Abstract
The wild type red fluorescent protein eqFP578 (from sea anemone Entacmaea quadricolor, λex = 552 nm, λem = 578 nm) and its bright far-red fluorescent variant Katushka (λex = 588 nm, λem = 635 nm) are characterized by the pronounced pH dependence of their fluorescence. The crystal structures of eqFP578f (eqFP578 with two point mutations improving the protein folding) and Katushka have been determined at the resolution ranging from 1.15 to 1.85 Å at two pH values, corresponding to low and high level of fluorescence. The observed extinguishing of fluorescence upon reducing pH in eqFP578f and Katushka has been shown to be accompanied by the opposite trans-cis and cis-trans chromophore isomerization, respectively. Asn143, Ser158, His197 and Ser143, Leu174, and Arg197 have been shown to stabilize the respective trans and cis fluorescent states of the chromophores in eqFP578f and Katushka at higher pH. The cis state has been suggested as being primarily responsible for the observed far-red shift of the emission maximum of Katushka relative to that of eqFP578f.
Keywords: fluorescent proteins, biomarkers, chromophore, gene-engineered variants, crystal structure
Introduction
Green fluorescent proteins (GFP) and GFP-like fluorescent proteins (FPs) have become important noninvasive tools for visualization and monitoring of the internal processes within cells or whole organisms, and the range of their application is continuously expanding. Utilization of FPs in cell biology, biotechnology, and biomedical studies enabled optical imaging of gene expression, measuring of cellular pH and ion concentration, monitoring of embryogenesis and inflammatory processes, tracking of proteins, etc.1–8 In recent years FP-based biomarkers have found important applications in the studies of various aspects of cancer.9–11
Proteins that emit red light, and especially those emitting in the far-red region, are of particular interest.6,12 The longer wavelength light is more advantageous for imaging applications. Its lower energy and the reduced absorption in tissues cause lesser damage to cells. The most favorable “optical window” for the visualization in living tissues is ∼650–1100 nm.13 Detection of fluorescence from proteins with emission peaks much shorter than 650 nm encounters the problem of interfering cellular autofluorescence. The emission with the wavelength larger than 1100 nm is absorbed by water. At present, the brightest red FPs have emission maxima too far from the preferred “optical window.” Besides, their excitation maxima are found in the range of 550–560 nm, where living tissues are almost opaque and the fluorescence of these proteins cannot be effectively excited (see Table I in Ref.6). Far-red fluorescent variants, HcRed, mPlum, and AQ143, crossing the 650 nm barrier, have been developed,14–16 but these proteins are characterized by low brightness relative to EGFP (<0.15), strongly limiting their practical application. Lately, an optimized far-red emitting variant RFP639 (λem = 639 nm), with enhanced relative brightness (0.39), has been reported.12
Table I.
Crystallographic Data and Refinement Statistics
| Protein | eqFP578f pH 5.5 (PDB_ID: 3PIB) | eqFP578f pH 4.0 (PDB_ID: 3PJB) | Katushka pH 8.5 (PDB_ID: 3PJ7) | Katushka pH 5.0 (PDB_ID: 3PJ5) |
|---|---|---|---|---|
| Crystallographic data | ||||
| Space group | I41 | P6122 | I41 | P6122 |
| Cell dimensions (Å, °) | a, b = 161.2, c = 75.7 | a, b = 104.7, c = 216.5 | a, b = 161.4, c = 74.5 | a, b = 104.4, c = 218.0 |
| Z/(Z') | 32 (4) | 24 (2) | 32 (4) | 24 (2) |
| Estimated solvent content (%) | 45 | 60 | 44 | 60 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Wavelength (Å) | 1.00 | 1.00 | 1.00 | 1.00 |
| Resolution range (Å) | 35.9–1.15 (1.19–1.15)a | 34.7–1.75 (1.82–1.75)a | 25.0–1.85 (1.93–1.85)a | 30.0–1.60 (1.66–1.60)a |
| Total reflections measured | 1,761,066 | 793,793 | 390,600 | 645,605 |
| Unique reflections observed | 314,476 | 69,631 | 78,120 | 88,439 |
| Multiplicity | 5.6 (4.7)a | 11.4 (11.6)a | 5.0 (5.0)a | 7.3 (7.2)a |
| I/σ (I) | 28.3 (2.4)a | 28.6 (4.4)a | 16.7 (2.6)a | 19.6 (2.4)a |
| Rmerge | 0.055 (0.488)a | 0.062 (0.562)a | 0.096 (0.655)a | 0.106 (0.719)a |
| Completeness (%) | 92.8 (59.9)a | 97.8 (94.8)a | 97.7 (97.1)a | 97.5 (97.0)a |
| Refinement statistics | ||||
| Non-H atoms in model | ||||
| Protein | 7,288 [4×(1-230) res] | 3,559 [2×(2-226) res] | 7,151 [4×(3-227) res] | 3,646 [2×(0-225) res] |
| Water | 721 | 340 | 463 | 288 |
| SO4 | 15 (3 ions) | |||
| Glycerol | 24 (4 mol) | 12 (2 mol) | 0 | 0 |
| Rworkb | 0.155 (99.0% data, F ≥ 0) | 0.177 (98.0% data, F ≥ 0) | 0.196 (97.9% data, F ≥ 0) | 0.187 (98.0% data, F ≥ 0) |
| Rfreeb | 0.177 (1.0% data, F ≥ 0) | 0.208 (2.0% data, F ≥ 0) | 0.252 (2.1% data, F ≥ 0) | 0.226 (2.0% data, F ≥ 0) |
| Mean B factor/(RMSD)c (Å2) | ||||
| Protein atoms | ||||
| main chain | 13.6 (0.9)c | 25.2 (2.0)c | 33.8 (0.9)c | 23.9 (0.7)c |
| side chain | 16.3 (2.0)c | 29.1 (3.5)c | 35.2 (2.1)c | 25.6 (1.6)c |
| Chromophore | 13.5 (3.3)c | 24.3 (4.3)c | 31.7 (1.4)c | 21.0 (2.4)c |
| Ramachandran statistics (%) | ||||
| Preferred/allowed/disallowed regions | 98.2/1.8/0.0 | 98/2.0/0.0 | 97.0/2.5 /0.5 | 97.0/2.5/0.5 |
Values in parentheses are given for the highest-resolution shells.
Percent of the data reserved for working and free sets.
Root mean square deviation.
Recently, more advanced bright far-red variant Katushka (λex = 588 nm, λem = 635 nm, and brightness = 0.70) has been successfully designed.6 This FP is derived from the wild type red fluorescent progenitor eqFP578 (Entacmaea quadricolor), with spectral maxima λex = 552 nm and λem = 578 nm. Katushka displays fast maturation and high photostability. It is significantly brighter than the spectrally close variant RFP63912 having identical chromophore-forming sequence Met63-Tyr64-Gly65. The fluorescence of Katushka and its progenitor eqFP578 is pH-dependent, with emission diminishing upon reducing pH. Compared with other far-red FPs, Katushka exhibits evident superiority for visualization in living tissues.
In this article, we present the results of crystallographic studies of the three-dimensional structures of eqFP578f at pH 4.0 and 5.5, as well as its far-red variant Katushka, at pH 5.0 and 8.5. eqFP578f is the spectrally identical variant of the wild type eqFP578 with two peripheral substitutions, Arg32Gly and Leu79Phe, which increase the maturation rate almost three times and notably improve folding efficiency. These four crystal structures were solved at resolution ranging from 1.15 to 1.85 Å. We have analyzed in detail the stereochemical features of these FPs that are responsible for the observed pH dependence of their fluorescence, and for the outstanding spectral characteristics of Katushka.
Results
pH dependence of spectral properties
The red wild type progenitor eqFP578f (λex = 552 nm and λem = 578 nm) and its far-red variant Katushka (λex = 588 nm and λem = 635 nm), both characterized by the chromophore forming triad Met63-Tyr64-Gly65, demonstrate pronounced dependence of fluorescence on pH [Fig. 1(a–c)]. The red fluorescence of eqFP578f gradually decreases at pH below ∼5.2, practically diminishing to zero at ∼3.2. Katushka shows lesser pH stability- its far-red fluorescence starts to decrease at pH below ∼8 and reaches zero at ∼4.5. The absorption spectra of eqF578f and Katushka at pH below certain level (∼4 and ∼5.5, respectively) show the decline of absorption at 552 and 588 nm and its raise at 380 and 450 nm, respectively [Fig. 1(d,e)]. The observed spectral variations at the extreme pH values presumably correlate with structural changes in the chromophore area.
Figure 1.
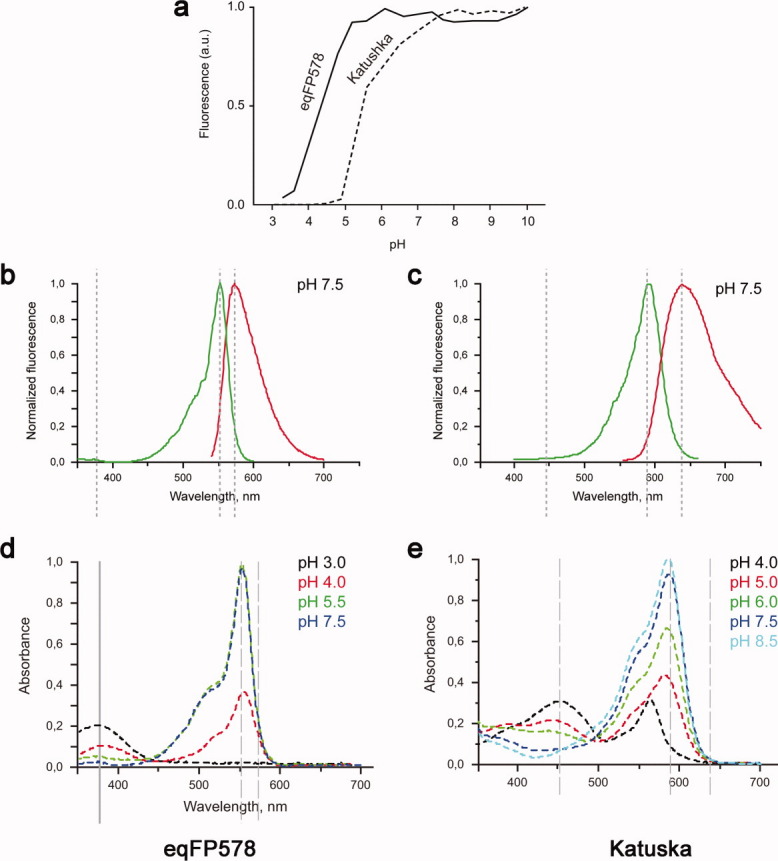
Spectroscopic data for eqFP578 and Katushka. pH dependence of the fluorescence for eqFP578 and Katushka (a). Excitation (in green) and emission (in red) spectra of eqFP578 (λex = 552 nm and λem = 578 nm) (b) and Katushka (λex = 558 nm and λem = 635 nm) (c) at pH 7.5. Absorbance at physiological and lower pH of eqFP578 (d), and Katushka (e).
Monomer and oligomer structure
The principal structural fold of the eqFP578f and Katushka at both high and low pH is an 11-stranded β-barrel, closed from both sides by loop caps. Chromophore moiety (matured from the sequence Met63-Tyr64-Gly65) is embedded in the middle of an internal α-helix that is wound along the β-barrel axis. Similar to TurboGFP and the FPs from Zoanthus,17,18 the β-barrel frames of these proteins show the presence of a pore, formed by the backbones of Trp140, Glu141, Ala142, His/Arg197, Arg/Lys198, and Leu199, with a chain of hydrogen bonded water molecules leading to the chromophore area. Evdokimov et al. suggested that this pore is essential for chromophore maturation, providing access for molecular oxygen.17 Two cis peptide bonds preceding Pro50 and Pro85 in the loop area have been detected. The high resolution structures enabled us to detect alternate stable conformations for a number of side chains.
The asymmetric units of the crystals of eqFP578f and Katushka grown at fluorescence-supporting higher pH (5.5 and 8.5, respectively) each contains a tetramer characterized by 222 point symmetry. The asymmetric units of the crystals of eqFP578f and Katushka at the fluorescence-quenching lower pH (4.0 and 5.0, respectively) contain only dimers which are, however, transformed to the conventional tetramers by the crystallographic symmetry operations. The irregular C-terminal tail of each monomer, starting from residue 222, is detached from the β-barrel side surface and sticks to cylindrical surface of the interacting counterpart in the tetrameric structure.
Fluorescent trans form of the eqFP578f chromophore at pH 5.5
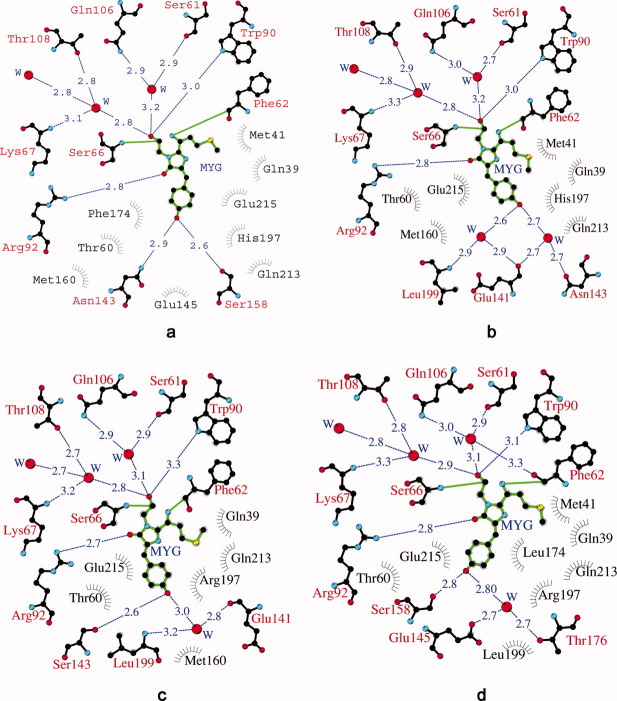
The chromophore of eqFP578f adopts a two-ring coplanar structure consisting of a five-membered imidazolone heterocycle with a p-hydroxybenzylidene substituent [Fig. 2(a)]. Analogously to the homologous red fluorescent protein eqFP611 with which it shares 76% sequence identity20 [Fig. 3], the distinguishing structural feature of the chromophore structure in eqFP578f at pH ≥ 5.5 is the atypical 100% trans conformation of the Tyr64 hydroxyphenyl group relative to Cα-N(64) bond. Similar to the other red and far-red fluorescent proteins,20–24 the first chromophore residue, Met63, is characterized by formation of N-acylimine partially double bond N = Cα, the sp2 hybridization of the corresponding Cα atom, and the cis configuration of the preceding peptide bond. An additional N-acylimine bond apparently extends the chromophore conjugated π-electronic system, resulting in a red shift in spectra. The trans chromophore with the immediate environment of ∼20 residues in eqFP578f provides at pH ≥ 5.5 bright red fluorescence [Figs. 1(b) and 4(a)]. This environment, including the catalytic Glu215 and Arg92, forms an extensive hydrogen bond network actively interacting with chromophore. A number of amino acids contribute strongly to stabilization of the trans isomer, and two hydrogen bonds are made between Tyr64 hydroxyl and the side chains of Asn143 and Ser158 [Fig. 4(a)]. Besides, the plane of the Phe64 hydroxyphenyl moiety is squeezed between the aromatic ring of Phe174, oriented perpendicularly, and the imidazole ring of His197, making the perfect π-stack (d ∼3.5 Å) with the phenolic ring.
Figure 2.
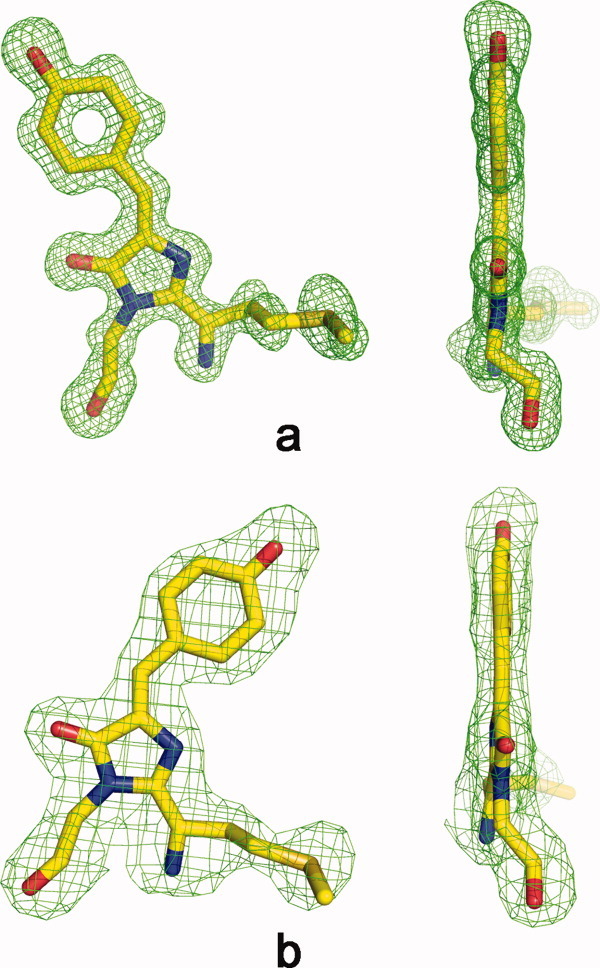
Front and side views of chromophores in fluorescent state in a Fo-Fc omit electron density (cutoff ρ = 3.0 σ). Trans conformation in eqFP578 at pH 5.5 (a) and cis conformation in Katushka at pH 8.5 (d). This figure was produced with PYMOL.19
Figure 3.
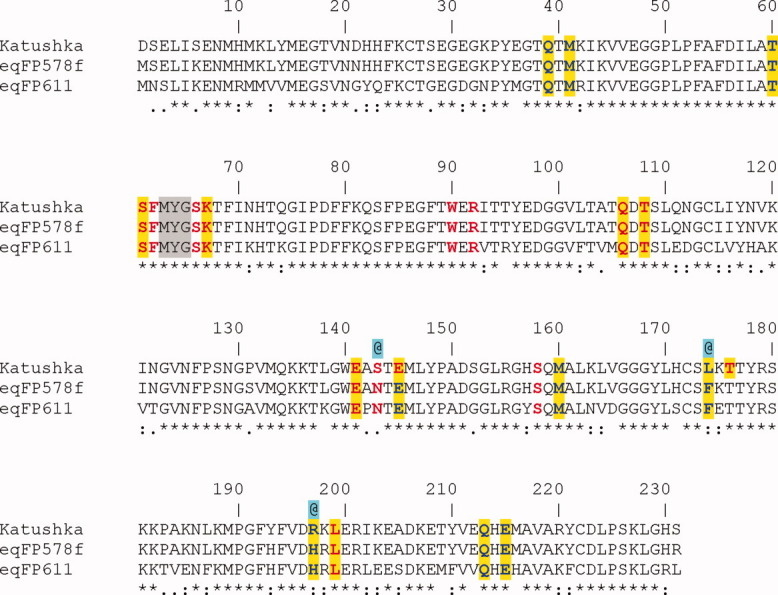
Sequence alignment displaying the residues interacting with the cis/trans chromophores. Red – direct H-bonding, Red (highlighted in yellow) – water mediated H-bonding, Blue – (highlighted in yellow) van der Waals contacts, @ – residues apparently responsible for the far-red shift in Katushka relative to eqFP578f and eqFP611, MYG – chromophore forming triad.
Figure 4.
A diagram illustrating the environment of chromophore in crystal structure. eqFP578: trans fluorescent state at pH 5.5 (a) and cis nonfluorescent state at pH 4.0 (b); Katushka: cis fluorescent state at pH 8.5 (c), and trans nonfluorescent state at pH 5.0 (d). Hydrogen bonds (≤ 3.3 Å) are shown as blue dashed lines, waters (W) as red spheres and van der Waals contacts (≤ 3.9 Å) as black “eyelashes.” This Figure was produced with LIGPLOT/HBPLUS.25,26
Nonfluorescent cis form of the eqFP578f chromophore at pH 4.0
Our data show that reducing pH causes gradual trans-cis isomerization of the eqFP578f chromophore, accompanied by extinguishing of fluorescence [Fig. 1(a)].
At pH 4.0, the crystal structure exhibits the presence of ∼80% cis and ∼20% trans chromophore isomers, compared with 100% trans population at pH 5.5. In contrast to the trans form, the cis bicyclic form is not fully planar. The deviation of the Tyr64 phenolic ring out of plane of the imidazolone ring is characterized by a ∼30° rotation around the Cβ-Cγ bond of Phe64. This state is stabilized by two proximal water molecules mediating the H-bond interactions with the Asn143 side chain and the backbones of Leu199 and Glu141 [Fig. 4(b)]. The trans-cis chromophore isomerization retains basically unchanged position of the chromophore imidazolone ring and the amino acid environmental structure in the chromophore area.
Similar to the close homolog eqFP611,12 the eqFP578f (76% of sequence identity with the former) was shown to remain essentially unaffected by lowering the pH from 8.0 to 5.0. It was suggested that the cis conformation of the red chromophore protects the acylimine bond from immediate reduction or hydrolysis.12 Below pH ∼4, the absorption spectra for eqFP578f and eqFP611 show the respective raise of the ∼370 and ∼380 nm bands at the expense of the ∼552 and ∼557 nm bands, suggesting reduction of the chromophore acylimine bond [Fig. 1(d)]. The 1.75 Å electron density of the eqFP578f at the borderline pH 4 does not show any evidence of the acylimine bond reduction. It is also possible that the local pH of the chromophore area might be somewhat higher than the pH in crystallization media.
Fluorescent cis form of the Katushka chromophore at pH 8.5
In contrast to eqFP578f, exhibiting bright red fluorescence at pH > 5 (λem = 578 nm) represented by the trans state of the chromophore, its engineered variant Katushka demonstrates bright far-red fluorescence at pH > 8 (λem = 635 nm), represented by chromophore in the 100% cis state [Figs. 1(c) and 2(b)]. In this state, the two rings of the chromophore are coplanar. Compared to the environment of the chromophore in eqFP578f, only three amino acids are different in Katushka (Ser143, Leu174, Arg197 instead of Asn143, Phe174, His197) (Fig. 3). At higher pH, these replacements favor of the cis state, with the Tyr64 phenolic ring flanked from both sides by the hydrophobic portions of the side chains of Arg197 and Thr60/Met160 (d ∼3.4–3.8 Å). In Katushka cis conformation of the chromophore is stabilized by two H-bonds between Phe64 hydroxyl and the side chain of Ser143 and a water molecule, mediating interactions with the backbones of Glu141 and Leu199 [Fig. 4(c)].
Nonfluorescent trans form of the Katushka chromophore at pH 5.0
In contrast to eqFP578, in Katushka the pH reduction causes the opposite cis-trans isomerization of the chromophore, accompanied by gradual loss of fluorescence. Below pH ∼5.5, the absorption spectrum of Katushka shows a reduction of 588 nm and the raise of 450 nm bands. The latter band corresponds to the protonated red chromophore [Fig. 1(e)]. At pH 5.0, the crystal structure of Katushka exhibits both trans and cis chromophore isomers in a ratio ∼80 and ∼20%. The appearance of a trans isomer at pH 5.0 correlates with the appearance of the second alternate conformation for the side chains of the chromophore proximal residues Ser158 and Arg197, in addition to those observed for the cis fluorescent state at higher pH 8.5. No other environmental structural differences between cis and trans states in the chromophore area have been found. The chromophore in the trans isomeric form exhibits almost planar bicyclic structure. It is stabilized by H-bonds between Phe64 hydroxyl and the side chain of Ser158 in the second alternate conformation and a water molecule, mediating interactions with the side chains of Glu145 and Thr176 [Fig. 4(d)]. The plane of the Phe64 phenolic ring resides between two positively charged guanidinium groups of the catalytic Arg92 side chain and the Arg197 side chain in the second alternate orientation (d ∼3.0–3.1 Å).
Discussion
The red progenitor eqFP578f demonstrates somewhat higher pH stability than its far-red variant Katushka. Its fluorescence gradually decreases at pH below ∼5.2, approaching zero at ∼3.2, compared with the corresponding values of ∼8 and ∼4.5 for Katushka [Fig. 1(a)]. Interestingly, the fluorescent states at higher pH are represented by different chromophore isomers, trans in eqFP578f and cis in Katushka. The fluorescent trans isomer in eqFP578f is stabilized by H-bonds between the hydroxyl of the chromophore phenolic ring and the side chains of Asn143 and Ser158 [Fig. 4(a)]. Additional stabilization is possibly provided by the imidazole ring of His197, making a perfect π-stack with the chromophore phenolic ring. The fluorescent cis isomer in Katushka is fixed by hydrogen bonding of the phenolic ring hydroxyl with the side chain of Ser143 (which replaced the Asn present in eqFP578f) and the proximal water molecule, mediating interactions with the backbones of Glu141 and Leu199 [Figs. 3 and 4(c)]. An additional indirect contribution may be provided by Arg197 and Leu174 (replacing respective His and Phe in eqFP578f), presumably destabilizing the trans chromophore state in favor of the cis state.
The proteins studied here demonstrate an opposite direction of isomerization upon decreasing pH – trans to cis in eqFP578f and cis to trans in Katushka. Increasing the acidity quenches charge-charge interactions mainly due to the contribution from protonation of the carboxylic groups, characterized by low pK values. Also, it presumably weakens the stabilizing H-bonds, enhancing the role of the electrostatic field in thermodynamic equilibrium of the two chromophore isomers. A significant change of the electrostatic field due to pH-induced protonation of the chromophore environment is presumably the main driving force of the chromophore isomerization.
The distinguishing feature of the fluorescent state of eqFP578f is the perfect π-stacking interaction of the trans chromophore phenolic ring with the imidazole ring of His197. It was proposed that such interactions with aromatic residues increase polarizability of the chromophore, resulting in the red shift of fluorescence.27 Corresponding observations on a number of selectively designed GFP variants support this proposal.28,29
Interestingly, the wt red eqFP578 (λex = 552 nm and λem = 578 nm) and the homologous wt far-red eqFP611 (76% sequence identity with eqFP578; λex = 559 nm, and λem = 611 nm), having identical chromophores (matured from Met63-Tyr64-Gly), residing in an identical nearest stereochemical environment (within ∼4 Å), exhibit significantly different fluorescence. Both structures are characterized by the RMSD values for all Cα atoms < 0.5 Å and exhibit strikingly similar arrangement of the side chains proximal to the chromophore. No evident structural reason explaining the red spectral shift of eqFP611 relative to eqFP578f has been found. Apparently, the combined effect of the charged residues from the second chromophore shell (d > 4 Å) contributes to the observed phenomenon. The electrostatic interactions, having long range nature (acting within ∼7 Å), might play the decisive role. Stereochemical analysis of the second shell drew our close attention to the residue at position 175, found next to the key Phe174 that interacts with the chromophore. This adjacent position, occupied by the positively charged Arg175 in eqFP578, is occupied by the negatively charged Glu175 in eqFP611. The side chains of these residues are oriented away from the chromophore area towards the intermonomer interface. However, the introduction of a single mutation Lys175Glu in the monomeric TagRFP; (the eqFP578 variant with 91% sequence identity and λem = 584 nm) did not result in the expected red shift of the emission maximum (unpublished results).
At physiological pH, the far-red variant Katushka exhibits fluorescence considerably shifted to the far-red spectral region (λex = 588 nm, λem = 635 nm) compared to eqFP578f and eqFP611. Our results indicate that the most critical residues responsible for the advanced spectral properties of Katushka (Ser143, Leu174, and Arg197) are located in the vicinity of chromophore. Indeed, during the process of designing Katushka from the wt eqFP578, we observed that two replacements, Asn143Ser and Phe174Leu, shifted the excitation maximum from 552 to 574 nm and the emission maximum from 578 to 602 nm. Further introduction of the His197Arg replacement, together with a limited number of mutations in the regions distant from the chromophore (the latter to improve the folding and aggregation properties) resulted in the final far-red variant Katushka.6 Moreover, these three replacements in TagRFP (λex = 588 nm and λem = 584 nm) were found to be sufficient to reach the far-red spectral characteristics similar to those in Katushka.6 The chromophore in Katushka adopts a conventional cis conformation, stabilized by H-bond with key residue Ser143, supported by contribution from water-mediated interactions with Glu141 and Leu191 [Fig. 4(c)]. Other key residues, Leu174 and Arg197, presumably destabilize the trans chromophore form in favor of cis. The cis conformation for the isolated chromophore was shown to be energetically more stable than the trans form.30 Therefore, we suggest that the significant red shift of the fluorescence emission from 578 nm in eqFP578f to 635 nm in Katushka is mainly caused by transformation of the chromophore trans form to the lower energy and consequently less strained cis form. For its excitation, the cis chromophore requires the lower energy (longer wavelength) light, which apparently provides the key contribution to the observed far-red spectral shift in the Katushka variant.
The importance of the Asn143Ser replacement in eqFP578 for the observed red spectral shift is supported by results of a mutational study of the homologous eqFP611.12,31 Similar to eqFP578, its fluorescent state is dependent on the 100% trans chromophore. The introduction of the Asn143Ser substitution in eqFP611 is accompanied by 80% trans-cis isomerization of the chromophore, with shift of the emission maximum from 611 nm to 630 nm. Addition of the second Ser158Cys substitution results in the variant RFP639 with 100% of cis chromophore and emission at 639 nm. Similar to Katushka, Ser143 in RFP639 stabilizes the cis chromophore state by H-bonding with the chromophore hydroxyl. Cys158 apparently provides additional support by weakening the corresponding H-bond with the trans chromophore and, thereby, destabilizing that state. An analogous replacement Ser158Cys in Katushka resulted in a variant with generally unchanged spectral characteristics, but exhibiting a significant portion of green fluorescence (unpublished data). In this context, it is worth to mention mKate, the monomeric far red variant of Katushka (91% of sequence homology; λem = 635 nm).32 Both proteins are characterized by the identical amino acid environment around their cis chromophores and practically identical 3D structure (RMSD value for all CA atoms < 0.5 Å). The mutation Ser158Ala in mKate left the spectral characteristics unchanged but increased the brightness of fluorescence by almost a factor of two.32 Here, breaking the corresponding H-bond with the chromophore in the potential nonfluorescent trans form indirectly stabilizes the fluorescent cis form.
Experimental Procedures
Expression, purification and characterization
For bacterial expression, PCR-amplified BamHI/ HindIII fragments encoding target proteins were cloned into the pQE30 vector (Qiagen). Proteins fused to the N-terminal polyhistidine tag were expressed in E. coli XL1 Blue strain (Invitrogen). The bacterial cultures were grown overnight at 37°C, and additionally incubated for 12 h at 25°C. No induction by IPTG was applied since promoter leakage was sufficient for effective expression. The cultures were centrifuged, the cell pellets resuspended in 20 mM Tris-HCl, 100 mM NaCl, pH 7.4 buffer and lysed by sonication. The recombinant proteins were purified using TALON metal-affinity resin (Clontech) followed by gel filtration on Superdex-200 (16/60) size exclusion column (Amersham).
Absorption spectra were recorded with a Beckman DU520 UV/VIS spectrophotometer. A Varian Cary Eclipse fluorescence spectrophotometer was used for measuring excitation-emission spectra. The pH titrations were performed by using a series of buffers in the range from 3.5 to 9.7. For each pH value, an aliquot of purified protein was diluted in the corresponding buffer solution, and the fluorescence brightness was estimated as a product of molar extinction coefficient and a fluorescence quantum yield after 12 h incubation at room temperature.
Crystallization and data collection
Before crystallization, the proteins were transferred to 10 mM Tris-HCl pH 7.5 100 mM NaCl 2.5 mM EDTA buffer and concentrated to ∼11 mg/mL. Initial searches for crystallization conditions were performed with Hampton Research and Emerald Biosystems crystallization kits using the Mosquito robotic system (TTP LabTech). Successful hits were further optimized manually. Crystals have been obtained under four different conditions. The eqFP578f_pH5.5 crystals have appeared from 25% w/v PEG 3350, 0.2-M ammonium acetate, and 0.1-M BIS-TRIS pH 5.5. The eqFP578f_pH4.0 was crystallized from 2.4-M ammonium sulfate, pH 4.0. Crystals of Katushka_pH8.5 were grown from 24% w/v PEG4000, 0.16-M MgCl2, 0.08 M TRIS hydrochloride pH 8.5, and 20% v/v glycerol. The Katushka_pH5.0 was crystallized from 1.5-M ammonium sulfate, pH 5.0, 25% v/v glycerol. All crystals have been obtained by hanging drop vapor diffusion method at 20°C. Typically, 1 μL of protein was mixed with 1μl of reservoir solution and equilibrated against 500 μL of reservoir solution. Crystal growth time varied from 10 to 20 days.
X-ray diffraction data were collected from single crystals flash-cooled in a 100 K nitrogen stream. Before cooling, the crystals of eqFP578f were transferred to a cryo-protecting solution containing 20% glycerol and 80% reservoir solution. Data were collected with a MAR300 CCD detector at the SER-CAT beamline 22-ID (Advanced Photon Source, Argonne National Laboratory, Argonne, IL) and were processed with HKL2000.33
Structure solution and crystallographic refinement
Crystal structures of eqFP578f and Katushka at higher pH (5.5 and 8.5, respectively) were solved by the molecular replacement method with MOLREP,34,35 using as a search model the coordinates of the closely homologous mKate monomer (PDB ID: 3BXA; Ref.32), without the chromophore. The refined coordinates of the eqFP578f and Katushka structures at higher pH with chromophore moiety removed were used to solve the corresponding structures at lower pH (4.0 and 5.5, respectively).
Structure refinement of eqFP578f was performed with PHENIX36 and that of Katushka with REFMAC5,37 alternating with manual correction of the model using COOT.38 Water molecules were located with PHENIX and ARP/wARP.36,39 The fractions of the cis and trans isomers of the chromophore were estimated from the occupancy coefficients determined with phenix.refine (within a general crystallographic refinement procedure), with the sum of occupancies for both isomers constrained to unity. Crystallographic data and refinement statistics are presented in Table I. Although the values of Rmerge were relatively high in the outermost shells for the Katushka data sets, the corresponding values of I/σ(I) indicated that these data were still significant. In all cases, the use of the complete experimental data sets for protein refinement improved the quality of the resulting electron density maps.
The electron density maps for all four structures were in good agreement with the corresponding amino acid sequences. The final models were characterized by clear electron density in the chromophore area. With one exception, no convincing density was observed for the N-terminal His-tag fragments introduced into the expressed constructs and used for protein purification. Only the eqFP578f structure at pH 5.5 contained a well-ordered His-tag fragment for monomer B. Besides the protein and water molecules, a single glycerol molecule (a component of the cryoprotectant solution) was located near each monomer in the eqFP578f structures at pH 5.5 and pH 4.0, whereas three sulfate ions (components of the crystallization solution) were located in the Katushka structure at pH 5.0. Structure validation was performed with COOT. The figures were prepared with LIGPLOT/HBPLUS25,26 and PYMOL.19
The coordinates and structure factors were deposited in the Protein Data Bank under accession codes 3PIB (eqFP578f_pH5.5), 3PJB (eqFP578f_pH4.0), 3PJ7 (Katushka_pH8.5), and 3PJ5 (Katushka_pH5.0).
Acknowledgments
We acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT), located at the Advanced Photon Source, Argonne National Laboratory. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U. S. Government.
References
- 1.Verkhusha VV, Lukyanov KA. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat Biotechnol. 2004;22:289–296. doi: 10.1038/nbt943. [DOI] [PubMed] [Google Scholar]
- 2.Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Zubova NN, Savitsky AP. Molecular cellular sensors created on the base of fluorescent proteins. Uspekhi Biol Khim (Rus) 2005;45:1–66. [Google Scholar]
- 4.Seitz G, Warmann SW, Fuchs J, Mau-Holzmann UA, Ruck P, Heitmann H, Hoffman RM, Mahrt J, Muller GA, Wessels JT. Visualization of xenotransplanted human rhabdomyosarcoma after transfection with red fluorescent protein. J Pediatr Surg. 2006;41:1369–1376. doi: 10.1016/j.jpedsurg.2006.04.039. [DOI] [PubMed] [Google Scholar]
- 5.Remington SJ. Fluorescent proteins: maturation, photochemistry and photophysics. Curr Opin Struct Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 7.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Shyy JY, Chien S. Fluorescence proteins, live-cell imaging, and mechanobiology: seeing is believing. Annu Rev Biomed Eng. 2008;10:1–38. doi: 10.1146/annurev.bioeng.010308.161731. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman RM. Advantages of multi-color fluorescent proteins for whole-body and in vivo cellular imaging. J Biomed Opt. 2005;10:41202–41210. doi: 10.1117/1.1992485. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman RM. In vivo cell biology of cancer cells visualized with fluorescent proteins. Curr Top Dev Biol. 2005;70:121–144. doi: 10.1016/S0070-2153(05)70006-5. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman RM. The multiple uses of fluorescent proteins to visualize cancer in vivo. Nat Rev Cancer. 2005;5:796–806. doi: 10.1038/nrc1717. [DOI] [PubMed] [Google Scholar]
- 12.Kredel S, Nienhaus K, Oswald F, Wolff M, Ivanchenko S, Cymer F, Jeromin A, Michel FJ, Spindler KD, Heilker R, Nienhaus GU, Wiedenmann J. Optimized and far-red-emitting variants of fluorescent protein eqFP611. Chem Biol. 2008;15:224–233. doi: 10.1016/j.chembiol.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Konig K. Multiphoton microscopy in life sciences. J Microsc. 2000;200:83–104. doi: 10.1046/j.1365-2818.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- 14.Gurskaya NG, Fradkov AF, Terskikh A, Matz MV, Labas YA, Martynov VI, Yanushevich YG, Lukyanov KA, Lukyanov SA. GFP-like chromoproteins as a source of far-red fluorescent proteins. FEBS Lett. 2001;507:16–20. doi: 10.1016/s0014-5793(01)02930-1. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci U S A. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shkrob MA, Yanushevich YG, Chudakov DM, Gurskaya NG, Labas YA, Poponov SY, Mudrik NN, Lukyanov S, Lukyanov KA. Far-red fluorescent proteins evolved from a blue chromoprotein from Actinia equina. Biochem J. 2005;392:649–654. doi: 10.1042/BJ20051314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evdokimov AG, Pokross ME, Egorov NS, Zaraisky AG, Yampolsky IV, Merzlyak EM, Shkoporov AN, Sander I, Lukyanov KA, Chudakov DM. Structural basis for the fast maturation of Arthropoda green fluorescent protein. EMBO Rep. 2006;7:1006–1012. doi: 10.1038/sj.embor.7400787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pletneva N, Pletnev V, Tikhonova T, Pakhomov AA, Popov V, Martynov VI, Wlodawer A, Dauter Z, Pletnev S. Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus. Acta Crystallogr D Biol Crystallogr. 2007;63:1082–1093. doi: 10.1107/S0907444907042461. [DOI] [PubMed] [Google Scholar]
- 19.DeLano WL. The PyMOL molecular graphics system. Palo Alto CA: DeLano Scientific; 2002. [Google Scholar]
- 20.Petersen J, Wilmann PG, Beddoe T, Oakley AJ, Devenish RJ, Prescott M, Rossjohn J. The 2.0-A crystal structure of eqFP611, a far red fluorescent protein from the sea anemone Entacmaea quadricolor. J Biol Chem. 2003;278:44626–44631. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- 21.Wall MA, Socolich M, Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat Struct Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 22.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Proc Natl Acad Sci U S A. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tubbs JL, Tainer JA, Getzoff ED. Crystallographic structures of Discosoma red fluorescent protein with immature and mature chromophores: linking peptide bond trans-cis isomerization and acylimine formation in chromophore maturation. Biochemistry. 2005;44:9833–9840. doi: 10.1021/bi0472907. [DOI] [PubMed] [Google Scholar]
- 24.Wilmann PG, Petersen J, Pettikiriarachchi A, Buckle AM, Smith SC, Olsen S, Perugini MA, Devenish RJ, Prescott M, Rossjohn J. The 2.1A crystal structure of the far-red fluorescent protein HcRed: inherent conformational flexibility of the chromophore. J Mol Biol. 2005;349:223–237. doi: 10.1016/j.jmb.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 25.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 26.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 27.Wachter RM, Elsliger MA, Kallio K, Hanson GT, Remington SJ. Structural basis of spectral shifts in the yellow-emission variants of green fluorescent protein. Structure. 1998;6:1267–1277. doi: 10.1016/s0969-2126(98)00127-0. [DOI] [PubMed] [Google Scholar]
- 28.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 29.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 30.Quillin ML, Anstrom DM, Shu X, O'Leary S, Kallio K, Chudakov DM, Remington SJ. Kindling fluorescent protein from Anemonia sulcata: dark-state structure at 1.38 A resolution. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 31.Nienhaus K, Nar H, Heilker R, Wiedenmann J, Nienhaus GU. Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611. J Am Chem Soc. 2008;130:12578–12579. doi: 10.1021/ja8046443. [DOI] [PubMed] [Google Scholar]
- 32.Pletnev S, Shcherbo D, Chudakov DM, Pletneva N, Merzlyak EM, Wlodawer A, Dauter Z, Pletnev V. A crystallographic study of bright far-red fluorescent protein mKate reveals pH-induced cis-trans isomerization of the chromophore. J Biol Chem. 2008;283:28980–28987. doi: 10.1074/jbc.M800599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project n4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 36.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Perrakis A, Sixma TK, Wilson KS, Lamzin VS. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D Biol Crystallogr. 1997;53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]