the mitochondria when more than 50% of the pixels of the carrier were mitochondria targeted positive (defined as green fluorescent values larger than 130). The fraction of mitochondrial targeted positive cells was calculated as follows;
where Nmt, Ntot represent the number of mitochondrial targeted positive cells and the total number of cells observed, respectively.
Investigation of the relationship between mitochondrial membrane fusion activity and cell viability. We prepared a graph showing mitochondrial membrane fusion activity (x-axis) and cell viability (y-axis). For the x-axis, we used data reported in a previous study,17 where we calculated mitochondrial fusion activities in terms of the cancellation of fluorescent resonance energy transfer (Supplementary Figure S2). For the y-axis, we prepared various DF-MITO-Porters unencapsulating DNase I and evaluated their cell viabilities as described below. HeLa cells (1 × 104 cells/well) were incubated in a 24-well plate (Corning, Corning, NY) with DMEM containing 10% fetal bovine serum, under 5% CO2/air at 37 °C for 24 hours. Sample-suspensions in 0.25 ml of serum-free DMEM (containing 12.5 µl of carriers) were added to the cells after washing the cells with phosphate-buffer saline (PBS (–)), followed by incubation under 5% CO2 at 37 °C for 3 hours. After replacing the medium with fresh DMEM containing 10% serum, the cells were incubated for a further 21 hours. The cells were then washed with PBS (–), and cell viability was measured using a Cell Proliferation Assay System with a Tetra Color ONE (Seikagaku Biobusiness, Tokyo, Japan), according to the instructions provided by the manufacturer. Briefly, a tetrazolium (WST-8) was added to a final volume of 275 µl/well. After 1-hour incubation, absorbance at 450 nm was measured by means of a Benchmark plus microplate reader (Bio-Rad Laboratories, Hercules, CA). Cell viability was calculated as follows;
where AT, AU represent the absorbance at 450 nm when cells were treated and untreated with samples, respectively.
Evaluation for mtDNA-levels after the mitochondrial delivery of DNase I. HeLa cells (2 × 105 cells/well) were seeded on a 6-well plate (Corning) with DMEM containing 10% fetal bovine serum, under 5% CO2/air at 37 °C for 24 hours. Sample-suspensions containing 6 µg of DNase I in 1 ml of serum-free DMEM were added to the cells after washing them with PBS (–), followed by incubation under 5% CO2 at 37 °C for 3 hours. After the addition of fresh DMEM containing 10% serum, the cells were incubated for a further 21 hours. The cells were then washed with PBS (–), trypsinized, suspended in DMEM with serum, precipitated by centrifugation (1,800g, 4 °C, 3 minutes). The pellets were washed with PBS (–) and precipitated by centrifugation (1,800g, 4 °C, 3 minutes). To evaluate the levels of mtDNA and nuclear DNA, total cellular DNA was purified from cell lysates by means of a GenElute Mammalian Genome DNA Miniprep kit (Sigma–Aldrich, St Louis, MO) and subjected to the PCR.
The PCR reaction mixture contained, in a total volume of 25 µl, 10 mmol/l of Tris-HCl (pH 8.3), 1.5 mmol/l of MgCl2, 50 mmol/l of KCl, 200 µmol/l of each one of the deoxynucleoside triphosphates, 0.5 µmol/l of primers ND6 (+) and ND6 (–) for mtDNA detection or primers β-actin (+) and β-actin (–) for nuclear DNA (as shown in Supplementary Table S2), and 10 ng of DNA obtained from HeLa cell lysates. After the addition of 0.625 U of Taq DNA polymerase (Finnzymes PCR Reagents; Thermo Fisher Scientific, Waltham, MA), the reaction mixture was first incubated at 94 °C for 2 minutes, then subjected to 30 cycles of 30 seconds at 94 °C, 30 seconds at 55 °C and 45 seconds at 72 °C, and finally to 10 minutes at 72 °C. Each 5 µl of PCR products were subjected to electrophoresis in 2% agarose gel in TAE (40 mmol/l Tris-HCl, 40 mmol/l acetic acid, 1 mmol/l EDTA, pH 8.0) at 100 V for 30 minutes. The DNA bands were visualized by UV after ethidium bromide staining.
For quantitative real-time PCR, each target, ND6 encoded on mtDNA, was amplified on the same plate with the reference, β-actin, with the SYBR Green Real-time PCR Master Mix (TOYOBO, Osaka, Japan) and the 7500 Real Time PCR System (Applied Biosystems, Foster City, CA), and the relative mtDNA amounts and range were determined. Briefly, we normalized each set of samples using the difference in threshold cycles (ΔCT) between the sample mtDNA and the housekeeping gene (β-actin): ΔCT = (ΔCT sample – ΔCT β-Actin). The calibrator sample (ΔCT calibration) was assigned as the sample with non-treatment cell. Relative mtDNA-levels were calculated by the expression 2−ΔΔCT where ΔΔCT= ΔCT sample (n) – ΔCT calibration (n). Each reaction was done, at least in duplicate. For the real-time PCR, two types of primers ND6 (+) and ND6 (–) and primers β-actin (+) and β-actin (–) were used as shown in Supplementary Table S2.
SUPPLEMENTARY MATERIAL Figure S1. Construction of the conventional MITO-Porter encapsulating DNase I. Figure S2. Summary of mitochondrial membrane fusion activities of R8-modified DOPE-LP. Table S1. Lipid composition and property of SUV. Table S2. Primers used in the PCR to evaluate the levels of mtDNA and nuclear DNA.
Acknowledgments
This work was supported, in part by, a Grant-in-Aid for Young Scientists (B) and Grants-in-Aid for Exploratory Research from the Ministry of Education, Culture, Sports, Science and Technology of Japanese Government (MEXT) and by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation, Japan (NIBIO). We also thank Milton Feather for his helpful advice in writing the manuscript.
Supplementary Material
Construction of the conventional MITO-Porter encapsulating DNase I.
Summary of mitochondrial membrane fusion activities of R8-modified DOPE-LP.
Lipid composition and property of SUV.
Primers used in the PCR to evaluate the levels of mtDNA and nuclear DNA.
REFERENCES
- Reeve AK, Krishnan KJ., and, Turnbull D. Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann N Y Acad Sci. 2008;1147:21–29. doi: 10.1196/annals.1427.016. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A., and, Weiner H. Delivery of drugs and macromolecules to mitochondria. Adv Drug Deliv Rev. 2007;59:729–738. doi: 10.1016/j.addr.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- Wallace DC. The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene. 2005;354:169–180. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Yamada Y., and, Harashima H. Mitochondrial drug delivery systems for macromolecule and their therapeutic application to mitochondrial diseases. Adv Drug Deliv Rev. 2008;60:1439–1462. doi: 10.1016/j.addr.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Clay Montier LL, Deng JJ., and, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomics. 2009;36:125–131. doi: 10.1016/S1673-8527(08)60099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J. Treatment of mitochondrial disorders . Eur J Paediatr Neurol. 2010;14:29–44. doi: 10.1016/j.ejpn.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Kyriakouli DS, Boesch P, Taylor RW., and, Lightowlers RN. Progress and prospects: gene therapy for mitochondrial DNA disease. Gene Ther. 2008;15:1017–1023. doi: 10.1038/gt.2008.91. [DOI] [PubMed] [Google Scholar]
- Flierl A, Jackson C, Cottrell B, Murdock D, Seibel P., and, Wallace DC. Targeted delivery of DNA to the mitochondrial compartment via import sequence-conjugated peptide nucleic acid. Mol Ther. 2003;7:550–557. doi: 10.1016/s1525-0016(03)00037-6. [DOI] [PubMed] [Google Scholar]
- Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- Endo T, Nakayama Y., and, Nakai M. Avidin fusion protein as a tool to generate a stable translocation intermediate spanning the mitochondrial membranes. J Biochem. 1995;118:753–759. doi: 10.1093/oxfordjournals.jbchem.a124976. [DOI] [PubMed] [Google Scholar]
- Esaki M, Kanamori T, Nishikawa S., and, Endo T. Two distinct mechanisms drive protein translocation across the mitochondrial outer membrane in the late step of the cytochrome b(2) import pathway. Proc Natl Acad Sci USA. 1999;96:11770–11775. doi: 10.1073/pnas.96.21.11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhler A, Ono H, Guiard B, Neupert W., and, Stuart RA. A novel intermediate on the import pathway of cytochrome b2 into mitochondria: evidence for conservative sorting. EMBO J. 1995;14:1349–1359. doi: 10.1002/j.1460-2075.1995.tb07121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil IA, Kogure K, Futaki S., and, Harashima H. High density of octaarginine stimulates macropinocytosis leading to efficient intracellular trafficking for gene expression. J Biol Chem. 2006;281:3544–3551. doi: 10.1074/jbc.M503202200. [DOI] [PubMed] [Google Scholar]
- Khalil IA, Kogure K, Futaki S, Hama S, Akita H, Ueno M.et al. (2007Octaarginine-modified multifunctional envelope-type nanoparticles for gene delivery Gene Ther 14682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Akita H, Kamiya H, Kogure K, Yamamoto T, Shinohara Y.et al. (2008MITO-Porter: A liposome-based carrier system for delivery of macromolecules into mitochondria via membrane fusion Biochim Biophys Acta 1778423–432. [DOI] [PubMed] [Google Scholar]
- Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K.et al. (2001Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery J Biol Chem 2765836–5840. [DOI] [PubMed] [Google Scholar]
- Nakase I, Niwa M, Takeuchi T, Sonomura K, Kawabata N, Koike Y.et al. (2004Cellular uptake of arginine-rich peptides: roles for macropinocytosis and actin rearrangement Mol Ther 101011–1022. [DOI] [PubMed] [Google Scholar]
- Joliot A., and, Prochiantz A. Transduction peptides: from technology to physiology. Nat Cell Biol. 2004;6:189–196. doi: 10.1038/ncb0304-189. [DOI] [PubMed] [Google Scholar]
- Schwarze SR, Ho A, Vocero-Akbani A., and, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- Yasuzaki Y, Yamada Y., and, Harashima H. Mitochondrial matrix delivery using MITO-Porter, a liposome-based carrier that specifies fusion with mitochondrial membranes. Biochem Biophys Res Commun. 2010;397:181–186. doi: 10.1016/j.bbrc.2010.05.070. [DOI] [PubMed] [Google Scholar]
- Asoh S, Ohsawa I, Mori T, Katsura K, Hiraide T, Katayama Y.et al. (2002Protection against ischemic brain injury by protein therapeutics Proc Natl Acad Sci USA 9917107–17112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gaizo V., and, Payne RM. A novel TAT-mitochondrial signal sequence fusion protein is processed, stays in mitochondria, and crosses the placenta. Mol Ther. 2003;7:720–730. doi: 10.1016/s1525-0016(03)00130-8. [DOI] [PubMed] [Google Scholar]
- Akita H, Kudo A, Minoura A, Yamaguti M, Khalil IA, Moriguchi R.et al. (2009Multi-layered nanoparticles for penetrating the endosome and nuclear membrane via a step-wise membrane fusion process Biomaterials 302940–2949. [DOI] [PubMed] [Google Scholar]
- Futaki S, Ohashi W, Suzuki T, Niwa M, Tanaka S, Ueda K.et al. (2001Stearylated arginine-rich peptides: a new class of transfection systems Bioconjug Chem 121005–1011. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Yamada Y., and, Harashima H. Efficient cytoplasmic protein delivery by means of a multifunctional envelope-type nano device. Biol Pharm Bull. 2007;30:758–762. doi: 10.1248/bpb.30.758. [DOI] [PubMed] [Google Scholar]
- El-Sayed A, Khalil IA, Kogure K, Futaki S., and, Harashima H. Octaarginine- and octalysine-modified nanoparticles have different modes of endosomal escape. J Biol Chem. 2008;283:23450–23461. doi: 10.1074/jbc.M709387200. [DOI] [PubMed] [Google Scholar]
- Kogure K, Moriguchi R, Sasaki K, Ueno M, Futaki S., and, Harashima H. Development of a non-viral multifunctional envelope-type nano device by a novel lipid film hydration method. J Control Release. 2004;98:317–323. doi: 10.1016/j.jconrel.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Kogure K, Akita H, Yamada Y., and, Harashima H. Multifunctional envelope-type nano device (MEND) as a non-viral gene delivery system. Adv Drug Deliv Rev. 2008;60:559–571. doi: 10.1016/j.addr.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Kogure K, Nakamura Y, Inoue K, Akita H, Nagatsugi F.et al. (2005Development of efficient packaging method of oligodeoxynucleotides by a condensed nano particle in lipid envelope structure Biol Pharm Bull 281939–1942. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kogure K, Yamada Y, Futaki S., and, Harashima H. Significant and prolonged antisense effect of a multifunctional envelope-type nano device encapsulating antisense oligodeoxynucleotide. J Pharm Pharmacol. 2006;58:431–437. doi: 10.1211/jpp.58.4.0002. [DOI] [PubMed] [Google Scholar]
- Homhuan A, Kogure K, Akaza H, Futaki S, Naka T, Fujita Y.et al. (2007New packaging method of mycobacterial cell wall using octaarginine-modified liposomes: enhanced uptake by and immunostimulatory activity of dendritic cells J Control Release 12060–69. [DOI] [PubMed] [Google Scholar]
- Lee RJ., and, Huang L. Folate-targeted, anionic liposome-entrapped polylysine-condensed DNA for tumor cell-specific gene transfer. J Biol Chem. 1996;271:8481–8487. doi: 10.1074/jbc.271.14.8481. [DOI] [PubMed] [Google Scholar]
- Akita H, Ito R, Khalil IA, Futaki S., and, Harashima H. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Mol Ther. 2004;9:443–451. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Hama S, Akita H, Ito R, Mizuguchi H, Hayakawa T., and, Harashima H. Quantitative comparison of intracellular trafficking and nuclear transcription between adenoviral and lipoplex systems. Mol Ther. 2006;13:786–794. doi: 10.1016/j.ymthe.2005.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Construction of the conventional MITO-Porter encapsulating DNase I.
Summary of mitochondrial membrane fusion activities of R8-modified DOPE-LP.
Lipid composition and property of SUV.
Primers used in the PCR to evaluate the levels of mtDNA and nuclear DNA.