Abstract
Post-transcriptional modifications of bases within the transfer RNAs (tRNA) anticodon significantly affect the decoding system. In bacteria and eukaryotes, uridines at the wobble position (U34) of some tRNAs are modified to 5-methyluridine derivatives (xm5U). These xm5U34-containing tRNAs read codons ending with A or G, whereas tRNAs with the unmodified U34 are able to read all four synonymous codons of a family box. In Escherichia coli (E.coli), the bifunctional enzyme MnmC catalyzes the two consecutive reactions that convert 5-carboxymethylaminomethyl uridine (cmnm5U) to 5-methylaminomethyl uridine (mnm5U). The C-terminal domain of MnmC (MnmC1) is responsible for the flavin adenine dinucleotide (FAD)-dependent deacetylation of cmnm5U to 5-aminomethyl uridine (nm5U), whereas the N-terminal domain (MnmC2) catalyzes the subsequent S-adenosyl-L-methionine-dependent methylation of nm5U, leading to the final product, mnm5U34. Here, we determined the crystal structure of E.coli MnmC containing FAD, at 3.0 Å resolution. The structure of the MnmC1 domain can be classified in the FAD-dependent glutathione reductase 2 structural family, including the glycine oxidase ThiO, whereas the MnmC2 domain adopts the canonical class I methyltransferase fold. A structural comparison with ThiO revealed the residues that may be involved in cmnm5U recognition, supporting previous mutational analyses. The catalytic sites of the two reactions are both surrounded by conserved basic residues for possible anticodon binding, and are located far away from each other, on opposite sides of the protein. These results suggest that, although the MnmC1 and MnmC2 domains are physically linked, they could catalyze the two consecutive reactions in a rather independent manner.
Keywords: tRNA modification enzyme, anticodon, wobble uridine, genetic code, two-codon set, decoding system, methyltransferase, FAD-dependent oxidoreductase, crystal structure
Introduction
Transfer RNAs (tRNAs) are heavily modified post-transcriptionally in all three domains of life. These modifications play critical roles for the fine-tuning of tRNA functions.1 Especially, the modification of the wobble position of the tRNA anticodon is known to affect the decoding system.2
The codon triplet in mRNA is decoded by the three anticodon bases in tRNA, at positions 34, 35, and 36. The interaction between the wobble position of tRNA (position 34) and the third base of the codon is relaxed, which enables tRNA to read more than one synonymous codon. The modification of the wobble base is used to either restrict or expand tRNA recognition in the decoding system.3–5 The uridine at the wobble position (U34) is often modified to 5-methyluridine derivatives (xm5U) in bacteria and eukaryotes, and the tRNA with xm5U34 is restricted to read the codons with A or G at the third position of the two-codon sets.2,3,5 In contrast, the tRNA with unmodified U34 can read all four synonymous codons of a family box in the genetic code.6–8
In Escherichia coli (E. coli), the C5 atom of uracil-34 is modified by a 5-methylaminomethyl group (mnm5U) in tRNAGln, tRNALys, tRNAGlu, tRNAArg, and tRNAGly.9–12 This modification is considered to be involved in restricting the codon recognition.13–16 The biosynthetic pathway leading to the addition of the methylaminomethyl group (mnm) on the C5 atom of uracil involves three consecutive steps [Fig. 1(A)]. The first step, performed by MnmE and GidA, forms 5-carboxymethylaminomethyl uridine (cmnm5U).12,17–21 The 5-cmnm group is then deacetylated to a 5-aminomethyl group, and subsequently methylated into the final product, methylaminomethyl uridine (mnm5U). These two last steps are performed by the bifunctional enzyme, MnmC,10,21–23 apparently without the accumulation of the intermediate product, aminomethyl uridine (nm5U).24 In the unique case of E. coli tRNALeu-4, the C5-atom of U34 remains hypomodified (cmnm5U34).25 Thiolation of the 2-position of uracil-34 (s2U, leading to mnm5s2U) is accomplished independently only in tRNA Gln, tRNALys, and tRNAGlu by several other proteins,26,27 whereas in tRNALeu-4, the 2′-hydroxyl group of the U-ribose is independently methylated (Um, leading to cmnm5Um)25 by the methyltransferase (MTase) TrmL.28
Figure 1.
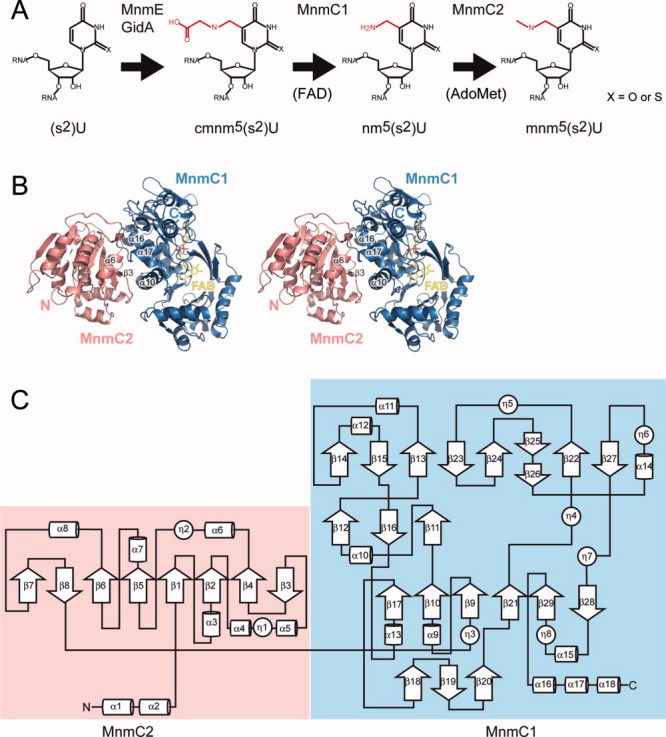
(A) Schematic representation of mnm5U biosynthesis in tRNA. X = O (in U) or S (in s2U). Modified positions are colored red; (B) Overall structure of MnmC with the FAD cofactor. The N-terminal domain (MnmC2) is colored salmon, and the C-terminal domain (MnmC1) is colored sky blue; and (C) Topology diagram of MnmC with secondary structure elements.
The deacetylation and subsequent methylation of the 5-cmnm group into the 5-mnm group of U34 [Fig. 1(A)] are each performed independently by distinct domains of MnmC.23 The C-terminal domain is responsible for the demodification reaction (MnmC1), and converts cmnm5U to nm5U in a flavin adenine dinucleotide (FAD)-dependent manner. The N-terminal domain catalyzes the methylation reaction (MnmC2) and changes nm5U to the final product mnm5U, using S-adenosyl-L-methionine (AdoMet). The sequence analysis revealed that the N-terminal MnmC2 domain belongs to the AdoMet-dependent class I MTase family, whereas the C-terminal MnmC1 domain is closely related to the FAD-dependent glycine/D-amino acid oxidases,22,23 within the glutathione reductase 2 (GR2) family.29
Several previous studies have examined the molecular functions and the enzymatic and biochemical properties of the individual domains of MnmC. However, little is known about how these fused domains cooperate in catalyzing the two consecutive reactions. Here, we report the crystal structure of MnmC bound with FAD, which provides new insights into its substrate recognition and catalytic mechanism.
Results
Overall structure
The crystal structure of MnmC consists of two globular domains [Fig. 1(B)]. The MnmC2 domain (residues 1-254 – E. coli numbering) [Figs. 1(C) and 2], which is responsible for the AdoMet-dependent methylation of the intermediate nm5U34, has a central region with the canonical secondary structure seen in the class I MTases.30 This domain is characterized by a seven-stranded β-sheet (β1-β6 and β8) sandwiched by two α-bundles (α3-α5 and α6-α8), and in MnmC it is extended by additional α-helices (α1 and α2 at the N-terminus and another β-strand (β7) in the central β-sheet [Fig. 1(C)].
Figure 2.
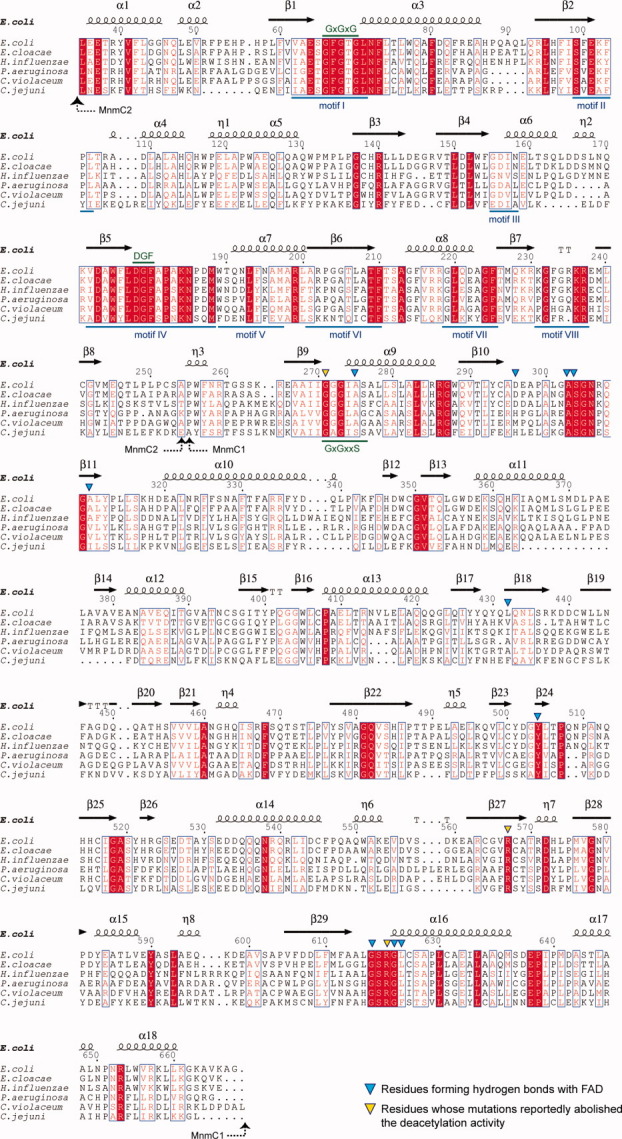
Sequence alignment of the MnmC family, created by using ESPript.31 The domain structures are indicated by dashed arrows. Numbering is adopted from the E. coli MnmC sequence. Conserved residues are shown in red. The highly conserved residues are shown in white within red-filled rectangles. The conserved motifs in the class I MTases are indicated by blue lines. The signature motif, GxGxG in motif I, the DGF sequence in motif IV, and the conserved GxGxxS motif in MnmC1 are indicated by green lines. Residues forming hydrogen bonds with FAD are indicated by blue triangles. Residues that interfere with the oxidation reaction23 are indicated by yellow triangles. GI numbers are given in parentheses: E. coli str. K-12 substr. W3110 (89109144), Enterobacter cloacae subsp. cloacae ATCC 13047 (296104010), Haemophilus influenzae 86-028NP (68249949), Pseudomonas aeruginosa PAO1 (15598652), Chromobacterium violaceum ATCC 12472 (34497397), Campylobacter jejuni subsp. jejuni NCTC 11168 (218562880).
As predicted from the sequence analysis,23 the structure of the MnmC1 domain (residues 255-668) [Figs. 1(C) and 2], which is responsible for the FAD-dependent deacetylation, bears a fold characteristic of the GR2 family.29 The general architecture consists of four β-sheets: a three-stranded antiparallel β-sheet (β18–β20), a six-stranded β-sheet (β17, β10, β9, β21, β29, and β28) flanked by an α-helix bundle (α9, α10, α13, and α15), and a three-stranded antiparallel β-sheet (β12, β16, and β11). In addition, the substrate binding domain consists of a mixed eight-stranded β-sheet (β14, β15, β13, β23, β24, β25-26, β22, and β27) flanked by the other α-bundle (α11, α12, and α14). The MnmC1 domain terminates with three α-helices (α16–α18) [Fig. 1(C)].
The overall conformation of the two domains is essentially identical among the six molecules in the asymmetric unit. The two domains interact with each other through β3 and α6 in the MnmC2 domain, and α10 α16, and α17 in the MnmC1 domain [Fig. 1(B)]. The interface between the two domains contains numerous hydrophilic residues.
N-terminal MnmC2 Domain
The structure of the MnmC2 domain is highly similar to that of a protein with unknown function (DUF752 from Aquifex aeolicus, PDBID=2E58) bound with AdoMet (RMSD of 1.73 Å for 177 Cα atoms). The structure revealed a cleft around the prospective AdoMet binding site (Fig. 3). A number of conserved basic residues surround the cleft, which is large enough to accommodate the anticodon loop of a tRNA (data not shown). To understand how the E. coli MnmC2 domain binds AdoMet, we superposed its structure with that of DUF752 [Fig. 4(A)]. The class I MTase contains the highly conserved GxGxG sequence (from Gly66 to Gly70 in MnmC), within the loop in motif I (Fig. 2).30 The superposed structures indicated that this motif interacts with the carboxypropyl moiety of AdoMet [Fig. 4(A)]. The mutation of the highly conserved Glu64 reportedly interfered with the AdoMet binding.23 The superposed model suggests that Glu64 may interact with the amino group in the methionine moiety of AdoMet. Many class I MTases use the conserved DPPY sequence in motif IV as a common substrate binding motif.30 However, the N-terminal domain of MnmC instead has the DGF sequence at the corresponding region. The first (Asp178) and third (Phe180) residues in this sequence are strictly conserved in the MnmC family, and the D178A and F180A substitutions reportedly abolished the methylation activity.23 In the structure of DUF752, Asp193 (corresponding to Asp178 of E. coli MnmC) interacts with the amino group of AdoMet, whereas Phe195 (corresponding to Phe180) forms hydrophobic interactions with both the ribose and donor methyl group.
Figure 3.
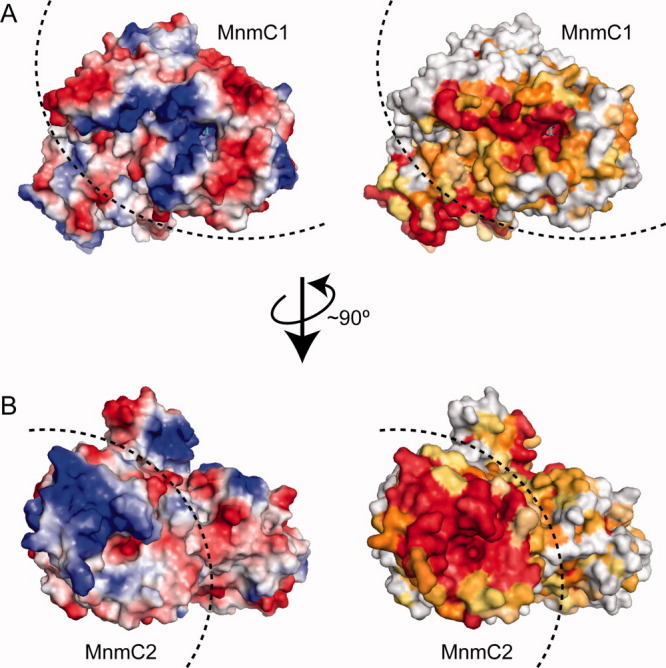
Surface representations of MnmC, from the FAD binding pocket side (A) and the AdoMet binding pocket side (B). Left: electrostatic surface potential representation, in which blue indicates positive charges and red indicates negative charges. Right: surface representation colored by conservation rate. The conservation rate was calculated using the ConSurf server.32–34 The highly conserved residues are colored red and the variable residues are colored white.
Figure 4.
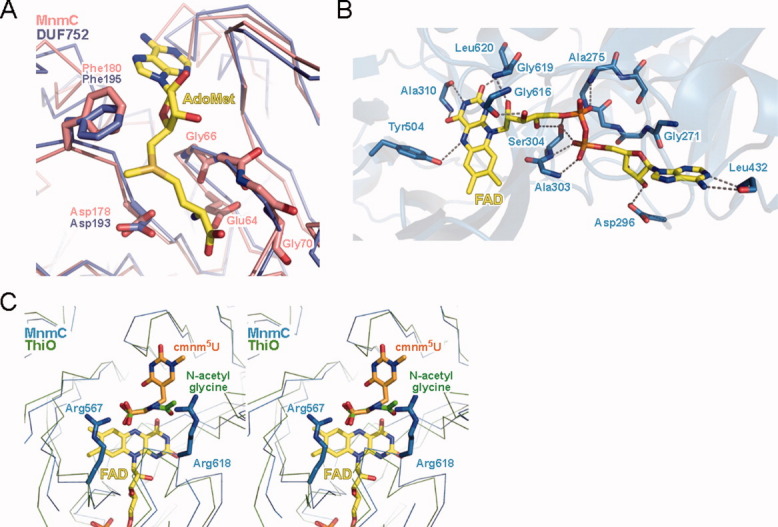
(A) Putative AdoMet binding site. The superposed structures of the MnmC2 domain (salmon) and DUF752 (blue) are shown in a Cα trace representation. The AdoMet molecule bound to DUF752 is shown in a stick representation; (B) FAD binding. The hydrogen bonds between the MnmC1 domain (sky blue) and FAD are indicated by dotted lines; and (C) cmnm5U binding model. The superposed structures of the MnmC1 domain (sky blue) and ThiO (green) are shown in a Cα trace representation.
C-terminal MnmC1 Domain
A DALI search revealed that the MnmC1 domain is homologous to the glycine oxidase ThiO [PDBID = 1NG3, Z score = 40.5] and the monomeric sarcosine oxidase [PDBID = 3M13, Z score = 39.7].35 The ThiO-containing structures that are homologous to MnmC1 are classified in the GR2 family. The substrates of ThiO and MnmC1 have similar chemical structures (glycine and carboxymethyl-amino group, respectively), and the reaction mechanism of MnmC1 was proposed based on that of ThiO.22
The MnmC1 domain bears some typical features of the GR2 family. The GR2 family contains the conserved GxGxxG motif, located in the loop connecting β9 and α9 in the Rossmann fold (Fig. 2), which functions to compensate for the charge of the pyrophosphate moiety of FAD.29 In the structure of MnmC, the corresponding GGGIAS (GxGxxS) sequence (from Gly271 to Ser276) is located adjacent to the pyrophosphate moiety of FAD, which is recognized by the amino groups of Ala275, Ala303, and Ser304, as well as the side chain of Ser304 [Fig. 4(B)]. The G271Q mutation reportedly decreases the oxidation activity, but does not affect the FAD-binding.23 The crystal structure revealed that Gly271 is located close to the adenosine moiety of FAD [Fig. 4(B)]. Thus, the substitution of the bulky glutamine for the glycine may disrupt the FAD binding site, thus interfering with the proper arrangement of the catalytic groups required for the reaction.
The flavin ring of FAD is recognized by hydrogen bonds with the backbones of Ala310 and Leu620, and the side chain of Tyr504. The three hydroxyl groups of the ribitol moiety interact with the backbones of Gly616 and Gly619, and the Ser304 side chain. The adenine base and the ribose 2′-OH group are recognized by Leu432 and Asp296, respectively [Fig. 4(B)].
Like the MnmC2 domain, the cavity around the FAD binding site of the MnmC1 domain also contains a number of conserved basic residues (Fig. 3), and its structure is complementary to that of the tRNA nucleotides, indicating the presence of the tRNA anticodon loop binding site.
Discussion
Substrate binding mechanism of MnmC1
The mutational analysis of MnmC1 revealed that the R567A mutation decreased the FAD-dependent oxidation, but did not interfere with the FAD binding (Fig. 2).23 This well conserved arginine residue is located in the vicinity of the flavin ring. Figure 4(C) shows the superposition model of MnmC1 and ThiO. The cmnm5U molecule is oriented so that the atoms of the terminal glycine moiety of its 5-cmnm group are superposed on the carboxyl carbon, Cα, and nitrogen atoms, respectively, of N-acetylglycine (the substrate analog) in the structure of ThiO. In the model, Arg567 is located near the terminal carboxyl group of cmnm5U, indicating that this residue is involved in the recognition of cmnm5U and the discrimination of mnm5U. The R618A substitution, which abolished the activity of MnmC, also resides near the FAD-binding site, suggesting that it may be involved in binding the anticodon loop of tRNA.
How are the two reactions coordinated?
tRNAs containing nm5(s2)U, the intermediate of the reactions catalyzed by MnmC, have never been detected in naturally occurring E. coli tRNAs.2,10 The nm5U-containing tRNAs can be produced only in vitro, when the reaction is conducted in the absence of AdoMet,24 or under experimental conditions favoring the incorporation of ammonium ions, instead of glycine, catalyzed by the GidA-MnmE complex.20 Obviously, the two consecutive MnmC- dependent enzymatic reactions are optimized in vivo, and thus no accumulation of the nm5U-containing tRNA intermediate is evident. One explanation could be that the two reactions occur without the release of the tRNA substrate from the enzyme. In this scenario, once bound to the enzyme, the portion of the tRNA anticodon containing nm5U34, produced by MnmC1, may simply flip (or channel) into the active site of MnmC2, where the free amino group of nm5U34 is rapidly methylated into mnm5U34. However, the present structure of bifunctional MnmC shows that the two catalytic cavities of the MnmC1 and MnmC2 domains face opposite sides of the protein (Fig. 3). We also observed the absence of extensive packing interactions between the MnmC monomers in the crystal. Instead, these observations favor an alternative reaction mechanism, in which the product of the first reaction is physically released from the MnmC1 domain before rebinding to the MnmC2 domain. The recent observation that purified recombinant E. coli bifunctional MnmC binds an nm5U-containing tRNA substrate more tightly than the cmnm5U-containing substrate (by about 10-fold)24 can partly explain the high efficiency of such an independent, two-step mechanism (‘assembly line’). Notably, when the two MnmC mutants possessing the catalytically dead MnmC2 or MnmC1 domain were mixed, partial recovery of the activity to produce mnm5U was observed, demonstrating that, at least in vitro, the two domains can indeed act independently.23
However, given the relatively hydrophilic nature of the interface between the two domains (this work), one cannot exclude the possibility that significant conformational changes within the bifunctional MnmC may also occur upon tRNA substrate binding and/or during the successive enzymatic reaction steps. A definitive understanding of MnmC's reaction mechanisms awaits the structural determination of its tRNA complex in multiple forms, where the wobble U34 of the anticodon loop is accommodated separately in the MnmC1 and MnmC2 domains. Finally, it is important to note that cmnm5U34 in naturally occurring E. coli tRNALeu-4 is never modified into mnm5U34, attesting to the fact that this particular tRNA does not interact with MnmC, probably due to the presence of the long-variable arm.
Materials and Methods
Purification and crystallization
The gene encoding MnmC (JW5380) from E. coli was cloned into pET-15b (Novagen) and expressed in the E. coli Rosetta2(DE3) strain (Novagen). The selenomethionine-labeled MnmC was expressed from the cloning vector pET-11b in E. coli strain B834(DE3) (Novagen). The cells were harvested and disrupted by sonication in 20 mM Tris-HCl buffer (pH 8.0), containing 300 mM NaCl and 2 mM DTT. The lysate was cleared by centrifugation for 30 min at 100,000 g. The supernatant was purified by a series of HiTrapQ, ResourceISO, MonoQ, and Superdex75 column chromatography steps (GE Healthcare Biosciences).
The crystals of MnmC were grown using the sitting-drop method. The best crystallization conditions employed a reservoir solution containing 100 mM Bis-Tris buffer (pH 5.5), containing 25% (w/v) PEG3350, 250 mM ammonium sulfate, and 10 mM hexamine cobalt(III) chloride. Diffraction quality crystals were obtained within 5 days.
X-ray data collection and structure determination
X-ray diffraction data were collected at the Photon Factory BL5A beamline and processed with the HKL2000 program suite. The structure was solved by the single wavelength anomalous dispersion method, using the selenomethionine-labeled crystal dataset. Selenium sites were located by using the program SHELX,36 and then used to calculate the initial phases with the program Phaser.37 Most of the model was automatically created by the program Phenix,38 and the structure was then manually corrected and refined by using the programs Coot39 and Phenix. Figures were prepared with the Pymol program (Schrödinger, LLC.). All data collection, phasing and refinement statistics are summarized in Table I. The PDB code is 3AWI.
Table I.
Data Collection, Phasing, and Refinement Statistics
| SeMet | Native | |
|---|---|---|
| Data collection | ||
| Cell dimensions (Å) | 92.1, 243.4, 176.4 | 92.1, 243.0, 175.5 |
| Wavelength (Å) | 0.97903 | 1 |
| Resolution (Å) | 50.0–3.00 (3.11–3.00)* | 50.0–3.00 (3.11–3.00)* |
| Rsym | 0.186 (0.870)* | 0.102 (0.336)* |
| Mean I/σ | 9.6 (1.5)* | 10.3 (3.1)* |
| Completeness (%) | 96.9 (94.7)* | 98.3 (94.0)* |
| Redundancy | 6.6 (5.6)* | 4.1 (4.0)* |
| Refinement | ||
| Resolution (Å) | 50–3.00 | |
| No. reflections | 146580 | |
| Rwork/Rfree | 0.205/0.251 | |
| Number of atoms | ||
| Protein/ligand/solvent | 29853/318/61 | |
| Average B-factors | ||
| Protein/ligand/solvent | 87.8/84.4/80.5 | |
| Ramachandran most favored/allowed (%) | 95.8/4.1 | |
| Rmsd bond length (Å)/angles (°) | 0.010/1.026 | |
Values in parentheses are for highest-resolution shell.
Acknowledgments
We thank Emiko Fusatomi and Rie Shibata for technical assistance during purification and crystallization. We also thank the staff of the beamline BL5A at the Photon Factory (Tsukuba, Japan) for their kind help in data collection. We are grateful to Drs. Henri Grosjean and Damien Brégeon for rewarding discussions on the modified nucleotides in tRNA.
References
- 1.Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 2.Björk GR, Hagervall TG, et al. Transfer RNA modification. In: Böck A, Curtis R III, Kaper JB, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington DC: ASM; 2008. Chap 4.6.2. [Google Scholar]
- 3.Grosjean H, de Crécy-Lagard V, Marck C. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010;584:252–264. doi: 10.1016/j.febslet.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 4.Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takai K, Yokoyama S. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 2003;31:6383–6391. doi: 10.1093/nar/gkg839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inagaki Y, Kojima A, Bessho Y, Hori H, Ohama T, Osawa S. Translation of synonymous codons in family boxes by Mycoplasma capricolum tRNAs with unmodified uridine or adenosine at the first anticodon position. J Mol Biol. 1995;251:486–492. doi: 10.1006/jmbi.1995.0450. [DOI] [PubMed] [Google Scholar]
- 7.de Crécy-Lagard V, Marck C, Brochier-Armanet C, Grosjean H. Comparative RNomics and modomics in Mollicutes: prediction of gene function and evolutionary implications. IUBMB Life. 2007;59:634–658. doi: 10.1080/15216540701604632. [DOI] [PubMed] [Google Scholar]
- 8.Ohama T, Inagaki Y, Bessho Y, Osawa S. Evolving genetic code. Proc Jpn Acad Ser B Phys Biol Sci. 2008;84:58–74. doi: 10.2183/pjab.84.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elseviers D, Petrullo LA, Gallagher PJ. Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 1984;12:3521–3534. doi: 10.1093/nar/12.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagervall TG, Edmonds CG, McCloskey JA, Björk GR. Transfer RNA(5-methylaminomethyl-2-thiouridine)-methyltransferase from Escherichia coli K-12 has two enzymatic activities. J Biol Chem. 1987;262:8488–8495. [PubMed] [Google Scholar]
- 11.Sakamoto K, Kawai G, Niimi T, Satoh T, Sekine M, Yamaizumi Z, Nishimura S, Miyazawa T, Yokoyama S. A modified uridine in the first position of the anticodon of a minor species of arginine tRNA, the argU gene product, from Escherichia coli. Eur J Biochem. 1993;216:369–375. doi: 10.1111/j.1432-1033.1993.tb18154.x. [DOI] [PubMed] [Google Scholar]
- 12.Yim L, Moukadiri I, Björk GR, Armengod ME. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli. Nucleic Acids Res. 2006;34:5892–5905. doi: 10.1093/nar/gkl752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama S, Watanabe T, Murao K, Ishikura H, Yamaizumi Z, Nishimura S, Miyazawa T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc Natl Acad Sci U S A. 1985;82:4905–4909. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sierzputowska-Gracz H, Sochacka E, Malkiewicz A, Kuo K, Gehrke CW, Agris PF. Chemistry and structure of modified uridines in the anticodon, wobble position of transfer RNA are determined by thiolation. J Am Chem Soc. 1987;109:7171–7177. [Google Scholar]
- 15.Krüger MK, Pedersen S, Hagervall TG, Sørensen MA. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J Mol Biol. 1998;284:621–631. doi: 10.1006/jmbi.1998.2196. [DOI] [PubMed] [Google Scholar]
- 16.Murphy FVt, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- 17.Brégeon D, Colot V, Radman M, Taddei F. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 2001;15:2295–2306. doi: 10.1101/gad.207701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer S, Wittinghofer A, Versées W. G-domain dimerization orchestrates the tRNA wobble modification reaction in the MnmE/GidA complex. J Mol Biol. 2009;392:910–922. doi: 10.1016/j.jmb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Shi R, Villarroya M, Ruiz-Partida R, Li Y, Proteau A, Prado S, Moukadiri I, Benitez-Paez A, Lomas R, Wagner J, Matte A, Velazquez-Campoy A, Armengod ME, Cygler M. Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme. J Bacteriol. 2009;191:7614–7619. doi: 10.1128/JB.00650-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moukadiri I, Prado S, Piera J, Velazquez-Campoy A, Björk GR, Armengod ME. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic Acids Res. 2009;37:7177–7193. doi: 10.1093/nar/gkp762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bessho Y, Yokoyama S. Enzymatic formation of 5-aminomethyl-uridine derivatives in tRNA. In: Grosjean H, editor. DNA and RNA modification enzymes. Austin: Landes Bioscience; 2009. pp. 409–425. [Google Scholar]
- 22.Bujnicki JM, Oudjama Y, Roovers M, Owczarek S, Caillet J, Droogmans L. Identification of a bifunctional enzyme MnmC involved in the biosynthesis of a hypermodified uridine in the wobble position of tRNA. RNA. 2004;10:1236–1242. doi: 10.1261/rna.7470904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roovers M, Oudjama Y, Kaminska KH, Purta E, Caillet J, Droogmans L, Bujnicki JM. Sequence-structure-function analysis of the bifunctional enzyme MnmC that catalyses the last two steps in the biosynthesis of hypermodified nucleoside mnm5s2U in tRNA. Proteins. 2008;71:2076–2085. doi: 10.1002/prot.21918. [DOI] [PubMed] [Google Scholar]
- 24.Pearson D, Carell T. Assay of both activities of the bifunctional tRNA-modifying enzyme MnmC reveals a kinetic basis for selective full modification of cmnm5s2U to mnm5s2U. Nucleic Acids Res. doi: 10.1093/nar/gkr071. (in press) [doi: 10.1093/nar/gkr1071] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horie N, Yamaizumi Z, Kuchino Y, Takai K, Goldman E, Miyazawa T, Nishimura S, Yokoyama S. Modified nucleosides in the first positions of the anticodons of tRNA(Leu)4 and tRNA(Leu)5 from Escherichia coli. Biochemistry. 1999;38:207–217. doi: 10.1021/bi981865g. [DOI] [PubMed] [Google Scholar]
- 26.Kambampati R, Lauhon CT. MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry. 2003;42:1109–1117. doi: 10.1021/bi026536+. [DOI] [PubMed] [Google Scholar]
- 27.Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol Cell. 2006;21:97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Benítez-Páez A, Villarroya M, Douthwaite S, Gabaldón T, Armengod ME. YibK is the 2′-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNALeu isoacceptors. RNA. 2010;16:2131–2143. doi: 10.1261/rna.2245910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dym O, Eisenberg D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001;10:1712–1728. doi: 10.1110/ps.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 32.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(Suppl):W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 35.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]


