Abstract
The mammalian target of rapamycin complex 1 (mTORC1) links the control of mRNA translation, cell growth, and metabolism to diverse stimuli. Inappropriate activation of mTORC1 can lead to cancer. Phorbol esters are naturally occurring products that act as potent tumor promoters. They activate isoforms of protein kinase C (PKCs) and stimulate the oncogenic MEK/ERK signaling cascade. They also activate mTORC1 signaling. Previous work indicated that mTORC1 activation by the phorbol ester PMA (phorbol 12-myristate 13-acetate) depends upon PKCs and may involve MEK. However, the precise mechanism(s) through which they activate mTORC1 remains unclear. Recent studies have implicated both the ERKs and the ERK-activated 90-kDa ribosomal S6 kinases (p90RSK) in activating mTORC1 signaling via phosphorylation of TSC2 (a regulator of mTORC1) and/or the mTORC1 component raptor. However, the relative importance of each of these kinases and phosphorylation events for the activation of mTORC1 signaling is unknown. The recent availability of MEK (PD184352) and p90RSK (BI-D1870) inhibitors of improved specificity allowed us to address the roles of these protein kinases in controlling mTORC1 in a variety of human and rodent cell types. In parallel, we used specific shRNAs against p90RSK1 and p90RSK2 to further test their roles in regulating mTORC1 signaling. Our data indicate that p90RSKs are dispensable for the activation of mTORC1 signaling by phorbol esters in all cell types tested. Our data also reveal striking diversity in the requirements for MEK/ERK in the control of mTORC1 between different cell types, pointing to additional signaling connections between phorbol esters and mTORC1, which do not involve MEK/ERK. This study provides important information for the design of efficient strategies to combat the hyperactivation of mTORC1 signaling by oncogenic pathways.
Keywords: MAP Kinases (MAPKs), mTOR Complex (mTORC), Phorbol Esters, Protein Kinase C (PKC), Rsk, Tuberous Sclerosis (Tsc)
Introduction
Signaling through mTORC1 (mammalian target of rapamycin complex 1)4 regulates numerous cellular functions and is implicated in disease states such as tissue hypertrophy and cancer. mTORC1 lies downstream of numerous proto-oncogenes such as epidermal growth factor receptor, Ras (rat sarcoma), PI3K (phosphatidylinositide 3-kinase), and PKB (protein kinase B, also referred to as Akt for acutely transforming retrovirus AKT8 in rodent T cell lymphoma) as well as tumor suppressors including PTEN (phosphatase and tensin homolog deleted on chromosome 10) and TSC1/TSC2 (tuberous sclerosis complex 1 and 2) (reviewed in Ref. 1)). Accordingly, mTORC1 signaling is highly active in cells lacking tumor suppressors such as PTEN, TSC1/TSC2, and moderately active in cells deficient for neurofibromin 1 or neurofibromin 2 (2–6). In view of this, there is currently a high level of interest in inhibiting mTORC1 signaling as a potential anti-cancer therapy (7). Some inhibitors of mTORC1, such as rapamycin derivatives, are already in use for cancer therapy (8, 9) but they have proved rather inefficient as a single anti-cancer therapy in the clinic (10). Inhibiting dysregulated signaling pathways upstream of mTORC1 provides an alternative way to reverse the hyperactivation of mTORC1 in cells containing oncogenic signaling proteins or mutated tumor suppressors. Understanding the signaling cascades impinging on mTORC1 is thus of paramount importance when targeting mTORC1 signaling.
mTORC1 signaling is activated upstream by amino acids via the Rag GTPases (11, 12) and hormones (e.g. insulin) through the PI3K/PKB/TSC1/TSC2 signaling cascade (reviewed in Ref. 1). The available evidence suggests that insulin activates mTORC1 signaling through the tuberous sclerosis complex (TSC1/TSC2), which acts as a GTPase-activator protein (GAP) for the small G-protein Rheb (Ras homolog enriched in brain) (1, 13–15). In its GTP-bound form, Rheb activates mTORC1 (16). Upon insulin stimulation, PKB/Akt phosphorylates TSC2 at several sites including Ser939, Ser981, Ser1130, Ser1132, and Thr1462 (17–19). Phosphorylation of Ser939/981 has been proposed to modulate mTORC1 signaling promoting the association of TSC2 with 14-3-3 proteins. This apparently sequesters TSC2 away from its membrane-bound binding partner (TSC1) and its substrate (Rheb) (20).
mTORC1 signaling is also activated by hypertrophic α1-adrenergic agonists (e.g. phenylephrine), which drive the growth of cardiomyocytes (leading to cardiac hypertrophy, a potentially lethal condition) (21) and tumor-promoting phorbol esters (e.g. phorbol myristate acetate, PMA) in human embryonic kidney (HEK) 293 cells (22–24). Unlike insulin, α1-adrenergic agonists and phorbol esters do not activate PKB/Akt but rather stimulate isoforms of protein kinase C (PKC), which are indispensable for the activation of mTORC1 signaling by these agents (25, 26). Similarly, it has been recently reported that epidermal growth factor (EGF) signals to mTORC1 through PKC and independently of PKB/Akt in glioma cells (27). How do PKCs relay their signal to mTORC1? PKC phosphorylates the proto-oncogene Raf-1 (28) to activate the classical MAP kinase (MEK/ERK/p90RSK) cascade and stimulate cell proliferation. Both ERKs and p90RSKs have been proposed to control mTORC1 signaling but which kinase relays the signal to mTORC1 remains a point of contention. One report suggests that this involves the direct phosphorylation of TSC2 by ERK (24), whereas others indicate that TSC2 is phosphorylated and inactivated by p90RSK (p90 ribosomal protein S6 kinases), which are activated by ERK (23, 29, 30). It has also been reported that p90RSKs phosphorylate raptor (a component of mTORC1) and thereby activate mTORC1 signaling (31). MEK/ERK signaling is also required for the activation of mTORC1 by α1-adrenergic stimuli in cardiomyocytes (21, 29). This involves upstream signaling through atypical PKC isoforms (25).
In this study, we sought to investigate the contributions of MEKs, ERKs, and p90RSK to the control of mTORC1 signaling by PMA. To this end, we have used two protein kinase inhibitors: PD184352 (also referred to as CI-1040), a MEK inhibitor (32, 33) and BI-D1870, an inhibitor of the p90RSK kinases (34). They were tested in multiple cell types that are frequently used to study the control of mTORC1 signaling. To complement this approach, we have also used short hairpin-mediated knockdown of p90RSK1 and p90RSK2 isoforms.
Our main finding is that p90RSKs are not required for mTORC1 activation by PMA in any of the various cell lines tested. Regarding the role of MEKs and ERKs in the control of mTORC1 by PMA, our pharmacological data indicate a possible role for MEK/ERK signaling in the control of mTORC1 in certain cell types (namely HEK293, HEK293E, and HEK293T) by PMA. In contrast, in other cell types (such as MCF-7 and NIH/3T3), mTORC1 signaling is partially or in some instances completely resistant to MEK/ERK inhibition, indicating the existence of additional signaling connections between PMA and mTORC1, which do not involve MEK or ERK.
Our findings contribute to the overall understanding of the signaling networks that control mTORC1. Because activation of MEK/ERK signaling is widespread in cancers, due to mutations in epidermal growth factor receptor, Ras, Raf, and neurofibromin 1/2, understanding how the classical MAPK pathway activates mTORC1 is important for the design of effective anti-cancer therapies against tumors resulting from hyperactivated mTORC1 signaling. In particular, our data reconcile seemingly discrepant reports on the control of mTORC1 via MEK/ERK and p90RSK, while also highlighting the surprising diversity/complexity of the regulatory mechanisms that control mTORC1.
EXPERIMENTAL PROCEDURES
Chemicals, Vectors, and Antisera
General laboratory chemicals were from Sigma and Fisher Scientific. Rapamycin and PD098059 were from Calbiochem. BI-D1870 was obtained from the Division of Signal Transduction Therapy, College of Life Sciences, University of Dundee (UK). Recombinant human insulin and PMA were purchased from Sigma. Recombinant human epidermal growth factor (number 13247-051) was purchased from Invitrogen. Protein G-Sepharose CL-4B was from GE Healthcare. BSA (fraction V) was from Roche Molecular Biochemicals. Collagenase (type II) was from Worthington (Edmonton, AB, Canada). Tissue culture reagents were bought from Wisent Inc. (St. Bruno, QC, Canada). The vectors encoding human FLAG-TSC1 and FLAG-TSC2 were generous gifts from Dr. Andrew Tee (Cardiff, UK) and have been described previously (35). Myc-tagged human raptor was a generous gift from Dr. David Sabatini (Whitehead Institute, Boston, MA). Lentivirus packaging vectors pLP1 (gag/pol), pLP2 (rev), and pLP/vesicular stomatitis virus-G (glycoprotein) were purchased from Invitrogen. Anti-phospho-Thr389 S6K (number 9234), anti-phospho-Thr308 PKB/Akt (number 2965), and anti-phospho-Ser473 PKB/Akt (number 9271), anti-phospho-Ser21/9 GSK3α/β (number 9331), anti-phospho-Thr202/Tyr204 p44/42 MAPK (ERK1/2) (number 9106), anti-phospho(Ser/Thr) Akt-substrate antisera (numbers 9611 and 9614), anti-phospho-Thr37/46 4E-BP1 (number 2855), anti-phospho-Ser65 4E-BP1 (number 9451), anti-phospho-Thr70 4E-BP1 (number 9455), anti-phospho-Ser235/236 S6 (number 2211), anti-phospho-Ser240/244 S6 (number 2215), anti-4E-BP1 (number 9452), anti-GSK3 (number 9332), anti-p44/42 MAPK (ERK1/ERK2) (number 9107), anti-PKB/Akt (number 4691), anti-RSK1 (number 9333), and anti-RSK2 (number 9340) were from Cell Signaling Technology (Danvers, MA). Anti-S6K (C-18, sc-230) antiserum was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-eIF4E (number 610270) was from BD Transduction Laboratories (Mississauga, ON, Canada). Anti-Myc (TAG003) was from BioShop Canada Inc. Anti-raptor (number 09-217) was obtained from Millipore (Billerica, MA).
Cell Isolation, Culture, and Treatments
ARVC were isolated from adult male Sprague-Dawley rats (250–300 g; Animal Care Centre, University of British Columbia) as described (21). After isolation, ARVC were washed and seeded onto laminin-coated tissue culture dishes (21). HEK293, HEK293E, HEK293T, and NIH/3T3 cells were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 100 μg/ml of streptomycin sulfate, 100 units/ml of penicillin G, 2 mm l-glutamine. MCF-7 cells were cultured in Roswell Park Memorial Institute-1640 (RPMI 1640) medium supplemented with 10% (v/v) FBS, 100 μg/ml of streptomycin sulfate, and 100 units/ml of penicillin G. HEK293 and HEK293T cells were transfected with cDNAs using the BES/CaCl2 method as described previously (36).
Short Hairpin RNAs, Generation of Lentivirus, and Transduction of HEK293T Cells
Lentiviral vectors for shRNA silencing of RSK1 and RSK2 and a scramble sequence were obtained from Sigma. The accession numbers and targeting regions for each shRNA used are listed in supplemental Table S1. Each shRNA vector was co-transfected into HEK293T cells with lentivirus packaging plasmids pLP1, pLP2, and pLP/vesicular stomatitis virus-G using Lipofectamine 2000 (Invitrogen). Supernatants containing viral particles were collected at 48 and 72 h post-transfection and filtered through a 0.45-μm nitrocellulose membrane. Filtered supernatants were subsequently applied to HEK293T target cells in the presence of Polybrene (5 μg/ml). Cells were incubated with virus for 24 h and then re-infected for a further 24-h period prior to selection with 5 μg/ml of puromycin (Sigma). Puromycin selection medium was prepared in DMEM supplemented with 10% (v/v) FBS, 100 μg/ml of streptomycin sulfate, and 100 units/ml of penicillin G. Cells were selected in puromycin for 3 days and then transferred to growth medium without puromycin for the experiments.
Cell Harvesting and Protein Immunoprecipitation
All cells were harvested in extraction buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 25 mm β-glycerophosphate, 2.5 mm sodium pyrophosphate, 1% (v/v) Triton X-100, 10 mm sodium fluoride, 1 mm sodium orthovanadate, 1 mm dithiothreitol, and 1× complete EDTA-free protease inhibitor mixture (04693132001), Roche Applied Science) adapted from Ref. 37 and protein concentrations were determined as described in Ref. 38. Raptor, MYC-raptor, FLAG-TSC1, and FLAG-TSC2 proteins were immunoprecipitated from HEK293, HEK293E, or HEK293T lysates typically by incubating 500 μg of protein with 30 μl (packed volume) of protein G-Sepharose and 5 to 10 μg of anti-FLAG, anti-Myc or anti-raptor antisera for 3 h at 4 °C. Immunoprecipitates were washed twice with lysis buffer. Beads were then resuspended in sample buffer containing 100 mm dithiothreitol (DTT) and boiled at 95 °C for 5 min.
Gel Electrophoresis and Western Blotting
SDS-PAGE and Western blotting were performed as described previously (39).
Assessment of Cell Proliferation Using the 5′-Bromo-2′-deoxyuridine 5 (BrdU) Colorimetric Assay in HEK293E and HEK293T Cells
HEK293E and HEK293T cells were seeded at 1 × 104 cells/well in a 96-well microtiter plate and allowed to adhere for 24 h in growth media at which point cells were treated with inhibitors for 3 days. Cells were then labeled with BrdU for 2 h and incubated with peroxidase-conjugated anti-BrdU (clone BMG, 6H8, Fab fragments) antibody as per the manufacturer's instructions (number 11647229001, Roche Applied Science). Absorbance was recorded at 450 nm using a Varioskan plate reader (Thermo Electron Corp.).
In Vivo 32P Radiolabeling and Two-dimensional Peptide Mapping
HEK293 cells were transfected with 4 μg of FLAG-TSC1 and 5 μg of FLAG-TSC2 in a 10-cm dish. Twenty-four hours following transfection, the cells were serum starved for 16 h and subsequently radiolabeled with [32P]orthophosphate as described previously (26, 36). The cells were then stimulated with 1 μm PMA for 25 min and harvested as described above. Two-dimensional peptide mapping was carried out as detailed previously (26, 36).
Microscopy
HEK293E and HEK293T cells were treated as described above and cell density was assessed by brightfield microscopy using the ×10 achroplan objective in an Axiovert 200M microscope (Zeiss).
RESULTS
BI-D1870, a Specific p90RSK Inhibitor, Blocks PMA-induced Phosphorylation of TSC2 but Not mTORC1 Activation in HEK293 Cells
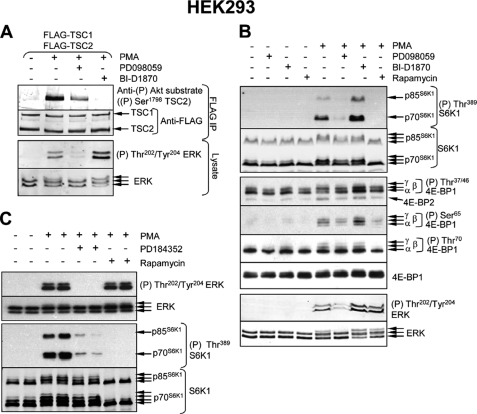
BI-D1870 was recently identified as a specific inhibitor of p90RSK (34). It potently inhibits all four p90RSK isoforms (p90RSK1–4) in vitro and in vivo. We set out to use this inhibitor to investigate the role of p90RSK in the activation of mTORC1. We first studied its ability to block PMA-induced TSC2 phosphorylation in vivo. FLAG-tagged TSC2 was expressed (with its partner, TSC1) in HEK293 cells. These cells were chosen as we have previously shown, using pharmacological inhibitors, that the phorbol ester PMA potently activates mTORC1 in this cell line via PKCs/MEKs (22, 26).
HEK293 cells were starved of serum and then treated with PMA in the presence or the absence of the allosteric MEK inhibitor PD098059 or the p90RSK inhibitor BI-D1870. PMA induced a pronounced increase in TSC2 phosphorylation as detected using the anti-phospho-Akt-substrate antisera (Fig. 1A). This antibody does not actually recognize the true PKB/Akt phosphorylation sites in TSC2 but instead detects phosphorylation at Ser1798, which is phosphorylated by p90RSK, as revealed by the observation that the reactivity with this antibody was lost when a TSC2(S1798A) mutant is used (29). We used 10 μm BI-D1870 as this concentration efficiently blocks p90RSK activity without interfering significantly with the catalytic activity of other kinases tested (34). Preincubation of HEK293 cells with 10 μm BI-D1870 completely prevented PMA-induced TSC2 phosphorylation at Ser1798 (Fig. 1A). These data support earlier findings that Ser1798 is indeed an intracellular substrate for p90RSK (23, 30).
FIGURE 1.
Inhibition of MEKs, but not p90RSK, impairs activation of mTORC1 signaling by phorbol ester in HEK293 cells. A, HEK293 cells were co-transfected with vectors encoding FLAG-TSC1 and FLAG-TSC2. Twenty-four hours later, the cells were starved of serum for 16 h and subsequently stimulated with 1 μm PMA for 25 min. In some instances, cells were pre-treated with 50 μm PD098059 (45 min) or 10 μm BI-D1870 (1 h), prior to stimulation with PMA. The cells were harvested and FLAG-TSC1 and FLAG-TSC2 were immunoprecipitated, followed by Western blot analysis as detailed in A, upper section. Lysates were also analyzed using anti-ERK and anti-phospho-ERK antisera (lower section). B, HEK293 cells were deprived of serum for 16 h and then stimulated with 1 μm PMA (25 min). Where indicated, cells were pre-treated with 50 μm PD098059 (1 h), 10 μm BI-D1870 (1 h), or 100 nm rapamycin (30 min), prior to stimulation with PMA. Cells were harvested and lysate samples were analyzed by SDS-PAGE and Western blot, using the indicated antisera. C, HEK293 cells were cultured to near confluence and subsequently starved of serum overnight, and treated with 10 μm PD184352 or 100 nm rapamycin (both for 1 h) prior to stimulation with 1 μm PMA for 25 min. Cells were harvested and lysates were analyzed by SDS-PAGE and Western blot using the indicated antisera.
The MEK inhibitor PD098059 partially inhibited the PMA-induced phosphorylation of Ser1798 in TSC2 (Fig. 1A), consistent with phosphorylation of this residue being mediated by p90RSK downstream of MEK/ERK signaling. We observed that PD098059 did not completely block the phosphorylation of ERKs either (Fig. 1A). The residual activity of MEK/ERK signaling in PD098059-treated cells presumably suffices for some activation of p90RSK, thereby explaining the observed residual Ser1798 phosphorylation (Fig. 1A). Interestingly, BI-D1870 treatment actually increased ERK1/2 phosphorylation, as indicated by a phosphospecific anti-ERK1/2 antibody and the reduced electrophoretic mobility of ERK1/2 in the gel (Fig. 1A). This is consistent with earlier data (34) that pointed to the existence of an inhibitory feedback loop involving p90RSK that normally limits ERK activation.
The fact that BI-D1870 completely blocks the p90RSK-mediated phosphorylation of TSC2 at Ser1798 allowed us to test whether such phosphorylation is required for the activation of mTORC1 by PMA. To assess this, we examined the phosphorylation of 4E-BP1 and S6K1, the two best characterized mTORC1 substrates. Two main bands were observed for phospho-S6K1, corresponding to the shorter (p70) and longer (p85) isoforms (Fig. 1B, top). PMA induced a marked increase in the phosphorylation of both S6K1 isoforms at the mTORC1 site (Thr389 in the shorter isoform; Fig. 1B) and induced a shift to the slower migrating species that is characteristic of S6K1 hyperphosphorylation. These effects were inhibited by PD098059, indicating that MEK activity is required for PMA-induced S6K1 phosphorylation, and blocked by rapamycin. However, BI-D1870 did not inhibit the effect of PMA on S6K1 phosphorylation. If anything, BI-D1870 actually potentiated it. This indicates that p90RSK activity is not required for PMA-induced S6K1 phosphorylation in HEK293 cells (Fig. 1B).
PMA also markedly increased the phosphorylation of 4E-BP1 at Ser65 (Fig. 1B, center). This was strongly inhibited by rapamycin, confirming that it requires mTORC1, and was moderately decreased by PD098059. In contrast, inhibiting p90RSK actually slightly increased the phosphorylation of Ser65 in 4E-BP1 (Fig. 1B). Collectively, these data indicate that activation of mTORC1 signaling by PMA in HEK293 cells involves MEK activity but not that of p90RSK, either through the phosphorylation of TSC2 (23, 30) or raptor (31).
To further assess the role of MEKs in the mTORC1 activation by PMA in HEK293 cells, we used an alternative MEK inhibitor, PD184352, which directly interferes with MEK activity (32). This compound blocked PMA-induced S6K1 phosphorylation in HEK293 cells (Fig. 1C) confirming the importance of MEK signaling for the activation of the mTORC1 pathway by phorbol esters in these cells.
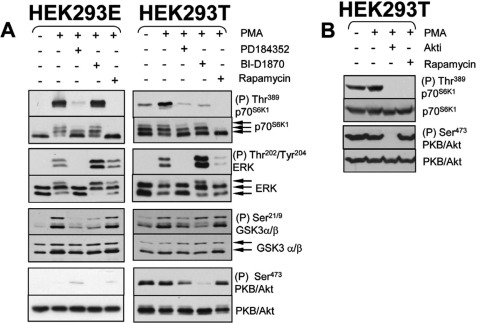
p90RSK Isoforms 1 and 2 Are Dispensable for mTORC1 Activation by Phorbol Esters and Growth Factors in HEK293E and HEK293T Cells
HEK293E cells are a variant HEK293 cell line that express the Epstein-Barr virus nuclear antigen 1 (EBNA1) and have often been used to study mTORC1 signaling (see for instance, Ref. 30). HEK293T cells (which express the SV40 large T antigen) are also widely used to study the control of mTORC1 signaling (40, 41). Therefore, we considered it important to assess the role of MEKs and p90RSK in the control of mTORC1 signaling in these HEK293 variants. PMA activated MEK and mTORC1 signaling in both cell types (Fig. 2A). In the case of HEK293E, p70S6K1 phosphorylation was completely dependent upon MEK signaling (it was blocked by PD184352) but was not reduced by BI-D1870 (Fig. 2A, left panel). BI-D1870 did, however, block p90RSK activity, as judged by the reduced phosphorylation of GSK3α/β at Ser21/9 in cells treated with this compound (Fig. 2A, left panel). PD184352 also blocked phosphorylation of GSK3α/β consistent with MEK/ERK functioning upstream of p90RSK (Fig. 2A, left panel). Taken together these data indicate that MEKs, and not p90RSK, are involved in mTORC1 activation by PMA in HEK293E cells. As observed in HEK293E cells, PMA also potently activated both ERK and mTORC1 signaling pathways in HEK293T cells (Fig. 2A, right panel). In the latter cell type, however, PMA-induced phosphorylation of S6K1 was blocked by either PD184352 or BI-D1870, suggesting that both MEK/ERK and p90RSK do play a role in the activation of mTORC1 by PMA in this setting.
FIGURE 2.
Differential regulation of mTORC1 signaling in HEK293E and HEK293T cells. A, HEK293E and HEK293T cells were propagated to near confluence at which point cells were starved of serum overnight. Cells were then stimulated with 1 μm PMA (25 min). Where indicated, cells were treated with 10 μm PD184352, 10 μm BI-D1870, or 100 nm rapamycin (all for 1 h) prior to stimulation. Samples were analyzed for MEK, p90RSK, mTORC1, and mTORC2 activity using antisera against phosphorylated ERK, GSK3α/β, p70S6K1, and PKB/Akt, respectively. B, HEK293T cells were cultured to near confluence and starved of serum overnight. Where indicated, cells were treated with 10 μm Akt inhibitor VIII (isozyme selective, Akt-I-1/2) or 100 nm rapamycin both for 1 h and then stimulated with 1 μm PMA for 25 min.
HEK293T cells (but not HEK293E cells) exhibit constitutively active PI3K/Akt signaling as shown by the elevated basal phosphorylation of PKB/Akt at Ser473 seen in serum-starved cells (Ref. 36 and Fig. 2A, left and right panels), likely as a consequence of the binding of the SV40 large T antigen to IRS-1 (42). We noted that in addition to blocking p90RSK activity, BI-D1870 markedly reduces basal phosphorylation of PKB/Akt in HEK293T cells: it is not clear why BI-D1870 exhibits this effect, but it is entirely consistent with the recent observation that BI-D1870 inhibited the insulin-induced phosphorylation of both Thr308 and Ser473 on PKB/Akt in 3T3-L1 adipocytes (43). Because PKB/Akt plays a prominent role in the activation of mTORC1 (reviewed in Ref. 1), it is possible that the inhibitory effects of the BI-D1870 compound on mTORC1 activity are mediated through inhibition of PKB/Akt rather than p90RSK. To test this possibility further we investigated whether Akt-I-1,2, a specific Akt inhibitor (44) impairs mTORC1 signaling. As shown in Fig. 2B, treatment of HEK293T cells with 10 μm Akt-I-1,2 abrogated p70S6K1 phosphorylation, indicating that PKB/Akt activity is indeed required for mTORC1 activation. Given that PKB/Akt activity is required for mTORC1, the inhibitory effect of BI-D1870 on mTORC1 may result from PKB/Akt inhibition, rather than inhibition of p90RSK.
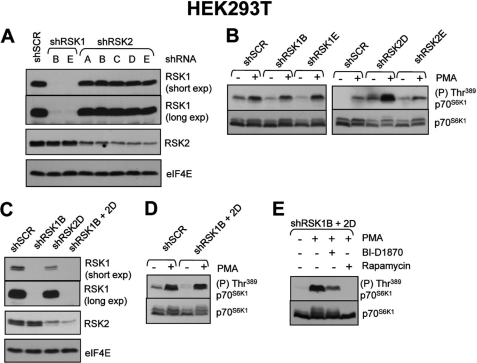
To further explore the roles of p90RSK in the activation of mTORC1 signaling, we adopted the complementary approach of “knocking down” their expression by shRNA in HEK293T cells. These cells express p90RSK1 and p90RSK2 (Fig. 3A), but p90RSK3 and p90RSK4 were not detected (data not shown). Multiple shRNAs were used against p90RSK1 and p90RSK2 to ensure that any detectable change in mTORC1 activation was the direct effect of the shRNA on its target mRNA. p90RSK1-directed shRNAs “B” and “E” each gave efficient knock-down of p90RSK1 without interfering with p90RSK2 protein levels (Fig. 3A). Conversely, each of the p90RSK2-directed shRNAs tested led to partial loss of p90RSK2 protein (but had no effect on p90RSK1 protein levels) (Fig. 3A). Knock-down of p90RSK1 with shRNAs B or E lowered basal p70S6K1 phosphorylation but did not impair PMA-induced phosphorylation of p70S6K1 (Fig. 3B). The ability of PMA to induce p70S6K1 phosphorylation in cells in which p90RSK1 had been knocked down with either shRNA B or E is comparable with that in HEK293T cells infected with scrambled (control) shRNA. Knockdown of p90RSK2 with shRNAs “D” or “E” did not block p70S6K1 phosphorylation either. On the contrary, it appeared to slightly increase p70S6K1 phosphorylation. p90RSK1 and p90RSK2 exhibit 95.6% identity at the amino acid level. It was therefore conceivable that the lack of effect of p90RSK1 knockdown on mTORC1 reflected a contribution from p90RSK2 to mTORC1 activation (or vice versa). Therefore, we transduced cells with effective shRNAs against p90RSK1 and p90RSK2 in combination. Combined knockdown of p90RSK1 and p90RSK2 was effective in depleting these enzymes (Fig. 3C), but again this did not impair the ability of PMA to induce the phosphorylation of p70S6K1 (Fig. 3D). In such cells, the PMA-induced phosphorylation of p70S6K1 was still sensitive to BI-D1870, suggesting that the inhibition by this compound reflects an effect on another target, e.g. as suggested above, PKB/Akt (Fig. 3E).
FIGURE 3.
RNA interference reveals that p90RSK isoforms 1 and 2 are not required for the activation of mTORC1 signaling in response to PMA. A, HEK293T were transduced with lentiviruses encoding short hairpin RNAs against p90RSK1, p90RSK2, or a scrambled sequence control. Transduced cells were subsequently selected with puromycin as described under “Experimental Procedures.” Once selected, cells were grown to near confluence in medium without puromycin and lysate samples were analyzed for knockdown efficiency using specific p90RSK1 and p90RSK2 antisera. Anti-eIF4E antisera was used to monitor loading. B, HEK293T cells prepared in A were used to assess mTORC1 activation. Cells were serum starved overnight and then stimulated with 1 μm PMA for 25 min. Lysate samples were analyzed for mTORC1 activation using S6K1 phosphospecific antisera. C, HEK293T cells were transduced with control shRNA, shRNA against p90RSK1, shRNA against p90RSK2, or shRNA against both p90RSK1 and p90RSK2. In the latter case, the cells were first infected with shRNA against p90RSK2 and then p90RSK1. Cells were selected using puromycin as described under “Experimental Procedures.” Level of knockdown was monitored as described in A. D, cells prepared in C were assessed for mTORC1 activation in response to 1 μm PMA (25 min) after overnight serum starvation. E, HEK293T cells prepared in C were grown to near confluence and subsequently deprived of serum overnight. Cells were then treated with 10 μm BI-D1870 or 100 nm rapamycin (each for 1 h) prior to stimulation with 1 μm PMA. Lysate samples were analyzed for mTORC1 activation using S6K1 phosphospecific antisera.
In addition to being activated by phorbol esters, mTORC1 is also stimulated by growth factors, such as EGF. EGF-mediated activation of mTORC1 depends upon PKCs, as recently shown in an elegant study by Fan and colleagues (27). We have, therefore, also tested whether p90RSK1 or p90RSK2 (downstream targets of the PKC/Raf/MEK/ERK pathway) are required for mTORC1 activation by EGF. As observed for PMA, p90RSK1 is dispensable for the activation of mTORC1 by EGF, whereas depletion of p90RSK2 by shRNA actually increased mTORC1 signaling (supplemental Fig. S1).
It has also been reported (31) that raptor phosphorylation by p90RSK positively regulates mTORC1 activity. Therefore, we also tested whether PMA, a potent activator of PKC/Raf/MEK/ERK/p90RSK signaling, elicited raptor phosphorylation in HEK293E or HEK293T cells. We were unable to observe any signal indicative of phosphorylation of endogenous raptor in response to PMA in either cell type using the commercially available anti-“phospho-PKB/Akt substrate” antisera (also used by others (31), (supplemental Fig. 2, A and B)). The study reporting raptor phosphorylation by p90RSK used ectopically expressed raptor. We, therefore, also analyzed the phosphorylation of overexpressed raptor. HEK293T cells were transfected with MYC-raptor and subsequently treated with the MAPK pathway activating stimuli: serum, PMA, or EGF. In our hands, we were unable to detect phosphorylation of MYC-raptor by p90RSKs using the reported phospho-PKB/Akt substrate antibodies under the conditions used (supplemental Fig. 2C). It remains possible, however, that the phosphorylation stoichiometry is low such that the raptor phosphorylation cannot be detected using these antibodies.
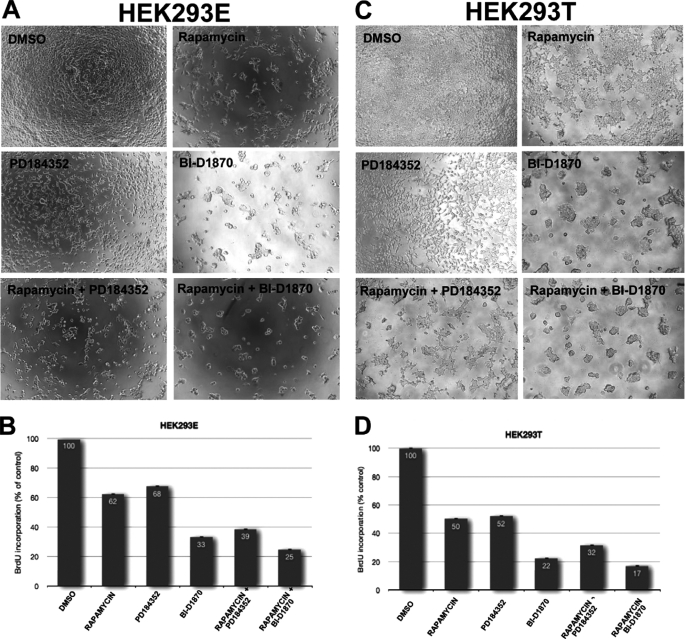
BI-D1870, PD184352, and Rapamycin Exert Additive Inhibitory Effects on the Proliferation of HEK293 Cell Types
MEK/ERK, p90RSK, and mTORC1 have all been previously linked to the control of the cell cycle and/or cell proliferation. For instance, the MEK inhibitor PD184352 induces G1 arrest in a variety of cells including Chinese hamster fibroblasts (CCl-39), melanoma (SKEMEL28) cells, lymphoid (Jurkat) cells, and blocks colony formation by acute myeloid leukemic (AML) cells deficient for the tumor suppressor neurofibromin 1 (45–48), whereas inhibition of mTORC1 by rapamycin effectively reduces the proliferation of numerous cancer cell types and in particular cell types that lack functional PTEN (see e.g. Refs. 2 and 49). p90RSK1 and p90RSK2 are also critical for proliferation of MCF-7 cells, as recently shown in an elegant study by Smith and colleagues (50), where treatment of MCF-7 cells with SL0101, a newly identified RSK inhibitor, blocked the cell cycle in G1. Having shown that MEK/ERK but not p90RSK regulate PMA-induced mTORC1 activation in HEK293 cell lines, we next sought to analyze the effect of MEK/ERK, p90RSK, or mTORC1 inhibition on the proliferation of these cell types. To this end, we tested the effects of rapamycin, BI-D1870, and PD184352 (alone or in combination) on the proliferation of HEK293E cells (Fig. 4, A and B). Effects on cell proliferation were assessed by visual inspection of phase-contrast micrographs (Fig. 4A) and quantified by BrdU incorporation (Fig. 4B). As shown in Fig. 4B, rapamycin and PD184352 inhibited BrdU incorporation by about 40 and 30%, respectively. Notably, a further decrease in the proliferation of HEK293E cells was observed when the two inhibitors were used in combination. These findings show that simultaneous blockade of the MEK/ERK/p90RSK and mTORC1 pathways has additive effects on cell proliferation, indicating, importantly, that MEK/ERK and mTORC1 provide independent inputs to the cell cycle. BI-D1870 decreased the proliferation of HEK293E cells (by ∼60%). As far as we are aware, these are the first data showing that BI-D1870 impairs cell proliferation. Notably, the simultaneous inhibition of p90RSK and mTORC1 with BI-D1870 and rapamycin gave rise to an additive inhibitory effect on proliferation (Fig. 4, A and B). Taken together these data indicate that the MEK/ERK/p90RSK and mTORC1 pathways exert independent effects on cell proliferation. Rapamycin, PD184352, or BI-D1870 had comparable anti-proliferative effects on HEK293T cells (cf. Fig. 4, C and D).
FIGURE 4.
PD184352, BI-D1870, and rapamycin inhibit the proliferation of HEK293E and HEK293T cells. A and B, HEK293E cells were propagated as described under “Experimental Procedures” and then treated with 10 μm PD184352, 10 μm BI-D1870, and/or 100 nm rapamycin for 72 h, at which point the cells were incubated with BrdU and incorporation into DNA was measured (B) as described under “Experimental Procedures” and according to the manufacturer's guidelines. Cell proliferation was also monitored by visual inspections of microphotographs (A). C and D, HEK293T cells were propagated, treated, and proliferation was measured as described in A and B.
Regulation of p70S6K1 Phosphorylation by PMA in Other Cell Types
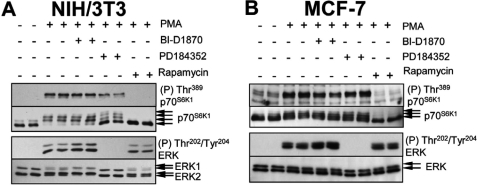
In view of the differing signaling connections in distinct cell types, we sough to extend our study to other cell lines. We chose human MCF-7 cells (a breast cancer-derived cell line) as well as the rodent NIH/3T3 cells (widely used in studies on cell transformation). In NIH/3T3 cells, MEK inhibition caused a small (but detectable) decrease in PMA-induced p70S6K1 phosphorylation (mobility shift; Fig. 5A), suggesting that this response requires signaling via MEKs. In contrast, BI-D1870 had no effect on the phosphorylation of p70S6K1 at Thr389 or on p70S6K1 mobility, indicating MEK signaling activates mTORC1 independently of p90RSK in these cells. In MCF-7 cells, PMA induced an mTORC1-dependent increase in p70S6K1 phosphorylation that was again independent of MEKs and p90RSK (Fig. 5B).
FIGURE 5.
MEKs and p90RSK are dispensable for activation of mTORC1 signaling in human (MCF-7) and rodent (NIH/3T3) cells. A and B, MCF-7 and NIH/3T3 cells were grown to near confluence and subsequently incubated with 100 nm rapamycin, 10 μm PD184352, or 10 μm BI-D1870 for 1 h followed by stimulation with 1 μm PMA for 25 min. Lysate samples (containing equal amounts of protein, typically 30–40 μg) were analyzed by SDS-PAGE and Western blot, using the indicated antisera.
PD098059, but Not BI-D1870, Reduces Phenylephrine-induced Phosphorylation of mTORC1 Targets in Cardiomyocytes
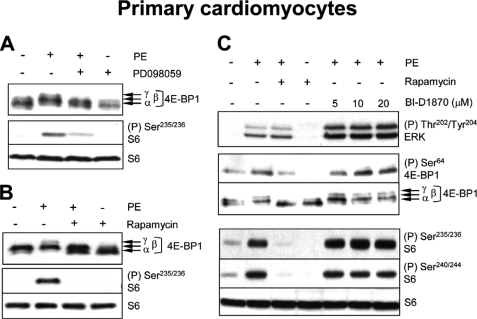
In addition to using a pharmacological agent such as PMA in transformed cells, we considered it important to study the control of mTORC1 by an agonist that activates ERK signaling via physiologically relevant receptors in primary cells. We have previously shown that the α1-adrenergic agonist phenylephrine (PE) activates mTORC1 signaling in ARVC and that this requires MEK (21, 51) and ERK (29). The activation of cardiomyocyte protein synthesis by α1-agonists is of particular interest, because it underlies cardiac hypertrophy, an important risk factor for heart failure (52). Both the PE-induced activation of protein synthesis (21) and pathological cardiac hypertrophy (53, 54) are dependent upon signaling through mTORC1.
PE activates S6K1 (21) and S6K2 (51) in ARVC, and induces the phosphorylation of 4E-BP1 (indicated by its shift to the slower-migrating, hyperphosphorylated γ-form; Fig. 6A) and S6 (assessed using the phospho-Ser235/236 antibody; Fig. 6A). These effects are decreased by inhibiting MEKs (using PD098059) and eliminated by rapamycin, demonstrating that PE-induced S6 phosphorylation is entirely dependent upon mTORC1 (Fig. 6, A and B). This implies that p90RSKs do not contribute directly to S6 phosphorylation here, greatly simplifying the interpretation of the data. In contrast, treatment of ARVC with BI-D1870 did not decrease the PE-induced phosphorylation of 4E-BP1, even at relatively high doses (20 μm) (Fig. 6C). Treatment of ARVC with BI-D1870 also failed to decrease PE-induced phosphorylation of S6 at all sites tested (Fig. 6C), which, as noted above, is completely dependent upon signaling through mTORC1. In fact, BI-D1870 tended to increase the phosphorylation of 4E-BP1 and S6, especially at the higher concentrations used. This likely reflects the enhanced activation of ERK seen under conditions of p90RSK inhibition (Fig. 6C). These findings provide strong evidence that MEK but not p90RSK activity is required for the activation of mTORC1 signaling by PE in primary cardiomyocytes.
FIGURE 6.
The p90RSK inhibitor, BI-D1870, does not impair PE-induced activation of mTORC1 signaling in ARVC. A–C, ARVC were treated with 50 μm PD098059 (30 min), 100 nm rapamycin (30 min), or varying concentrations of BI-D1870 (1 h), prior to stimulation with 10 μm PE (1 h). Lysate samples (containing equal amounts of protein, typically 30–40 μg) were analyzed by SDS-PAGE/Western blot, using the indicated antisera.
PMA Induces Only Weak Phosphorylation of Multiple Sites on TSC2 in HEK293 Cells
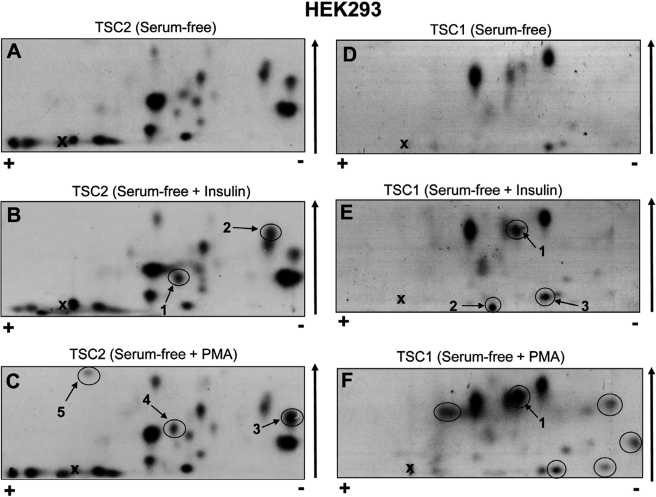
The diverse regulation of mTORC1 signaling prompted us to examine the phosphorylation of TSC2 in vivo in greater detail. Next we studied whether PMA activates mTORC1 signaling through the phosphorylation of alternative sites on TSC2. To this end, HEK293 cells were transfected with FLAG-tagged TSC2 (and its binding partner FLAG-TSC1). The transfected cells were subsequently starved of serum to decrease the phosphorylation of any sites stimulated by e.g. insulin, mitogens, or growth factors, and then metabolically labeled with [32P]orthophosphate. In some instances, cells were then treated with insulin or PMA. Radiolabeled TSC1 and TSC2 were immunoprecipitated, digested with trypsin, and the resulting peptides resolved by two-dimensional mapping.
The complex pattern of the two-dimensional peptide map (Fig. 7A) reveals that TSC2 is phosphorylated at several sites even under serum-starved conditions. Two major and 16 minor species were observed. In response to insulin, only two species showed a slight increase in radiolabeling (peptides 1 and 2, arrows in Fig. 7B). Remarkably, the intensities of the insulin-induced labeling of peptides 1 and 2 are much lower than those of the major basal species observed in serum-starved cells. Radioactivity incorporation into the stimulated sites, if phosphorylated stoichiometrically, is expected to be similar or higher than that of the basal sites (because it will involve new phosphorylation by radiolabeled phosphate, whereas basal sites may already be (perhaps partially) phosphorylated with unlabeled phosphate). The lower extent of labeling of the peptides that respond to insulin is thus probably quite substoichiometric begging the question: how can the insulin-induced phosphorylation of only a subpopulation of TSC2 trigger the inhibition of the entire TSC2 pool in the cell?
FIGURE 7.
TSC1 and TSC2 are each phosphorylated at multiple sites in vivo. A–F, HEK293 cells were co-transfected with vectors for FLAG-TSC1 and FLAG-TSC2. Twenty-four hours later, cells were starved of serum for 16 h and then metabolically labeled with [32P]orthophosphate for 4 h. In some instances, cells were subsequently stimulated with either 100 nm insulin for 25 min (B and E) or 1 μm PMA for 25 min (C and F). Cells were harvested and radiolabeled FLAG-TSC1/TSC2 were immunoprecipitated. Immunoprecipitated proteins were resolved by SDS-PAGE. Radiolabeled FLAG-TSC1 and -TSC2 were excised from the gel and subjected to digestion with trypsin. Peptides were separated by two-dimensional peptide mapping and visualized by autoradiography. Positive and negative electrodes are noted by the plus (+) and minus (−) signs. Arrow denotes the direction of the chromatography. The cross marks the origin. Labeled peptides discussed in the main text are circled.
Like insulin, PMA elicited only a minor increase in the phosphorylation of TSC2 (Fig. 7C). Three minor species (Fig. 7C, arrows 3, 4, and 5) appear to increase in the maps from PMA-treated cells (cf. Fig. 7, A and C). Given the substoichiometric levels of phosphorylation of peptides 3, 4, and 5, it is possible that phosphorylation of multiple (as opposed to a single) sites accounts for the ability of PMA to activate mTORC1 signaling.
TSC1 Also Undergoes PMA-stimulated Phosphorylation in Vivo
While analyzing the phosphorylation of TSC2, it became clear that TSC1 is also weakly phosphorylated in serum-starved conditions. We therefore asked whether PMA affected the phosphorylation of TSC1. To this end, we isolated FLAG-TSC1 (from the same radiolabeled HEK293 cells that had been analyzed for TSC2 phosphorylation) and subjected TSC1 to tryptic digestion and two-dimensional peptide map (Fig. 7, D–F). In samples from serum-starved cells, two major and several minor phosphopeptides were observed (Fig. 7D). Peptide maps were also prepared from TSC1 isolated from insulin- or PMA-treated cells (Fig. 7, E and F). PMA caused a marked increase in the phosphorylation of one species (denoted 1 in Fig. 7F), which became as strongly labeled as the main basal species. PMA also caused the appearance of five minor phosphopeptides (circled in Fig. 7F), which alone, as argued above, are unlikely to lead to substantial inactivation of the TSC1/TSC2 complex. Although to a lesser extent than PMA, insulin also induced the phosphorylation of peptide 1 (Fig. 7E) and the appearance or strengthening of two others (numbered 2 and 3 in Fig. 7E) although their labeling remained below the levels of the major basal phosphopeptides. Importantly, to our knowledge these data are the first to show that TSC1, like TSC2, undergoes phosphorylation in living cells in a regulated manner. Additional work, beyond the scope of this study, is required to confirm whether phosphorylation of TSC1 plays a regulatory role in the control of mTORC1 pathway by agents that activate the PKC/Raf/MEK/ERK signaling cascade.
DISCUSSION
The ability of phorbol esters and growth factors to activate mTORC1 signaling is well documented (see for example, Refs. 22 and 24). Several mechanisms have been proposed to explain this, including phosphorylation of TSC2 and raptor by ERKs and p90RSK (23, 24, 31, 31). However, their relative contribution to mTORC1 signaling is not known. In this study we have used both pharmacological inhibitors and RNA interference to evaluate the relative contribution of MEK, ERKs, and p90RSK to the control of mTORC1 in a variety of immortalized cell types widely used to study mTORC1 signaling in the context of cancer, as well as in primary cardiac muscle cells used as a model of cardiac hypertrophy.
Our study confirms and extends earlier findings that phorbol esters potently activate mTORC1 (in all cell types tested). We also show that MEK/ERK are likely required for the activation of mTORC1 signaling by phorbol esters in HEK293 cell lines. Our conclusion that MEK/ERK play a role in the control of mTORC1 in these cell types relies exclusively on data obtained with two pharmacological inhibitors (PD098059 and PD184352) with distinct modes of action. Although the data obtained with these two inhibitors are concordant and supports a role for MEK/ERK in the control of mTORC1, future work (involving shRNA experiments) is still required to categorically implicate MEK/ERK in the control of mTORC1 signaling by phorbol esters in HEK293 cell types.
Interestingly, we observe that certain cell lines (e.g. NIH/3T3) exhibit only partial sensitivity to MEK inhibitors, whereas in others (e.g. MCF-7) mTORC1 signaling is completely refractory to MEK/ERK inhibition. Our studies also show that the requirement of MEK/ERK for mTORC1 activation depends not only on the cell type but also on the stimulus used. For instance, the MEK inhibitor, PD184352, potently inhibits EGF-induced mTORC1 signaling in MCF-7 cells5 but fails to block PMA-mediated mTORC1 activation in the same cell type. The intricate signaling mechanisms underlying such diverse cellular responses are not known. Considerable additional work, beyond the scope of this study, is required to explain the differing dependences on MEK/ERK signaling for mTORC1 activation in response to different stimuli and in distinct cell types.
Because MEK signaling switches on p90RSK and mTORC1 signaling is reduced upon MEK inhibition in HEK293 cells, it remained possible that MEKs regulated mTORC1 signaling via p90RSK in these cell types, as proposed earlier (31). To address this possibility we made use of BI-D1870, a novel p90RSK inhibitor, as well as shRNA-mediated knockdown of p90RSK1 and/or p90RSK2. We observed that BI-D1870 did not impair the activation of mTORC1 signaling in any of the various cell types tested, with the exception of HEK293T cells. Knockdown of p90RSK isoforms 1 and 2 in HEK293T cells failed to reprise the effect of BI-D1870. We postulate that the inhibitory effect of BI-D1870 likely arises from its inhibitory effect on Akt, which itself regulates mTORC1 signaling. The inhibitory effect of this compound on Akt signaling was also shown in 3T3-L1 adipocytes (43). Our pharmacological data also show that p90RSK are dispensable for that activation of mTORC1 by α1-agonists (PE).
Having shown that MEK/ERK are likely required (whereas p90RSK are dispensable) for mTORC1 activation in HEK293 cells, we turned our attention to how MEK/ERK may exert their control over mTORC1 in these cell types. ERKs have been previously implicated in the control of mTORC1 through the phosphorylation of Ser664 on TSC2 (24). Our data show that TSC2 is already heavily phosphorylated under serum starvation conditions (where mTORC1 signaling is switched off) and PMA elicits only small increases in the phosphorylation of TSC2. These findings suggest the existence of additional modes of regulation, beside TSC2 phosphorylation. A possible additional point of regulation is TSC1, which we now show to be phosphorylated in response to PMA.
The upstream control of mTORC1 by PMA and PE is clearly complex, involving multiple regulatory inputs. Given the significance of dysregulated mTORC1 signaling in cancer, further studies are needed to complete our understanding of the mechanisms by which oncogenic signals that turn on the PKC/Raf/MEK signaling cascade activate mTORC1. Encouragingly, the observation that MEK/ERK regulate mTORC1 signaling in some cell lines indicates that MEK inhibitors may prove useful in reversing the hyperactivation of mTORC1 signaling (even if only in certain cell types) in disease states, such as cancer, where mTORC1 activity is frequently dysregulated. Although, p90RSK are dispensable for mTORC1 activation, inhibition of the former enzymes may also be of therapeutic value in the cancer setting. This notion is supported by our findings that BI-D1870 potently inhibits the proliferation of HEK293E cells.
Supplementary Material
Acknowledgments
A special thanks Dr. Ivan Topisirovic for helpful discussions and Dr. Maritza Jaramillo and Dr. Polen Sean for critical reading of the manuscript.
This work was supported in part by grants from the University of British Columbia, the Heart and Stroke Foundation of British Columbia and the Yukon, the British Heart Foundation, and Canadian Institutes of Health Research (all to C. G. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
B. Fonseca and C. G. Proud, unpublished data.
- mTORC1
- mammalian target of rapamycin complex 1
- Akt
- acutely transforming retrovirus AKT8 in rodent T cell lymphoma
- PKB
- protein kinase B
- PMA
- phorbol 12-myristate 13-acetate
- S6K1
- S6 kinase 1
- TSC1/2
- tuberous sclerosis 1/2
- ARVC
- adult rat ventricular cardiomyocytes
- MCF-7
- Michigan Cancer Foundation line 7
- PTEN
- phosphatase and tensin homolog deleted on chromosome 10
- p90RSK
- ribosomal S6 kinase protein of 90 kDa
- Rheb
- Ras homolog enriched in brain
- raptor
- regulatory associated protein of mTOR
- PE
- phenylephrine
- BES
- 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid.
REFERENCES
- 1. Huang J., Manning B. D. (2008) Biochem. J. 412, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neshat M. S., Mellinghoff I. K., Tran C., Stiles B., Thomas G., Petersen R., Frost P., Gibbons J. J., Wu H., Sawyers C. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10314–10319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kwiatkowski D. J., Zhang H., Bandura J. L., Heiberger K. M., Glogauer M., el-Hashemite N., Onda H. (2002) Hum. Mol. Genet. 11, 525–534 [DOI] [PubMed] [Google Scholar]
- 4. Onda H., Crino P. B., Zhang H., Murphey R. D., Rastelli L., Gould Rothberg B. E., Kwiatkowski D. J. (2002) Mol. Cell. Neurosci. 21, 561–574 [DOI] [PubMed] [Google Scholar]
- 5. Johannessen C. M., Reczek E. E., James M. F., Brems H., Legius E., Cichowski K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8573–8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. James M. F., Han S., Polizzano C., Plotkin S. R., Manning B. D., Stemmer-Rachamimov A. O., Gusella J. F., Ramesh V. (2009) Mol. Cell. Biol. 29, 4250–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gibbons J. J., Abraham R. T., Yu K. (2009) Semin. Oncol. 36, S3–S17 [DOI] [PubMed] [Google Scholar]
- 8. Easton J. B., Houghton P. J. (2006) Oncogene 25, 6436–6446 [DOI] [PubMed] [Google Scholar]
- 9. Faivre S., Kroemer G., Raymond E. (2006) Nat. Rev. Drug Discov. 5, 671–688 [DOI] [PubMed] [Google Scholar]
- 10. Guertin D. A., Sabatini D. M. (2009) Sci. Signal. 2, pe24. [DOI] [PubMed] [Google Scholar]
- 11. Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008) Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. L. (2008) Nat. Cell. Biol. 10, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y., Corradetti M. N., Inoki K., Guan K. L. (2004) Trends Biochem. Sci. 29, 32–38 [DOI] [PubMed] [Google Scholar]
- 14. Corradetti M. N., Guan K. L. (2006) Oncogene 25, 6347–6360 [DOI] [PubMed] [Google Scholar]
- 15. Manning B. D., Cantley L. C. (2003) Trends Biochem. Sci. 28, 573–576 [DOI] [PubMed] [Google Scholar]
- 16. Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. (2005) Curr. Biol. 15, 702–713 [DOI] [PubMed] [Google Scholar]
- 17. Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. (2002) Mol. Cell 10, 151–162 [DOI] [PubMed] [Google Scholar]
- 18. Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002) Nat. Cell. Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 19. Dan H. C., Sun M., Yang L., Feldman R. I., Sui X. M., Ou C. C., Nellist M., Yeung R. S., Halley D. J., Nicosia S. V., Pledger W. J., Cheng J. Q. (2002) J. Biol. Chem. 277, 35364–35370 [DOI] [PubMed] [Google Scholar]
- 20. Cai S. L., Tee A. R., Short J. D., Bergeron J. M., Kim J., Shen J., Guo R., Johnson C. L., Kiguchi K., Walker C. L. (2006) J. Cell Biol. 173, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang L., Proud C. G. (2002) Circ. Res. 91, 821–829 [DOI] [PubMed] [Google Scholar]
- 22. Herbert T. P., Kilhams G. R., Batty I. H., Proud C. G. (2000) J. Biol. Chem. 275, 11249–11256 [DOI] [PubMed] [Google Scholar]
- 23. Roux P. P., Ballif B. A., Anjum R., Gygi S. P., Blenis J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13489–13494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P. P. (2005) Cell 121, 179–193 [DOI] [PubMed] [Google Scholar]
- 25. Wang L., Rolfe M., Proud C. G. (2003) Biochem. J. 373, 603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fonseca B. D., Lee V. H., Proud C. G. (2008) Biochem. J. 411, 141–149 [DOI] [PubMed] [Google Scholar]
- 27. Fan Q. W., Cheng C., Knight Z. A., Haas-Kogan D., Stokoe D., James C. D., McCormick F., Shokat K. M., Weiss W. A. (2009) Sci. Signal. 2, ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kolch W., Heidecker G., Kochs G., Hummel R., Vahidi H., Mischak H., Finkenzeller G., Marmé D., Rapp U. R. (1993) Nature 364, 249–252 [DOI] [PubMed] [Google Scholar]
- 29. Rolfe M., McLeod L. E., Pratt P. F., Proud C. G. (2005) Biochem. J. 388, 973–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ballif B. A., Roux P. P., Gerber S. A., MacKeigan J. P., Blenis J., Gygi S. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carrière A., Cargnello M., Julien L. A., Gao H., Bonneil E., Thibault P., Roux P. P. (2008) Curr. Biol. 18, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 32. Sebolt-Leopold J. S., Dudley D. T., Herrera R., Van Becelaere K., Wiland A., Gowan R. C., Tecle H., Barrett S. D., Bridges A., Przybranowski S., Leopold W. R., Saltiel A. R. (1999) Nat. Med. 5, 810–816 [DOI] [PubMed] [Google Scholar]
- 33. Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sapkota G. P., Cummings L., Newell F. S., Armstrong C., Bain J., Frodin M., Grauert M., Hoffmann M., Schnapp G., Steegmaier M., Cohen P., Alessi D. R. (2007) Biochem. J. 401, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tee A. R., Fingar D. C., Manning B. D., Kwiatkowski D. J., Cantley L. C., Blenis J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fonseca B. D., Smith E. M., Lee V. H., MacKintosh C., Proud C. G. (2007) J. Biol. Chem. 282, 24514–24524 [DOI] [PubMed] [Google Scholar]
- 37. Tee A. R., Proud C. G. (2000) Oncogene 19, 3021–3031 [DOI] [PubMed] [Google Scholar]
- 38. Bradford M. M. (1976) Anal. Biochem. 77, 248–254 [DOI] [PubMed] [Google Scholar]
- 39. Scheper G. C., Parra J. L., Wilson M., van Kollenburg B., Vertegaal A. C., Han Z. G., Proud C. G. (2003) Mol. Cell. Biol. 23, 5692–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 41. Kim D. H., Sarbassov D. D., Ali S. M., Latek R. R., Guntur K. V., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2003) Mol. Cell 11, 895–904 [DOI] [PubMed] [Google Scholar]
- 42. Yu Y., Kudchodkar S. B., Alwine J. C. (2005) J. Virol. 79, 6882–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen S., Mackintosh C. (2009) Cell Signal. 21, 1984–1993 [DOI] [PubMed] [Google Scholar]
- 44. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Squires M. S., Nixon P. M., Cook S. J. (2002) Biochem. J. 366, 673–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meng X. W., Chandra J., Loegering D., Van Becelaere K., Kottke T. J., Gore S. D., Karp J. E., Sebolt-Leopold J., Kaufmann S. H. (2003) J. Biol. Chem. 278, 47326–47339 [DOI] [PubMed] [Google Scholar]
- 47. Solit D. B., Garraway L. A., Pratilas C. A., Sawai A., Getz G., Basso A., Ye Q., Lobo J. M., She Y., Osman I., Golub T. R., Sebolt-Leopold J., Sellers W. R., Rosen N. (2006) Nature 439, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lauchle J. O., Kim D., Le D. T., Akagi K., Crone M., Krisman K., Warner K., Bonifas J. M., Li Q., Coakley K. M., Diaz-Flores E., Gorman M., Przybranowski S., Tran M., Kogan S. C., Roose J. P., Copeland N. G., Jenkins N. A., Parada L., Wolff L., Sebolt-Leopold J., Shannon K. (2009) Nature 461, 411–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Podsypanina K., Lee R. T., Politis C., Hennessy I., Crane A., Puc J., Neshat M., Wang H., Yang L., Gibbons J., Frost P., Dreisbach V., Blenis J., Gaciong Z., Fisher P., Sawyers C., Hedrick-Ellenson L., Parsons R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10320–10325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith J. A., Poteet-Smith C. E., Xu Y., Errington T. M., Hecht S. M., Lannigan D. A. (2005) Cancer Res. 65, 1027–1034 [PubMed] [Google Scholar]
- 51. Wang L., Gout I., Proud C. G. (2001) J. Biol. Chem. 276, 32670–32677 [DOI] [PubMed] [Google Scholar]
- 52. Hannan R. D., Jenkins A., Jenkins A. K., Brandenburger Y. (2003) Clin. Exp. Pharmacol. Physiol. 30, 517–527 [DOI] [PubMed] [Google Scholar]
- 53. McMullen J. R., Sherwood M. C., Tarnavski O., Zhang L., Dorfman A. L., Shioi T., Izumo S. (2004) Circulation 109, 3050–3055 [DOI] [PubMed] [Google Scholar]
- 54. Shioi T., McMullen J. R., Tarnavski O., Converso K., Sherwood M. C., Manning W. J., Izumo S. (2003) Circulation 107, 1664–1670 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.