A complex of KAP1 and HP1 is needed to tether AID to the H3K9me3-marked donor switch region during CSR.
Abstract
Immunoglobulin class switch recombination (CSR) is initiated by double-stranded DNA breaks (DSBs) in switch regions triggered by activation-induced cytidine deaminase (AID). Although CSR correlates with epigenetic modifications at the IgH locus, the relationship between these modifications and AID remains unknown. In this study, we show that during CSR, AID forms a complex with KAP1 (KRAB domain–associated protein 1) and HP1 (heterochromatin protein 1) that is tethered to the donor switch region (Sμ) bearing H3K9me3 (trimethylated histone H3 at lysine 9) in vivo. Furthermore, in vivo disruption of this complex results in impaired AID recruitment to Sμ, inefficient DSB formation, and a concomitant defect in CSR but not in somatic hypermutation. We propose that KAP1 and HP1 tether AID to H3K9me3 residues at the donor switch region, thus providing a mechanism linking AID to epigenetic modifications during CSR.
During immune responses, B cells diversify their Ig genes by somatic hypermutation (SHM) and class switch recombination (CSR). SHM modifies antibody affinities by introducing mutations in the variable region of heavy (IgH) and light chain genes (Di Noia and Neuberger, 2007). CSR modulates antibody effector functions by replacing the antibody isotype expressed through a DNA recombination reaction that joins two switch regions (Chaudhuri et al., 2007). SHM and CSR are initiated by the deamination of cytosines to uracils in single-stranded DNA mediated by activation-induced cytidine deaminase (AID; Petersen-Mahrt et al., 2002; Chaudhuri et al., 2007; Di Noia and Neuberger, 2007). The resulting dU:dG mismatches are differentially processed to generate mutations in variable regions during SHM or double-stranded DNA breaks (DSBs) in switch regions during CSR (Chaudhuri et al., 2007; Di Noia and Neuberger, 2007). AID appears to find its targets by its binding to Spt5 and RNA polymerase II (Pavri et al., 2010) and distinguishes between variable and switch regions by its association with proteins like RPA, PKA, 14-3-3, or by the formation of higher-order DNA structures in switch regions (Chaudhuri et al., 2007; Vuong et al., 2009; Xu et al., 2010; Yamane et al., 2011). AID itself also contributes to this choice, as N-terminal mutations in AID result in normal CSR but defective SHM (Shinkura et al., 2004; Wei et al., 2011) and conversely C-terminal truncations in AID result in normal SHM but defective CSR (Chaudhuri et al., 2007; Kracker et al., 2010). The latter phenotype is also observed in mice deficient for DNA damage response (DDR) components (Ramiro et al., 2007) and in a subset of hyper IgM patients with a CSR-specific defect (Kracker et al., 2010). It has been proposed that this domain associates with CSR-specific factors that could be required for targeting AID to switch regions (Ta et al., 2003) or for recombination downstream of DSB formation either by mediating RNA editing (Doi et al., 2009; Nonaka et al., 2009) or by recruiting factors that promote efficient DNA repair (Barreto et al., 2003; Ta et al., 2003; Ito et al., 2004; Shinkura et al., 2004; Kracker et al., 2010). In addition, epigenetic modifications at the IgH locus, including histone H3 trimethylation at lysine 9 (H3K9me3), have been suggested to target the CSR machinery to switch regions (Wang et al., 2006, 2009; Chowdhury et al., 2008; Kuang et al., 2009). However, a direct causal link between these modifications and AID function in CSR has not been established.
RESULTS
AID associates with KAP1 (KRAB domain–associated protein 1)
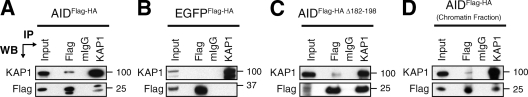
To identify nuclear proteins associating with AID, we performed sequential immunoprecipitations followed by mass spectrometry identification using nuclear extracts prepared from CH12 B cells expressing tagged AID (AIDFlag-HA; Fig. S1). As negative controls, we used cell lines expressing the tags alone (Flag-HA) or an irrelevant tagged protein (EGFPFlag-HA). Among the identified proteins specifically coprecipitating with AID but not with the negative controls, we found known AID partners, DNA-PKCS (Wu et al., 2005) and CRM1 (Brar et al., 2004; Ito et al., 2004; McBride et al., 2004; Geisberger et al., 2009), proteins previously implicated in CSR, Ikaros (Sellars et al., 2009), MSH2 (Chaudhuri et al., 2007), and MSH6 (Chaudhuri et al., 2007), and proteins with no known function in SHM or CSR, like KAP1 (Fig. S1). KAP1 (also known as TRIM28, KRIP1, or Tif1β) was selected for analysis because it has been previously implicated as an effector of the DDR (Ziv et al., 2006). KAP1 is also a transcriptional corepressor (Cammas et al., 2000) that associates with members of the HP1 (heterochromatin protein 1) family to participate in chromatin packaging and heterochromatin formation (Nielsen et al., 1999). The association between AID and KAP1 was confirmed by reciprocal immunoprecipitations and Western blotting (Fig. 1 A) and was specific, as KAP1 did not coprecipitate with EGFPFlag-HA (Fig. 1 B), despite relatively higher expression of EGFPFlag-HA (Fig. S1). To determine whether AID associates with KAP1 through its C terminus, we performed immunoprecipitation and Western blot experiments using extracts prepared from CH12 cells expressing an AID C-terminal truncation mutant (AIDFlag-HA Δ182–198). Although the expression level of AIDFlag-HA Δ182–198 is significantly lower than AIDFlag-HA or EGFPFlag-HA (Fig. S1), we found that C-terminal deletion in AID did not disrupt its association with KAP1 (Fig. 1 C). To determine whether the association between AID and KAP1 can take place within chromatin, we performed a coimmunoprecipitation experiment on chromatin fractions prepared from CH12 B cells expressing tagged AIDFlag-HA. We found that chromatin-bound AID reciprocally coimmunoprecipitated with KAP1 (Fig. 1 D). We conclude that endogenous KAP1 associates with nuclear and chromatin-bound tagged AID through interactions that do not require its C-terminal domain.
Figure 1.
KAP1 associates with AID. (A–C) Nuclear extracts prepared from CH12 cells stably expressing AIDFlag-HA (A), EGFPFlag-HA (B), or AIDFlag-HA Δ182–198 (C) were immunoprecipitated and blotted with anti-KAP1 and anti-Flag antibodies. Because of the lower expression levels of AIDFlag-HA Δ182–198 when compared with AIDFlag-HA or EGFPFlag-HA (Fig. S1), loading for Western blot (WB) analysis was adjusted accordingly in C. (D) Chromatin fractions prepared from CH12 cells expressing AIDFlag-HA were immunoprecipitated and blotted with anti-KAP1 and anti-Flag antibodies. Input control represents 1% of material used for immunoprecipitation (IP). Molecular mass markers in kilodaltons are indicated. Data are representative of five independent experiments.
KAP1 is required for efficient CSR but is dispensable for SHM
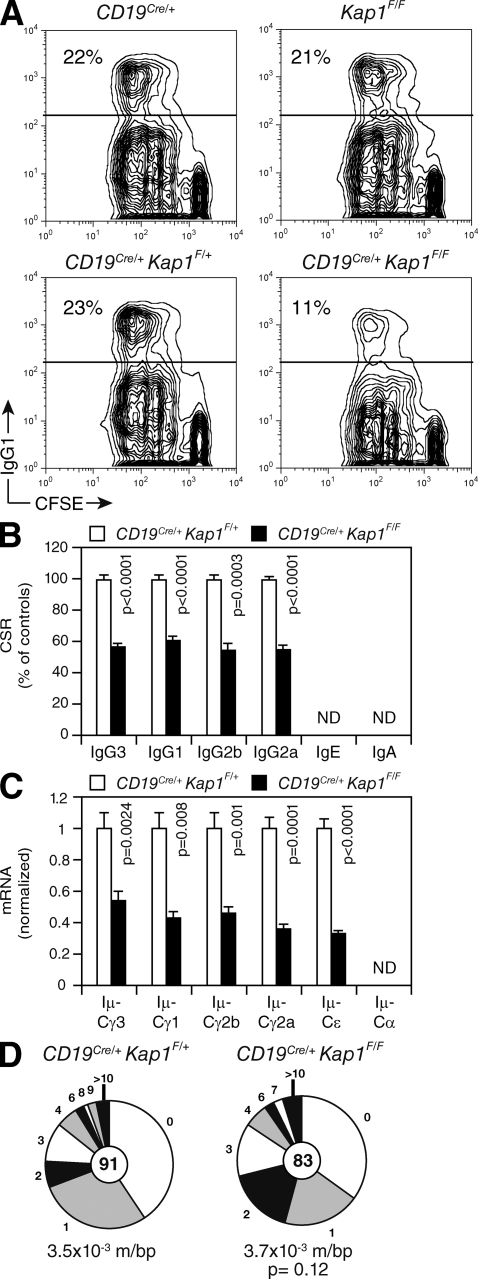
To determine whether KAP1 plays a role in CSR and SHM, we bred Kap1 floxed mice (Kap1F/F; Cammas et al., 2000) with CD19Cre/+ transgenic mice (Rickert et al., 1997) to inactivate Kap1 in developing B cells. Despite efficient Cre-mediated deletion, no differences were observed in cellularity in the bone marrow or the spleen (not depicted), and B cell development appeared to be unaffected (Fig. S2). To determine whether CSR is affected by Kap1 deficiency, we cultured in vitro CFSE-labeled splenic B cells isolated from CD19Cre/+Kap1F/F and control mice under conditions that induce CSR to various isotypes. We found that Kap1 deficiency had no effect on proliferation (Fig. 2 A) or survival (not depicted). However, Kap1 deficiency resulted in a 40–50% reduction in CSR to all isotypes tested (Fig. 2, A–C). Consistent with this, the level of postrecombination Iμ-CH transcripts, which appear only after recombination, was significantly reduced in CD19Cre/+Kap1F/F B cells (Fig. 2 C and Fig. S3). We conclude that Kap1 deficiency results in a B cell–intrinsic defect in CSR that is independent of abnormalities in survival or proliferation.
Figure 2.
KAP1 is required for efficient CSR but is dispensable for SHM. (A) IgG1 surface expression and CFSE dilution by flow cytometry in CD19Cre/+, Kap1F/F, CD19Cre/+Kap1F/+, and CD19Cre/+Kap1F/F B cells stimulated with LPS + IL-4 for 72 h. The percentage of switched cells is indicated in each plot. (B) Percentage (+SD) of CSR after 72 h in CD19Cre/+Kap1F/F relative to CD19Cre/+Kap1F/+ for the different isotypes tested (CSR to IgA and IgE could not be determined). CSR in CD19Cre/+Kap1F/+ B cells was set to 100%. Data are from five independent experiments. (C) qRT-PCR for postswitch (Iμ-CH) transcripts in CD19Cre/+Kap1F/F relative to CD19Cre/+Kap1F/+ stimulated B cells (72 h). Expression is normalized to Cd79b expression and is presented relative to the expression in CD19Cre/+Kap1F/+ B cells, set as 1. Data are representative of three experiments. Error bars indicate SD. (D) JH4 intron sequences from germinal center B cells (B220+Fas+GL-7+) sorted from the lymph nodes of immunized CD19Cre/+Kap1F/F and CD19Cre/+Kap1F/+ mice were analyzed for mutations. Segment sizes are proportional to the number of sequences bearing the indicated mutations. Mutation frequency (mutations/base pair [m/bp]) and number of sequences analyzed (in center) are indicated. Sequences were obtained from four independent experiments. (B–D) P-values were determined by the Student’s t test.
To determine whether SHM is also affected by Kap1 deficiency, we immunized CD19Cre/+Kap1F/F and control mice with NP-CGG, sorted germinal center B cells from the lymph nodes, and analyzed JH4 intron sequences for mutations (Jolly et al., 1997). We did not find significant differences in mutation frequency (Fig. 2 D; P = 0.12), distribution, or pattern (Fig. S4) between sequences obtained from CD19Cre/+Kap1F/F (n = 83) and control (n = 91) animals. We conclude that KAP1 is involved in mediating CSR but appears to be dispensable for SHM.
The function of KAP1 in CSR is upstream of DSB resolution
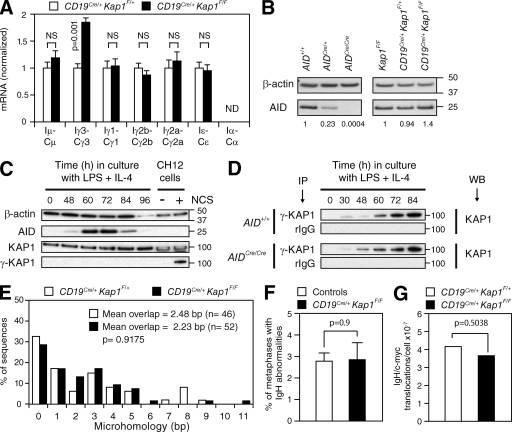
CSR is dependent on transcription (Chaudhuri et al., 2007), and KAP1 is a transcriptional corepressor (Cammas et al., 2000). To determine whether switch region transcription is affected by Kap1 deficiency, we measured by quantitative RT-PCR (qRT-PCR) the level of switch region germline transcripts in activated B cells. We found no significant reduction in the level of these transcripts in Kap1-deficient B cells (Fig. 3 A and Fig. S3). However, occasional increases in the levels of γ3, γ1, γ2b, and γ2a transcripts were found (Fig. 3 A and Fig. S3). This is similar to what is observed in Atm−/−, H2ax−/−, or 53bp1−/− B cells in which CSR is reduced despite normal or increased levels of switch region transcripts (Reina-San-Martin et al., 2003, 2004; Ward et al., 2004). We conclude that switch regions continue to be efficiently transcribed in Kap1-deficient B cells and that switch regions are accessible for DNA deamination by AID. Therefore, the function of KAP1 is downstream of switch region transcription.
Figure 3.
KAP1 functions downstream of switch region transcription and upstream of DSB formation. (A) qRT-PCR for germline (IH-CH) transcripts in CD19Cre/+Kap1F/F relative to CD19Cre/+Kap1F/+ stimulated B cells (72 h). Expression is normalized to Cd79b and relative to CD19Cre/+Kap1F/+ B cells, set as 1. P-values were determined by the Student’s t test. Data are from three independent experiments. Error bars indicate SD. (B) Western blot for AID and β-actin in Kap1F/F, CD19Cre/+Kap1F/+, CD19Cre/+Kap1F/F, AID+/+, AIDCre/+, and AID-deficient (AIDCre/Cre) B cells. Numbers below the panel reflect the intensity of bands representing AID protein levels relative to controls after normalization to β-actin. Molecular mass markers in kilodaltons are indicated. Data are representative of three independent experiments. (C) KAP1 phosphorylation (γ-KAP1) by Western blot in wild-type B cells cultured with LPS + IL-4. As a positive control, CH12 cells treated with neocarzinostatin (NCS). (D) γ-KAP1 was immunoprecipitated from activated AID+/+ and AID-deficient (AIDCre/Cre) B cells and blotted for KAP1. Data are representative of two independent experiments. IP, immunoprecipitation; WB, Western blot. (E) Percentage of Sμ/Sγ3 switch junction sequences with indicated nucleotide overlap from CD19Cre/+Kap1F/+ and CD19Cre/+Kap1F/F LPS-stimulated B cells (72 h) from five independent experiments. Mean length of overlap in base pairs and the number of sequences analyzed (n) is indicated. (F) Quantification of IgH locus breaks as determined by IgH-FISH on metaphases prepared from control (n = 780) and CD19Cre/+Kap1F/F (n = 593) B cells cultured for 3 d with anti-CD40 and IL-4. Mean (+SEM) is shown. Data are from two independent experiments (Table S1). (E and F) P-values were determined by the Mann-Whitney test. (G) Frequency of IgH/c-myc translocations in CD19Cre/+Kap1F/F and CD19Cre/+Kap1F/+ B cells determined by long-range PCR and Southern blot in three independent experiments. The p-value was determined by the one-tailed exact Fisher’s test.
To determine whether Kap1 deficiency affects AID expression, we measured the level of AID protein in activated B cells by Western blot. For comparison, we used AID-deficient (AIDCre/Cre) B cells, which were obtained from mice bearing a targeted insertion of the CRE recombinase cDNA into AID’s exon 1 (Robbiani et al., 2008) and which displayed the same phenotype as AID−/− mice (Muramatsu et al., 2000). Reduced levels in AID protein were found in AIDCre/+ B cells (Fig. 3 B), as has been reported for AID+/− B cells (Sernández et al., 2008). In contrast, Kap1 deficiency had no deleterious effect on AID protein levels (Fig. 3 B). Consistent with this, retroviral overexpression of full-length AID in Kap1-deficient B cells did not rescue CSR, whereas it did in AID-deficient B cells (Fig. S5). As expected, overexpression of C-terminal truncation AID mutants had no effect on CSR in either AID- or in Kap1-deficient B cells (Fig. S5). We conclude that defective CSR in Kap1-deficient B cells is not caused by reduced levels of AID and that KAP1 functions downstream of AID expression.
KAP1 is phosphorylated at serine 824 (γ-KAP1) by the ATM kinase and accumulates in γ-H2AX–containing foci in response to DNA damage (Ziv et al., 2006). Furthermore, inactivation of DDR components (i.e., ATM and H2AX) results in defective CSR (Ramiro et al., 2007). To determine whether KAP1 is phosphorylated during CSR, we assayed for γ-KAP1 by Western blot. KAP1 phosphorylation was readily detected in control cells exposed to the DSB-inducing chemical neocarzinostatin (Fig. 3 C). Despite robust levels of AID expression, γ-KAP1 was not detected in activated B cells (Fig. 3 C), indicating that KAP1 phosphorylation, if any, is below detection threshold. To determine whether a small fraction of KAP1 is phosphorylated during CSR, we immunoprecipitated γ-KAP1 and blotted for KAP1 (Fig. 3 D). Although γ-KAP1 was detected under these conditions, KAP1 phosphorylation was not dependent on AID expression (Fig. 3 D). We conclude that the role of KAP1 in CSR is independent of its phosphorylation status at serine 824.
CSR requires the generation of DSBs in switch regions and their resolution by nonhomologous end joining (NHEJ; Yan et al., 2007). To determine whether CSR junctions are affected by Kap1 deficiency, we compared IgG3 CSR junctions from CD19Cre/+Kap1F/F (n = 52) and control (n = 46) B cells. Sequence analysis revealed no significant differences (P = 0.9175) in the amount of microhomology at the junctions between groups (Fig. 3 E). We conclude that CSR junctions are unaffected by Kap1 deficiency and that DNA ends are repaired normally by NHEJ.
Inactivation of core NHEJ or DDR components results in global and IgH-specific genomic instability (Franco et al., 2006; Yan et al., 2007). To determine whether Kap1 deficiency results in the accumulation of unresolved DNA breaks triggered by AID in switch regions, we performed two-color IgH-specific fluorescence in situ hybridization (IgH-FISH; Franco et al., 2006; Yan et al., 2007) on metaphase spreads prepared from CD19Cre/+Kap1F/F (n = 593) and control (n = 780) activated B cells. Contrary to Atm−/− or Xrcc4−/− B cells (Yan et al., 2007), we found no significant increase (P = 0.9) in the percentage of metaphases with abnormalities in Kap1-deficient B cells (Fig. 3 F and Table S1). Consistent with this, we found that Kap1 disruption did not result in increased frequency of AID-dependent IgH/c-myc translocations (Fig. 3 G). We conclude that in the absence of KAP1, DSBs generated at switch regions are efficiently repaired and do not lead to global or IgH-specific genomic instability. Therefore, KAP1 functions upstream of the AID-induced DNA breaks that initiate CSR.
AID binding to Sμ is impaired by Kap1 deficiency
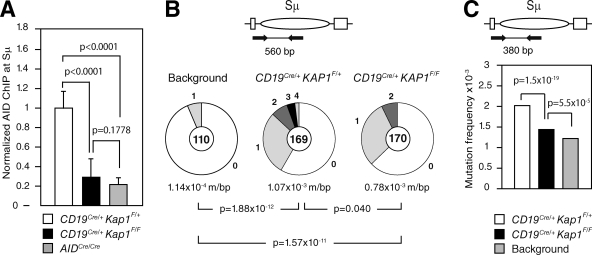
To determine whether AID recruitment to the Sμ switch region is dependent on KAP1, we performed chromatin immunoprecipitation (ChIP) analysis using an anti-AID antibody (Pavri et al., 2010) on chromatin prepared from CD19Cre/+Kap1F/+, CD19Cre/+Kap1F/F, and AIDCre/Cre activated B cells (Fig. 4 A). We found that AID occupancy at the Sμ switch region was significantly reduced (P < 0.0001) in CD19Cre/+Kap1F/F when compared with CD19Cre/+Kap1F/+ B cells (Fig. 4 A). AID-ChIP signal in CD19Cre/+Kap1F/F B cells relative to controls ranged between 20 and 50% (depending on the experiment; not depicted) and was specific, as only background levels of immunoprecipitated AID protein were observed at Sμ in AIDCre/Cre B cells. We conclude that AID binding to the donor switch region is impaired in Kap1-deficient B cells.
Figure 4.
AID binding to Sμ is impaired by Kap1 deficiency and results in reduced levels of DNA damage. (A) ChIP analysis for AID occupancy at the Sμ switch region in CD19Cre/+Kap1F/+, CD19Cre/+Kap1F/F, and AIDCre/Cre B cells cultured in vitro with LPS + IL-4 for 60 h. Normalized AID-ChIP data from three experiments assayed with two different primer sets are shown. For each sample, AID-ChIP values were normalized to the input control and subtracted from the bead-only negative control. AID-ChIP signal in CD19Cre/+Kap1F/+ B cells was assigned an arbitrary value of 1. P-values were determined by the one-tailed Student’s t test. Error bars indicate SD. (B and C) Mutation analysis in the 5′ end of Sμ in CD19Cre/+Kap1F/F and CD19Cre/+Kap1F/+ LPS + IL-4–stimulated B cells (72 h) as determined by Sanger sequencing (B) or HTS (C). Data are from five and four independent experiments, respectively. Background mutation frequency was determined on tail genomic DNA. Pie charts in B are as in Fig. 2. P-values were determined by the two-tailed Student’s t test.
Impaired AID binding results in reduced levels of DNA damage at the Sμ
To determine whether impaired AID recruitment in Kap1-deficient B cells translates into reduced levels of DNA damage, we measured the mutation frequency in the 5′ end of Sμ by cloning and sequencing. This assay has been used to assess the level of AID-induced DNA damage in switch regions (Barreto et al., 2003; Guikema et al., 2007). We found that the proportion of mutated sequences and the mutation frequency were reduced (P = 0.040) in CD19Cre/+Kap1F/F (F = 0.78 × 10−3 mutations/bp; n = 170) when compared with controls (F = 1.07 × 10−3 mutations/bp; n = 169; Fig. 4 B). A significant reduction (P = 1.5 × 10−19) in mutation frequency in the 5′ end of Sμ was also found in Kap1-deficient B cells when sequences were analyzed in a larger dataset by high-throughput amplicon sequencing (HTS; Fig. 4 C). We conclude that impaired AID recruitment to Sμ results in reduced levels of DNA damage and consequently in defective CSR.
AID forms a complex with KAP1 and HP1 that binds H3K9me3
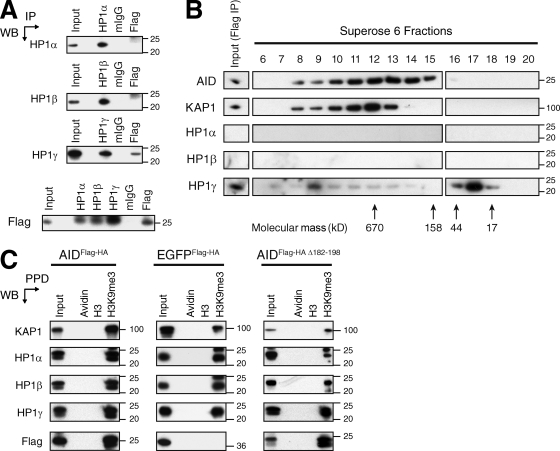
Epigenetic modifications at the IgH locus have been suggested to target the CSR machinery to switch regions by relaxing chromatin or by providing binding motifs for factors involved in CSR (Odegard et al., 2005; Wang et al., 2006; Fraenkel et al., 2007; Chowdhury et al., 2008; Kuang et al., 2009; Wang et al., 2009). Among these, H3K9me3, which is usually associated with silent genes and heterochromatin, was found at transcribed donor and acceptor switch regions (Chowdhury et al., 2008; Kuang et al., 2009). We hypothesized that AID could be tethered to transcribed switch regions bearing this modification through the association between AID, KAP1, and HP1 and the ability of the latter to bind H3K9me3 (Bannister et al., 2001). To determine whether AID also associates with HP1, we performed reciprocal coimmunoprecipitation experiments using nuclear extracts prepared from cells expressing AIDFlag-HA and antibodies specific for the different HP1 isoforms. Although AIDFlag-HA coprecipitated with all HP1 isoforms, only HP1-γ reciprocally coprecipitated AIDFlag-HA (Fig. 5 A). To determine whether KAP1, HP1, and AID exist in a complex, we fractionated Flag immunoprecipitates by gel filtration chromatography using a Superose 6 column and assayed the different fractions for AID, KAP1, and HP1 isoforms by Western blot (Fig. 5 B). We found that AIDFlag-HA, KAP1, and HP1-γ coelute in fractions corresponding to a complex of ∼670 kD (Fig. 5 B). However, the majority of HP1-γ was eluted in low molecular weight fractions (Fig. 5 B), indicating that only a small fraction of the complex contains HP1-γ or that the in vitro association of HP1-γ within the complex is weaker. HP1-α and HP1-β were not detected (Fig. 5 B). To determine whether the AID–KAP1–HP1 complex is able to bind H3K9me3 residues, we performed peptide pull-down experiments using nuclear extracts from cells expressing AIDFlag-HA, EGFPFlag-HA, or AIDFlag-HA Δ182–198 and biotinylated histone H3 peptides either unmodified or trimethylated at lysine 9. We found that the H3K9me3 peptide precipitated KAP1, all the HP1 isoforms, and AIDFlag-HA (Fig. 5 C). Precipitation of AIDFlag-HA was specific, as this peptide did not precipitate EGFPFlag-HA (Fig. 5 C). The H3K9me3 peptide also precipitated AIDFlag-HA Δ182–198 (Fig. 5 C), indicating that the C-terminal domain of AID is not required for the recognition of H3K9me3 residues by the AID–KAP1–HP1 complex. We conclude that KAP1, HP1, and AID exist in a complex that binds H3K9me3 in vitro. Therefore, this modification could serve as a tethering motif for a complex formed by AID, KAP1, and HP1 during CSR in vivo.
Figure 5.
AID forms a complex with KAP1 and HP1 that binds H3K9me3. (A) Nuclear extracts prepared from CH12 cells expressing AIDFlag-HA were immunoprecipitated and blotted with anti–HP1-α, anti–HP1-β, anti–HP1-γ, and anti-Flag antibodies. Data are representative of three independent experiments. (B) Flag immunoprecipitates eluted with the Flag peptide were fractionated with a Superose 6 gel filtration column. The indicated fractions were analyzed by Western blot using antibodies specific for KAP1, HP1-α, HP1-β, HP1-γ, and AID. Arrows indicate the elution position of calibration proteins of known molecular mass. Data are representative of two independent experiments. (C) Peptide pull-downs (PPD) using biotinylated unmodified (H3) or modified H3 peptides (H3K9me3) and avidin-agarose beads (Avidin) on nuclear extracts prepared from CH12 cells stably expressing AIDFlag-HA, EGFPFlag-HA, or AIDFlag-HA Δ182–198. Precipitated proteins were separated by SDS-PAGE and blotted with antibodies specific for KAP1, HP1-α, HP1-β, HP1-γ, and Flag. Data are representative of three independent experiments. (A–C) Molecular mass markers in kilodaltons are indicated. IP, immunoprecipitation; WB, Western blot.
H3K9me3 and KAP1 mark the donor switch region before and during CSR
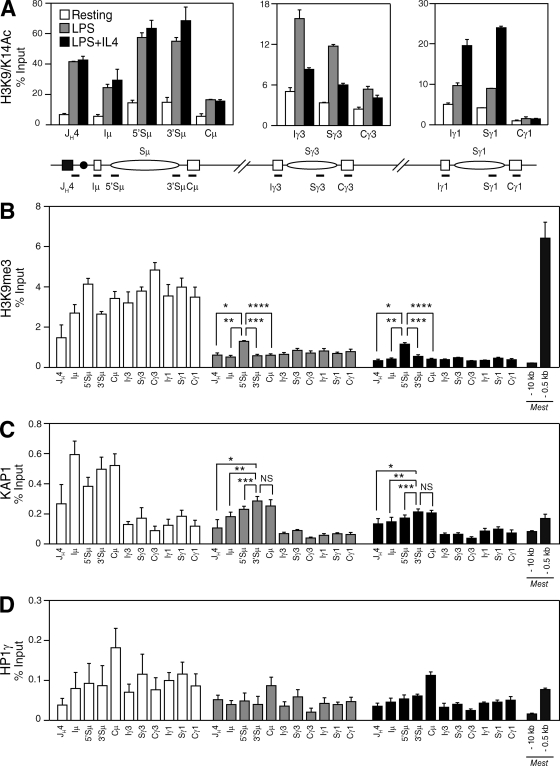
To determine whether KAP1 and HP1 are recruited to switch regions during CSR in vivo and whether this correlates with histone modifications associated with the IgH locus, we performed ChIP experiments on resting and activated control and CD19Cre/+Kap1F/F B cells (Fig. 6 and Figs. S6 and S7). Consistent with a previous study (Wang et al., 2009), we found that histone H3 associated with the pair of recombining switch regions is acetylated at lysine 9 and 14 (H3K9/K14Ac) in B cells undergoing CSR (Fig. 6 A) and that this correlated with switch region transcription (Fig. 3 A). Consistent with normal levels of germline transcripts in Kap1-deficient B cells (Fig. 3 A), we found that H3K9/K14Ac was not affected by Kap1 deficiency (Fig. S6). Although H3K9me3 has been reported to be induced at the donor and acceptor switch regions during CSR (Chowdhury et al., 2008; Kuang et al., 2009), we found that H3K9me3 was already present at the IgH locus before recombination in resting B cells (Fig. 6 B). Upon stimulation, the level of H3K9me3 was substantially reduced throughout the locus but was specifically retained over the donor switch region (Sμ; Fig. 6 B and Table S2) when compared with sequences located immediately downstream of the variable region (JH4), the I exons, the CH exons, or regions surrounding the Mest promoter, which were included as positive and negative controls (Riclet et al., 2009). Surprisingly, H3K9me3 retention was not observed at Sγ3 when cells were stimulated to switch to IgG3 with LPS, nor at Sγ1 when cells were stimulated to switch to IgG1 with LPS + IL-4 (Fig. 6 B). The H3K9me3 pattern was similar in Kap1-deficient B cells (Fig. S6 and Table S3), indicating that this modification is not dependent on KAP1. Interestingly, we found that KAP1, together with H3K9me3, is recruited to the IgH locus in resting B cells. However, KAP1 binding was predominant over Sμ when compared with downstream regions (Fig. 6 C). Upon stimulation, KAP1 was released from the locus but was preferentially retained over Sμ (Fig. 6 C). KAP1 recruitment was specific as no significant ChIP signal above background was found in Kap1-deficient B cells (not depicted). HP1-γ binding differed from H3K9me3 and KAP1 in that it was distributed throughout the locus in resting and activated B cells (Fig. 6 D, Fig. S7, and Table S4). Increased HP1-γ binding at Cμ in activated B cells was observed (Fig. 6 D) but was not reproducible (Fig. S7). We conclude that H3K9me3, HP1-γ, and KAP1 are present at the IgH locus as part of a heterochromatic signature in resting B cells and that H3K9me3 and KAP1 specifically mark the donor switch region (Sμ) before and during CSR.
Figure 6.
H3K9me3 and KAP1 mark the donor switch region (Sμ) before and during CSR. (A–D) ChIP analysis performed on chromatin prepared from resting or LPS-stimulated (48 h) and LPS + IL-4–stimulated (48 h) B cells obtained from CD19Cre/+Kap1F/+ mice using antibodies specific for H3K9/K14Ac (A), H3K9me3 (B), KAP1 (C), and HP1-γ (D). Real-time quantitative PCR was performed by using primer pairs specific for JH4, Iμ, Sμ, Cμ, Iγ3, Sγ3, Cγ3, Iγ1, Sγ1, and Cγ1. Regions surrounding the Mest promoter (−10 kb and −0.5 kb) were used as negative and positive controls, respectively (Riclet et al., 2009). Fold change over control IgG is expressed as a percentage of input. Mean (+SD) of triplicate samples is shown. P-values were determined by the one-tailed Student’s t test (H3K9me3, LPS: *, P = 0.0003; **, P < 0.0001; ***, P < 0.0001; ****, P < 0.0001; H3K9me3, LPS + IL-4: *, P < 0.0001; **, P = 0.0001; ***, P = 0.0004; ****, P < 0.0001; KAP1, LPS: *, P = 0.0040; **, P = 0.0065; ***, P = 0.0289; KAP1, LPS + IL-4: *, P = 0.0134; **, P = 0.0170; ***, P = 0.0346). See Tables S2 and S5 for detailed statistical analysis and primer sequences, respectively. Data are representative of four independent experiments (see Fig. S7 for an additional experiment).
The in vivo KAP1–HP1 association is required for efficient AID binding at Sμ and CSR
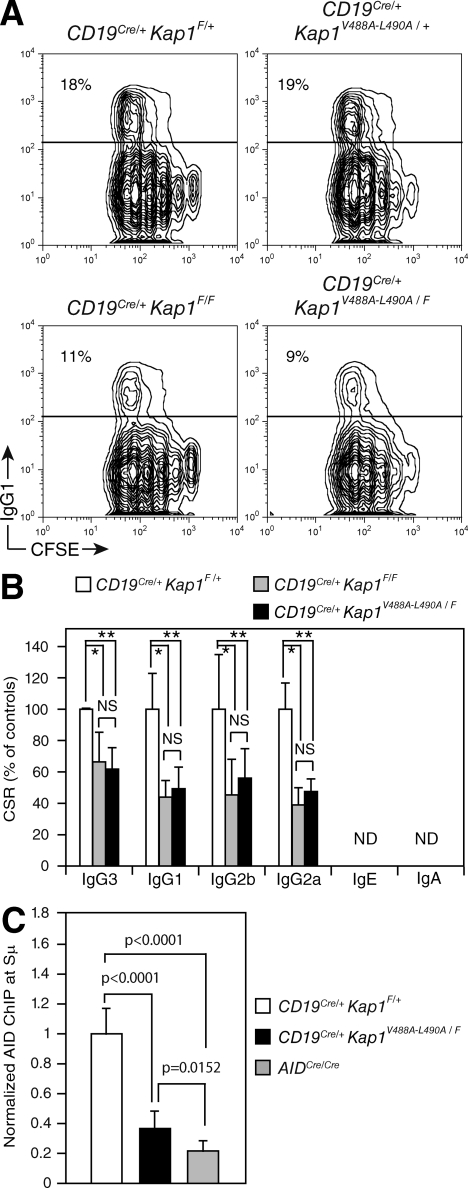
To determine whether the in vivo association between KAP1 and HP1 is required to sustain CSR, we bred CD19Cre/+Kap1F/F mice with knockin mice (Kap1V488A-L490A; Herzog et al., 2011) expressing a mutated KAP1 protein that is unable to associate with the HP1 proteins (Cammas et al., 2004) to generate CD19Cre/+Kap1V488A-L490A/F mice. Although B cells isolated from CD19Cre/+Kap1V488A-L490A/+ mice (expressing a wild-type KAP1 and a mutant protein) underwent CSR at wild-type frequencies (similar to CD19Cre/+Kap1F/+), B cells isolated from CD19Cre/+Kap1V488A-L490A/F mice (expressing only the KAP1 mutant protein, which cannot associate with HP1; Fig. S2) displayed the same CSR defect observed in CD19Cre/+Kap1F/F B cells (Fig. 7, A and B). We conclude that the in vivo association between KAP1 and HP1 is required to sustain efficient CSR.
Figure 7.
The in vivo association between KAP1 and HP1 is required for efficient CSR. (A) IgG1 cell surface expression in CD19Cre/+Kap1F/+, CD19Cre/+ Kap1V488A-L490A/+, CD19Cre/+Kap1F/F, and CD19Cre/+ Kap1V488A-L490A/F CFSE-labeled B cells stimulated with LPS + IL-4 for 3 d. The percentage of switched cells is indicated in each plot. (B) Percentage (+SD) of CSR in CD19Cre/+Kap1F/+, CD19Cre/+Kap1F/F, and CD19Cre/+ Kap1V488A-L490A/F relative to CD19Cre/+Kap1F/+. CSR in CD19Cre/+Kap1F/+ B cells was set to 100%. Data are from five independent experiments. P-values were determined by the one-tailed Student’s t test (*, CD19Cre/+Kap1F/+ vs. CD19Cre/+Kap1F/F: IgG3, P = 0.0154; IgG1, P = 0.0035; IgG2b, P = 0.0264; IgG2a, P = 0.001; **, CD19Cre/+Kap1F/+ vs. CD19Cre/+Kap1V488A-L490A/F: IgG3, P = 0.0017; IgG1, P = 0.0036; IgG2b, P = 0.0276; IgG2a, P = 0.0004). (C) ChIP analysis for AID occupancy at the Sμ switch region in CD19Cre/+Kap1F/+, CD19Cre/+Kap1V488A-L490A/F, and AIDCre/Cre B cells cultured in vitro with LPS + IL-4 for 60 h. Normalized AID-ChIP data from three experiments assayed with two different primer sets are shown. For each sample, AID-ChIP values were normalized to the input control and subtracted from the bead-only negative control. AID-ChIP signal in CD19Cre/+Kap1F/+ B cells was assigned an arbitrary value of 1. P-values were determined by the one-tailed Student’s t test. Error bars indicate SD.
To determine whether AID binding to Sμ is dependent on the interaction between KAP1 and HP1, we performed AID-ChIP experiments on chromatin prepared from CD19Cre/+Kap1F/+, CD19Cre/+Kap1V488A-L490A/F, and AIDCre/Cre activated B cells (Fig. 7 C). We found that AID occupancy at the Sμ switch region was significantly reduced (P < 0.0001) in CD19Cre/+Kap1V488A-L490A/F when compared with CD19Cre/+Kap1F/+ B cells (Fig. 7 C). The reduction in AID retention at Sμ was similar to that observed in Kap1-deficient B cells (Fig. 4 A). We conclude that the in vivo association between KAP1 and HP1 is required to mediate the efficient binding of AID to the donor switch region and to sustain CSR and that a complex formed by AID, KAP1, and HP1 is tethered to the transcribed donor switch region bearing the H3K9me3 mark in vivo.
DISCUSSION
We have found that AID forms a complex with KAP1 and HP1 and that Kap1 inactivation results in a CSR-specific defect that phenocopies mice deficient for components of the DDR (i.e., defective CSR and normal SHM; Ramiro et al., 2007). Because KAP1 has been reported to be an effector of the DDR downstream of ATM (Ziv et al., 2006) and because Kap1 inactivation results in a CSR-specific defect similar to Atm−/− mice (Lumsden et al., 2004; Reina-San-Martin et al., 2004), we hypothesized that the function of KAP1 in CSR is to respond to AID-induced DNA damage. However, we found that the role of KAP1 in CSR is independent of its phosphorylation status at serine 824 and that its inactivation does not result in defective DNA repair or increased genomic instability. Therefore, KAP1 does not appear to participate in the repair of AID-induced DNA damage during CSR.
Impaired CSR in the absence of KAP1 could be explained by a global reactivation of genes normally silenced by a KAP1-dependent mechanism (Nielsen et al., 1999) or by a substantial deregulation of chromatin structure (Ziv et al., 2006) at the IgH locus. However, this is unlikely as the transcriptional repressor activity of KAP1 requires its association with HP1 (Sripathy et al., 2006), and we show that the expression of a KAP1 mutant protein that cannot associate with HP1 (Cammas et al., 2004) was unable to restore CSR to wild-type levels. Furthermore, no significant alterations in switch region transcription were observed because of Kap1 deficiency. We believe that the long-range interactions between switch region promoters and IgH enhancers, which are controlled by transcription (Wuerffel et al., 2007), are most likely not affected and that the global three-dimensional structure of the IgH locus is not altered by Kap1 deficiency. The local access of AID to the switch region is most likely not affected in the absence of KAP1 because AID targeting to switch regions appears to be dependent on its association with Spt5 and RNA polymerase II (Pavri et al., 2010) and because Kap1 inactivation reduces but does not abolish the recruitment of AID to Sμ.
We pinpoint the role of KAP1 in CSR to a step that facilitates AID recruitment and the subsequent generation of DNA damage at Sμ. We find a substantial reduction in AID recruitment that is in contrast with the 50% reduction in CSR that we observe in Kap1-deficient B cells. This might relate to differences in the immunoprecipitation efficiency between experiments. As the AID-ChIP signal in Kap1-deficient B cells (relative to controls) ranged between 20 and 50% depending on the experiment, we think that it might be underestimated.
We show that AID forms a complex with KAP1 and HP1 that selectively recognizes H3K9me3 in vitro. We also show that H3K9me3, KAP1, and HP1 are recruited to the IgH locus in resting B cells and that upon activation H3K9me3 and KAP1 are released but retained over Sμ. With respect to the pattern of H3K9me3, our results differ from a previous study (Kuang et al., 2009) as we find that this modification is not inducible. Although it is difficult to pinpoint the reason for this discrepancy, it most likely relates to technical issues such as differences in culture conditions, time points assayed, detection methods, etc. Nevertheless, the specific enrichment of KAP1 binding at Sμ that we observe suggests that H3K9me3 can serve as a recognition motif for an AID–KAP1–HP1 complex in vivo at the donor switch region during CSR. Although we cannot provide an explanation as to why KAP1 recruitment expands up to Cμ or why it is not recruited to the acceptor switch regions, this suggests that additional factors and/or histone modifications may be required to limit the range of AID recruitment and/or to modulate the efficiency of its retention. As we did not observe recruitment of KAP1 at Sγ3 or Sγ1, it is possible that KAP1 deficiency has no impact in the tethering efficiency of AID to acceptor switch regions, the efficiency of cytidine deamination, and the subsequent generation of DSBs. As DSB formation at acceptor switch regions has been suggested to be a rate-limiting step in CSR (Reina-San-Martin et al., 2003; Schrader et al., 2003), it might explain why the impairment in CSR in the absence of KAP1 is not more profound.
H3K9me3 is usually associated with heterochromatin formation and gene silencing. However, increasing evidence suggests additional roles for heterochromatin (Grewal and Jia, 2007), for example in recruiting factors that facilitate access of RNA polymerase II to heterochromatic regions (Zofall and Grewal, 2006) or by acting as a recruitment platform for factors regulating long-range chromosomal interactions (Jia et al., 2004). The presence of HP1-γ throughout the IgH locus before and during CSR is reminiscent of the mating-type switching in the fission yeast Schizosaccharomyces pombe, in which heterochromatin promotes the spreading of the recombination-promoting complex across the mating locus and imposes structural constraints that are important for the choice of the recombination donor site (Jia et al., 2004). Indeed, H3K9me3 has been shown to be present, along with HP1-γ, at actively transcribed genes in mammalian cells (Vakoc et al., 2005) and at recombining switch regions in mouse and human B cells (Chowdhury et al., 2008; Kuang et al., 2009). Strikingly, we demonstrate that the in vivo KAP1–HP1 association is required for efficient AID binding at Sμ and to sustain CSR by expressing a KAP1 mutant that cannot associate with HP1 (in a Kap1-deficient background).
Our observations are consistent with a model in which AID forms a complex with KAP1 and HP1 that is tethered to transcribed switch regions bearing the H3K9me3 mark. There is precedent for a model based on the recognition of a modified histone, as recognition of hypermethylated histone H3 at lysine 4 by the PHD domain of RAG2 promotes efficient V(D)J recombination (Liu et al., 2007; Matthews et al., 2007). In our model, H3K9me3 and associated KAP1 and HP1-γ mark the Sμ region before recombination. Upon stimulation, these marks are retained over the Sμ and provide a docking motif for AID tethering in vivo. AID would then be retained in close proximity to its substrate by KAP1, HP1, and H3K9me3, thus increasing the probability of cytidine deamination, the efficiency of DSB formation, and consequently of CSR. Our results thus provide a mechanism linking AID to epigenetic modifications associated with the IgH locus during CSR.
MATERIALS AND METHODS
Mice.
Kap1F/F (Cammas et al., 2000), CD19Cre/+ (provided by K. Rajewsky, Harvard Medical School, Boston, MA; Rickert et al., 1997), Kap1V488A-L490A/+ (Herzog et al., 2011), and AIDCre/Cre (Robbiani et al., 2008) mice were on a C57BL/6 background and were bred and maintained under specific pathogen-free conditions. Age-matched littermates (8–12 wk old) were used in all experiments. All animal work was performed under protocols approved by the Direction des Services Vétérinaires du Bas-Rhin, France (Authorization no. 67–343).
Splenic B cell purification, CSR assays, and two-color IgH-FISH.
Resting splenic B cells were isolated, cultured in vitro with LPS (CSR to IgG3 and IgG2b), LPS + IL-4 (CSR to IgG1 and IgE), or LPS + IFN-γ (CSR to IgG2a), and assayed for CSR as described previously (Robert et al., 2009). Switch transcripts, switch junctions, and IgH/c-myc translocations were analyzed as described previously (Robert et al., 2009). Metaphase spreads were prepared and subjected to two-color IgH-FISH as described previously (Franco et al., 2006).
Nuclear extracts and tandem affinity purification.
Nuclear extracts and chromatin fractions were prepared from 12 × 109 cells using standard techniques. 20 mg of clarified extracts was taken into immunoprecipitation buffer (IP-300: 20 mM Tris, pH 7.9, 300 mM KCl, 20% glycerol, 0.25 mM EDTA, 0.125 mM EGTA, 0.5 mM PMSF, 1 mM DTT, 0.025% NP-40, 1× protease inhibitor cocktail [Roche], and 100 U/ml Benzonase [EMD]) and precleared with mouse IgG and protein G–agarose (GE Healthcare) for 1 h at 4°C. Flag M2 agarose beads (100 µl, 50% slurry; Sigma-Aldrich) were added and incubated overnight at 4°C. Immune complexes were eluted three times with 100 µl of 0.2 mg/ml Flag peptide (30 min at 4°C). Elutions were pooled and subjected to immunoprecipitation with anti-HA agarose beads (40 µl, 50% slurry; 4 h at 4°C; Roche). Immune complexes were eluted twice with 40 µl of 0.1 M glycine, pH 2.4 (10 min), or by overnight incubation with 40 µl of 0.6 mg/ml HA peptide. Proteins were fractionated by SDS-PAGE in 4–12% gradient gels (Invitrogen) and stained with Coomassie blue (G250; Bio-Rad Laboratories) before processing for mass spectrometry.
Mass spectrometry analysis.
Whole lanes from one-dimensional electrophoresis were systematically excised in ∼5-mm bands. Proteins in bands were subjected to in-gel reduction, alkylation, and trypsin digestion. Peptides were dried, resuspended in 0.1% formic acid, and injected into the nano liquid chromatography (Ultimate 3000; Dionex) coupled with a linear ion trap mass spectrometer equipped with a nanoelectrospray source (LTQ XL ETD; Thermo Fisher Scientific). Tandem mass spectrometry spectra were recorded in the data-dependent mode on the five most intense ions observed in mass spectrometry scan with collision-induced dissociation and electron transfer dissociation fragmentation. Peptides selected for tandem mass spectrometry acquisition were then dynamically excluded for 30 s. Protein ID was performed using Proteome Discoverer 1.1 (Thermo Fisher Scientific) and SEQUEST (Thermo Fisher Scientific) searching against the mouse Swiss-Prot database (version 57.9) with the following fixed parameters: precursor mass tolerance of ±0.5 D, product ion mass tolerance of ±0.8 D, and two missed cleavages, and carbamidomethylation of cysteine as fixed modification and methionine oxidation as variable modification. SEQUEST results were filtered with Xcorr versus charge state 1.5-1, 1.6-2, 3-3, 3.2-4.
SHM analysis.
Germinal center B cells (B220+Fas+GL-7+) were sorted from the lymph nodes of NP-CGG–immunized mice and analyzed for mutation in JH4 intron sequences (Jolly et al., 1997) with SHMTool (Maccarthy et al., 2009).
Real-time quantitative PCR.
RNA and cDNA were prepared using standard techniques. Quantitative PCR was performed using SYBR green JumpStart Taq ReadyMix (Sigma-Aldrich) or QuantiTect SYBR green PCR kit (QIAGEN). Approximately 3 ng cDNA or 3–5 × 106 cell equivalents of de–cross-linked chromatin were run (in triplicate) and analyzed on a LightCycler 480 (Roche). Transcript or DNA quantities were calculated relative to standard curves and normalized to CD79b transcripts or input DNA. Gene of interest/CD79b or region of interest/input DNA ratios were averaged across experiments and normalized to the appropriate controls.
Sμ mutation analysis.
The 5′ end of Sμ was amplified by PCR using Pfu Turbo as described previously (Reina-San-Martin et al., 2004) or using Platinum Taq polymerase (Invitrogen) and bar-coded primers (Table S5) for HTS. HTS amplification conditions were as follows: 30 cycles of 94°C (15 s), 60°C (15 s), and 68°C (1 min). PCR products were purified with AmPure beads (Agencourt), quantified using Quan-iT Pico Green (Invitrogen), and mixed in equimolar ratios before sequencing with a 454 GS FLX sequencer (Roche; GATC). Sequences were aligned with Lasergene (DNASTAR) and analyzed with SHMTool (Maccarthy et al., 2009).
Peptide pull-downs.
Avidin-conjugated agarose beads (Thermo Fisher Scientific) were coupled to biotinylated unmodified or modified H3 peptides (A1RTKQTARK9[me3]STGGKAPRKQLATK23-biotin; letters in bold indicate the modified amino acid, and the type of modification is in brackets) for 3 h at 25°C with rotation, washed in PBS 1× NP-40 0.1% and resuspended as a 50% slurry. 5 mg of nuclear extracts was precleared with avidin-agarose beads and incubated overnight with peptide-bound resin (40 µl, 50% slurry). Resin was washed and resuspended in 20 µl of Laemmli buffer. Proteins were fractionated by SDS-PAGE and analyzed by Western blot.
ChIP.
The protocol was adapted from Millipore (http://www.millipore.com/userguides/tech1/mcproto407). In brief, 3 × 107 resting or stimulated B cells were cross-linked at 37°C for 10 min in 5 ml PBS/0.5% BSA with 1% formaldehyde. The reaction was quenched with 0.125 M glycine. After lysis, chromatin was sonicated to 0.5–1 kb using a Vibracell 75043 (Thermo Fisher Scientific). After 2× dilution in ChIP buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, and 167 mM NaCl), chromatin was precleared by rotating for 2 h at 4°C with 80 µl of 50% protein A/G slurry (0.2 mg/ml sheared salmon sperm DNA, 0.5 mg/ml BSA, and 50% protein A/G; GE Healthcare). 0.3–0.5 × 106 cell equivalents were saved as input, and 3–5 × 106 cell equivalents were incubated overnight with specific or control antibodies (Table S6). Immune complexes were precipitated by the addition of protein A/G or Dynabeads protein A (for AID) and processed according to the Millipore protocol.
Gel filtration chromatography.
20 mg of nuclear extract was immunoprecipitated with Flag M2–agarose beads in IP-300 buffer, washed, and eluted with 0.2 mg/ml Flag peptide in 100 µl. Eluted proteins were fractionated using an micro Superose 6 PC 3.2/30 column (ÄKTA). 100 µl of proteins in each fraction was analyzed by Western blot.
Online supplemental material.
Fig. S1 shows Western blot analysis of nuclear and cytoplasmic extracts prepared from CH12 cells expressing AIDFlag-HA, Western blot analysis of nuclear extracts from CH12 cells expressing AIDFlag-HA, EGFPFlag-HA, and AIDFlag-HA Δ182–198, SDS-PAGE (silver stain) analysis of proteins eluted after tandem affinity purification, and the KAP1 peptides identified by mass spectrometry. Fig. S2 shows that Kap1 deficiency has no major impact on B cell development. Fig. S3 displays two additional experiments showing that Kap1 deficiency results in reduced levels of postrecombination transcripts without decreasing germline transcription. Fig. S4 shows that Kap1 deficiency has no effect on the frequency of germinal center B cells and the distribution or pattern of somatic mutations. Fig. S5 shows that AID overexpression in Kap1-deficient B cells does not rescue CSR. Fig. S6 displays two independent experiments showing that Kap1 deficiency does not alter the pattern of H3K9me3 or H3K9/K14Ac marks at the IgH locus. Fig. S7 displays an additional ChIP experiment (corresponding to Fig. 6) showing that H3K9me3 and KAP1 mark the donor switch region (Sμ) before and during CSR. Table S1 shows the two-color IgH-FISH analysis. Tables S2, S3, and S4, included as separate PDF files, show the statistical analyses corresponding to the ChIP experiments in Fig. 6, Fig. S6, and Fig. S7, respectively. Table S5 shows the primers used in this study. Table S6 shows the antibodies used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110118/DC1.
Acknowledgments
We thank members of the Losson and Reina-San-Martin laboratories for discussions and Evi Soutoglou and Olivier Pourquié for comments on the manuscript. We also thank Klaus Rajewsky for CD19Cre mice, Michel Nussenzweig for the anti-AID antibody for ChIP analysis, Anna Gazumyan for advice on AID-ChIP, Raphaël Riclet for advice on H3K9me3, KAP1, and HP1 ChIP, Manuela Argentini and Frank Ruffenach for assistance in mass spectrometry analysis, Isabelle Billas for advice on gel filtration chromatography, Sonia Beroud for assistance with switch junction analysis, Claudine Ebel for assistance with cell sorting, Pascal Eberling for peptide synthesis, and Michael Gendron for animal care and immunizations.
B.P. Jeevan-Raj was supported by fellowships from the Association pour la Recherche sur le Cancer (ARC) and the Fondation pour la Recherche Médicale. F.W. Alt is an investigator of the Howard Hughes Medical Institute. This work was supported by grants to B. Reina-San-Martin from the Agence Nationale pour la Recherche (ANR-07-MIME-004-01), the ARC, and the Institut National de la Santé et de la Recherche Médicale (INSERM). B. Reina-San-Martin is an AVENIR-INSERM young investigator.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AID
- activation-induced cytidine deaminase
- ChIP
- chromatin immunoprecipitation
- CSR
- class switch recombination
- DDR
- DNA damage response
- DSB
- double-stranded DNA break
- HTS
- high-throughput amplicon sequencing
- IgH-FISH
- IgH-specific fluorescence in situ hybridization
- NHEJ
- nonhomologous end joining
- qRT-PCR
- quantitative RT-PCR
- SHM
- somatic hypermutation
References
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 410:120–124 10.1038/35065138 [DOI] [PubMed] [Google Scholar]
- Barreto V., Reina-San-Martin B., Ramiro A.R., McBride K.M., Nussenzweig M.C. 2003. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell. 12:501–508 10.1016/S1097-2765(03)00309-5 [DOI] [PubMed] [Google Scholar]
- Brar S.S., Watson M., Diaz M. 2004. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J. Biol. Chem. 279:26395–26401 10.1074/jbc.M403503200 [DOI] [PubMed] [Google Scholar]
- Cammas F., Mark M., Dollé P., Dierich A., Chambon P., Losson R. 2000. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development. 127:2955–2963 [DOI] [PubMed] [Google Scholar]
- Cammas F., Herzog M., Lerouge T., Chambon P., Losson R. 2004. Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev. 18:2147–2160 10.1101/gad.302904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Basu U., Zarrin A., Yan C., Franco S., Perlot T., Vuong B., Wang J., Phan R.T., Datta A., et al. 2007. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 94:157–214 10.1016/S0065-2776(06)94006-1 [DOI] [PubMed] [Google Scholar]
- Chowdhury M., Forouhi O., Dayal S., McCloskey N., Gould H.J., Felsenfeld G., Fear D.J. 2008. Analysis of intergenic transcription and histone modification across the human immunoglobulin heavy-chain locus. Proc. Natl. Acad. Sci. USA. 105:15872–15877 10.1073/pnas.0808462105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J.M., Neuberger M.S. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22 10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Doi T., Kato L., Ito S., Shinkura R., Wei M., Nagaoka H., Wang J., Honjo T. 2009. The C-terminal region of activation-induced cytidine deaminase is responsible for a recombination function other than DNA cleavage in class switch recombination. Proc. Natl. Acad. Sci. USA. 106:2758–2763 10.1073/pnas.0813253106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel S., Mostoslavsky R., Novobrantseva T.I., Pelanda R., Chaudhuri J., Esposito G., Jung S., Alt F.W., Rajewsky K., Cedar H., Bergman Y. 2007. Allelic ‘choice’ governs somatic hypermutation in vivo at the immunoglobulin kappa-chain locus. Nat. Immunol. 8:715–722 10.1038/ni1476 [DOI] [PubMed] [Google Scholar]
- Franco S., Gostissa M., Zha S., Lombard D.B., Murphy M.M., Zarrin A.A., Yan C., Tepsuporn S., Morales J.C., Adams M.M., et al. 2006. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 21:201–214 10.1016/j.molcel.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Geisberger R., Rada C., Neuberger M.S. 2009. The stability of AID and its function in class-switching are critically sensitive to the identity of its nuclear-export sequence. Proc. Natl. Acad. Sci. USA. 106:6736–6741 10.1073/pnas.0810808106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I., Jia S. 2007. Heterochromatin revisited. Nat. Rev. Genet. 8:35–46 10.1038/nrg2008 [DOI] [PubMed] [Google Scholar]
- Guikema J.E., Linehan E.K., Tsuchimoto D., Nakabeppu Y., Strauss P.R., Stavnezer J., Schrader C.E. 2007. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J. Exp. Med. 204:3017–3026 10.1084/jem.20071289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog M., Wendling O., Guillou F., Chambon P., Mark M., Losson R., Cammas F. 2011. TIF1β association with HP1 is essential for post-gastrulation development, but not for Sertoli cell functions during spermatogenesis. Dev. Biol. 350:548–558 10.1016/j.ydbio.2010.12.014 [DOI] [PubMed] [Google Scholar]
- Ito S., Nagaoka H., Shinkura R., Begum N., Muramatsu M., Nakata M., Honjo T. 2004. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc. Natl. Acad. Sci. USA. 101:1975–1980 10.1073/pnas.0307335101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S., Yamada T., Grewal S.I. 2004. Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell. 119:469–480 10.1016/j.cell.2004.10.020 [DOI] [PubMed] [Google Scholar]
- Jolly C.J., Klix N., Neuberger M.S. 1997. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res. 25:1913–1919 10.1093/nar/25.10.1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracker S., Gardes P., Mazerolles F., Durandy A. 2010. Immunoglobulin class switch recombination deficiencies. Clin. Immunol. 135:193–203 10.1016/j.clim.2010.01.012 [DOI] [PubMed] [Google Scholar]
- Kuang F.L., Luo Z., Scharff M.D. 2009. H3 trimethyl K9 and H3 acetyl K9 chromatin modifications are associated with class switch recombination. Proc. Natl. Acad. Sci. USA. 106:5288–5293 10.1073/pnas.0901368106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Subrahmanyam R., Chakraborty T., Sen R., Desiderio S. 2007. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 27:561–571 10.1016/j.immuni.2007.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden J.M., McCarty T., Petiniot L.K., Shen R., Barlow C., Wynn T.A., Morse H.C., III, Gearhart P.J., Wynshaw-Boris A., Max E.E., Hodes R.J. 2004. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J. Exp. Med. 200:1111–1121 10.1084/jem.20041074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccarthy T., Roa S., Scharff M.D., Bergman A. 2009. SHMTool: a webserver for comparative analysis of somatic hypermutation datasets. DNA Repair (Amst.). 8:137–141 10.1016/j.dnarep.2008.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews A.G., Kuo A.J., Ramón-Maiques S., Han S., Champagne K.S., Ivanov D., Gallardo M., Carney D., Cheung P., Ciccone D.N., et al. 2007. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 450:1106–1110 10.1038/nature06431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride K.M., Barreto V., Ramiro A.R., Stavropoulos P., Nussenzweig M.C. 2004. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199:1235–1244 10.1084/jem.20040373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Nielsen A.L., Ortiz J.A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R. 1999. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 18:6385–6395 10.1093/emboj/18.22.6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T., Doi T., Toyoshima T., Muramatsu M., Honjo T., Kinoshita K. 2009. Carboxy-terminal domain of AID required for its mRNA complex formation in vivo. Proc. Natl. Acad. Sci. USA. 106:2747–2751 10.1073/pnas.0812957106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard V.H., Kim S.T., Anderson S.M., Shlomchik M.J., Schatz D.G. 2005. Histone modifications associated with somatic hypermutation. Immunity. 23:101–110 10.1016/j.immuni.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., Reina San-Martin B., Barreto V., et al. 2010. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 143:122–133 10.1016/j.cell.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt S.K., Harris R.S., Neuberger M.S. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–103 10.1038/nature00862 [DOI] [PubMed] [Google Scholar]
- Ramiro A., Reina San-Martin B., McBride K., Jankovic M., Barreto V., Nussenzweig A., Nussenzweig M.C. 2007. The role of activation-induced deaminase in antibody diversification and chromosome translocations. Adv. Immunol. 94:75–107 10.1016/S0065-2776(06)94003-6 [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R.F., Nussenzweig A., Nussenzweig M.C. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197:1767–1778 10.1084/jem.20030569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Chen H.T., Nussenzweig A., Nussenzweig M.C. 2004. ATM is required for efficient recombination between immunoglobulin switch regions. J. Exp. Med. 200:1103–1110 10.1084/jem.20041162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert R.C., Roes J., Rajewsky K. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25:1317–1318 10.1093/nar/25.6.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riclet R., Chendeb M., Vonesch J.L., Koczan D., Thiesen H.J., Losson R., Cammas F. 2009. Disruption of the interaction between transcriptional intermediary factor 1beta and heterochromatin protein 1 leads to a switch from DNA hyper- to hypomethylation and H3K9 to H3K27 trimethylation on the MEST promoter correlating with gene reactivation. Mol. Biol. Cell. 20:296–305 10.1091/mbc.E08-05-0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Bothmer A., Callen E., Reina-San-Martin B., Dorsett Y., Difilippantonio S., Bolland D.J., Chen H.T., Corcoran A.E., Nussenzweig A., Nussenzweig M.C. 2008. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 135:1028–1038 10.1016/j.cell.2008.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert I., Dantzer F., Reina-San-Martin B. 2009. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J. Exp. Med. 206:1047–1056 10.1084/jem.20082468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C.E., Bradley S.P., Vardo J., Mochegova S.N., Flanagan E., Stavnezer J. 2003. Mutations occur in the Ig Smu region but rarely in Sgamma regions prior to class switch recombination. EMBO J. 22:5893–5903 10.1093/emboj/cdg550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellars M., Reina-San-Martin B., Kastner P., Chan S. 2009. Ikaros controls isotype selection during immunoglobulin class switch recombination. J. Exp. Med. 206:1073–1087 10.1084/jem.20082311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sernández I.V., de Yébenes V.G., Dorsett Y., Ramiro A.R. 2008. Haploinsufficiency of activation-induced deaminase for antibody diversification and chromosome translocations both in vitro and in vivo. PLoS ONE. 3:e3927 10.1371/journal.pone.0003927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R., Ito S., Begum N.A., Nagaoka H., Muramatsu M., Kinoshita K., Sakakibara Y., Hijikata H., Honjo T. 2004. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 5:707–712 10.1038/ni1086 [DOI] [PubMed] [Google Scholar]
- Sripathy S.P., Stevens J., Schultz D.C. 2006. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 26:8623–8638 10.1128/MCB.00487-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta V.T., Nagaoka H., Catalan N., Durandy A., Fischer A., Imai K., Nonoyama S., Tashiro J., Ikegawa M., Ito S., et al. 2003. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat. Immunol. 4:843–848 10.1038/ni964 [DOI] [PubMed] [Google Scholar]
- Vakoc C.R., Mandat S.A., Olenchock B.A., Blobel G.A. 2005. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 19:381–391 10.1016/j.molcel.2005.06.011 [DOI] [PubMed] [Google Scholar]
- Vuong B.Q., Lee M., Kabir S., Irimia C., Macchiarulo S., McKnight G.S., Chaudhuri J. 2009. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat. Immunol. 10:420–426 10.1038/ni.1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Whang N., Wuerffel R., Kenter A.L. 2006. AID-dependent histone acetylation is detected in immunoglobulin S regions. J. Exp. Med. 203:215–226 10.1084/jem.20051774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wuerffel R., Feldman S., Khamlichi A.A., Kenter A.L. 2009. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J. Exp. Med. 206:1817–1830 10.1084/jem.20081678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Reina-San-Martin B., Olaru A., Minn K., Tamada K., Lau J.S., Cascalho M., Chen L., Nussenzweig A., Livak F., et al. 2004. 53BP1 is required for class switch recombination. J. Cell Biol. 165:459–464 10.1083/jcb.200403021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M., Shinkura R., Doi Y., Maruya M., Fagarasan S., Honjo T. 2011. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat. Immunol. 12:264–270 10.1038/ni.1991 [DOI] [PubMed] [Google Scholar]
- Wu X., Geraldes P., Platt J.L., Cascalho M. 2005. The double-edged sword of activation-induced cytidine deaminase. J. Immunol. 174:934–941 [DOI] [PubMed] [Google Scholar]
- Wuerffel R., Wang L., Grigera F., Manis J., Selsing E., Perlot T., Alt F.W., Cogne M., Pinaud E., Kenter A.L. 2007. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 27:711–722 10.1016/j.immuni.2007.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Fulop Z., Wu G., Pone E.J., Zhang J., Mai T., Thomas L.M., Al-Qahtani A., White C.A., Park S.R., et al. 2010. 14-3-3 adaptor proteins recruit AID to 5′-AGCT-3′-rich switch regions for class switch recombination. Nat. Struct. Mol. Biol. 17:1124–1135 10.1038/nsmb.1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A., Resch W., Kuo N., Kuchen S., Li Z., Sun H.W., Robbiani D.F., McBride K., Nussenzweig M.C., Casellas R. 2011. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat. Immunol. 12:62–69 10.1038/ni.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.T., Boboila C., Souza E.K., Franco S., Hickernell T.R., Murphy M., Gumaste S., Geyer M., Zarrin A.A., Manis J.P., et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 449:478–482 10.1038/nature06020 [DOI] [PubMed] [Google Scholar]
- Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8:870–876 10.1038/ncb1446 [DOI] [PubMed] [Google Scholar]
- Zofall M., Grewal S.I. 2006. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell. 22:681–692 10.1016/j.molcel.2006.05.010 [DOI] [PubMed] [Google Scholar]