Abstract
A new approach has been developed to probe the structural properties of membrane peptides and proteins using the pulsed electron paramagnetic resonance technique of electron spin echo envelope modulation (ESEEM) spectroscopy and the α-helical M2δ subunit of the acetylcholine receptor incorporated into phospholipid bicelles. To demonstrate the practicality of this method, a cysteine-mutated nitroxide spin label (SL) is positioned 1, 2, 3, and 4 residues away from a fully deuterated Val side chain (denoted i + 1 to i + 4). The characteristic periodicity of the α-helical structure gives rise to a unique pattern in the ESEEM spectra. In the i + 1 and i + 2 samples, the 2H nuclei are too far away to be detected. However, with the 3.6 residue per turn pattern of an α-helix, the i + 3 and i + 4 samples reveal a strong signal from the 2H nuclei of the Val side chain. Modeling studies verify these data suggesting that the closest 2H-labeled Val to SL distance would in fact be expected in the i + 3 and i + 4 samples. This technique is very advantageous, because it provides pertinent qualitative structural information on an inherently difficult system like membrane proteins in a short period of time (minutes) with small amounts of protein (μg).
Keywords: ESEEM, spin label, solid phase peptide synthesis
Introduction
New approaches and technologies are needed to more efficiently probe the structural properties of membrane proteins. Biophysical techniques such as solid-state NMR spectroscopy are powerful and provide a wealth of information but can be expensive and time consuming in the preparation of samples and data collection. This work highlights the application of the pulsed electron paramagnetic resonance (EPR) technique of electron spin echo envelope modulation (ESEEM) spectroscopy to directly and efficiently probe the secondary structure of a membrane peptide. The information obtained from this ESEEM approach can be used qualitatively to determine the secondary structure of the protein segment and also in a quantitative manner to measure relative distances between a nitroxide spin label (SL) and deuterium nuclei.
Membrane proteins play vital roles as receptors for communication, channels for transportation, drug binding targets, and enzymes to catalyze reactions within the cell membrane.1 Limited structural information exists for membrane proteins due to their inherent hydrophobic nature, poor over expression yields, and lack of high quality crystals.2,3 Thus, it is very challenging to glean pertinent structural information with traditional biophysical techniques such as solution NMR and X-ray crystallography.4 Because of these limitations, other biophysical techniques need to be developed to study the structures of membrane proteins. EPR spectroscopy can be used to study specific questions about the structure and dynamics of biological samples in a fast and efficient manner, when compared to NMR techniques.4
Site-directed spin labeling (SDSL) coupled with EPR spectroscopy has been used to obtain pertinent structural and dynamic information on membrane proteins.4–6 For these SDSL experiments, a nitroxide SL is covalently attached to a cysteine residue at a site of interest. The distance between two spin labels (SL–SL) can be measured with EPR spectroscopy. For these dual-labeled samples, continuous wave electron paramagnetic resonance (CW-EPR) dipolar line broadening studies have been utilized to determine medium range distances between 8 and 20 Å5. Pulsed EPR spectroscopy techniques such as Pulsed Electron-Electron Double Resonance (PELDOR)/Double Electron Electron Resonance (DEER) have also measured longer range distances of 20–70 Å.7 Shorter distances can be measured with the pulsed EPR ESEEM technique.8–13 ESEEM spectroscopy observes NMR transitions indirectly by the way of EPR spectroscopy (EPR-detected NMR) through an electron spin that is weakly coupled to a nearby NMR active nucleus.12–15 The three-pulse ESEEM (π/2–τ–π/2–T–π/2) technique can detect distances between a SL and a 2H (i = 1) nucleus, where the modulation depth (k) produced by a weakly dipolar-coupled nucleus is scaled by r−6. If the 2H nucleus and the SL are in close (< 8 Å) proximity, the 2H modulation can be observed in the time domain data, and a Fourier transformation of the ESEEM data will reveal a peak at or near the 2H Larmor frequency. ESEEM spectroscopy has been used to probe the ligand coordination sphere of metalloenzymes, protein solvent accessibility, and membrane protein depth.16–18 For the first time, we demonstrate that SDSL ESEEM can be used to directly probe the secondary structure of a protein and/or distances between SLs and amino acid side chains.
The secondary structure of an α-helix is periodic and distance measurements can be easily modeled. A typical α-helix has a turn periodicity of 3.6 amino acid units. The distance from the beginning to the end of the turn in the α-helix is 5.4 Å. Taking this into account, every three or four residues in a α-helical segment should have the minimum distance between the side chain residues, assuming that the helix is straight.
We hypothesize that, by using SDSL coupled with ESEEM spectroscopy, the secondary structure of an α-helical membrane peptide can be probed by detecting 2H modulation between a 2H-labeled amino acid and a nearby spin-labeled Cys. The M2δ subunit of the acetylcholine receptor serves as an ideal α-helical model transmembrane peptide to test this conjecture.19,20
Results
To completely map out the α-helical content of the M2δ peptide, four different peptides were designed by fixing the 2H-labeled Val d8 at position 15 (i), and varying the (1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)methanesulfonate (MTSL) spin label (X) at 4 successive positions (i + 1 to i + 4). The sequence for the peptides are as follows: i + 1 (EKMSTAISVXLA QAVFLLLTSQR), i + 2(EKMSTAISVLXAQAVFL LLTSQR), i + 3 (EKMSTAISVLLXQAVFLLLTSQR), and i + 4 (EKMSTAISVLLAXAVFLLLTSQR). The Val d8 is shown in boldface as V, and the SL is shown as X. Additionally, a control sample was prepared such that Val 15 was not 2H-labeled (Fig. 1, black) at position i + 3 (EKMSTAISVLLXQAVFLLLTSQR). Figure 1 shows three-pulse ESEEM data of the M2δ peptide incorporated into unoriented 1,2-Dimyristoyl-sn-Glycero-3-Phosphocholine (DMPC)/1,2-Dihexanoyl-sn-Glycero-3-Phosphocholine (DHPC) lipid bicelles with a 2H-labeled Val d8 and a SL three residues away (i + 3) at two different τ values. From the ESEEM data collected, both τ values provided high-quality time domain and FT ESEEM data. However, 2H modulation was optimized, and the proton modulation was effectively suppressed with τ equal to 200 ns, when compared to the τ 120 ns data. The optimal τ values can vary depending on the field and frequency that the data was collected at.
Figure 1.
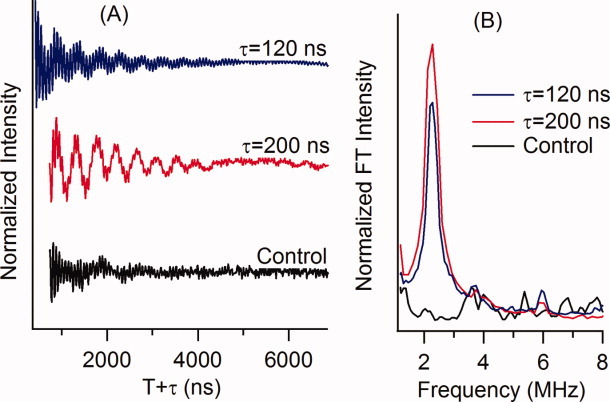
(A) Three-pulse ESEEM time-domain spectra of M2δ in a lipid bicelle. The top is i + 3 2H-labeled Val 15 at τ = 120 ns (blue), the middle is i + 3 2H-labeled Val 15 at τ = 200 ns (red), and the bottom is the nondeuterated control sample at τ = 200 ns (Black). (B) Frequency domain where red illustrates the i + 3 sample, blue illustrates the i + 3 sample, and black the control.
Low-frequency 2H modulation in the time-domain data in Figure 1(A) (red and blue, 2H Val) is clearly evident in the 2H-labeled Val sample when compared to the control (black, 1H Val) sample. The cross-term averaged FT data [Fig. 1(B)] reveal a large well-resolved peak centered at 2.3 MHz originating from weakly coupled 2H nuclei. 2H modulation is not detected in the control sample. The ESEEM spectra unequivocally demonstrate that we can detect the dipolar interaction between the SL and the 2H-labeled Val d8 3 residues away. If the distance between all of the 2H nuclei (Val d8) and the SL is greater than ∼8 Å, no 2H modulation or peak would be observed.21,17 However, if any 2H-SL distances are closer than ∼8 Å, a peak will be observed at the 2H Larmor frequency.
Figure 2 shows three-pulse ESEEM data for all four successive positions (i + 1 to i + 4) of the M2δ peptide. 2H modulation is observed in the time domain and a peak centered at the 2H Larmor frequency for the i + 3 and i + 4 M2δ 2H-labeled Val 15 samples, and not for the i + 1 and i + 2 positions.
Figure 2.
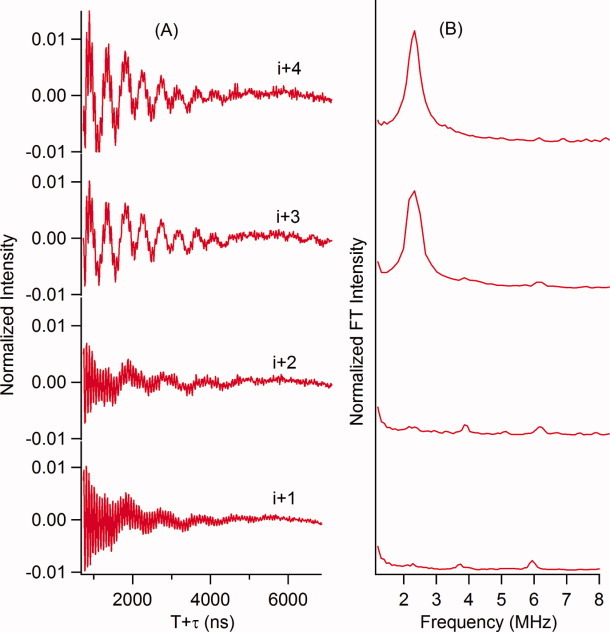
Three-pulsed ESEEM experimental data with a τ = 200 ns of the i + 1 through the i + 4 2H-labeled Val15 M2δ lipid bilayer samples. (A) Time domain and (B) frequency domain.
Molecular modeling and molecular dynamics studies were conducted on the α-helical M2δ peptide to estimate distance ranges between the N–O bond on the SL and 2H-labeled Val nuclei at the i + 1, i + 2, i + 3, and i + 4 positions.22,23 2H-SL distance ranges for the i + 1 (8–12 Å) and i + 2 (11–15 Å) positions were found to be outside the ESEEM detection range of ∼8 Å. However, 2H-SL distances for the i + 3 (7–11 Å) and i + 4 (7–11 Å) positions were found within the ESEEM detection range.
Discussion
Typically, ESEEM data can be simulated and theoretically give a distance between a 2H label and the MTSL spin label. In the current set of experiments, however, it is challenging to glean a single 2H-SL distance. Broad distributions of distances are observed due to a large number of possible orientations between the SL and multiple 2H nuclei on the Val sidechain. The corresponding distance distribution is due largely to many allowed orientations of the MTSL and the 2H-labled valine residue. MTSL is known to exhibit three torsional angle rotations about the χ1, χ2, and χ3 closest to the Cα. Additionally, two free torsional angle rotations are present about the χ4, χ5 angles.24 In the case of valine, two additional rotations are present. One mode of rotation is about the two Cα–Cβ χ1 torsional angles, which correspond to the rotation of the (CD3) methyl groups. The second mode of rotation is about the χ2 torsional angle.25,26 As the sample is frozen at 80 K for the ESEEM measurements, the eight 2H nuclei on the Val side chain are distributed over a range of distances with respect to the SL. The broad distance ranges makes it difficult to measure a single 2H distance via ESEEM simulation with only the modulation depth.14,27 The 2H modulation observed in the ESEEM spectra are most likely dominated by the shorter 2H-SL distances (< 8 Å) and the longer range distances are not easily detected (modulation depth scales as 1/r6). The 2H modulation and 2H peak intensity in Figure 2 are slightly larger for the i + 4 position when compared to the i + 3. This matches well with MD simulations on the M2δ peptide, which reveal a slight increase in the population of 2H-SL distances below the 8 Å ESEEM detection limit. For the i + 3 and i + 4 positions of a helical structure, the modulation depth can vary slightly depending on the local environment and conformation of the SL, dynamic properties of the SL and side chain, and type of 2H-labeled side chain.
The application of SDSL ESEEM to probe the secondary structural properties of membrane proteins is comparable to solid-state NMR spectroscopic techniques.20,28,29 The rotational echo double resonance (REDOR) NMR technique measures dipolar couplings between NMR active nuclei, such as 13C and 15N.29–31 REDOR solid-state NMR spectra coupled with spectral simulations can be used to probe the α-helical content of 13C/15N-labeled membrane proteins and peptides. These experiments require mg quantities of isotopically labeled protein and a significant amount of instrument time (multiple days and up to weeks) for sufficient signal-to-noise (S/N). In sharp contrast, the ESEEM spectra in this work yielded high-quality data in less than an hour with as little as 35 μg of protein sample.
This efficient ESEEM spectroscopic technique does not provide the same high-resolution structural information obtained from NMR spectroscopy or X-ray crystallography but can provide very important qualitative secondary structural information on membrane proteins systems or other biological systems of unknown structure. For SDSL EPR researchers, this approach will provide additional tools to probe the structures of biological systems. The MD studies are consistent with the presence and absence of 2H modulation for all positions in Figure 2. Similar modeling studies were conducted for a β-strand. 2H-SL distance ranges for the i + 1 (6–11 Å) and i + 2 (7–12 Å) positions were found to be within the ESEEM detection range, and outside the 2H-SL ESEEM detection range for the i + 3 (9–14 Å) and i + 4 (14–19 Å) positions. Random coil distances were found to be similar to the β-strand but varied depending on the structure. Thus, this ESEEM approach should be valuable for distinguishing between an α-helix and a β-strand. 2H modulation would be detected at the i + 3 and i + 4 positions for an α-helix, whereas modulation would not be detected at the i + 1 and i + 2 positions. For a β-strand, 2H modulation would be detected at the opposite positions i + 1 and i + 2 and not at the i + 3 and i + 4 positions. This work explores the helical content of the M2δ peptide with ESEEM spectroscopy. Future ESEEM experiments using this approach will study the structure of a β-strand and additional 2H sidechain labels.
For the first time, this work illustrates that the pulsed EPR ESEEM technique can be used in a manner to probe the secondary structural properties of membrane peptides and proteins. ESEEM spectroscopy can be used to both confirm the presence of an α-helix and provide pertinent inter side chain structural information. One of the greatest advantages of this technique is the short data acquisition time and small amount of protein needed to obtain pertinent structural information of an inherently difficult system like membrane proteins. This ESEEM technique also lends itself to probing the structural properties of larger membrane proteins and water-soluble proteins using an over expression system coupled with deuterium-labeled amino acids introduced into a minimal media. There are no size restrictions using this novel ESEEM approach, when compared to analogous NMR experiments.
Materials and Methods
The M2δ peptides were synthesized on a CEM microwave solid phase synthesizer using Fmoc-chemistry. The peptides were cleaved from the solid support and purified via reverse phase high performance liquid chromatography (HPLC) as described previously.30 After purification, the peptides were labeled with MTSL overnight at room temperature and repurified via HPLC using the same purification conditions.30,31 The purity of the peptides was confirmed to be over 95% pure by mass spectrometry. The peptides were incorporated into DMPC/DHPC (3.5/1) lipid bicelles.30 Bicelles were used in these experiments, because they serve as an excellent membrane mimic that yield high-quality pulsed EPR data. Comparable data could be obtained with proteoliposomes using a larger multimembrane spanning α-helix membrane protein (data not shown).
CW-EPR X-Band (∼9 GHz) spectroscopy was performed to check final spin concentration (∼200 μM) by double integration. A Bruker ELEXSYS E580 was used to collect all three-pulse ESEEM data using τ values of 120 and 200 ns. A standard Bruker X-band MS3 split-ring resonator was used for the pulsed experiments. All samples were performed under the same experimental parameters at 80 K with a microwave frequency of ∼9.269 GHz for all of the samples with a starting T of 300 ns for the experiments with τ = 120 ns, a starting T of 532 ns for experiments with τ = 200 ns and 512 points in 12 ns increments. Both τ values provided high-quality time-domain and FT ESEEM data. However, 2H modulation was optimized, and the proton modulation was effectively suppressed with τ equal to 200 ns, when compared to the τ 120 ns data. The optimal τ values can vary depending on the field and frequency that the data was collected at. The ESEEM data were fit to an exponential decay curve, which was normalized to 1, according to the literature.14 The exponential fit was then subtracted from the experimental spectrum as shown in Figures 1 and 2 before cross-term averaged Fourier Transformation.32
The molecular modeling was performed using nanoscale molecular dynamics (NAMD) with the molecular graphics software VMD.22,23 The structure of AchR M2δ peptide was obtained from the solution NMR coordinates (PDB entry: 1EQ8). The Cys mutants were created at i + 1, i + 2, i + 3, and i + 4 positions using VMD, a MTSSL nitroxide spin-robe attached by using CHARMM force-field topology files incorporated in NAMD, where, i represents the 15th residue, on the peptide. The molecular dynamics simulations were collected out to 100 ps at room temperature using Langevin dynamics under NAMD. The trajectory data for every 1000 fs were recorded. The possible distance distribution for each deuterium and SL was obtained from the analysis of the trajectory data file using VMD.
Acknowledgments
The authors greatly thank Dr. Kurt Warncke and his research group for helpful discussions and support with the ESEEM simulations. The authors also thank Drs. Jimmy Feix and Ralph Weber (Bruker BioSpin) for consultation about spin labeling and ESEEM measurements.
Glossary
Abbreviations
- ACHR
acetylcholine receptor
- ESEEM
electron spin echo envelope modulation
References
- 1.McLuskey K, Roszak AW, Zhu Y, Isaacs NW. Crystal structures of all-alpha type membrane proteins. Eur Biophys J Biophy. 2010;39:723–755. doi: 10.1007/s00249-009-0546-6. [DOI] [PubMed] [Google Scholar]
- 2.Bordag N, Keller S. alpha-Helical transmembrane peptides: A “Divide and Conquer” approach to membrane proteins. Chem Phys Lipids. 2010;163:1–26. doi: 10.1016/j.chemphyslip.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Huang C, Mohanty S. Challenging the limit: NMR assignment of a 31 kDa helical membrane protein. J Am Chem Soc. 2010;132:3662–3663. doi: 10.1021/ja100078z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander N, Bortolus M, Al-Mestarihi A, Mchaourab H, Meilerl J. De novo high-resolution protein structure determination from sparse spin-labeling EPR data. Structure. 2008;16:181–195. doi: 10.1016/j.str.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klug CS, Feix JB. Methods and applications of site-directed spin labeling EPR spectroscopy. Method Cell Biol. 2008;84:617–658. doi: 10.1016/S0091-679X(07)84020-9. [DOI] [PubMed] [Google Scholar]
- 6.Zou P, Mchaourab HS. Increased sensitivity and extended range of distance measurements in spin-labeled membrane proteins: Q-band double electron-electron resonance and nanoscale bilayers. Biophys J. 2010;98:L18–L20. doi: 10.1016/j.bpj.2009.12.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeschke G. Determination of the nanostructure of polymer materials by electron paramagnetic resonance spectroscopy. Macromol Rapid Commun. 2002;23:227–246. [Google Scholar]
- 8.Lakshmi KV, Brudvig GW. Pulsed electron paramagnetic resonance methods for macromolecular structure determination. Curr Opin Struc Biol. 2001;11:523–531. doi: 10.1016/s0959-440x(00)00242-6. [DOI] [PubMed] [Google Scholar]
- 9.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Milov AD, Samoliov RI, Shubin AA, Gorbunova EY, Mustaeva LG, Ovchinnikova TV, Raap J, Tsvetkov YD. Self-aggregation and orientation of the ion channel-forming zervamicin IIA in the membranes of ePC vesicles studied by cw EPR and ESEEM spectroscopy. Appl Magn Reson. 2010;38:75–84. [Google Scholar]
- 11.Lorigan GA, Britt RD, Kim JH, Hille R. Electron spin echo envelope modulation spectroscopy of the molybdenum center of xanthine oxidase. BBA-Bioenergetics. 1994;1185:284–294. doi: 10.1016/0005-2728(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 12.Force DA, Randall DW, Lorigan GA, Clemens KL, Britt RD. ESEEM studies of alcohol binding to the manganese cluster of the oxygen evolving complex of Photosystem II. J Am Chem Soc. 1998;120:13321–13333. [Google Scholar]
- 13.Mims WB. Envelope modulation in spin-echo experiments. Phys Rev B. 1972;5:2409–2419. [Google Scholar]
- 14.Heming M, Narayana M, Kevan L. Analysis of nuclear quadrupole interaction effects in electron spin-echo modulation spectra by second-order perturbation methods. J Chem Phys. 1985;83:1478–1484. [Google Scholar]
- 15.Kevan L, Schwartz RN. Time domain electron spin resonance. New York: John Wiley and Sons; 1979. [Google Scholar]
- 16.Shubin AA, Dikanov SA. Numerical analysis of the influence of nuclear quadrupole interaction on modulation effects in electron spin echo from deuterium nuclei in disordered systems. J Magn Reson. 1985;64:185–193. [Google Scholar]
- 17.Cieslak JA, Focia PJ, Gross A. Electron spin-echo envelope modulation (ESEEM) reveals water and phosphate interactions with the KcsA potassium channel. Biochemistry. 2010;49:1486–1494. doi: 10.1021/bi9016523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romanelli M, Ristori S, Martini G, Kang YS, Kevan L. Electron spin echo study of doxyl spin probes in micellar systems of ammonium perfluorooctanoate. J Phys Chem. 1994;98:2125–2128. [Google Scholar]
- 19.Kim J, McNamee MG. Topological disposition of Cys 222 in the alpha-subunit of nicotinic acetylcholine receptor analyzed by fluorescence-quenching and electron paramagnetic resonance measurements. Biochemistry. 1998;37:4680–4686. doi: 10.1021/bi972666k. [DOI] [PubMed] [Google Scholar]
- 20.Inbaraj JJ, Cardon TB, Laryukhin M, Grosser SM, Lorigan GA. Determining the topology of integral membrane peptides using EPR spectroscopy. J Am Chem Soc. 2006;128:9549–9554. doi: 10.1021/ja0622204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carmieli R, Papo N, Zimmermann H, Potapov A, Shai Y, Goldfarb D. Utilizing ESEEM spectroscopy to locate the position of specific regions of membrane-active peptides within model membranes. Biophys J. 2006;90:492–505. doi: 10.1529/biophysj.105.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 23.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Compt Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fanucci G, Cafiso D. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:1–10. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Kinsey RA, Kintanar A, Tsai MD, Smith RL, Janes N, Oldfield EA. First observation of amino acid side chain dynamics in membrane proteins using high field deuterium nuclear magnetic resonance spectroscopy. J Biol Chem. 1981;256:4146–4149. [PubMed] [Google Scholar]
- 26.Liu W, Crocker E, Siminovitch DJ, Smith SO. Role of side-chain conformational entropy in transmembrane helix dimerization of glycophorin A. Biophys J. 2003;84:1263–1271. doi: 10.1016/S0006-3495(03)74941-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun L, Hernandez-Guzman J, Warncke K. OPTESIM, a versatile toolbox for numerical simulation of electron spin echo envelope modulation (ESEEM) that features hybrid optimization and statistical assessment of parameters. J Magn Reson. 2009;200:21–28. doi: 10.1016/j.jmr.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howell SC, Mesleh MF, Opella SJ. NMR structure determination of a membrane protein with two transmembrane helices in micelles: MerF of the bacterial mercury detoxification system. Biochemistry. 2005;44:5196–5206. doi: 10.1021/bi048095v. [DOI] [PubMed] [Google Scholar]
- 29.Chu S, Coey AT, Lorigan GA. Solid-state H-2 and N-15 NMR studies of side-chain and backbone dynamics of phospholamban in lipid bilayers: investigation of the N27A mutation. BBA-Biomembranes. 2010;1798:210–215. doi: 10.1016/j.bbamem.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayo DJ, Inbaraj JJ, Subbaraman N, Grosser SM, Chan CA, Lorigan GA. Comparing the structural topology of integral and peripheral membrane proteins utilizing electron paramagnetic resonance spectroscopy. J Am Chem Soc. 2008;130:9656–9657. doi: 10.1021/ja803590w. [DOI] [PubMed] [Google Scholar]
- 31.Bhargava K, Feix JB. Membrane binding, structure, and localization of cecropin-mellitin hybrid peptides: a site-directed spin-labeling study. Biophys J. 2004;86:329–336. doi: 10.1016/S0006-3495(04)74108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoll S, Britt RD. General and efficient simulation of pulse EPR spectra. Phys Chem. 2009;11:6614–6625. doi: 10.1039/b907277b. [DOI] [PubMed] [Google Scholar]


