Abstract
HMG-CoA reductase (HMGR) catalyzes a rate-limiting step in sterol biosynthesis and is a key control point in the feedback inhibition that regulates this pathway. Through the action of the membrane protein Insig, HMGR synthesis and degradation are regulated to maintain sterol homeostasis. The fission yeast Schizosaccharomyces pombe encodes homologs of HMGR and Insig called hmg1+ and ins1+, respectively. In contrast to the mammalian system, Ins1 regulates Hmg1 by a nondegradative mechanism involving phosphorylation of the Hmg1 active site. Here, we investigate the role of the Ins1-Hmg1 system in coupling glucose sensing to regulation of sterol biosynthesis. We show that Ins1-dependent Hmg1 phosphorylation is strongly induced in response to glucose withdrawal and that HMGR activity is correspondingly reduced. We also find that inability to activate Hmg1 phosphorylation under nutrient limiting conditions results in overaccumulation of sterol pathway intermediates. Furthermore, we show that regulation of Hmg1 phosphorylation requires the protein phosphatase 2A-related phosphatase Ppe1 and its regulator Sds23. These results describe a mechanism by which cells tune the rate of sterol synthesis to match nutrient availability.
Keywords: AMP-activated Kinase (AMPK), Glucose, Phosphatase, PP2A, Protein Phosphorylation, Sterol, Yeast Metabolism, HMG-CoA Reductase, Insig, Schizosaccharomyces pombe
Introduction
Sterol synthesis is a highly conserved and tightly regulated metabolic pathway that maintains homeostasis through multiple systems of feedback inhibition (1, 2). HMG-CoA reductase (HMGR),2 an eight-span integral membrane protein that resides in the endoplasmic reticulum, catalyzes a rate-limiting step in this pathway, and is regulated by mechanisms including transcription, translation, degradation, and phosphorylation (3). Aspects of this regulation require Insigs, a family of six-span endoplasmic reticulum-resident integral membrane proteins. Insigs control three of these regulatory systems in different eukaryotes: transcription and degradation in mammals (4), degradation in budding yeast (5), and phosphorylation in the fission yeast Schizosaccharomyces pombe (6).
In mammalian cells, Insig negatively regulates HMGR transcription by suppressing activation of the membrane-bound transcription factor sterol regulatory element-binding protein under sterol-replete conditions (4). Insig can also promote degradation of HMGR in the presence of 24,25-dihydrolanosterol, which stimulates binding between Insig and the HMGR transmembrane domain (2). This interaction allows Insig to recruit gp78, a ubiquitin E3 ligase, to HMGR, resulting in ubiquitination of HMGR and proteasomal degradation.
In addition to controlling HMGR activity by regulating transcription and protein degradation, mammalian cells also control HMGR activity by phosphorylation. A high AMP:ATP ratio activates the AMP-activated protein kinase (AMPK), which phosphorylates a conserved serine in the HMGR active site (7, 8). This modification, which does not require Insig (9), reversibly decreases HMGR activity and allows the cell to conserve energy in response to metabolic stress (10). The enzyme primarily responsible for dephosphorylating HMGR is protein phosphatase 2A (PP2A) (3). PP2A is a heterotrimeric complex consisting of a structural subunit, a regulatory subunit, and a catalytic subunit, referred to as A, B, and C respectively. PP2A phosphatases achieve substrate specificity by incorporating interchangeable B subunits and participate in a wide variety of cellular processes (11). S. pombe encodes three PP2A-related phosphatases: ppa1+, ppa2+, and ppe1+. Unlike mammals, fission yeast PP2As are regulated by Sds23, a cystathionine β-synthase domain-containing protein. Sds23 physically interacts with Ppa1, Ppa2, and Ppe1 and inhibits the phosphatase activity of these enzymes in vitro (12).
S. pombe encodes single homologs of HMGR and Insig called Hmg1 and Ins1, respectively. As in the mammalian system, Hmg1 and Ins1 form a complex, but in contrast to mammals Ins1 binding does not stimulate Hmg1 degradation. Instead, Ins1 binding stimulates phosphorylation of serine 1024 and threonine 1028 in the Hmg1 catalytic site, attenuating enzyme activity (6). Hmg1 phosphorylation is induced in response to hypertonic stress and growth in minimal medium, and this signaling pathway requires the stress-activated MAP kinase Sty1.
In the current study, we investigated the role of glucose in regulating sterol biosynthesis. Our results show that Ins1-dependent Hmg1 phosphorylation is strongly induced when cells are deprived of glucose and that this response occurs in the absence of AMPK. Cells that are unable to phosphorylate Hmg1 inappropriately accumulate sterol pathway intermediates as the cells enter stationary phase. Furthermore, we find that the PP2A-related phosphatase Ppe1 and its regulator Sds23 are required for this nutrient regulation of Hmg1. This study advances our understanding of how sterol synthesis is regulated in S. pombe and describes a new mechanism by which cells match their anabolism to nutrient availability.
EXPERIMENTAL PROCEDURES
We obtained yeast extract from Fisher; oligonucleotides (supplemental Table 1) from Integrated DNA Technologies; [14C[HMG-CoA (55 Ci/mol) from American Radiolabeled Chemicals; NADPH from Roche; horseradish peroxidase-conjugated, affinity-purified donkey anti-rabbit and anti-mouse IgG from Jackson ImmunoResearch; protein A beads from RepliGen; prestained protein standards from Bio-Rad; glucose from Fisher; sucrose from J. T. Baker; and galactose and sorbitol from Sigma. Protease inhibitors (10 μg/ml leupeptin, 5 μg/ml pepstatin A, 1 mm PMSF) were used at the indicated concentrations and obtained from Sigma. Standard genetic manipulations and molecular biology techniques were performed as described previously (13).
Strains and Media
Wild-type haploid S. pombe KGY425 and derived strains were grown to log phase at 30 °C in YES medium (5 g/liter yeast extract plus 30 g/liter glucose and supplements, 225 mg/liter each of uracil, adenine, leucine, histidine, and lysine) unless otherwise indicated (14). Glucose-free medium was identical to YES medium except for the absence of glucose. Strains used in this study are described in supplemental Table 2 (15). sds23Δ, ppa1Δ, ppa2Δ, and ppe1Δ strains were constructed by homologous recombination using oligonucleotides oJB506/oJB507, oJB500/oJB501, oJB502/oJB503, and oJB504/oJB505, respectively (16).
Antibodies
Rabbit polyclonal antisera recognizing Hmg1 (amino acids 35–204), Ins1 (amino acids 1–80), and phosphoserine 1024 of Hmg1 were generated as described previously (6). Monoclonal antibody recognizing Hmg1 phosphothreonine 1028 was obtained from Cell Signaling (2321) and used according to the manufacturer's instructions.
Hmg1 Immunoprecipitation
Hmg1 immunoprecipitation was conducted as described previously (6). Briefly, cells (1 × 108) were lysed by vortexing with glass beads in lysis buffer (6 mm Na2HPO4, 4 mm NaH2PO4, pH 7.2, 1% (v/v) IGEPAL CA-630, 150 mm NaCl, 4 mm EDTA, 50 mm NaF, 0.3 mm Na3VO4) containing protease inhibitors. The lysate was cleared by centrifugation at 20,000 × g for 10 min, and the resulting supernatant was subjected to immunoprecipitation using 5 μg of anti-Hmg1 IgG and 20 μl of protein A beads in 1 ml of lysis buffer. Beads were washed three times in 1 ml of lysis buffer, and purified protein was eluted and assayed by SDS-PAGE and immunoblotting.
HMG-CoA Reductase Activity Assay
The HMGR activity assay was performed as described previously (6). Briefly, yeast microsomes (3.75 μg) were incubated with 2 mm NADPH and 0.24 mm [14C]HMG-CoA in 50 μl of assay buffer (68.4 mm Na2HPO4, 31.6 mm NaH2PO4, 150 mm NaCl, 1 mm EDTA, 50 mm NaF, 10 mm DTT, pH 7.2) plus protease inhibitors at 30 °C. Reactions were stopped by adding 12.5 μl of 5 n HCl, and insoluble material was removed by centrifugation at 16,000 × g for 2 min. 10 μl of the resulting supernatant was applied to Silica Gel 60 F254 TLC plates (Merck) which were developed in 1:1 acetone/benzene. 14C-Labeled mevalonate was scraped from plates and quantified by liquid scintillation counting.
Sterol Analysis
Gas chromatography was performed as described previously (17). Briefly, cells (1 × 108) were incubated at 75 °C for 2 h in a mixture of 4.5 ml of 60% (w/v) KOH and 9 ml of methanol. Sterols were extracted with petroleum ether, which was evaporated under a stream of N2 gas. Sterols were resuspended in heptane and analyzed by gas chromatography.
RESULTS
Glucose Regulates Hmg1 Phosphorylation
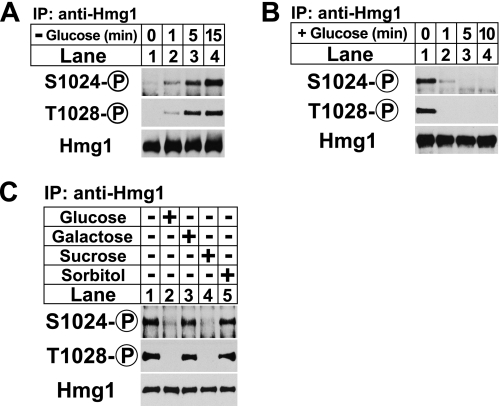
To determine whether Hmg1 phosphorylation was regulated by the presence of a carbon source, we grew wild-type S. pombe cells in rich YES medium containing the standard concentration of glucose (30 g/liter) and then transferred the cells to glucose-free medium for 0, 1, 5, and 15 min. Detergent-solubilized extracts were subjected to anti-Hmg1 immunoprecipitation, and the bound fraction was analyzed by SDS-PAGE followed by immunoblotting with phosphospecific antibodies to Hmg1 amino acid residues serine 1024 and threonine 1028 (6). Phosphorylation of Hmg1 Ser-1024 and Thr-1028 was visible within 1 min of glucose deprivation (Fig. 1A, lane 2), increasing at 5 and 15 min (Fig. 1A, lanes 3 and 4). Total Hmg1 levels remained unchanged (Fig. 1A, lower panel). We next sought to determine whether glucose readdition suppressed Hmg1 phosphorylation induced by low glucose. Hmg1 phosphorylation was induced by growing wild-type cells in glucose-free medium for 30 min, and then the cells were shifted to YES medium. We found that Hmg1 phosphorylation was suppressed by 1 min (Fig. 1B).
FIGURE 1.
Glucose regulates Hmg1 phosphorylation. A, wild-type cells growing exponentially in YES medium were collected by centrifugation and resuspended in glucose-free medium. Samples were taken at the indicated times, and Hmg1 immunoprecipitates (IP) were blotted using anti-Hmg1 IgG or Hmg1 phosphospecific antibodies. B, wild-type cells were grown to exponential phase overnight in rich YES medium, collected by centrifugation, and shifted to glucose-free medium for 30 min. At time 0, cells were again collected by centrifugation and transferred to glucose-rich medium. Samples were taken at the indicated times and processed as in A. C, wild-type cells growing exponentially in YES medium were collected by centrifugation and resuspended in glucose-free medium for 30 min. The indicated carbon sources (30 g/liter) were added for 15 min. Cells were harvested by centrifugation and processed as in A.
We showed previously that osmotic stress induced Hmg1 phosphorylation (6). Because of this we next asked whether the ability to suppress Hmg1 phosphorylation was specific to glucose or whether any osmolyte could produce the same effect. To test this we grew wild-type cells for 30 min in the absence of glucose, then added different osmolytes for 15 min and assayed Hmg1 phosphorylation. As expected, glucose suppressed Hmg1 phosphorylation (Fig. 1C, lanes 1 and 2). Sucrose, a disaccharide that is converted to glucose and fructose by the secreted invertase Inv1 (18, 19), also suppressed Hmg1 phosphorylation (Fig. 1C, lane 4). Neither sorbitol nor galactose, an isomer of glucose that cannot be used as a carbon source (20), suppressed Hmg1 phosphorylation (Fig. 1C, lanes 3 and 5). Collectively, these data show that Hmg1 phosphorylation can be induced by glucose deprivation and suppressed by glucose addition and that this phosphorylation is not due to changes in osmotic pressure.
Low Glucose Suppresses Hmg1 Activity by Stimulating Enzyme Phosphorylation
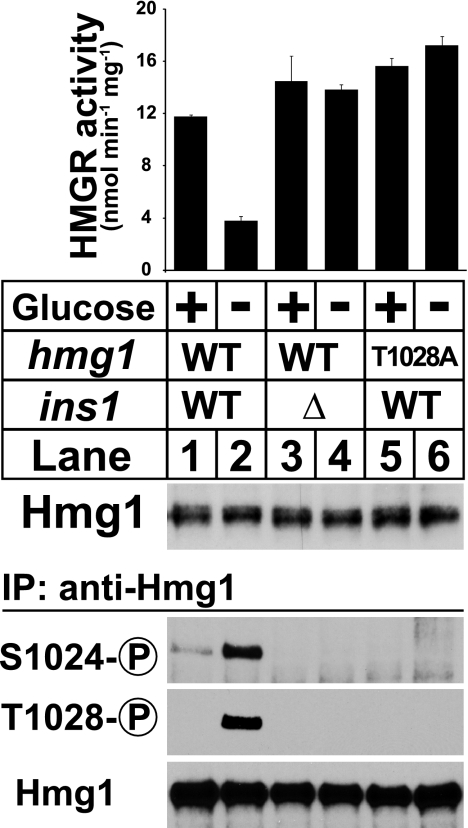
Previous work in our laboratory showed that phosphorylation of Hmg1 residues Ser-1024 and Thr-1028 suppresses activity and that this phosphorylation requires both the presence of Thr-1028 and the endoplasmic reticulum-resident integral membrane protein Ins1 (6). To test whether glucose deprivation suppresses Hmg1 activity through Ins1-dependent phosphorylation, we measured Hmg1 activity in microsomes from wild-type cells, ins1Δ cells, and hmg1-T1028A cells, each grown in the presence or absence of glucose for 30 min. Glucose deprivation suppressed Hmg1 activity 3-fold (Fig. 2, lanes 1 and 2). Suppression of Hmg1 activity required ins1+ and T1028 (Fig. 2, lanes 3–6). Consistent with previous results, Hmg1 phosphorylation required both Ins1 and Thr-1028 (Fig. 2, lower panel). These results indicate that glucose deprivation suppresses Hmg1 activity by stimulating Ins1-dependent phosphorylation of the enzyme active site.
FIGURE 2.
Low glucose suppresses Hmg1 activity by stimulating enzyme phosphorylation. Wild-type cells, ins1Δ cells, and hmg1-T1028A cells were grown in YES medium overnight to exponential phase, collected by centrifugation, and transferred to either YES medium or glucose-free medium for 30 min. Microsomes were prepared, assayed for Hmg1 activity, and immunoblotted with anti-Hmg1 IgG. Data are the average of three technical replicates; error bars show S.E. Whole cell lysates were also prepared, from which Hmg1 was immunoprecipitated (IP) and analyzed by immunoblotting with anti-Hmg1 IgG or Hmg1 phosphospecific antibodies.
Ins1 Is Required for Sterol Homeostasis in Stationary Phase
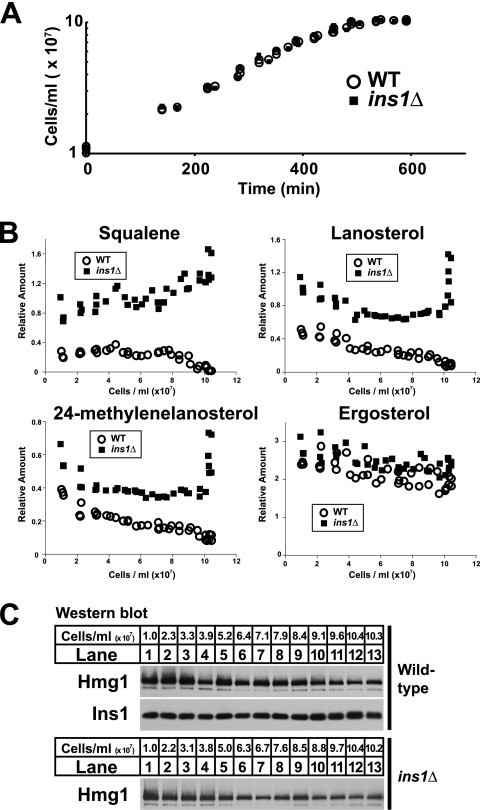
We reasoned that a mechanism for responding to glucose deprivation may be needed when exponentially growing cells begin to exhaust available glucose. Thus, we tested whether sterol homeostasis was altered upon entry to stationary phase in cells lacking the ability to phosphorylate Hmg1. We measured both cell growth and levels of sterol pathway intermediates in cultures of wild-type and ins1Δ cells. Although no difference was observed in growth rate between the two strains (Fig. 3A), substantial differences were observed in sterol pathway intermediates (Fig. 3B). Squalene, lanosterol, and 24-methylenelanosterol levels approached zero in wild-type cells as growth stopped (Fig. 3B, open circles). Conversely, ins1Δ cells accumulated all three intermediates (Fig. 3B, closed squares). Ergosterol, the end product of the pathway, was similar between the two strains. Observed differences were not due to levels of Hmg1, which were similar between wild-type and ins1Δ cells and decreased slightly as cell growth slowed (Fig. 3C). This experiment shows that Ins1 is required to prevent inappropriate accumulation of sterol pathway intermediates as cells transition from exponential growth into stationary phase.
FIGURE 3.
Ins1 is required for sterol homeostasis in stationary phase. A, growth of wild-type and ins1Δ cells in YES medium was monitored by A600 nm. B and C, cells grown to the indicated density were assayed by gas chromatography for levels of squalene, lanosterol, 24-methylenelanosterol, and ergosterol (B), and by immunoblotting for Hmg1 and Ins1 (C). Aggregate data from three biological replicates are shown.
Hmg1 Is Phosphorylated in Stationary Phase Due to Low Glucose
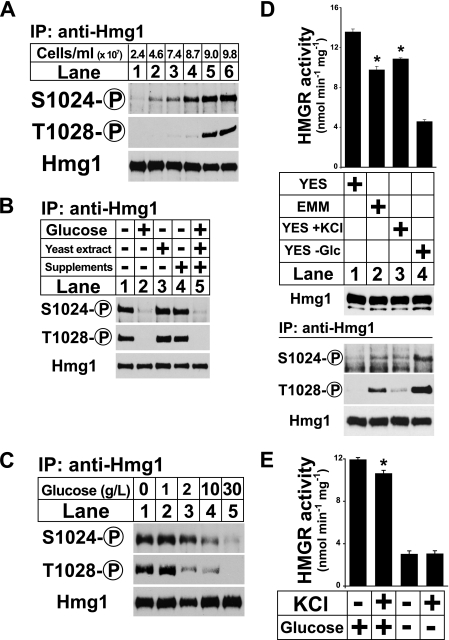
To investigate whether Hmg1 phosphorylation is induced by the gradual depletion of glucose that occurs as a culture reaches saturation, we monitored Hmg1 phosphorylation in a culture of wild-type cells as the cell density increased. Hmg1 phosphorylation increased in proportion to cell density and plateaued shortly before the cells stopped growing (Fig. 4A). We next tested whether Hmg1 phosphorylation in stationary phase resulted from the depletion of glucose or another component of the growth medium. We grew wild-type cells to saturation and then added either nothing or the individual components of YES medium: 30 g/liter glucose, 5 g/liter yeast extract, supplements (225 mg/liter each adenine, uracil, leucine, lysine, and histidine), or all three (Fig. 4B). After 15 min cells were harvested and assayed for Hmg1 phosphorylation. As expected, addition of glucose or complete YES medium suppressed Hmg1 phosphorylation (Fig. 4B, lanes 1, 2, and 5). Neither yeast extract nor supplements were able to suppress Hmg1 phosphorylation (Fig. 4B, lanes 3 and 4). Thus, glucose is sufficient to suppress stationary phase Hmg1 phosphorylation.
FIGURE 4.
Hmg1 is phosphorylated in stationary phase due to low glucose. A, wild-type cells were grown in YES medium and harvested by centrifugation at the indicated cell densities. Hmg1 immunoprecipitates were blotted using anti-Hmg1 IgG or Hmg1 phosphospecific antibodies. B, wild-type cells (1 × 108) grown in YES medium to early stationary phase were treated with 30 g/liter glucose, 5 g/liter yeast extract, and/or 1 × supplements (see “Experimental Procedures”) as indicated. Samples were harvested after 15 min and processed as in A. C, wild-type cells were grown in YES medium, harvested by centrifugation, and resuspended in medium containing different concentrations of glucose as indicated. Samples were harvested after 15 min and processed as in A. D, wild-type cells were grow in YES medium overnight to exponential phase, collected by centrifugation, and transferred to either YES medium, EMM minimal medium, YES medium containing 0.6 m KCl, or glucose-free medium for 30 min. Microsomes were prepared, assayed for Hmg1 activity, and immunoblotted with anti-Hmg1 IgG. Data are the average of three technical replicates; error bars show S.E. Asterisk indicates significant difference from YES plus glucose condition (p < 0.001). Whole cell lysates were also prepared, from which Hmg1 was immunoprecipitated and analyzed by immunoblotting with anti-Hmg1 IgG or Hmg1 phosphospecific antibodies. E, wild-type cells were grown overnight in YES to exponential phase, collected by centrifugation, and resuspended in YES medium, YES containing 0.6 m KCl, glucose-free medium, or glucose-free medium containing 0.6 m KCl for 30 min. Microsomes were prepared and assayed for Hmg1 activity. Data are the average of three technical replicates; error bars show S.E. Asterisk indicates significant difference from YES plus glucose condition (p < 0.01).
To determine the glucose concentration at which Hmg1 phosphorylation is induced, we grew wild-type cells for 15 min in a range of glucose concentrations and assayed Hmg1 phosphorylation (Fig. 4C). Hmg1 phosphorylation was induced at 10 g/liter glucose (Fig. 4C, lane 4) and was maximally induced at glucose concentrations <1 g/liter (0.1% w/v), a typical glucose concentration for S. pombe cultures entering stationary phase (Fig. 4C, lanes 2 and 3) (21, 22). These data show that Hmg1 phosphorylation is negatively regulated by glucose concentration.
Because we have shown previously that Hmg1 phosphorylation is activated by growth in minimal medium and by osmotic stress (6), we wanted to compare the effects of these stimuli with that of low glucose. To make this comparison, we grew wild-type cells in YES medium, EMM (minimal) medium, YES medium containing 0.6 m KCl, and low glucose medium for 30 min. We assayed Hmg1 protein levels, enzyme activity, and phosphorylation. Microsomal Hmg1 protein was equal in all four samples (Fig. 4D, middle panel). Consistent with our previous study (6), Hmg1 phosphorylation was increased for cells grown in EMM and 0.6 m KCl (Fig. 4D, bottom panel, lanes and 3). Correspondingly, Hmg1 activity was decreased to a small, but significant extent. By comparison, low glucose medium decreased Hmg1 activity 3-fold, consistent with the high level of Hmg1 phosphorylation observed (Fig. 4D, lane 4). This experiment shows that low glucose induces enzyme phosphorylation and suppresses Hmg1 activity to a greater magnitude than previously observed for minimal medium or osmotic stress.
Because osmotic stress and low glucose both suppress Hmg1 activity, we next asked whether the effects were additive. To answer this question, we grew wild-type cells for 30 min in YES medium, 0.6 m KCl, low glucose medium, or low glucose medium containing 0.6 m KCl. KCl-induced osmotic stress reduced Hmg1 activity by a small but statistically significant amount (Fig. 4E, first two bars), whereas low glucose greatly decreased Hmg1 activity (Fig. 4E, third bar). When low glucose and osmotic stress were applied simultaneously, no further decrease in Hmg1 activity was observed (Fig. 4E, fourth bar). This result indicates that osmotic stress cannot further suppress Hmg1 activity in low glucose conditions.
AMPK Is Not Essential for Low Glucose Regulation of Hmg1
Although our previous study identified the stress-activated MAP kinase Sty1 as an essential factor for Hmg1 phosphorylation induced by osmotic stress or growth in minimal medium (6), we found no role for Sty1 in low glucose phosphorylation of Hmg1 (supplemental Fig. S1). Therefore, we sought to identify a different kinase involved in low glucose Hmg1 phosphorylation.
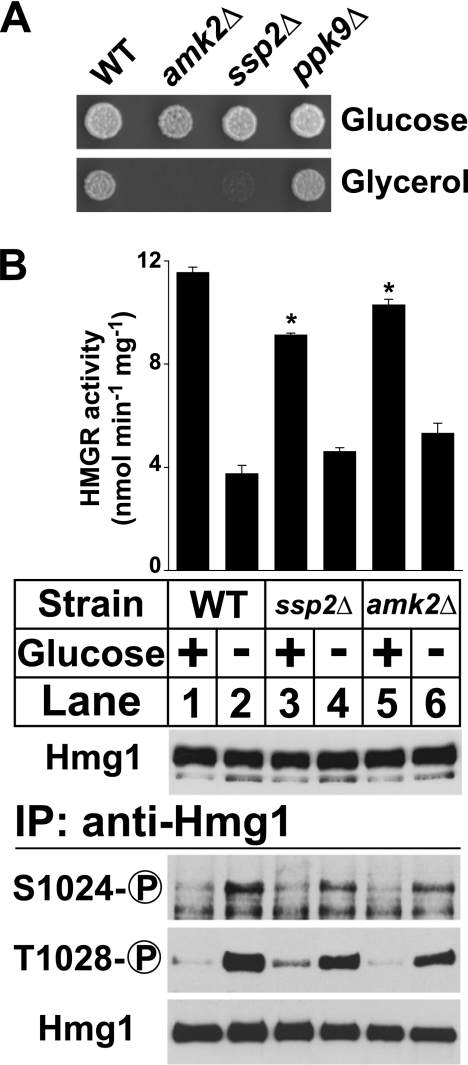
Mammalian HMG-CoA reductase is phosphorylated by AMPK when the intracellular AMP:ATP ratio increases (10). AMPK consists of β and γ regulatory subunits and one α catalytic subunit, which have structurally conserved homologs in S. pombe (12, 23, 24). To determine which genes function as the S. pombe AMPK, we reasoned that cells lacking AMPK would be unable to grow on nonfermentable carbon sources such as glycerol, consistent with the phenotype of Saccharomyces cerevisiae AMPK mutants (25). We tested strains lacking the sole candidate β subunit (amk2+) and two candidate α subunits (ssp2+ and ppk9+) for growth on glucose and glycerol (Fig. 5A). amk2+ and ssp2+ were required for growth on glycerol; ppk9+ was not. This result suggests that amk2+ and ssp2+ are the functional β and α subunits of S. pombe AMPK, respectively.
FIGURE 5.
AMPK is not essential for low glucose regulation of Hmg1. A, wild-type and mutant strains lacking amk2+, ssp2+, or ppk9+ (1000 cells) were grown at 30 °C on YES medium or YES medium containing glycerol (3% (v/v)) instead of glucose. B, wild-type, ssp2Δ, and amk2Δ cells growing exponentially in YES medium were harvested by centrifugation, resuspended in either fresh YES medium or glucose-free medium, and harvested after 30 min. Microsomes were prepared, assayed for Hmg1 activity, and immunoblotted with anti-Hmg1 IgG. Data are the average of three technical replicates; error bars show S.E. Asterisk indicates significant difference from wild-type cells plus glucose (p < 0.01). Whole cell lysates were also prepared, from which Hmg1 was immunoprecipitated and analyzed by immunoblotting with anti-Hmg1 IgG or Hmg1 phosphospecific antibodies.
To test whether S. pombe AMPK is required for low glucose Hmg1 phosphorylation, we grew wild-type cells and cells lacking either the AMPK α catalytic subunit ssp2+ or the sole β regulatory subunit amk2+ in the presence or absence of glucose for 30 min and assayed Hmg1 activity and phosphorylation. Both wild-type cells and cells lacking AMPK subunits induced Hmg1 phosphorylation under low glucose and proportionally decreased Hmg1 activity (Fig. 5B). We observed two differences between wild-type and AMPK mutant strains. First, the AMPK mutant strains showed a small decrease in low glucose Hmg1 phosphorylation and corresponding increase in Hmg1 activity relative to wild-type cells. Second, the AMPK mutant strains showed a small but consistent decrease in HMGR activity in high glucose conditions and corresponding small increase in Hmg1 phosphorylation. Despite differences between wild-type and AMPK mutant strains, the results as a whole show that low glucose Hmg1 phosphorylation does not require S. pombe AMPK.
Low Glucose Hmg1 Phosphorylation Requires the PP2A-related Phosphatase Ppe1 and Its Regulator Sds23
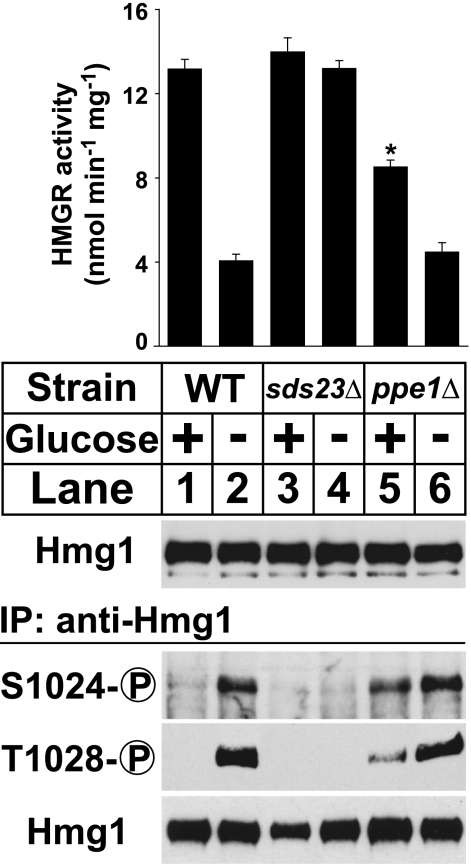
Sds23 is a suppressor of PP2A-related phosphatase activity that is required for cell division in low glucose conditions (12). To examine the role of Sds23 in low glucose Hmg1 phosphorylation, we grew wild-type and sds23Δ cells in the presence or absence of glucose for 30 min and assayed Hmg1 phosphorylation. As expected, glucose deprivation induced Hmg1 phosphorylation in wild-type cells (Fig. 6, lower panel, lanes 1 and 2). However, cells lacking sds23+ showed no Hmg1 phosphorylation (Fig. 6, lower panel, lanes 3 and 4), indicating that sds23+ is required for low glucose Hmg1 phosphorylation. Correspondingly, cells lacking sds23+ showed constitutively high Hmg1 activity (Fig. 6, upper panel, lanes 2 and 3). This shows that Sds23 is required for low glucose phosphoregulation of Hmg1.
FIGURE 6.
Low glucose Hmg1 phosphorylation requires the PP2A-related phosphatase Ppe1 and its regulator Sds23. Wild-type, sds23Δ, and ppe1Δ cells growing exponentially in YES medium were harvested by centrifugation and resuspended in either fresh YES medium or glucose-free medium for 30 min. Microsomes were prepared, assayed for Hmg1 activity, and immunoblotted with anti-Hmg1 IgG. Data are the average of three technical replicates; error bars show S.E. Asterisk indicates significant difference from YES plus glucose condition (p < 0.001). Whole cell lysates were also prepared, from which Hmg1 was immunoprecipitated and analyzed by immunoblotting with anti-Hmg1 IgG or Hmg1 phosphospecific antibodies.
Because Sds23 is a negative regulator of PP2A-related phosphatases (12), we hypothesized that cells lacking the PP2A-related phosphatase might show constitutive Hmg1 phosphorylation. The S. pombe genome encodes three PP2A-related phosphatase catalytic subunits: ppa1+, ppa2+, and ppe1+. Two of these, ppa1+ and ppa2+, are not individually required to regulate low glucose Hmg1 phosphorylation (supplemental Fig. S2). To test whether ppe1Δ cells show constitutive Hmg1 phosphorylation, we assayed low glucose Hmg1 phosphorylation and enzyme activity in a strain lacking ppe1+. Cells lacking ppe1+ showed increased Hmg1 Ser-1024 and Thr-1028 phosphorylation in the presence of glucose (Fig. 6, lower panel, lanes 5 and 6). This result shows that ppe1+ is required to decrease Hmg1 phosphorylation in high glucose conditions. Consistent with this, Hmg1 activity in the presence of glucose was significantly reduced in ppe1Δ cells (Fig. 6, upper panel, lane 5). Taken together, our results show that low glucose phosphoregulation of Hmg1 requires the PP2A-related phosphatase Ppe1 and its regulator Sds23.
DISCUSSION
Ins1 regulates Hmg1 by a nondegradative mechanism, promoting phosphorylation of the Hmg1 catalytic domain to regulate enzyme activity (6). In this study we identified a glucose signaling pathway that utilizes Ins1-dependent phosphoregulation of Hmg1 to control sterol synthesis. Depriving S. pombe cells of glucose induces Ins1-dependent phosphorylation of the Hmg1 catalytic domain, suppressing enzyme activity (Fig. 2). Adding glucose suppresses Hmg1 phosphorylation, restoring enzyme activity. When the ability to phosphorylate Hmg1 is removed, S. pombe cells are unable to down-regulate sterol synthesis upon entering stationary phase, and consequently sterol intermediates accumulate.
Mammalian cells down-regulate HMGR activity in response to glucose starvation by AMPK-mediated phosphorylation; AMPK phosphorylates a conserved serine in the HMGR catalytic domain that corresponds to Hmg1 Ser-1024. AMPK regulation of HMGR does not require mammalian Insig (9). In contrast, phosphoregulation of Hmg1 requires Ins1. Although deletion of S. pombe AMPK has a subtle effect on Hmg1 phosphorylation, it is not absolutely required. (Fig. 5B). Thus, both mammals and fission yeast regulate sterol synthesis under low glucose by controlling phosphorylation of a conserved site in the HMGR catalytic domain, but the mechanisms by which this is accomplished are different.
The PP2A-related phosphatase Ppe1 and its upstream regulator Sds23 are required for glucose regulation of Hmg1 phosphorylation (Fig. 6). Although cells lacking Sds23 are unable to phosphorylate Hmg1, cells lacking Ppe1 show constitutive Hmg1 phosphorylation. This is consistent with Sds23 acting as a negative regulator of Ppe1 (12). Mammalian HMGR is also regulated by PP2A (26). Although Hmg1 regulation can occur in the absence of AMPK (Fig. 5B), it is interesting to note that S. pombe and mammalian HMGR are both regulated by cystathionine β-synthase domain-containing proteins: Sds23 and the AMPK γ subunit, respectively. Cystathionine β-synthase domains bind adenosine nucleotides such as AMP and ATP and are proposed to act as metabolic sensors (27, 28). Thus it is tempting to speculate that Sds23 may sense changes in the AMP:ATP ratio to regulate sterol synthesis in a way analogous to that of AMPK in mammalian cells.
Ppe1 is necessary for complete dephosphorylation of Hmg1 Ser-1024 and Thr-1028 in the presence of glucose and the corresponding increase in HMGR activity (Fig. 6). However, even in cells lacking Ppe1, glucose causes some decrease in Hmg1 phosphorylation (Fig. 6, lanes 5 and 6). Likewise, there remains a glucose-dependent increase in Hmg1 activity in ppe1Δ cells. This residual effect may be due to two other PP2A-related phosphatases, Ppa1 and Ppa2, which do not individually affect Hmg1 phosphorylation (supplemental Fig. S2). However, we were unable to test this possibility directly because of synthetic lethal interactions among ppa1+, ppa2+, and ppe1+ (29).
In a previous study, we found that osmotic stress activates Hmg1 phosphorylation (6). Here, we compared Hmg1 phosphoregulation in osmotic stress and low glucose. We found that low glucose induces Hmg1 phosphorylation and suppresses Hmg1 activity to a greater extent than osmotic stress (Fig. 4D). When we measured Hmg1 activity in low glucose with simultaneous osmotic stress, we observed no further decrease in enzyme activity compared with low glucose alone (Fig. 4E). This result suggests that low glucose causes maximal inhibition of Hmg1 and that osmotic stress plays a comparatively minor role in control of Hmg1.
We showed previously that the stress-responsive MAP kinase Sty1 is required for Hmg1 phosphorylation in response to growth in a minimal medium (6). Although the current report also describes an increase in Hmg1 phosphorylation upon nutrient limitation, there are three key differences between these findings. First, the minimal medium-induced response requires Sty1, whereas the low glucose-induced response does not (supplemental Fig. S1). Second, Sty1-dependent phosphorylation of Hmg1 is suppressed by yeast extract, whereas the glucose-dependent phosphorylation of Hmg1 is not (Fig. 4B) (6). Third, low glucose causes an increase in Hmg1 phosphorylation and decrease in Hmg1 activity that is of greater magnitude than osmotic stress or minimal medium (Fig. 4D). These findings demonstrate that multiple signaling pathways converge on Hmg1 through the common mechanism of Ins1-dependent phosphorylation, allowing the cell to regulate sterol synthesis in response to multiple and diverse environmental stimuli.
In this study, we show that S. pombe cells regulate sterol synthesis in response to changing glucose levels. Our findings support a model in which glucose withdrawal activates Sds23, which in turn suppresses the activity of the PP2A-related phosphatase Ppe1. As a result, Ins1-dependent phosphorylation of the Hmg1 catalytic domain increases, suppressing HMGR activity and inhibiting sterol synthesis in response to decreased glucose availability. This regulatory mechanism may function to limit flow of carbon into the sterol biosynthetic pathway when cell growth is arrested upon entry into stationary phase. Current studies are focused on identifying the low glucose Hmg1 kinase and understanding the interplay between this kinase and the Sds23-Ppe1 pathway.
Supplementary Material
Acknowledgments
We thank Shan Zhao and Emerson Stewart for excellent technical assistance and Jianhua Liu (Genome Institute of Singapore) for yeast strains.
This work was supported, in whole or in part, by National Institutes of Health Grant HL-077588.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. S1 and S2.
- HMGR
- HMG-CoA reductase
- AMPK
- AMP-activated protein kinase
- PP2A
- protein phosphatase 2A.
REFERENCES
- 1. Goldstein J. L., Brown M. S. (1990) Nature 343, 425–430 [DOI] [PubMed] [Google Scholar]
- 2. Jo Y., Debose-Boyd R. A. (2010) Crit. Rev. Biochem. Mol. Biol. 45, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Friesen J. A., Rodwell V. W. (2004) Genome Biol. 5, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 5. Flury I., Garza R., Shearer A., Rosen J., Cronin S., Hampton R. Y. (2005) EMBO J. 24, 3917–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burg J. S., Powell D. W., Chai R., Hughes A. L., Link A. J., Espenshade P. J. (2008) Cell Metab. 8, 522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Omkumar R. V., Darnay B. G., Rodwell V. W. (1994) J. Biol. Chem. 269, 6810–6814 [PubMed] [Google Scholar]
- 8. Sato R., Goldstein J. L., Brown M. S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9261–9265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Engelking L. J., Liang G., Hammer R. E., Takaishi K., Kuriyama H., Evers B. M., Li W. P., Horton J. D., Goldstein J. L., Brown M. S. (2005) J. Clin. Invest. 115, 2489–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hardie D. G., Hawley S. A., Scott J. W. (2006) J. Physiol 574, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Virshup D. M. (2000) Curr. Opin. Cell Biol. 12, 180–185 [DOI] [PubMed] [Google Scholar]
- 12. Hanyu Y., Imai K. K., Kawasaki Y., Nakamura T., Nakaseko Y., Nagao K., Kokubu A., Ebe M., Fujisawa A., Hayashi T., Obuse C., Yanagida M. (2009) Genes Cells 14, 539–554 [DOI] [PubMed] [Google Scholar]
- 13. Hughes A. L., Todd B. L., Espenshade P. J. (2005) Cell 120, 831–842 [DOI] [PubMed] [Google Scholar]
- 14. Burke J. D., Gould K. L. (1994) Mol. Gen. Genet. 242, 169–176 [DOI] [PubMed] [Google Scholar]
- 15. Bimbó A., Jia Y., Poh S. L., Karuturi R. K., den Elzen N., Peng X., Zheng L., O'Connell M., Liu E. T., Balasubramanian M. K., Liu J. (2005) Eukaryot. Cell 4, 799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998) Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 17. Hughes A. L., Lee C. Y., Bien C. M., Espenshade P. J. (2007) J. Biol. Chem. 282, 24388–24396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mitchison J. M., Creanor J. (1969) J. Cell Sci. 5, 373–391 [DOI] [PubMed] [Google Scholar]
- 19. Moreno S., Ruíz T., Sánchez Y., Villanueva J. R., Rodríguez L. (1985) Arch. Microbiol. 142, 370–374 [DOI] [PubMed] [Google Scholar]
- 20. Matsuzawa T., Fujita Y., Tanaka N., Tohda H., Itadani A., Takegawa K. (2011) J. Biosci. Bioeng. 111, 158–166 [DOI] [PubMed] [Google Scholar]
- 21. Pluskal T., Hayashi T., Saitoh S., Fujisawa A., Yanagida M. (2011) FEBS J. 278, 1299–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zuin A., Carmona M., Morales-Ivorra I., Gabrielli N., Vivancos A. P., Ayté J., Hidalgo E. (2010) EMBO J. 29, 981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hardie D. G. (2003) Endocrinology 144, 5179–5183 [DOI] [PubMed] [Google Scholar]
- 24. Townley R., Shapiro L. (2007) Science 315, 1726–1729 [DOI] [PubMed] [Google Scholar]
- 25. Hardie D. G. (2007) Nat. Rev. Mol. Cell Biol. 8, 774–785 [DOI] [PubMed] [Google Scholar]
- 26. Ingebritsen T. S., Blair J., Guy P., Witters L., Hardie D. G. (1983) Eur. J. Biochem. 132, 275–281 [DOI] [PubMed] [Google Scholar]
- 27. Scott J. W., Hawley S. A., Green K. A., Anis M., Stewart G., Scullion G. A., Norman D. G., Hardie D. G. (2004) J. Clin. Invest. 113, 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ignoul S., Eggermont J. (2005) Am. J. Physiol. Cell Physiol. 289, C1369–1378 [DOI] [PubMed] [Google Scholar]
- 29. Shimanuki M., Kinoshita N., Ohkura H., Yoshida T., Toda T., Yanagida M. (1993) Mol. Biol. Cell 4, 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.