Abstract
EphA2 receptor tyrosine kinase and the human cytoplasmic protein tyrosine phosphatase (HCPTP) are overexpressed in a number of epithelial cancers. Overexpressed EphA2 in these cancers shows a significant decrease in phosphotyrosine content which results in suppression of receptor signaling and endocytosis and an increase in metastatic potential. The decreased phosphotyrosine content of EphA2 has been associated with decreased contact with its ligand, ephrin A1 and dephosphorylation by HCPTP. Potential specificity of the two HCPTP variants for tyrosines on EphA2 has not been investigated. We have used a mass spectrometry assay to measure relative rates of dephosphorylation for the two HCPTP variants at phosphotyrosine sites associated with control of the EphA2 kinase activity or interaction with downstream targets. Our results suggest that although both variants dephosphorylate the EphA2 receptor, the rate and specificity of dephosphorylation for specific tyrosines are different for HCPTP-A and HCPTP-B. The SAM domain tyrosine Y960 which has been implicated in downstream PI3K signaling is dephosphorylated exclusively by HCPTP-B. The activation loop tyrosine (Y772) which directly controls kinase activity is dephosphorylated about six times faster by HCPTP-A. In contrast, the juxtamembrane tyrosines (Y575, Y588 and Y594) which are implicated in both control of kinase activity and downstream signaling are dephosphorylated by both variants with similar rates. This difference in preference for dephosphorylation sites on EphA2 not only illuminates the different roles of the two variants of the phosphatase in EphA2 signaling, but also explains why both HCPTP variants are highly conserved in most mammals.
Keywords: EphA2 receptor tyrosine kinase, human cytoplasmic protein tyrosine phosphatase, phosphotyrosine signaling, selected reaction monitoring
Introduction
Overexpression of the EphA2 receptor tyrosine kinase has been implicated in aberrant cell signaling in a variety of epithelial cancers such as lung, breast, colon, and prostate. 1–3 In transformed cells, endocytic recycling of the EphA2 receptors appears to be interrupted, leaving these receptors clustered in ruffles on the plasma membrane. 4 EphA2 receptors in transformed cells show a significant decrease in their phosphotyrosine content, which has been correlated with increased tumor malignancy and metastatic potential 5 6 stimulating investigation of EphA2 as a potential target for cancer chemotherapy. 2 6 The decrease in phosphotyrosine content has been proposed to be due to decreased contact of EphA2 with its ligand, ephrin A1, as well as dephosphorylation by the simultaneously overexpressed human cytoplasmic protein tyrosine phosphatase (HCPTP).
EphA2 belongs to the largest subfamily of receptor tyrosine kinases, the Eph family, which recognize an equally diverse set of ephrin ligands displayed on adjacent cells. The family is comprised of 16 known receptors with nine known ephrin ligands. 7 8 The Eph receptors are transmembrane proteins with multidomain extracellular and cytoplasmic regions connected by a single transmembrane helix. EphA2 is activated on binding to ephrinA1 expressed on the membrane of an adjacent cell, producing an Eph–ephrin heterodimer at the cell junction, which dimerizes to form a heterotetramer. 8 Activation increases phosphorylation on the cytoplasmic tyrosines, which results in downstream signaling with subsequent endocytosis and degradation of the receptor. 6 9 Work with tumor-based models suggests roles for EphA2 signaling in the regulation of cell growth, maintenance of cell–cell contacts, survival, migration, and angiogenesis. 1
Previous experiments have shown a correlation between expression levels of HCPTP and EphA2 and the extent of EphA2 phosphorylation in transformed epithelial cells. 9–11 More direct evidence for interaction between HCPTP and EphA2 receptors in the cell was demonstrated by coimmunoprecipitation from nontransformed (MCF-10Aneo) and transformed (MCF-7, SK-BR-3, MDA-MB-435, and MDA-MB-231) mammary epithelial cells. 9 Overexpression of HCPTP was also shown to be sufficient to confer transformation on nontransformed epithelial cells; treatment with sodium orthovanadate reversed the cells from a transformed to a nontransformed phenotype. Increased recruitment of HCPTP to overexpressed EphA2 was shown to inhibit p190 Rho GAP activity, which in turn promoted destabilization of adherens junctions. 12 These studies, coupled with the demonstration that HCPTP efficiently dephosphorylates EphA2 in vitro, led to the conclusion that the oncogenic activity of HCPTP was directly linked to altered EphA2 function in vivo. 9
The cytoplasmic region of EphA2 has 17 tyrosine residues, many of which are functionally phosphorylated in the cell. Fang et al. 13 overexpressed murine EphA2 in COS7 cells and EphA2-null murine pulmonary microvascular endothelial cells and showed that tyrosines Y588, Y594, Y735, and Y772 were phosphorylated on activation by ephrin-A1. A large-scale kinome analysis by Oppermann et al. 14 on three cancer cell lines identified phosphorylation on Y575, Y588, Y594, Y628, Y772, Y791, and Y960.
Functions in EphA2 signaling have been proposed for several of these tyrosines. Y772 is the putative activation loop tyrosine in the kinase domain, although it is not required for kinase activity of the receptor. 1 Y735 has been implicated in phosphatidylinositol 3-kinase (PI3K) signaling and is the site for EphA2 interaction with the p85 regulatory subunit of PI3K. 13 Y588 and Y594 are located in the juxtamembrane (JM) region and are implicated in regulation and autoinhibition of the kinase, in addition to providing a binding site for Vav GEFs. 13 15 Y960 in the sterile alpha motif (SAM) domain has been proposed as part of the interaction interface between EphA2 and Ship2. 16 Most of these tyrosines are well conserved within the Eph family, except Y575 in the juxtamembrane region and Y960 in the SAM domain, both of which are unique to EphA2.
Humans have two functional splice variants of HCPTP, designated HCPTP-A and HCPTP–B. 17 These two variants share a high percentage of identity (87.3%) with a variable region of 34 amino acids from residues 40–73. Differences in amino acids in this variable region have been shown to change the sensitivity of the enzyme to inhibitors and are proposed to contribute to substrate specificity. 18 19 Also, the variants show a 25-fold difference in enzymatic activity at physiological pH. 20
Although it has long been postulated that HCPTP-A and HCPTP-B might have a different subset of physiological substrates, 18 21 22 the only physiological study of the interactions of HCPTP and EphA2 did not differentiate between the two splice variants. To test for differences in specificity between HCPTP variants, we have investigated the tyrosines that are the preferential targets of this phosphatase using novel quantitative selected reaction monitoring mass spectrometry (SRM-MS)-based methods and a recombinant autophosphorylated cytoplasmic region of EphA2. The results of our experiments show that HCPTP-A and HCPTP-B exhibit distinct specificities for EphA2 phosphotyrosines, which may provide a rationale for selective inhibitors necessary to investigate how HCPTP targets EphA2 in transformed cells.
Results
Phosphopeptide mapping of a recombinant cytoplasmic domain of EphA2 (EphA2 C0)
EphA2 C0, the cytoplasmic region of EphA2, has been expressed in E. coli. 23 This recombinant protein, containing all the structural elements from the juxtamembrane domain to the C-terminal PDZ binding motif, has kinase activity and is isolated in an autophosphorylated state. EphA2 C0 contains 17 tyrosines that are potential sites of phosphorylation. In an initial experiment, we analyzed the recombinant EphA2 C0 by liquid chromatography tandem mass spectrometric (LC-MSMS) to identify which tyrosine residues are autophosphorylated in this construct. Purified recombinant EphA2 C0 was digested with trypsin and AspN to achieve 100% sequence coverage. The tryptic digest was sufficient for detecting 15 of the 17 tyrosines. The remaining two tyrosines, Y813 and Y818, required the AspN digest for detection.
Of the 17 tyrosines in EphA2 C0, autophosphorylation was unambiguously detected on nine tyrosines (Table I and Supporting Information Fig. S1). All three tyrosines of the flexible juxtamembrane region (Y575, Y588, and Y594) were phosphorylated. Of the 11 kinase domain tyrosines, phosphorylation was detected on Y628, Y694, Y735, and Y772. Two of the three SAM domain tyrosines (Y921 and Y960) were also phosphorylated. Supporting Information Figure S1 shows the MSMS spectra for peptides containing all nine detected phosphotyrosines.
Table I.
Phosphopeptide Mapping of EphA2 C0 by LC-MSMS
| Tyrosine | Domains | Peptide sequencesa |
|---|---|---|
| 575 | Jm | 569-QSPEDV[Y]FSK−578 |
| 588b | 587-T[Y]VDPHTYEDPNQAVLK−603 | |
| 594b | 587-TYVDPHT[Y]EDPNQAVLK−603 | |
| 628 | Kinase | 618-VIGAGEFGEV[Y]KGMLK−633 |
| 694 | 685-YKPMMIITE[Y]MENGALDK−702 | |
| 735 | 729-YLANMN[Y]VHR−738 | |
| 772 | 763-VLEDDPEAT[Y]TTSGGK−778 | |
| 921 | SAM | 918-MQQ[Y]TEHFMAAGYTAIEK−935 |
| 960 | 958-IA[Y]SLLGLK−966 |
Peptide sequences represent the tryptic peptides with phosphorylated tyrosines indicated within brackets detected by mass spectrometry.
Tyrosines 588 and 594 are present on the same tryptic peptide and the doubly phosphorylated and both singly phosphorylated forms were detected.
Earlier studies have only looked at global EphA2 phosphorylation using anti-pY antibodies, 1 9 23 but results from two recent studies confirm that most of the phosphotyrosines observed in recombinant EphA2 C0 are also phosphorylated in cellular systems. Fang et al. 13 used LC-MSMS analysis to look at phosphorylated tyrosines in EphA2; their results suggest that Y588, Y594, Y735, and Y772 are autophosphorylated in vascular endothelial cells. Our phosphopeptide mapping results suggest that in addition to the tyrosines reported by Fang et al. Y575 (which is unique to EphA2), Y628 and Y694 in the kinase domain and Y921 and Y960 in the SAM domain can also be autophosphorylated. Y575, Y628, Y694, Y921, and Y960 have been reported to be phosphorylated in large-scale kinome analysis, 14 24 but to date, there has been no evidence that these sites are autophosphorylated by EphA2.
Global dephosphorylation of EphA2 C0 by HCPTP-A and HCPTP-B
Previous experiments have established that HCPTP can dephosphorylate EphA2 in a time and concentration dependent manner. 9 This study used recombinant HCPTP to dephosphorylate immunoprecipitated EphA2 from MDA-MB231 cells but did not identify the variant of the phosphatase. To determine if both variants of HCPTP are capable of dephosphorylating EphA2 C0, dephosphorylation reactions were performed in the presence of 1 mM EDTA to prevent the autophosphorylation of dephosphorylated tyrosines. As expected, both variants were able to dephosphorylate EphA2 C0 (Fig. 1). EphA2 C0 is substantially dephosphorylated by HCPTP-A within 10 min, based on loss of the slower mobility band, which correlates with the decrease in phosphotyrosine content. 25 By 60 min, the slow mobility band is almost invisible. However, some residual phosphotyrosine content, evident from the anti-pY blot, remains even after 240 min. In contrast to HCPTP-A, the rate of dephosphorylation of EphA2 C0 by HCPTP-B appears to be distinctly slower. The anti-pY immunoblot of EphA2 C0 dephosphorylation by HCPTP-B shows only a slight decrease in pY content at the 60-min timepoint. There appears to be substantial pY content even after 240 min; however, the increase in the faster mobility band visible in the Coomassie stained gel over time clearly indicates that some dephosphorylation is occurring. We conclude that both variants of HCPTP are capable of dephosphorylating EphA2 C0, although there is a striking difference in the efficiency of global dephosphorylation.
Figure 1.
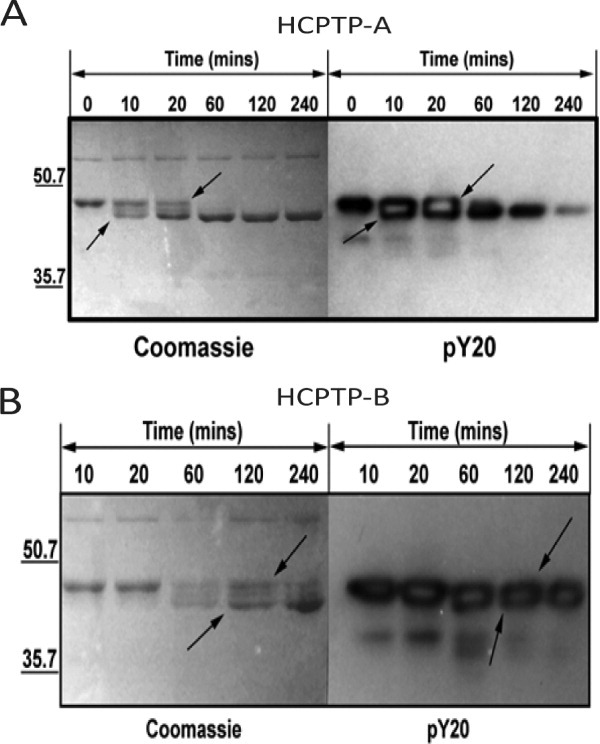
Comparison of global EphA2 C0 dephosphorylation catalyzed by HCPTP-A and HCPTP-B. Aliquots drawn at various time points were resolved on 12% SDS PAGE and either stained with Coomassie blue or probed with an anti-phosphotyrosine antibody (pY20). (A) Dephosphorylation of EphA2 C0 by HCPTP-A. (B) Dephosphorylation of EphA2 C0 by HCPTP-B. The increase in mobility of EphA2 C0 upon dephosphorylation is indicated by arrows.
Dephosphorylation of EphA2 phosphopeptides by HCPTP variants
Having established that EphA2 C0 is a substrate for both HCPTP variants, we then designed experiments to identify specific tyrosines that are dephosphorylated by each variant and to estimate their relative rates of dephosphorylation. We initially looked at dephosphorylation of EphA2 phosphopeptides by the two HCPTP variants. We tested two peptide substrates, VLEDDPEAT(pY772)TTSGGK and VRLPGHQKRIA(pY960)SLLGLKDQ, representing the activation loop and SAM domain tyrosines. At physiological pH, both variants have very low rates of dephosphorylation for these peptides (Supporting Information Fig. S2), preventing determination of steady-state kinetic parameters for these in vitro substrates. However, by comparing relative reaction rates, we found that the variants do not appear to discriminate between peptide substrates. In fact, the rates of dephosphorylation of the peptide substrates are not distinguishable from the dephosphorylation rate of phosphotyrosine by itself. An examination of the two structures of the HCPTP variants 18 22 reveals that the variable region forms a loop that is proximal to but not part of the active site of the phosphatase. This implies that the specificity of these two enzymes may be coded in a three-dimensional substrate that can interact with residues both in the variable region and in the active site.
SRM-MS method development and label free quantification
To more thoroughly explore potential differences between the two variants, we used autophosphorylated EphA2 C0 as an intact physiological substrate and monitored dephosphorylation of individual tyrosines using selected reaction monitoring mass spectrometry (SRM-MS) combined with a label-free quantification strategy. 26 We developed and optimized SRM method parameters for detection of five of the tyrosines of EphA2 in regions with proposed important functions in EphA2 signaling and endocytosis: Y575, Y588, Y594 (juxtamembrane region), Y772 (activation loop), and Y960 (SAM domain) in EphA2 C0 (Supporting Information Table SI). Tryptic peptides containing each of these tyrosines can be detected and quantified in a single SRM-MS experiment. SRM was used (1) to allow individual quantification of Y588 and Y594, which are present on the same tryptic peptide, 26 (2) to provide specificity for the phosphorylated Y960 peptide, which is isobaric with another prominent EphA2 ion (Supporting Information Fig. S3), and (3) to begin developing the methods necessary for future analysis of endogenous EphA2 phosphorylation from cell extracts. In addition to the phosphorylated peptides and their unmodified cognates, three other peptides within EphA2 C0 that do not contain any tyrosines were monitored as standard peptides used for normalization (Supporting Information Table SI). These methods cover five of the nine autophosphorylated tyrosines in EphA2 that are distributed across all three structural domains in the cytoplasmic region. This represents a good starting point to explore site specificity of the HCPTP variants that target EphA2 in vivo.
SRM traces for phosphorylated peptides monitored in one phosphatase assay at 0, 30, and 60 min are shown in Figure S4(A–C). Figure S4(D) shows the SRM traces for the standard peptides monitored at the same three timepoints. The average variation in measured peak area among the standard peptides was 7.2%. The stoichiometry of each autophosphorylation site monitored was first calculated as described. 26 27 The starting concentration of each phosphorylation site was then calculated from the stoichiometry data (Table II). First-order rate parameters, half-life and initial rate of dephosphorylation were determined by fitting the data obtained from the SRM dephosphorylation experiments and stoichiometry calculations with a first-order exponential equation.
Table II.
Measurement of Phosphorylation Stoichiometry for Each Tyrosine Monitored Using SRM-MS
| EphA2 Peptidesa | Peptide nomenclatureb | Measured Stoichiometry (%) | pY Content [S0] (μM) |
|---|---|---|---|
| QSPEDV[Y575]FSK | pY575 | 70 ± 5 | 7.0 |
| T[Y588]VDPHT[Y594]EDPNQAVLK | ppY588Y594 | 47 ± 10 | 4.7 |
| T[Y588]VDPHTY594EDPNQAVLK | pY588Y584 | 17 ± 4 | 1.7 |
| TY588VDPHT[Y594]EDPNQAVLK | Y588pY594 | 29 ± 7 | 2.9 |
| VLEDDPEAT[Y772]TTSGGK | pY772 | 80 ± 2 | 8.0 |
| IA[Y960]SLLGLK | pY960 | 8 ± 2 | 0.8 |
Phosphorylated tyrosines are indicated within brackets in the peptide sequence.
The peptides are referred to by this naming system in all figures and tables.
Dephosphorylation of EphA2 C0 by HCPTP-A
EphA2 C0 was reacted with HCPTP-A and samples were withdrawn over time for quantitative analysis of the phosphorylation state at each of the five tyrosines. Dephosphorylation assays were conducted at physiological pH. Figure 2(A) shows the relative percent phosphorylation for the three singly phosphorylated (Y772, Y575, and Y960) and the doubly phosphorylated (Y588Y594) peptide over time. Figure 2(B) shows dephosphorylation data for the two neighboring phosphorylation sites in the juxtamembrane region, Y588 and Y594. As Y588 and Y594 are present on the same tryptic peptide, three different phosphorylated peptides, the doubly phosphorylated peptide and the two singly phosphorylated peptides, are monitored for these tyrosines. Table III and Supporting Information Figure S5(A) list the exponential fit parameters and rate of dephosphorylation for each phosphotyrosine monitored. Overall, the activation loop pY772 (t1/2 = 5.8 min) and juxtamembrane region ppY588Y594 (t1/2 = 7.4 min) peptides are dephosphorylated by HCPTP-A at a faster rate than the other tyrosines monitored. In the first 10–15 min of the reaction, we observed a transient accumulation of the singly phosphorylated peptide pY588Y594 [Fig. 2(B)]. This would occur if pY594 is dephosphorylated faster than pY588 in the doubly phosphorylated ppY588Y594 peptide, generating the singly phosphorylated pY588Y594 peptide as product faster than it is depleted. This suggests that pY594 is preferred as a substrate over pY588 by HCPTP-A. pY575, the unique tyrosine site in the juxtamembrane region, is dephosphorylated by HCPTP-A more slowly (t1/2 = 34.3 min), and there is no observed dephosphorylation of the SAM domain tyrosine, pY960.
Figure 2.
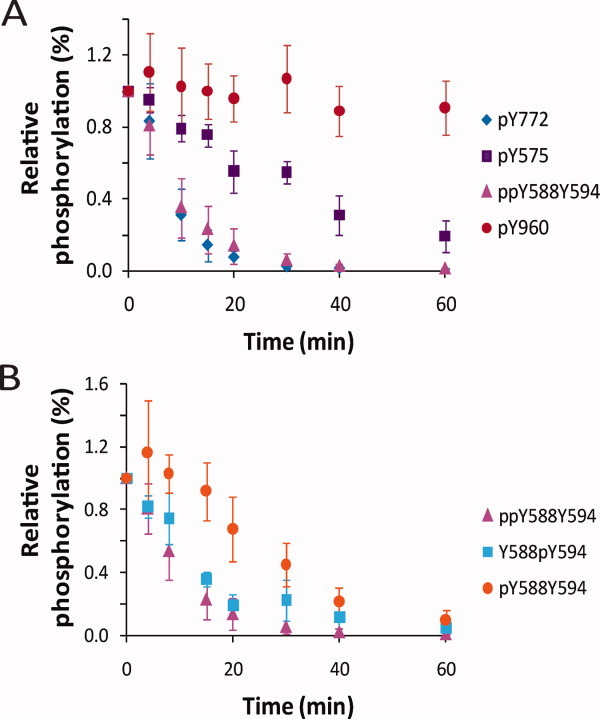
Dephosphorylation of EphA2 C0 by HCPTP-A. Samples drawn at various time points were subjected to SRM-MS analysis. The percentage phosphorylation for the sample at 0 min was adjusted to 100%, and the percent phosphorylation over time is plotted relative to 0 min. (A) A comparison of the dephosphorylation profiles for pY772, ppY588Y594, pY575 and pY960. (B) A comparison of the dephosphorylation profiles for the three phosphorylated forms of the peptide containing both pY588 and pY594. All experiments were repeated at least three times and the mean and SD from the mean are shown.
Table III.
First-order Rate Parameters (Fit to the First-order Exponential Equation [S] = [S0]e−kt) and Initial Rate of Dephosphorylation for Each Tyrosine by HCPTP-A and HCPTP-B
| k (min−1) | t1/2 (min) | Initial rate (10% decrease in [S]) (pmol min−1 μg−1 HCPTP) | ||||
|---|---|---|---|---|---|---|
| Tyrosine | A | B | A | B | A | B |
| pY575 | 0.02 ± 0.001 | 0.02 ± 0.001 | 34.3 | 45.4 | 0.8 | 0.4 |
| ppY588Y594 | 0.09 ± 0.007 | 0.11 ± 0.01 | 7.4 | 6.8 | 2.2 | 1.5 |
| pY588Y594a | 0.04 ± 0.004 | 0.04 ± 0.002 | 16.2 | 18.3 | 0.4 | 0.2 |
| Y588pY594 | 0.05 ± 0.002 | 0.06 ± 0.001 | 13.8 | 10.7 | 0.8 | 0.5 |
| pY772 | 0.12 ± 0.02 | 0.04 ± 0.002 | 5.8 | 19.4 | 4.9 | 0.8 |
| pY960 | — | 0.04 ± 0.002 | — | 18.3 | — | 0.1 |
For pY588Y594 the rate was fit from 10–60 min as this substrate increases for the first 10 min of the reaction.
Dephosphorylation of EphA2 C0 by HCPTP-B
Figures 3(A,B) show results from the same analysis as above performed with HCPTP-B. Table III and Supporting Information Figure S5(B) list the exponential fit parameters and rate of dephosphorylation for each phosphotyrosine monitored. For this variant, the doubly phosphorylated juxtamembrane peptide containing ppY588Y594 (t1/2 = 6.8 min) seems to be the most efficient substrate [Fig. 3(A)]. HCPTP-B also shows a similar preference for dephosphorylation of pY594 on the doubly phosphorylated peptide, demonstrated by the initial increase in the singly phosphorylated pY588Y594 peptide [Fig. 3(B)]. The rate of dephosphorylation of pY772 (t1/2 = 19.4), the activation loop tyrosine, is modest, comparable to the rate of dephosphorylation of the SAM domain pY960 (t1/2 = 18.3 min). pY575 is the slowest substrate for HCPTP-B. The most striking contrast between the two variants in these results is that pY960 shows no reactivity with HCPTP-A but is dephosphorylated with a rate similar to the other tyrosines by HCPTP-B. The amount of substrate was decreased 10-fold and the dephosphorylation assay was repeated with HCPTP-A to verify that dephosphorylation of pY960 was not inhibited due to an excess of other preferred substrates. The results clearly showed that pY960 was not a substrate for HCPTP-A and was only targeted by HCPTP-B.
Figure 3.
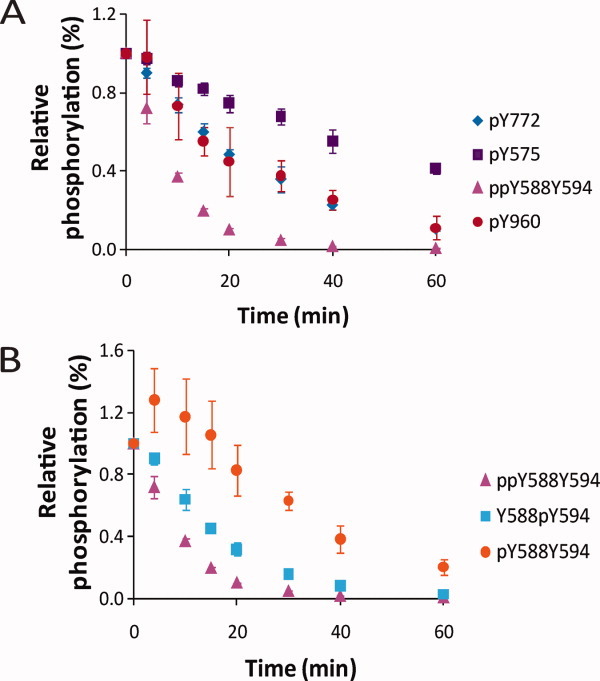
Dephosphorylation of EphA2 C0 by HCPTP-B. Samples drawn at various timepoints were subjected to SRM-MS analysis. The percentage phosphorylation for the sample at 0 min was adjusted to 100%, and the percent phosphorylation over time is plotted relative to 0 min. (A) A comparison of the dephosphorylation profiles for pY772, ppY588Y594, pY575, and pY960. (B) A comparison of the dephosphorylation profiles for the three phosphorylated forms of the peptide containing both pY588 and pY594. All experiments were repeated at least three times and the mean and SD from the mean are shown.
Preferential dephosphorylation of the juxtamembrane peptide at pY594
We observed that both HCPTP variants appeared to preferentially dephosphorylate Y594 on the doubly phosphorylated juxtamembrane peptide. To confirm this result, we decreased the amount of enzyme 10-fold and repeated the dephosphorylation assay. The results of this experiment for HCPTP-A are shown in Figure 4. Under these conditions, the difference between the dephosphorylation rates for Y588 and Y594 are exaggerated and a clear increase in pY588Y594 can be observed. Similar results were seen when the amount of either HCPTP variant was decreased. These results replicate our earlier observation that the pY594 site on the doubly phosphorylated peptide is dephosphorylated at a faster rate by HCPTP and indicate a clear preference of the phosphatase for this site.
Figure 4.
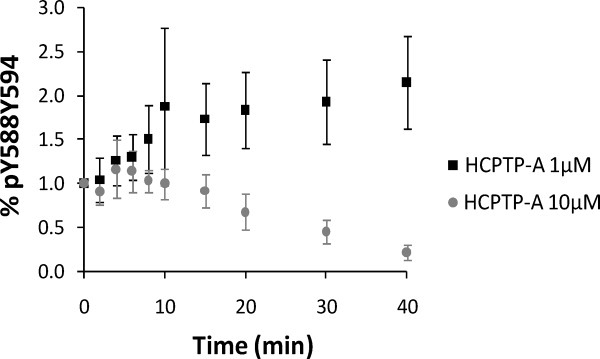
pY588Y594 builds up as a result of dephosphorylation at Y594 on the doubly phosphorylated peptide ppY588Y594 as the reaction proceeds. 10 μM EphA2 C0 was treated with 10 μM HCPTP-A (C0 + A 10:10) or with 1 μM HCPTP-A (C0 + A 10:1) and the SRM-MS data for pY588Y594 was analyzed at various timepoints. The mean and standard deviation of three independent replicates are plotted.
Discussion
The oncogenic potential of HCPTP has been proposed to result from the dephosphorylation of tyrosines involved in both the kinase activity of EphA2 and downstream signaling that leads to endocytosis and degradation of this receptor. 9–11 The link shown between overexpression of EphA2 and HCPTP, EphA2 signaling and the metastatic potential of tumor cells makes both of these oncogenic proteins attractive candidates for drug discovery in cancer. 2 6 20 28 Understanding the specificity of the HCPTP variants expands this research and may provide a way to more specifically target the functions of these ubiquitous PTPases.
Our study has revealed a clear difference in substrate specificity for the A and B variants of HCPTP using its physiological substrate, the cytoplasmic domain of EphA2. To achieve this result we used a novel, highly sensitive SRM mass spectrometric method that can follow individual phosphorylated tyrosine sites on the EphA2 receptor. This method allows us to determine the stoichiometry of phosphorylation and the rate of dephosphorylation of individual sites on an intact protein with multiple phosphorylated tyrosines, where each pY site is recognized differently by the HCPTP variants. This approach has proven to be extremely useful where in vitro experiments to differentiate the roles of these highly identical (87.3%) HCPTP splice variants will guide the more challenging in vivo experiments, as development of specific antibodies and siRNA for these closely related proteins is difficult. Importantly, differences in apparent specificity stemming from assays with EphA2 C0 and synthetic peptides have clearly shown the need to use biologically relevant substrates to determine physiological specificity.
The initial phosphorylation mapping studies were done on a soluble EphA2 cytoplasmic domain, parallel to similar mapping studies of EphB2 and EphA3. 29 30 Our study demonstrates that EphA2 can autophosphorylate nine of the 17 tyrosines in its cytoplasmic region. All these sites have also been observed as phosphorylated in vivo. 13 14 24 Fang et al. reported that the juxtamembrane Y588 and Y594 and the activation loop Y772 appeared to be the major phosphorylation sites on EphA2, when the receptor was overexpressed in vascular endothelial cells. Our SRM-MS-based stoichiometry calculations with EphA2 C0 also suggest that these sites are efficiently autophosphorylated (Table I).
The EphA2 phosphopeptide mapping data suggests that this receptor can also be autophosphorylated on five other sites. Of the five additional sites, Y575 and Y960 are both unique to EphA2 and are not conserved among the other Eph receptors. Our observation of autophosphorylation on these tyrosines suggests that they may serve as docking sites for interacting proteins that are specific to EphA2 function. Y960 has been proposed as part of the interaction interface between EphA2 and Ship2 16; Ship2 regulates EphA2 endocytosis through PI3K-dependent Rac1 activation. 31 Y575 is autophosphorylated as efficiently as nearby Y588 and Y594 (Table I) and could also potentially contribute to the regulation of juxtamembrane domain function in vivo.
The major goal of this research was to investigate specificity in the HCPTP variants. Among the differences, we found that HCPTP-A was far more active toward the activation loop tyrosine in the kinase domain (Y772) than HCPTP-B. Y772 is dephosphorylated about six times faster by HCPTP-A compared to HCPTP-B, and appears to be a primary target for this variant. In contrast, HCPTP-B was most active toward the peptide containing juxtamembrane tyrosines, with a preference for Y594. Both HCPTP-A and HCPTP-B dephosphorylate the doubly phosphorylated juxtamembrane peptide (ppY588Y594) with comparable rates, as might be expected for its role as a regulatory region. Where previous studies have not distinguished the accessibility of these two tyrosines, our results have revealed a significant difference in the order in which the two conserved juxtamembrane tyrosines Y588 and Y594 are dephosphorylated. The singly phosphorylated form pY588Y594 transiently accumulates early in the reaction with both HCPTP variants, demonstrating that the phosphate on Y594 is preferentially removed from the doubly phosphorylated peptide ppY588Y594.
The most striking difference between the two variants was revealed in their activity toward the SAM domain tyrosines. HCPTP-A showed no detectable activity toward pY960 in the SAM domain of EphA2, whereas this site was dephosphorylated at a rate similar to other sites by HCPTP-B. As the SAM domain has been implicated in protein–protein interactions that lead to EphA2 endocytosis, HCPTP-B could play a unique role in regulating this process.
Evidence from other studies may illuminate the roles of the two HCPTP variants in EphA2 signaling (Fig. 5). Mutagenesis and kinase activity assays of tyrosines corresponding to Y588, Y594, and Y772 in EphA2 and other ephrin receptors have indicated that these tyrosines directly regulate the catalytic activity of the receptor by a dual component mechanism. 15 Fang et al. showed Y588F inhibited and Y594F abolished ligand-stimulated kinase activity, confirming the importance of the juxtamembrane region in control of EphA2 function. 13 The evidence for interaction of these two molecules in vivo suggests that dephosphorylation on EphA2 at Y772 and Y588/Y594 by the HCPTP variants may lead to the inhibition of the autokinase activity of EphA2, resulting in the hypophosphorylated state of this receptor in cancer.
Figure 5.
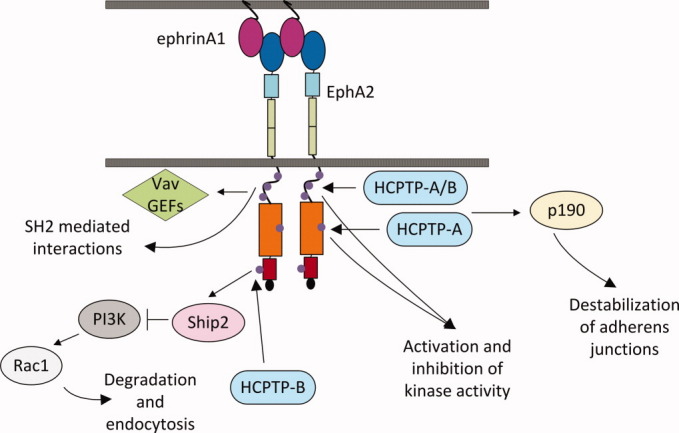
Specificity of HCPTP variants towards EphA2 tyrosines with linked pathways and functions. The juxtamembrane region is shown as an unstructured loop with three phosphorylated tyrosines (Y575, Y588, and Y594) represented by purple circles. The juxtamembrane tyrosines play a role in activation and inhibition of kinase activity, interact with Vav GEFs, and participate in other SH2 mediated reactions. The rate of dephosphorylation of the juxtamembrane by HCPTP variants is comparable. The kinase domain is shown in orange with the activation loop tyrosine (Y772) marked as a purple circle. Y772 is the preferred site for dephosphorylation by HCPTP-A. The SAM domain of EphA2 (red) has been shown to interact with the SAM domain of Ship2 and regulate EphA2 endocytosis through PI3K dependent Rac1 activation. The SAM domain tyrosine, Y960 is a target for dephosphorylation only by HCPTP-B.
In vivo experiments with Eph receptor mutants have shown that HCPTP dephosphorylation of the juxtamembrane tyrosines could have an even more complicated effect on downstream signaling. In EphB2, a Y610F mutation (Y594 cognate) produces a critical defect in the response of neuronal cells to ephrin stimulation, where Y606F (Y588 cognate) showed relatively small changes. 15 29 In contrast, an experiment involving mutants of EphA2 juxtamembrane tyrosines indicates that Y588 has a much more critical role than Y594 in controlling the incorporation of epithelial cells in vasculature and, so, would presumably have a greater effect in tumor angiogenesis. 13 These experiments suggest that Y588 and Y594 have multiples roles in vivo, so that the differential (or sequential) targeting by HCPTP variants may produce a nuanced effect in cellular transformation and metastasis.
Dephosphorylation of the EphA2 receptor tyrosines by these HCPTP variants may also affect downstream signaling by modifying the interface with interacting signaling proteins (Fig. 5). Experiments have linked the EphA2 receptor to the Rac1 pathway through interactions between the SH2 domains of Vav GEFs with the juxtamembrane region of EphA2, modulated by phosphorylated Y588 and Y594. 13 32 The interaction between the SAM domains of EphA2 and Ship2, which occurs at an interface containing Y960, suggests a role for this tyrosine in EphA2 endocytosis, also linked to Rac1 activation. Differential activity of the HCPTP variants toward these phosphorylated tyrosines may, therefore, be involved in the balance of EphA2 signaling that is linked to metastatic cell migration and tumor vascularization in cancer.
Different roles open the possibility of modulating specific HCPTP activities in the cell. Mutagenesis studies of EphA2, HCPTP-A and HCPTP-B will help us to determine a consensus about substrates selection for these two variants. These experiments will also help to identify residues in HCPTP-A and HCPTP-B that contribute to substrate specificity, which should allow us to design inhibitors that exclusively target one variant and more finely tune the response of cells to potential inhibitors of HCPTP designed to control metastatic transformation in cancer.
Methods
Purification of HCPTP-A and HCPTP-B
Both variants were purified as described previously. 17 The wild-type proteins were expressed in BL21(DE3) cells and purified using a combination of ion-exchange chromatography on a SP Sepharose™ fast flow (FF) column followed by secondary purification on a Sephacryl size exclusion column in pH 4.8, 10 mM sodium acetate, 150 mM NaCl and stored at 4°C. Enzymatic activity was monitored via a single point quenched product reaction with the substrate analog p-nitrophenyl phosphate as described previously. 20
Purification of the EphA2 C0–cytoplasmic region of EphA2
EphA2 C0 is a construct of the entire cytoplasmic region of EphA2 (aa 565–976, 48 kDa). This construct includes the juxtamembrane region, the kinase domain, the SAM domain, and the PDZ binding motif. The recombinant protein was purified as described by Zabell et al. 23 with the following modifications. EphA2 C0 was expressed as a fusion protein with an N-terminal 6HIS tag, a thioredoxin tag, and Factor Xa cleavage site using the pET32 Xa/LIC (Novagen) vector in BL21(DE3) OrigamiB cells. The cells were grown in terrific broth containing 50 μg mL−1 ampicillin, 15 μg mL−1 kanamycin and 12.5 μg mL−1 tetracycline at 37°C to a OD600 of 0.8 and then induced with 1 μM IPTG at 18°C for 16 h. The protein was purified using Ni-affinity chromatography in pH 7.4, 10 mM sodium phosphate, and 500 mM sodium chloride using an imidazole gradient. The fusion tags were cleaved after dialysis with Factor Xa (Novagen) at room temperature for 5 h using 1unit of Factor Xa per 50-μg of protein. This was followed by reverse purification over a Ni-affinity column. The flow through from the reverse purification containing EphA2 C0 was collected, dialyzed in pH 7.4, 10 mM Tris-HCl, 150 mM sodium chloride, 1 mM EDTA, concentrated to 1 mg mL−1, and stored at –80°C.
Dephosphorylation of EphA2 C0 by HCPTP
All dephosphorylation assays were performed in pH7.4, 10 mM Tris, 150 mM NaCl and 1 mM EDTA at 30°C. Total reaction volume was maintained at 200 μL, and the reaction was initiated by mixing the EphA2 C0 and HCPTP-A or B. EphA2 C0 (10 μM) was treated with 10 μM of HCPTP-A variant and for the B-variant 20 μM of enzyme was used. An aliquot (10 μL) of the reaction mixture at 0, 2, 4, 6, 8, 10, 20, 30, 40, and 60 min was withdrawn and 1 mM sodium orthovanadate (2.5 μL of 5 mM stock) was added to each sample to stop the dephosphorylation reaction for SRM analysis. For immunoblot analysis, 1 μM EphA2 C0 was mixed with 10 μM HCPTP, and the reaction was stopped by adding SDS PAGE loading buffer.
In-solution digests
Trypsin (7.5 μL of 20 μg mL−1; Sequence grade, Sigma) in 50 mM ammonium bicarbonate was directly added to each of the timecourse dephosphorylation samples and incubated at 37°C overnight (16 h). The samples were then diluted in an equal volume of 6% acetonitrile and 2% formic acid in water and immediately used for mass spectrometry analysis.
Phosphorylation site mapping
Peptides extracted from in gel digestion were resuspended in 5% acetonitrile, 1% formic acid immediately before LC-MSMS analysis. Peptides were separated on an Agilent 1100 capillary HPLC system using Zorbax C18 trap and 75-μm × 150-mm capillary columns. Peptides were eluted with a gradient of increasing acetonitrile in 0.1% formic acid at 300 nL min−1 and injected into a Thermo Scientific LTQ-orbitrap using a nanoelectrospray source. Peptides were initially identified by automated database searching using the Sorcerer program (Sage-N Research) All spectra from putative phosphopeptides were then interpreted manually to confirm correct peptide identification and localization of phosphorylated residues.
Selected reaction monitoring mass spectrometry
SRM was performed on an Agilent 1100 nanoflow HPLC coupled to an Agilent 6410 QQQ MS equipped with a chip cube. Peptide samples were separated on ProtID-Chip-150 (II) (Agilent) that has a 40-nL trapping column and a 150-mm analytical column, both packed with Zorbax-SB C-18. An acetonitrile gradient of 3–35% in 6 min, followed by a 90% acetonitrile wash for 2 min and reequilibration for 5 min was used for all LC-SRM runs. Fragmentor voltage and collision energy parameters were manually optimized for each of the 13 target peptides. Two transitions were programmed for each peptide with 20 ms dwell times. Q1 and Q3 resolutions were set to wide (full width at half maximum, FWHM = 1.2 μ).
Label-free quantification
The phosphorylation level at each site was quantified from SRM data using an isotope label-free system. 26 27 The quantification required normalization of phosphopeptides and their unmodified cognates using a collection of other tryptic peptides from EphA2 that are not subject to tyrosine phosphorylation. The area under the peak for each SRM transition was calculated using MassHunter quantitation software (Agilent). Normalization of phosphopeptide/unmodified peptide pairs using signals from other abundant EphA2 C0 peptides corrects for systematic variability in the samples. The mean signal intensities for the standard peptides were calculated. Percent deviation from the mean of the area for each timepoint was used to adjust the area under the curve for each phosphopeptide and its unmodified pair at every timepoint. The relative change in phosphorylation for each phosphopeptide was determined and plotted versus time. The initial stoichiometries and starting concentrations of each phosphorylation site were calculated as described from both SRM and selected ion monitoring data. 26 Rate constants and half life for each monitored phosphotyrosine were determined by fitting the dephosphorylation data to the first-order equation [S] = [S0]e−kt, where [S] is the concentration of the substrate at time t, [S0] is the initial concentration of the substrate and k is the rate constant, using Origin Pro 8.0 (Origin Lab Corporation).
Acknowledgments
The authors thank the Purdue Proteomics facility for the use of their equipment and the Purdue Center for Cancer Research for general research support. D.B., L.N.P., K.T.H., M.C.H., and C.V.S. designed the research, D.B. and L.N.P. performed the research, D.B. analyzed the data, D. B., M.C.H., and C.V.S. wrote the manuscript.
Supplementary material
References
- 1.Walker-Daniels J, Hess A, Hendrix M, Kinch M. Differential regulation of EphA2 in normal and malignant cells. Am J Pathol. 2003;162:1037–1042. doi: 10.1016/S0002-9440(10)63899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ireton RC, Chen J. EphA2 receptor tyrosine kinase as a promising target for cancer therapeutics. Curr Cancer Drug Targets. 2005;5:149–157. doi: 10.2174/1568009053765780. [DOI] [PubMed] [Google Scholar]
- 3.Kinch MS, Carles-Kinch K. Overexpression and functional alterations of the EphA2 tyrosine kinase in cancer. Clin Exp Metastasis. 2003;20:59–68. doi: 10.1023/a:1022546620495. [DOI] [PubMed] [Google Scholar]
- 4.Zantek ND, Azimi M, Fedor-Chaiken M, Wang B, Brackenbury R, Kinch MS. E-Cadherin regulates the function of the EphA2 receptor tyrosine kinase 1. Cell Growth Differ. 1999;10:629–638. [PubMed] [Google Scholar]
- 5.Zelinski DP, Zantek ND, Stewart JC, Irizarry AR, Kinch MS. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 2001;61:2301–2306. [PubMed] [Google Scholar]
- 6.Carles-Kinch K, Kilpatrick KE, Stewart JC, Kinch MS. Antibody targeting of the EphA2 tyrosine kinase inhibits malignant cell behavior. Cancer Res. 2002;62:2840–2847. [PubMed] [Google Scholar]
- 7.Zhou R. The Eph family receptors and ligands. Pharmacol Ther. 1998;77:151–181. doi: 10.1016/s0163-7258(97)00112-5. [DOI] [PubMed] [Google Scholar]
- 8.Himanen JP, Nikolov DB. Eph signaling: a structural view. Trends Neurosci. 2003;26:46–51. doi: 10.1016/s0166-2236(02)00005-x. [DOI] [PubMed] [Google Scholar]
- 9.Kikawa KD, Vidale DR, Van Etten RL, Kinch MS. Regulation of the EphA2 kinase by the low molecular weight tyrosine phosphatase induces transformation. J Biol Chem. 2002;277:39274–39279. doi: 10.1074/jbc.M207127200. [DOI] [PubMed] [Google Scholar]
- 10.Chiarugi P, Taddei ML, Schiavone N, Papucci L, Giannoni E, Fiaschi T, Capaccioli S, Raugei G, Ramponi G. LMW-PTP is a positive regulator of tumor onset and growth. Oncogene. 2004;23:3905–3914. doi: 10.1038/sj.onc.1207508. [DOI] [PubMed] [Google Scholar]
- 11.Parri M, Buricchi F, Taddei ML, Giannoni E, Raugei G, Ramponi G, Chiarugi P. EphrinA1 repulsive response is regulated by an EphA2 tyrosine phosphatase. J Biol Chem. 2005;280:34008–34018. doi: 10.1074/jbc.M502879200. [DOI] [PubMed] [Google Scholar]
- 12.Fang WB, Ireton RC, Zhuang G, Takahashi T, Reynolds A, Chen J. Overexpression of EPHA2 receptor destabilizes adherens junctions via a RhoA-dependent mechanism. J Cell Sci. 2008;121:358–368. doi: 10.1242/jcs.017145. [DOI] [PubMed] [Google Scholar]
- 13.Fang WB, Brantley-Sieders DM, Hwang Y, Ham A-JL, Chen J. Identification and functional analysis of phosphorylated tyrosine residues within EphA2 receptor tyrosine kinase. J Biol Chem. 2008;283:16017–16026. doi: 10.1074/jbc.M709934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Kéri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binns KL, Taylor PP, Sicheri F, Pawson T, Holland SJ. Phosphorylation of tyrosine residues in the kinase domain and juxtamembrane region regulates the biological and catalytic activities of Eph receptors. Mol Cell Biol. 2000;20:4791–4805. doi: 10.1128/mcb.20.13.4791-4805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leone M, Cellitti J, Pellecchia M. NMR studies of a heterotypic Sam-Sam domain association: the interaction between the lipid phosphatase Ship2 and the EphA2 receptor. Biochemistry. 2008;47:12721–12728. doi: 10.1021/bi801713f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wo YY, McCormack AL, Shabanowitz J, Hunt DF, Davis JP, Mitchell GL, Van Etten RL. Sequencing, cloning, and expression of human red cell-type acid phosphatase, a cytoplasmic phosphotyrosyl protein phosphatase. J Biol Chem. 1992;267:10856–10865. [PubMed] [Google Scholar]
- 18.Zabell APR, Schroff AD, Jr, Bain BE, Van Etten RL, Wiest O, Stauffacher CV. Crystal structure of the human B-form low molecular weight phosphotyrosyl phosphatase at 1.6 Å resolution. J Biol Chem. 2006;281:6520–6527. doi: 10.1074/jbc.M506285200. [DOI] [PubMed] [Google Scholar]
- 19.Cirri P, Fiaschi T, Chiarugi P, Camici G, Manao G, Raugei G, Ramponi G. The molecular basis of the differing kinetic behavior of the two low molecular mass phosphotyrosine protein phosphatase isoforms. J Biol Chem. 1996;271:2604–2607. doi: 10.1074/jbc.271.5.2604. [DOI] [PubMed] [Google Scholar]
- 20.Homan K, Balasubramaniam D, Zabell A, Wiest O, Helquist P, Stauffacher C. Identification of novel inhibitors for a low molecular weight protein tyrosine phosphatase via virtual screening. Bioorg Med Chem. 2010;18:5449–5456. doi: 10.1016/j.bmc.2010.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang ZY, Dixon JE. Protein tyrosine phosphatases: mechanism of catalysis and substrate specificity. Adv Enzymol Relat Areas Mol Biol. 1994;68:1–36. doi: 10.1002/9780470123140.ch1. [DOI] [PubMed] [Google Scholar]
- 22.Zhang M, Stauffacher CV, Lin D, Van Etten RL. Crystal structure of a human low molecular weight phosphotyrosyl phosphatase. Implications for substrate specificity. J Biol Chem. 1998;273:21714–21720. doi: 10.1074/jbc.273.34.21714. [DOI] [PubMed] [Google Scholar]
- 23.Zabell KM, Laurence JS, Kinch MS, Knapp DW, Stauffacher CV. Expression and purification of the intact cytoplasmic domain of the human ephrin receptor A2 tyrosine kinase in Escherichia coli. Protein Expr Purif. 2005;47:210–216. doi: 10.1016/j.pep.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 24.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, MacNeill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu T-L, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 25.Wegener AD, Jones LR. Phosphorylation-induced mobility shift in phospholamban in sodium dodecyl sulfate-polyacrylamide gels. Evidence for a protein structure consisting of multiple identical phosphorylatable subunits. J Biol Chem. 1984;259:1834–1841. [PubMed] [Google Scholar]
- 26.Balasubramaniam D, Eissler C, Stauffacher C, Hall M. Use of selected reaction monitoring data for label free quantification of protein modification stoichiometry. Proteomics. 2010;10:4301–4305. doi: 10.1002/pmic.201000232. [DOI] [PubMed] [Google Scholar]
- 27.Steen H, Jebanathirajah JA, Springer M, Kirschner MW. Stable isotope-free relative and absolute quantitation of protein phosphorylation stoichiometry by MS. Proc Natl Acad Sci USA. 2005;102:3948–3953. doi: 10.1073/pnas.0409536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koolpe M, Dail M, Pasquale EB. An ephrin mimetic peptide that selectively targets the EphA2 receptor. J Biol Chem. 2002;277:46974–46979. doi: 10.1074/jbc.M208495200. [DOI] [PubMed] [Google Scholar]
- 29.Kalo MS, Yu H-H, Pasquale EB. In vivo tyrosine phosphorylation sites of activated Ephrin-B1 and EphB2 from neural tissue. J Biol Chem. 2001;276:38940–38948. doi: 10.1074/jbc.M105815200. [DOI] [PubMed] [Google Scholar]
- 30.Lawrenson ID, Wimmer-Kleikamp SH, Lock P, Schoenwaelder SM, Down M, Boyd AW, Alewood PF, Lackmann M. Ephrin-A5 induces rounding, blebbing and de-adhesion of EphA3-expressing 293T and melanoma cells by CrkII and Rho-mediated signalling. J Cell Sci. 2002;115:1059–1072. doi: 10.1242/jcs.115.5.1059. [DOI] [PubMed] [Google Scholar]
- 31.Zhuang G, Hunter S, Hwang Y, Chen J. Regulation of EphA2 receptor endocytosis by SHIP2 lipid phosphatase via phosphatidylinositol 3-kinase-dependent Rac1 activation. J Biol Chem. 2007;282:2683–2694. doi: 10.1074/jbc.M608509200. [DOI] [PubMed] [Google Scholar]
- 32.Hunter SG, Zhuang G, Brantley-Sieders D, Swat W, Cowan CW, Chen J. Essential role of Vav family guanine nucleotide exchange factors in EphA receptor-mediated angiogenesis. Mol Cell Biol. 2006;26:4830–4842. doi: 10.1128/MCB.02215-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


