Abstract
Ste4 is the β subunit of a heterotrimeric G protein that mediates mating responses in Saccharomyces cerevisiae. Here we show that Ste4 undergoes ubiquitination in response to pheromone stimulation. Ubiquitination of Ste4 is dependent on the E3 ligase Rsp5. Disrupting the activity of Rsp5 abolishes ubiquitination of Ste4 in vivo, and recombinant Rsp5 is capable of ubiquitinating Ste4 in vitro. We find also that Lys-340 is a major ubiquitination site on Ste4, as pheromone-induced ubiquitination of the protein is prevented when this residue is mutated to an arginine. Functionally, ubiquitination does not appear to regulate the stability of Ste4, as blocking ubiquitination has no apparent effect on either the abundance or the half-life of the protein. However, when presented with a concentration gradient of pheromone, Ste4K340R mutant cells polarize significantly faster than wild-type cells, indicating that ubiquitination limits pheromone-directed polarized growth. Together, these findings reveal a novel stimulus-dependent posttranslational modification of a Gβ subunit, establish Ste4 as a new substrate of the E3 ligase Rsp5, and demonstrate a role for G protein ubiquitination in cell polarization.
Keywords: Cellular Regulation, G Proteins, MAP Kinases (MAPKs), Signal Transduction, Ubiquitination
Introduction
G proteins are membrane-associated signal transducers that transmit signals from cell surface receptors to intracellular effectors. Without stimulation, G proteins exist as a heterotrimer of α, β, and γ subunits. G protein-coupled receptor activation leads to GDP release, GTP binding, and dissociation of Gα-GTP from the Gβγ subunits. Depending on the system, either Gα or Gβγ or both transmit the signal and activate downstream effectors (1, 2).
Multiple mechanisms exist to ensure that G protein signaling is tightly controlled (2, 3). For instance, the cell surface expression of receptors is regulated via feedback phosphorylation and internalization (4). The duration of G protein activation is limited by regulators of G protein signaling (RGS protein), which enhance the GTPase activity of Gα and promote reassociation of the heterotrimer and G protein deactivation (5). Likewise, the activity of most effector kinases, such as mitogen-activated protein kinases, is regulated by the action of phosphatases (6, 7).
The ubiquitin system is now recognized as an important regulator of cell function (8–10). The process of ubiquitination is catalyzed by a cascade of enzymes composed of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-ligating enzyme or ligase (E3) (8, 9). In most studied examples, ubiquitin is conjugated to the ϵ-amino group of lysine via the formation of an isopeptide bond (9). Less commonly, the free amino terminus of a target protein is used for conjugation (11). Specificity of ubiquitination is afforded by the large number of E2 and E3 enzymes, which bind protein substrates with a high degree of selectivity (8, 12). Most substrates are modified with a polyubiquitin chain, which targets the substrates for degradation by the 26 S proteasome (8). Some proteins, especially those that are membrane-associated, are mono-ubiquitinated (a single ubiquitin moiety instead of a polyubiquitin chain is attached to a lysine residue on the substrate) (13). Distinct from polyubiquitination, monoubiquitination typically does not lead to proteasome-dependent proteolysis of the protein. Rather, it acts as a signal to regulate intracellular protein trafficking and/or serves as a mechanism to modulate protein-protein interactions (14–16). In this regard, monoubiquitination could function much like phosphorylation to affect protein activity.
Evidence is emerging that ubiquitination is involved in the regulation of G protein signaling (17, 18). In mammalian cells, several heterotrimeric G protein subunits, including Gα and Gγ but not Gβ, are thought to be regulated by the ubiquitination pathway (17, 18). Among those, direct ubiquitination has been observed for Gαs (19), Gγ2 (20), and transducin Gγ (21). However, the ubiquitination site in these proteins has not been identified, and the mechanism and functional consequence of these ubiquitination events are not fully understood (18).
The G protein pathway is highly conserved across evolution, allowing the study of fundamental mechanisms of G protein signaling using simple model organisms such as the Baker's yeast Saccharomyces cerevisiae (22). Indeed, many principles of G protein signaling were initially discovered through studying the mating pathway in yeast (23). As with other G protein signaling pathways, activation of the yeast mating pathway is initiated by a ligand (i.e. the mating pheromone) binding to a cell surface receptor Ste2/Ste3, which in turn promotes GTP binding to the Gα subunit Gpa1. GTP triggers dissociation of Gpa1 from the Gβγ subunits Ste4/Ste18 (3, 24). Free Ste4/Ste18 transmits the signal, leading to activation of multiple downstream effectors including Far1 (a cyclin-dependent kinase inhibitor), Cdc24 (exchange factor for a small GTPase Cdc42), and a MAP kinase cascade comprised of Ste20, Ste11, Ste7, and Fus3 (3, 23). Activation of Far1 and the MAP kinase cascade results in growth arrest at G1 and transcription of genes required for mating (3).
Ste4 also regulates polarized cell growth via interactions with Cdc24 and Far1 (3, 25). When cells are treated with pheromone, they reorient their cytoskeleton and initiate polarized growth toward the highest concentration of pheromone, leading to the formation of a “shmoo” morphology (26, 27). It is possible that this behavior allows the yeast to mate with the best partner available because such yeast cells may release the strongest mating signal. It has been suggested that Ste4 may play a role in sensing the pheromone gradient, but direct evidence is lacking (28).
Despite the pivotal roles of Ste4 in activating a multitude of effectors that are responsible for all aspects of the pheromone response, Ste4 is not an abundant protein. In fact, earlier work suggested that Ste4 is the limiting component in the receptor/G protein complex. Estimates from a large scale quantitative immunoblotting study indicated that the number of Ste4 molecules (∼2050/cell) is much lower than either Gα Gpa1 (∼9920/cell) or Gγ Ste18 (∼5550/cell) (29). Moreover, as little as 2-fold overexpression of Ste4 (but not Gpa1 and Ste18) is sufficient to yield full activation of the pathway (30). Given the limiting abundance of Ste4 and its crucial roles in pheromone signaling, it is likely that a battery of mechanisms may exist to regulate its activity to ensure accurate cellular responses to pheromone treatment.
In this study, we examined the potential role of the ubiquitination pathway in the regulation of Ste4. We find that Ste4 is monoubiquitinated and that ubiquitination is stimulated by pheromone treatment. Through genetic and biochemical analysis, we identify Rsp5, a homologous to the E6-AP carboxyl terminus type E3 ligase, as the enzyme responsible for Ste4 ubiquitination. We find also that lysine 340 in Ste4 serves as a major ubiquitination site. Finally, we find that blocking Ste4 ubiquitination alters the rate of polarized growth triggered by pheromone stimulation. Together, this study reveals a novel stimulus-dependent modification of the G protein β subunit required for proper cell polarization.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
Standard methods for the growth, maintenance, and transformation of yeast and bacteria and for the manipulation of DNA were used throughout. The yeast S. cerevisiae strains used in this study are BY4741 (MATa leu2Δ met15Δ his3Δ ura3Δ), BY4741-derived mutants lacking STE4, FUS3, or KSS1 (Research Genetics, Huntsville, AL), MYY290 (MATa leu2 his3 ura3), and MYY808 (MATa leu2 his3 ura3 rsp5–1, a temperature-sensitive mutant) (from Dr. Michael Yaffe, University of California at San Diego) (31), doxycycline-sensitive yeast strains (MATa URA3::CMV-tTA his3–1 leu2–0 met15–0) (Open Biosystems) (32), YPH499 (MATa ura3–52 lys2–801am ade2–101oc trp1-Δ63 his3-Δ200 leu2-Δ1), and YPH499-derived mutants lacking STE4 (ste4::KanMX, this work).
Expression plasmids used in this study that have been described previously are pYES-His-8-Ubiquitin (33), pYES-GAL-STE4-FLAG (34), YCp-STE4, and the YCp-STE4-derived mutants STE4Δ310–346 and STE4T320A/S335A (from Dr. David Stone, University of Illinois at Chicago) (35). YCp-STE4K340R was generated by site-directed mutagenesis using YCp-STE4 as the template. Mutagenesis primers were 5′-CAA TCG CCA CAA ACT TTA AGA TCA ACA AGC TCA AGC TAT CTA GAC-3′ and 5′-GTC TAG ATA GCT TGA GCT TGT TGA TCT TAA AGT TTG TGG CGA TTG-3′. pRS306-STE4K340R was constructed by subcloning the STE4 open reading frame plus 1000 base pairs of upstream promoter sequence and 472 base pairs of downstream sequence from YCp-STE4K340R into the EcoRI/NotI sites of pRS306. The PCR primers used were 5′-AAG GAA AAA AGC GGC CGC ACA GAA ATA TTT GAA ATA TAT TTC C-3′ and 5′-CTA GGA ATT CAA ATT CAG GCA TTT TTG AAA TTA CC-3′. The resulting plasmid was linearized with StuI and integrated at the URA3 locus of YPH499-derived mutants lacking STE4. The transformants were selected on SCD-Ura, and the integrations were confirmed by immunoblotting of Ste4 protein.
pRS315-His-8-Ubiquitin (CEN/ARS, LEU2, GAL1 promoter, CYC1 terminator) was constructed by subcloning the GAL1-His-8-Ubiquitin-CYC1 fragment from pYES-His-8-Ubiquitin to the SpeI site of pRS315. The PCR primers used were 5′-GGA CTA GTA CGG ATT AGA AG-3′ and 5′-GGA CTA GTG CCG ATT CAT TAA TGC AGG GC-3′. For construction of pYES-RSP5-FLAG, a triple-FLAG epitope tag was placed at the C terminus of Rsp5 (RSP5-FLAG) by PCR amplification and subcloning into the pYES2.1/V5-His-TOPO (2 μm, URA3, GAL1 promoter, CYC1 terminator) (Invitrogen). PCR primers were 5′-CCC AAG CTT CCA GAA TGC CTT CAT CCA TAT CCG TC-3′, and 5′-TTA CTT GTC ATC GTC ATC TTT ATA ATC CTT GTC ATC GTC ATC TTT ATA ATC CTT GTC ATC GTC ATC TTT ATA ATC CCC AAG CTT TTC TTG ACC AAA CCC TAT GG-3′. The plasmid pDS30 (FUS1-GFP reporter) was from Daria Siekhaus, University of California, Berkeley (36).
Signaling and Degradation Bioassays
The pheromone-induced phosphorylation of Fus3 and Kss1 were monitored by immunoblotting of whole cell extracts with antibodies that specifically recognize phosphorylated Fus3 and Kss1 (9101, phosphor-p44/42 MAPK antibody from Cell Signaling Technology, Inc.). Specificity of detection was established using fus3Δ and kss1Δ cell extracts as negative controls. The pheromone-dependent transcription assay using the green fluorescent protein reporter (FUS1-GFP) has been described previously (30, 37). To monitor the loss of Ste4 over time, early log cell cultures were treated with 3 μm α-factor for 60 min, followed by cycloheximide (10 μg/ml in 0.1% ethanol, final concentrations) for up to 2 h. Growth was stopped by the addition of 10 mm NaN3 and transfer to an ice bath. Cell extracts were prepared via TCA precipitation, separated by 8% SDS-PAGE, and transferred to a nitrocellulose membrane (Bio-Rad). The membrane was probed with antibodies to Ste4 at 1:2000 (from Duane Jenness, University of Massachusetts), and Pgk1 at 1:75,000 (from Jeremy Thorner, University of California, Berkeley, CA). Immunoreactive species were visualized by enhanced chemiluminescence detection (Pierce) of horseradish peroxidase-conjugated anti-rabbit IgG (Bio-Rad) or anti-goat IgG (Santa Cruz Biotechnology, Inc.). Specificity of detection was established using ste4Δ cell extract as a negative control.
Isolation of Ste4 Conjugated to His-tagged Ubiquitin
Ubiquitination of Ste4 was detected by purifying His-tagged ubiquitin conjugates as described previously (33), followed by immunoblotting with antibodies that were raised against Ste4. Briefly, wild-type or ste4Δ mutant cells were transformed with either pYES-His-8-Ubiquitin or vector and were grown in synthetic galactose medium to early log phase. Upon treatment with pheromone (3 μm) for the indicated time, about 60 A600 cells from each culture were harvested by centrifugation. Extracts were prepared for metal affinity purification of His-tagged ubiquitin and conjugates by methods that were adapted from Muratani et al. (38). Each cell pellet was suspended in 650 μl of buffer A2 (6 m guanidine-HCl, 100 mm Na2HPO4/NaH2PO4 (pH 8.0), 10 mm imidazole, 250 mm NaCl, 0.5% Nonidet P-40, 2 mm N-ethylmaleimide, and 1 pellet of complete EDTA-free protease inhibitor (Roche) for every 50 ml of buffer). Suspensions were subjected to eight cycles of glass bead vortex homogenization of 30 s each. The lysates were solubilized by mixing at 4 °C for 1 h and clarified by two rounds of centrifugation at a speed of 12,000 × g for 5 min and 25 min at 4 °C. The resulting supernatants were incubated with TALON Superflow metal affinity resin (BD Biosciences, Clontech) for 2 h at room temperature with rocking. Following this incubation, the resin was washed twice with buffer A2, twice with buffer A2/T2 (1 volume of buffer A2 and 3 volumes of buffer T2 (50 mm Na2HPO4/NaH2PO4 (pH 8.0), 250 mm NaCl, 20 mm imidazole, 0.5% Nonidet P-40), and twice with buffer T2. Resin was then washed with buffer T2 containing 50 mm histidine to reduce the retention of proteins that nonspecifically bind to the resin. Finally, 8His-ubiquitin and any conjugated proteins were eluted with 2× SDS-PAGE loading buffer. Samples of the metal affinity-purified proteins and total extract were resolved by 7.5% SDS-PAGE and transferred to nitrocellulose. Detection of Ste4 was carried out with anti-Ste4 antibodies followed by goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology, Inc.) at 1:6000. Visualization of HRP-conjugated complexes was accomplished by the enhanced chemiluminescence Reagent Plus from PerkinElmer Life Sciences.
Immunoprecipitation
The association of Ste4 and Rsp5 was examined by immunoprecipitation of FLAG-tagged Rsp5 and immunoblotting with anti-Ste4 antibodies. Cells transformed with pYES-RSP5-FLAG or empty vector were grown to early log phase, treated as indicated with 3 μm pheromone for 60 min, harvested by centrifugation, and resuspended in 550 μl of lysis (FLAG/Triton) buffer (50 mm NaPO4 (pH 7.5), 400 mm NaCl, 0.1% Triton X-100, 10% glycerol, 0.5 mm dithiothreitol, 25 mm NaF, 25 mm glycerophosphate, 1 mm sodium orthovanadate, 10 mm N-ethylmaleimide, 5 mm phenylmethylsulfonyl fluoride, and one pellet of complete EDTA-free protease inhibitor mixture (Roche) for every 50 ml of buffer). This and all subsequent manipulations were carried out at 4 °C. Cells were subjected to glass bead vortex homogenization for 30 s, repeated eight times, and centrifuged twice at 6000 × g for 5 min and 25 min. Lysates were incubated for 2 h with a bead volume of 10 μl of anti-FLAG M2 affinity resin (Sigma) equilibrated in lysis buffer. Immunoprecipitates were collected by centrifugation at 1000 × g for 30 s, and pellets were washed with 1 ml of lysis buffer for 3 min and repeated four times before final resuspension in 30 μl of 2× SDS-PAGE loading buffer. Each sample was resolved by 7.5% SDS-PAGE and immunoblotting with anti-Ste4 antibodies at 1:1000 or anti-FLAG monoclonal antibodies at 1:2000.
Purification of Ste4
FLAG-tagged Ste4 was purified from the ste4Δ cells transformed with plasmid pYES-GAL-STE4-FLAG. Expression of Ste4 was induced by switching early log phase cells to galactose-containing media for 8 h. Briefly, cells were grown in glucose-containing medium to A600 0.8, harvested by centrifugation, resuspended in galactose-containing medium to A600 0.3, and grown for an additional 8 h to induce the expression of Ste4-FLAG. After treatment with 3 μm pheromone for 60 min, cells were harvested by centrifugation and resuspended in lysis (FLAG/Triton) buffer. Ste4-FLAG was immunoprecipitated using anti-FLAG M2 resin. The immunoprecipitates were collected by centrifugation at 1000 × g for 30 s, and the pellet was washed four times with lysis buffer (10 ml for the first wash and 1 ml for the remaining three washes), four times with 1 ml of 1× ubiquitination buffer (25 mm Tris (pH 7.5), 50 mm NaCl, 4 mm MgCl2), and was stored in glycerol-containing 1× ubiquitination buffer (25 mm Tris (pH 7.5), 50 mm NaCl, 4 mm MgCl2, 30% glycerol) prior to the ubiquitination reaction.
In Vitro Ubiquitination of Ste4 by Rsp5
In vitro ubiquitination reactions were conducted in compact reaction columns (USB Corp.) with Ste4-FLAG immobilized on M2 FLAG affinity beads. (Note: We observed that elution with FLAG/Triton buffer prior to the ubiquitination reaction inhibited Rsp5 catalytic activity). Briefly, 15 μl of bead-immobilized Ste4-FLAG representing 15% of a single affinity purification from a 3,600 A cell pellet was prewashed with 0.7 ml 1× ubiquitin assay buffer (25 mm Tris (pH 7.5), 50 mm NaCl, 4 mm MgCl2) to remove the glycerol storage buffer. Ubiquitination assay components were then added to the beads, including (unless otherwise indicated) 1 μm human E1-activating enzyme Uba1 (Boston Biochem), 1 μm yeast E2-conjugating enzyme Ubc5b, 0.2 μm yeast E3 ligase Rsp5 (or the catalytically inactive mutant Rsp5C777A), 5 μm ubiquitin, and 4.8 mm ATP (Sigma-Aldrich), resulting in a 25 μl final reaction volume (beads plus liquid). Reactions were allowed to proceed for 30 min at room temperature and then quenched by addition of SDS (to 2% final) and heated to 95 °C for 2 min followed by dilution with 6× SDS-PAGE loading buffer containing 100 mm DTT. Ubiquitination of Ste4-FLAG was detected by 10% SDS-PAGE and immunoblotting with anti-FLAG antibody as described above.
Polarized Growth
Cells were grown to A600 0.2, treated with dimethyl sulfoxide (1%, final concentration) for 1.5 h before the addition of nocodazole (15 μg/ml in 1% dimethyl sulfoxide, final concentrations) for 2.5–3 h. Cultures were visualized by microscopy to confirm G2/M arrest. The cells were then loaded into a microfluidics device as described previously (39), and the nocodazole was washed out for 20 min prior to stimulation with 150 nm α factor. Note that the concentration of α factor required to give a good response in liquid culture is much higher because of accumulation of Bar1 and degradation of the α factor. Cells were imaged at 2-min intervals for 2 h. For each cell, the time at which the first sign of polarized growth detectable in the differential interference contrast image was recorded and plotted as a function of time.
RESULTS
Ste4 is the β subunit of a heterotrimeric G protein that mediates mating responses in S. cerevisiae. Earlier studies indicate that Ste4 represents a crucial component in the pathway, and its activity must be tightly regulated (30, 40). Growing evidence suggests that ubiquitination is involved in the regulation of G protein signaling (17, 18). Thus, we considered the possibility that Ste4 may be regulated via ubiquitination.
Pheromone Stimulation Induces Ubiquitination of Ste4
To examine whether Ste4 undergoes ubiquitination, we exploited an affinity purification method for isolating ubiquitin-conjugated proteins (38). Cells were transformed with a plasmid that expresses poly-His-tagged ubiquitin (His-8-Ubi)2 from the GAL1 promoter or an empty vector as a control. Cells were treated with mating pheromone α factor over time and collected at the indicated time points. Extracts from collected cell pellets were prepared for affinity purification. The purifications were conducted under denaturing conditions to dissociate Ste4 from other ubiquitinated proteins that happen to bind Ste4 via non-covalent interactions. Samples of the extracts and purified proteins were resolved by SDS-PAGE and analyzed by immunoblotting.
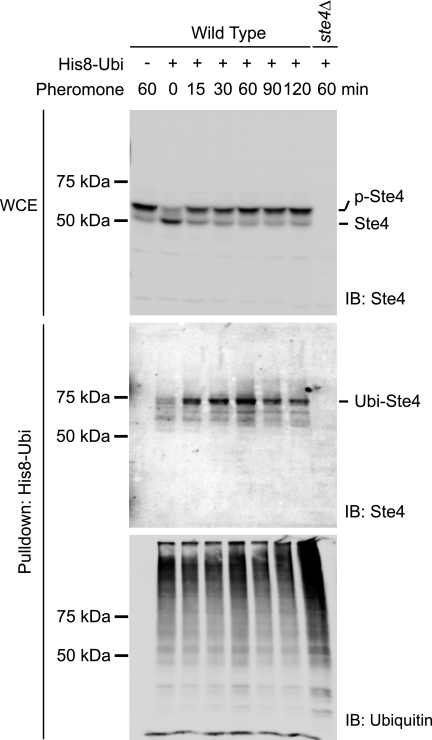
Immunoblot analysis of whole-cell lysates specifically identified Ste4, as this species is clearly absent in the ste4Δ mutant (Fig. 1, top panel). The pheromone-induced mobility-shift of Ste4 is because of phosphorylation, as documented previously (35, 41). The population of His-8-Ubi-conjugated proteins specifically isolated by affinity purification is shown on the anti-ubiquitin blot (Fig. 1, bottom panel). Immunoblotting of the affinity-purified His-8-Ubi-conjugated proteins with anti-Ste4 antibodies revealed a discrete band of about 70 kDa (Fig. 1, center panel). The intensity of the 70-kDa band was significantly increased in the pheromone-treated samples, peaking 60 min after pheromone addition. The 70-kDa band as well as the minor species around it were completely absent from the vector and ste4Δ control samples (Fig. 1, center panel), indicating that they represent ubiquitinated Ste4. Because the majority of ubiquitinated Ste4 exists as a discrete band rather than a ladder of high molecular weight bands, it is very likely that Ste4 is modified with a single ubiquitin moiety. Taken together, this analysis shows that Ste4 is monoubiquitinated and that ubiquitination of Ste4 is promoted by pheromone treatment.
FIGURE 1.
Ste4 undergoes ubiquitination in response to pheromone stimulation. Wild-type or ste4Δ cells were transformed with either empty vector or a plasmid expressing His-8-tagged ubiquitin (His8-Ubi). Early log phase cells were treated with 3 μm pheromone for the indicated time (min). His-8-Ubi-conjugated proteins were purified from whole cell lysates under denaturing conditions. The level of Ste4 in the whole cell extracts (WCE) prior to purification was detected by immunoblotting (IB) with anti-Ste4 antibodies (top panel). The purified samples were analyzed by immunoblotting with anti-Ste4 antibodies (center panel) and anti-ubiquitin antibodies (bottom panel). p-Ste4, phosphorylated Ste4; Ubi-Ste4, ubiquitinated Ste4.
Ubiquitination of Ste4 May Be Regulated via Its Prior Phosphorylation
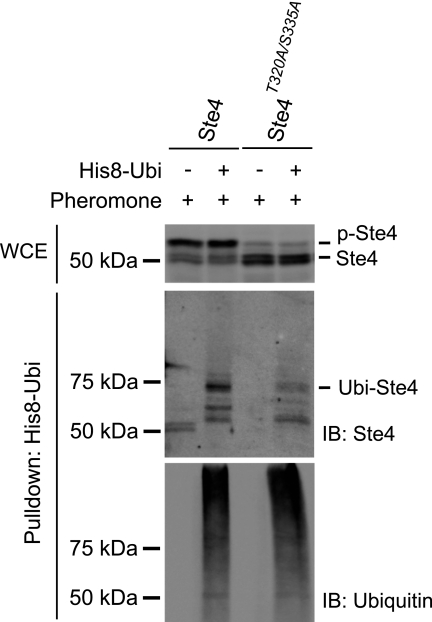
Next, we investigated potential mechanisms that may regulate Ste4 ubiquitination. Phosphorylation is a common mechanism that targets signaling proteins for ubiquitination (9, 42). Ste4 is rapidly phosphorylated following pheromone stimulation (Fig. 1, top panel) (35, 41), raising the possibility that there may be a mechanistic link between these two modifications. The precise phosphorylation sites of Ste4, as well as the kinase responsible, have not been definitively identified (3). Thus, we are not able to directly test whether phosphorylation is required for Ste4 ubiquitination. However, previous work did map Ste4 phosphorylation to a small region encompassing residues 310 to 346. Phosphorylation is completely blocked in a mutant form of Ste4 that lacks this region (Ste4Δ310–346) (41). A mutagenesis analysis of serine and threonine residues within this region suggested that phosphorylation of Ste4 occurs on residues Thr-320 and Ser-335, as phosphorylation is significantly impaired in the Ste4T320A/S335A mutant (35). To test whether these residues are required for Ste4 ubiquitination, we examined the ubiquitination status of the Ste4T320A/S335A mutant. As shown in Fig. 2, the Ste4T320A/S335A mutant displays a substantial decrease in the level of ubiquitination. The remaining residual ubiquitination might be due to residual phosphorylation of the mutant (Fig. 2). These data suggest that phosphorylation is required for Ste4 ubiquitination.
FIGURE 2.
Phosphorylation may be required for Ste4 ubiquitination. Cells expressing either wild-type Ste4 or Ste4T322A/S335A were transformed with either an empty vector or a plasmid expressing His-8-tagged ubiquitin (His8-Ubi). Early log phase cells were treated with 3 μm pheromone for 60 min. His-8-Ubi-conjugated proteins were purified from whole cell lysates under denaturing conditions. The level of Ste4 in the whole cell extracts (WCE) prior to purification was detected by immunoblotting (IB) with anti-Ste4 antibodies (top panel). The purified samples were analyzed by immunoblotting with anti-Ste4 antibodies (center panel) and anti-ubiquitin antibodies (bottom panel). p-Ste4, phosphorylated Ste4; Ubi-Ste4, ubiquitinated Ste4.
Ubiquitination of Ste4 Is Dependent on the E3 Ligase Rsp5
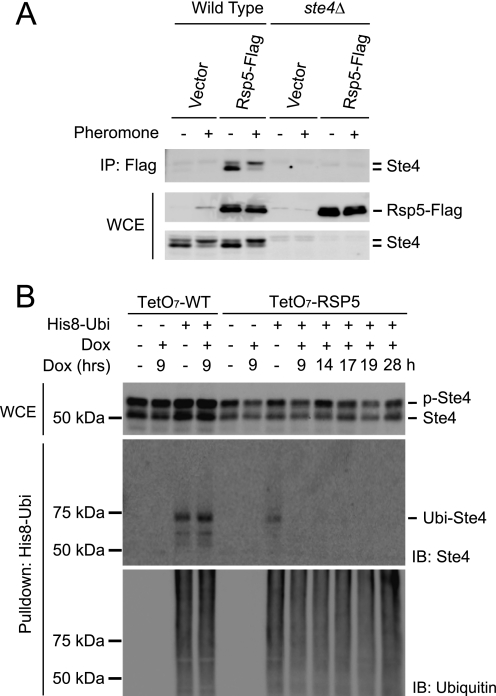
Monoubiquitination of many plasma membrane-associated proteins, including Ste2 and Gpa1, is catalyzed by Rsp5, a HECT-type E3 ubiquitin ligase (43, 44). One unique feature of Rsp5 is that it can form strong interactions with its substrates (45). Thus, to test whether Rsp5 may be the ligase responsible for Ste4 ubiquitination, we first examined whether there is any detectable interaction between Ste4 and Rsp5. For this purpose, FLAG-tagged Rsp5 was expressed in either wild-type or the ste4Δ mutant and was purified via immunoprecipitation from cultures treated or not treated with mating pheromone. Copurification of Ste4 with Rsp5 was examined via immunoblotting of the purified samples with anti-Ste4 antibodies. As shown in Fig. 3A, Ste4 indeed copurifies with Rsp5.
FIGURE 3.
Rsp5 is required for Ste4 ubiquitination in vivo. A, Ste4 coimmunoprecipitates with Rsp5. Wild-type or ste4Δ cells were transformed with either an empty vector or a plasmid expressing FLAG-tagged Rsp5 (Rsp5-FLAG). Early log phase cells were treated with 3 μm pheromone for 1 h. FLAG-tagged Rsp5 was immunoprecipitated (IP) using anti-FLAG M2 resin, and the copurified Ste4 was detected by immunoblotting of the immunoprecipitates (IP: Flag) using anti-Ste4 antibodies. The relative level of input Ste4 and Rsp5-FLAG in the whole cell extracts (WCE) prior to purification is shown in the lower panels. B, turning off RSP5 expression abolishes Ste4 ubiquitination. Wild-type (TetO7-WT) or TetO7-RSP5 (RSP5 under the control of doxcyclin-regulatable promoter) cells were transformed with either an empty vector or a plasmid expressing His-8-tagged ubiquitin (His8-Ubi). Early log phase cells were treated or not treated with 60 μg/ml doxycycline (Dox) for the indicated time (h), followed by treatment of 3 μm pheromone for 1 h. His-8-Ubi-conjugated proteins were purified from whole cell lysates under denaturing conditions. The level of Ste4 in the whole cell extracts (WCE) prior to purification was detected by immunoblotting (IB) with anti-Ste4 antibodies (top panel). The purified samples were analyzed by immunoblotting with anti-Ste4 antibodies (center panel) and anti-ubiquitin antibodies (bottom panel). p-Ste4, phosphorylated Ste4; Ubi-Ste4, ubiquitinated Ste4.
Having detected interactions between Rsp5 and Ste4, we asked whether Rsp5 is required for in vivo ubiquitination of Ste4. RSP5 is an essential gene. To determine the consequences of Rsp5 depletion, we used a strain in which the promoter of the RSP5 gene has been replaced with a tetracyclin-regulatable promoter (32). In this strain, addition of doxycycline will turn off transcription of the RSP5 gene. As shown in Fig. 3B, ubiquitination of Ste4 is completely blocked after turning off the expression of the RSP5 gene. Ubiquitination of Ste4 is also blocked in a mutant strain that harbors a temperature-sensitive allele of RSP5 (rsp5–1), grown at the restrictive temperature (31) (supplemental Fig. 1). These data indicate that Rsp5 is required for the ubiquitination of Ste4 in vivo.
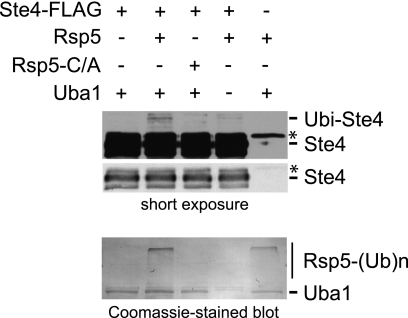
The above analysis clearly shows that Rsp5 is necessary for ubiquitination of Ste4. To determine whether Rsp5 is sufficient for Ste4 ubiquitination, we conducted an in vitro ubiquitination assay using purified recombinant Rsp5 in the presence of E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, ubiquitin, and ATP. As shown in Fig. 4, reconstitution of the ubiquitinating enzymes with purified Ste4 resulted in the formation of monoubiquitinated Ste4. Importantly, exclusion of E1 (Uba1) or incubation with a catalytically inactive form of Rsp5 (Rsp5C777A) does not result in the production of monoubiquitinated Ste4 (Fig. 4). These data show that Rsp5 is sufficient to ubiquitinate Ste4.
FIGURE 4.
Rsp5 ubiquitinates Ste4 in vitro. Bead-immobilized Ste4-FLAG was incubated with Rsp5 or catalytically inactive Rsp5 (Rsp5-C/A) in the absence or presence of E1 activating enzyme (Uba1). All reactions contained Ubc5b, ATP, and ubiquitin. Reactions were stopped after 30 min with 2% SDS. Proteins were separated by 10% SDS-PAGE, transferred to nitrocellulose membranes, and blotted with M2 anti-FLAG antibody and goat-anti-mouse-HRP secondary antibody. Monoubiquitination slows the electrophoretic mobility of Ste4-FLAG by the approximate mass of ubiquitin (∼8 kDa). Long (top panel) and short exposures (center panel) are shown. After immunoblotting, the nitrocellulose blot was stained with Coomassie to show Rsp5 autoubiquitination and Uba1 loading (bottom panel). *, antibody heavy chain. Because we observed that FLAG peptide elution buffer inhibited Rsp5 catalytic activity, we conducted in vitro ubiquitination with bead-immobilized Ste4-FLAG, which may also reduce reaction efficiency.
Lys340 Is Required for Pheromone-induced Ubiquitination of Ste4
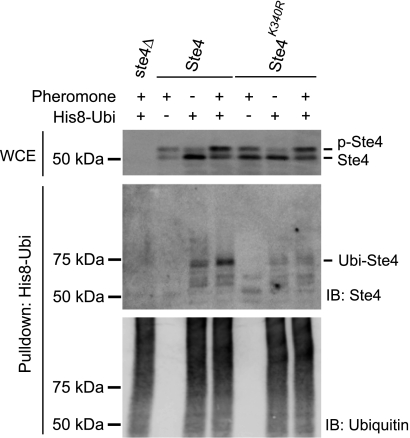
Next, we sought to identify the site of Ste4 ubiquitination. For this purpose, we made use of a recently developed bioinformatics tool (UbPred) that identifies lysine residues likely to serve as an ubiquitination site (46). Analyzing the Ste4 sequence with this tool revealed lysine 340 as the highest probability ubiquitination site among all 22 lysine residues in the protein. To test whether Lys-340 indeed serves as an ubiquitination site, we mutated it to an arginine residue, expressed the mutant from an integrated gene, and examined its ubiquitination status. Although basal ubiquitination is still detectable in the Ste4K340R mutant, pheromone-induced ubiquitination is nearly completely abolished (Fig. 5). Therefore, our data indicate that Lys-340 is primarily responsible for pheromone-induced ubiquitination of Ste4.
FIGURE 5.
Lys-340 is required for pheromone-induced ubiquitination of Ste4. Cells expressing either wild-type Ste4 or Ste4K340R were transformed with either an empty vector or plasmids expressing His-8-tagged ubiquitin (His8-Ubi). Early log phase cells were treated with 3 μm pheromone for 60 min. His-8-Ubi-conjugated proteins were purified from whole cell lysates under denaturing conditions. The level of Ste4 in the whole cell extracts (WCE) prior to purification was detected by immunoblotting (IB) with anti-Ste4 antibodies (top panel). The purified samples were analyzed by immunoblotting with anti-Ste4 antibodies (center panel) and anti-ubiquitin antibodies (bottom panel). p-Ste4, phosphorylated Ste4; Ubi-Ste4, ubiquitinated Ste4.
Functional Consequences of Ste4 Ubiquitination
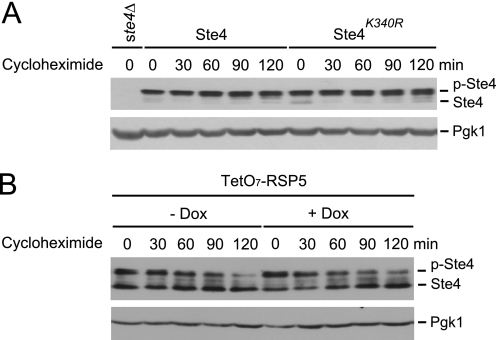
Having identified the site of ubiquitination and the enzyme responsible, we next sought to determine the functional consequences of ubiquitination. Because ubiquitination typically leads to protein degradation, we first investigated whether ubiquitination regulates the stability of Ste4. For this purpose, we added the protein synthesis inhibitor cycloheximide to pheromone-treated cells either expressing wild-type Ste4 or the ubiquitination site mutant and monitored the protein levels for up to 2 h. As shown in Fig. 6A, there is no obvious difference between the stability of the wild type and that of Ste4K340R. Similarly, blocking ubiquitination of Ste4 by switching off the expression of RSP5 did not appear to alter the stability of Ste4 (Fig. 6B). On the basis of these data, we conclude that ubiquitination does not alter the stability of Ste4 protein.
FIGURE 6.
Blocking ubiquitination does not alter the stability of Ste4. A, cells expressing either wild-type Ste4 or Ste4K340R were grown to early log phase, treated with 3 μm pheromone for 1 h, and followed by treatment with the protein synthesis inhibitor cycloheximide for the indicated times. The level of Ste4 and Ste4K340R was detected by immunoblotting (IB). B, early log TetO7-RSP5 (RSP5 under the control of doxycycline-regulatable promoter) cells were first not-treated (-Dox) or treated with doxycycline (+Dox) for 9 h (to switch off RSP5 expression in the TetO7-RSP5 cells). Cells were then treated with 3 μm pheromone for 1 h, followed by treatment with the protein synthesis inhibitor cycloheximide for the indicated times. The abundance of Ste4 was detected by immunoblotting (IB). Equal loading of each lane was confirmed by immunoblotting with anti-Pgk1. Note that Ste4 is dephosphorylated more prominently in the TetO7-RSP5 strain, which resulted in the appearance of two bands.
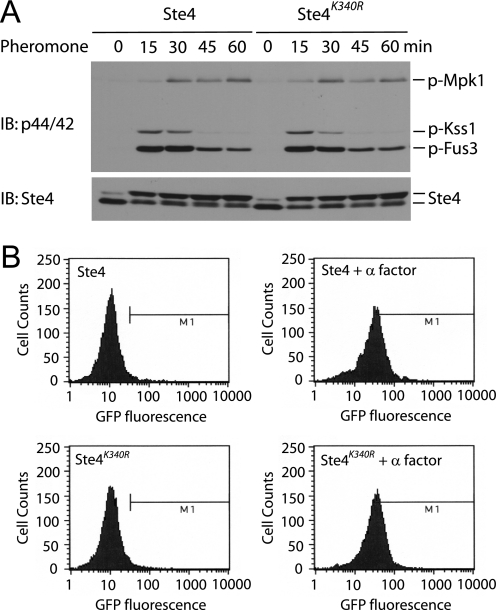
Ubiquitination, and in particular monoubiquitination, can in some cases alter protein function. Thus, we next investigated whether ubiquitination of Ste4 affects its ability to transmit the pheromone signal. Upon stimulation, Ste4 activates several effectors leading to highly coordinated mating responses, including growth arrest, new gene transcription, and morphological changes (3, 27). Ste4 activates a MAP kinase cascade by recruiting the MAP kinase scaffold (Ste5) and the associated kinase complex (Ste11/Ste7/Fus3) to the plasma membrane and facilitating its activation by Ste20 (3, 47). To determine whether ubiquitination of Ste4 plays any role in regulating the magnitude and/or duration of MAPK activation, we compared the activation status of Fus3 in wild-type cells versus the Ste4K340R mutant treated with the mating pheromone α factor over time. Fus3 activity was monitored by immunoblotting with an antibody directed against the dually phosphorylated form of the protein (Thr-202/Tyr-204) (p44/42) (48, 49). As shown in Fig. 7A, no obvious difference in either the magnitude or duration of Fus3 activation was detected between wild-type and the Ste4K340R mutant. Consistent with this result, the Ste4K340R mutant displayed the same level of pheromone-induced activation of gene transcription as measured by the FUS1-GFP reporter (50, 51) (Fig. 7B). This analysis indicates that Ste4 ubiquitination plays no apparent role in regulating the MAP kinase cascade.
FIGURE 7.
Ubiquitination of Ste4 does not regulate the magnitude and duration of pheromone-induced activation of MAP kinases. A, whole cell extracts were prepared from the wild type and the Ste4K340R mutant treated with 3 μm pheromone α factor for the indicated time, resolved by 10% SDS-PAGE, and probed with anti-phospho-p44/42 (top panel) or anti-Ste4 (bottom panel) antibodies. p-Mpk1, p-Kss1, and p-Fus3 denote phosphorylated and thus activated Mpk1, Kss1, and Fus3. B, pheromone-dependent transcriptional induction was measured following transformation of the above cells with a pheromone-responsive FUS1 promoter-GFP reporter. Cells were then treated with 3 μm α-factor for 90 min, and the resulting fluorescence in each cell was monitored by cell sorting. Pathway activation results in an increase in cells with >40 fluorescence units of activity (as indicated by an M1 bar on each graph).
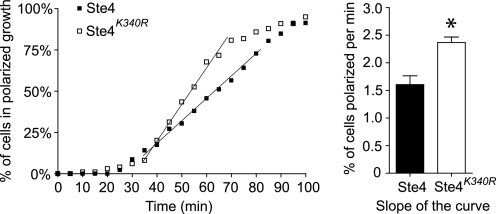
In addition to activating a MAP kinase cascade via binding Ste5 and Ste20, Ste4 also regulates polarized cell growth via interactions with Cdc24 and Far1 (3, 25). When cells are treated with pheromone, they reorient their cytoskeleton and initiate polarized growth toward the highest concentration of pheromone, leading to the formation of a shmoo morphology (26, 27). To determine whether ubiquitination of Ste4 may regulate this process, we compared the polarized cell growth of the wild type and the Ste4K340R mutant via live cell microscopy. To ensure consistent Ste4 expression from cell to cell, the Ste4K340R mutant was integrated to the genome. To minimize the potential effects of the cell cycle, the cells were synchronized to M phase via nocodazole treatment before pheromone stimulation. To maintain a consistent environment with constant replenishment of media and pheromone, washout of nocodozole and treatment with pheromone was done in a microfluidic chamber (39). As shown in Fig. 8, cells start to undergo polarized growth after 30 min of pheromone treatment. By 80 min of treatment, the majority of cells display polarized growth. However, the kinetics of polarized growth displayed by the wild type and Ste4K340R mutant are very different. On average, the population of Ste4K340R mutants polarizes significantly faster than the wild type (Fig. 8). Such a behavior of the Ste4K340R mutant indicates that ubiquitination regulates the process of pheromone-triggered polarized growth.
FIGURE 8.
Ubiquitination of Ste4 limits pheromone-induced polarized growth. Cells expressing either wild-type Ste4 or Ste4K340R were grown to A600 0.2 and treated with dimethyl sulfoxide (1%, final concentration) for 1.5 h before the addition of nocodazole (15 μg/ml in 1% dimethyl sulfoxide, final concentrations) for 2.5–3 h. Cultures were visualized by microscopy to confirm G2/M arrest. The cells were then loaded into a microfluidics device, and the nocodazole was washed out for 20 min prior to stimulation with 150 nm α factor. Cells were imaged at 5 min intervals for 2 h. For each cell, the time at which the first sign of polarized growth detectable in the differential interference contrast image was recorded and plotted as a function of time. The slope was taken from the linear portion of each curve, and the average slopes from three independent experiments are shown in the bar graph (right panel). The difference between Ste4 and Ste4K340R was statistically analyzed by t test. *, p < 0.05). Error bars, mean ± S.E.
DISCUSSION
Heterotrimeric G proteins are molecular switches that control many important biological processes. Thus, a clear understanding of the mechanisms that regulate G proteins is essential for revealing new disease mechanisms and for designing novel approaches for pharmaceutical intervention. In this report, we showed that the Gβ protein undergoes ubiquitination in response to pheromone stimulation. To our knowledge, this is the first demonstration of stimulus-dependent ubiquitination of a G protein subunit in any system.
Our findings show that Rsp5 is responsible for Ste4 ubiquitination. First, either turning off Rsp5 expression (the TET-RSP5 strain in the presence of doxycycline) or disrupting Rsp5 function (the temperature-sensitive rsp5–1 mutant) completely abolishes Ste4 ubiquitination in vivo. Second, Ste4 physically interacts with Rsp5 in yeast. Third, Rsp5 is capable of ubiquitinating Ste4 in vitro. Notably, earlier studies showed that Rsp5 is also responsible for the ubiquitination of Ste2 (receptor) (44) and the monoubiquitination (but not the polyubiquitination) of Gpa1 (Gα) (43). Thus, Ste4 joins a growing list of signaling proteins that serve as substrates of Rsp5. It would be interesting to examine whether there is any coordination among these ubiquitination events. Given the easily detectable interaction between Rsp5 and Ste4, an attractive model is that Ste4 may serve as an adaptor protein (in addition to being a substrate) for Rsp5 to facilitate its ubiquitination of other membrane-associated proteins. However, monoubiquitination of Gpa1 is apparently not dependent on Rsp5/Ste4 interaction because similar level of mono-ubiquitinated Gpa1 is present in the wild type versus a ste4Δ mutant3.
Although polyubiquitination is commonly regarded as a degradation signal, monoubiquitination appears to have a distinct role in protein regulation (13, 52). In accord with this view, monoubiquitination does not appear to modulate Ste4 stability. Blocking ubiquitination of Ste4, either via mutating its major ubiquitination site (Lys-340) or turning off the expression of its ligase Rsp5, has no obvious effect on either the abundance or the stability of Ste4. In addition, the abundance of Ste4 is not discernibly decreased in response to pheromone treatment, a condition that markedly induces Ste4 ubiquitination (Fig. 1). This is in marked contrast with both Ste2 and Gpa1. In both cases, ubiquitination by Rsp5 serves to target the proteins for internalization and eventual degradation in the vacuole (43, 44).
Thus, although Ste4 is a critical determinant of the intensity and duration of downstream MAP kinase signaling (30, 40), ubiquitination does not appear to alter the abundance of Ste4 and thereby the intensity of downstream MAP kinase signaling. Rather, ubiquitination of Ste4 appears to have a very specific role in regulating the timing of polarized cell growth in response to pheromone stimulation. Compared with the wild type, the point mutation (Ste4K340R) that blocks ubiquitination allows cells to polarize significantly faster. On average, the mutant cells initiate polarized growth 8–10 min earlier than the wild type upon stimulation by pheromone. Further investigation will be needed to determine how ubiquitination of Ste4 selectively regulates one effector (cell polarity machinery) but not the other (i.e. a MAP kinase cascade).
Growing evidence indicates that ubiquitination, especially monoubiquitination, can modulate protein-protein interactions, which consequently can lead to a very specific outcome (14). A prominent example is K-Ras, a small GTPase that controls cell differentiation and growth. It has been shown that a very small fraction of K-Ras is modified by mono- or diubiquitin (53). Interestingly, ubiquitinated K-Ras has substantially increased affinity toward some but not all of its downstream effectors (53). Consequently, blocking ubiquitination of K-Ras impacts some but not all of the cellular responses controlled by the protein. In the case of Ste4, it is entirely possible that ubiquitination may serve to regulate its interaction with downstream effectors such as Far1, Cdc24, Ste5, and Ste20. However, it should be pointed out that the difference of binding could be subtle, dynamic, and limited to certain subcellular locations. Therefore, typical coimmunoprecipitation may fail to capture those potential differences.
How is ubiquitination of Ste4 itself regulated? Pheromone stimulation induces both phosphorylation and ubiquitination of Ste4. In agreement with a model that phosphorylation targets Ste4 for ubiquitination, a mutant (Ste4T320A/S335A) that significantly diminishes phosphorylation of Ste4 also substantially abolishes ubiquitination. It is interesting to note that the major ubiquitination site Lys-340 is adjacent to the putative phosphorylation sites (Thr-320 and Ser-335), raising a possibility that phosphorylation may lead to a conformational change that helps to expose the lysine residue to receive an activated ubiquitin from Rsp5. However, in consideration of this model, it is important to keep in mind that Thr-320 and Ser-335 were implicated as possible phosphorylation sites on the basis of their requirement for the phosphorylation-induced mobility shift (35). Thus, it remains possible that the Ste4T320A/S335A mutant may reduce phosphorylation through an indirect mechanism.
In summary, our data clearly demonstrate that the Gβ subunit Ste4 undergoes pheromone-dependent ubiquitination and that Rsp5 is responsible for this modification. Ubiquitination of Ste4 occurs primarily on a single lysine residue. Significantly, ubiquitination of Ste4 selectively regulates some (e.g. polarized cell growth) but not other aspects of pheromone responses. Together, these findings reveal a new mechanism for regulating signal transmission mediated by G proteins.
Supplementary Material
Acknowledgments
We thank Drs. David E. Stone, Michael P. Yaffe, and Jeremy Thorner for generously providing strains, plasmids, and antibodies, and Laura M. Hall for preliminary analysis of cell polarization.
This work was supported, in whole or in part, by National Institutes of Health Grants R15GM093313-01 (to Y. W.), K99 GM094533-01 (to M. T.), and R01 GM073180 (to H. G. D.). This work was also supported by American Heart Association Scientist Development Grant 0635271N (to Y. W.) and a research grant from Sigma Xi (to M. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
M. Zhu and Y. Wang, unpublished observation.
- His-8-Ubi
- His-8-tagged ubiquitin.
REFERENCES
- 1. Sprang S. R. (1997) Annu. Rev. Biochem. 66, 639–678 [DOI] [PubMed] [Google Scholar]
- 2. Cabrera-Vera T. M., Vanhauwe J., Thomas T. O., Medkova M., Preininger A., Mazzoni M. R., Hamm H. E. (2003) Endocr. Rev. 24, 765–781 [DOI] [PubMed] [Google Scholar]
- 3. Dohlman H. G., Thorner J. W. (2001) Annu. Rev. Biochem. 70, 703–754 [DOI] [PubMed] [Google Scholar]
- 4. Pierce K. L., Premont R. T., Lefkowitz R. J. (2002) Nat. Rev. Mol. Cell Biol. 3, 639–650 [DOI] [PubMed] [Google Scholar]
- 5. Dohlman H. G., Thorner J. (1997) J. Biol. Chem. 272, 3871–3874 [DOI] [PubMed] [Google Scholar]
- 6. Johnson G. L., Lapadat R. (2002) Science 298, 1911–1912 [DOI] [PubMed] [Google Scholar]
- 7. Martín H., Flández M., Nombela C., Molina M. (2005) Mol. Microbiol. 58, 6–16 [DOI] [PubMed] [Google Scholar]
- 8. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 9. Pickart C. M., Eddins M. J. (2004) Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 10. Ciechanover A., Finley D., Varshavsky A. (1984) J. Cell. Biochem. 24, 27–53 [DOI] [PubMed] [Google Scholar]
- 11. Ciechanover A., Ben-Saadon R. (2004) Trends Cell Biol. 14, 103–106 [DOI] [PubMed] [Google Scholar]
- 12. Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 13. Hicke L. (2001) Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 14. Chen Z. J., Sun L. J. (2009) Mol. Cell 33, 275–286 [DOI] [PubMed] [Google Scholar]
- 15. Sigismund S., Polo S., Di Fiore P. P. (2004) Curr. Top Microbiol. Immunol. 286, 149–185 [DOI] [PubMed] [Google Scholar]
- 16. Schnell J. D., Hicke L. (2003) J. Biol. Chem. 278, 35857–35860 [DOI] [PubMed] [Google Scholar]
- 17. Wang Y., Dohlman H. G. (2006) Circ. Res. 99, 1305–1314 [DOI] [PubMed] [Google Scholar]
- 18. Shenoy S. K. (2007) Circ. Res. 100, 1142–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Naviglio S., Pagano M., Romano M., Sorrentino A., Fusco A., Illiano F., Chiosi E., Spina A., Illiano G. (2004) Cell. Signal. 16, 1229–1237 [DOI] [PubMed] [Google Scholar]
- 20. Hamilton M. H., Cook L. A., McRackan T. R., Schey K. L., Hildebrandt J. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5081–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Obin M., Lee B. Y., Meinke G., Bohm A., Lee R. H., Gaudet R., Hopp J. A., Arshavsky V. Y., Willardson B. M., Taylor A. (2002) J. Biol. Chem. 277, 44566–44575 [DOI] [PubMed] [Google Scholar]
- 22. Dohlman H. G., Thorner J., Caron M. G., Lefkowitz R. J. (1991) Annu. Rev. Biochem. 60, 653–688 [DOI] [PubMed] [Google Scholar]
- 23. Wang Y., Dohlman H. G. (2004) Science 306, 1508–1509 [DOI] [PubMed] [Google Scholar]
- 24. Bardwell L. (2005) Peptides 26, 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bar E. E., Ellicott A. T., Stone D. E. (2003) J. Biol. Chem. 278, 21798–21804 [DOI] [PubMed] [Google Scholar]
- 26. Madden K., Snyder M. (1998) Annu. Rev. Microbiol. 52, 687–744 [DOI] [PubMed] [Google Scholar]
- 27. Dohlman H. G. (2002) Annu. Rev. Physiol. 64, 129–152 [DOI] [PubMed] [Google Scholar]
- 28. Suchkov D. V., DeFlorio R., Draper E., Ismael A., Sukumar M., Arkowitz R., Stone D. E. (2010) Mol. Biol. Cell 21, 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 30. Hao N., Yildirim N., Wang Y., Elston T. C., Dohlman H. G. (2003) J. Biol. Chem. 278, 46506–46515 [DOI] [PubMed] [Google Scholar]
- 31. Fisk H. A., Yaffe M. P. (1999) J. Cell Biol. 145, 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mnaimneh S., Davierwala A. P., Haynes J., Moffat J., Peng W. T., Zhang W., Yang X., Pootoolal J., Chua G., Lopez A., Trochesset M., Morse D., Krogan N. J., Hiley S. L., Li Z., Morris Q., Grigull J., Mitsakakis N., Roberts C. J., Greenblatt J. F., Boone C., Kaiser C. A., Andrews B. J., Hughes T. R. (2004) Cell 118, 31–44 [DOI] [PubMed] [Google Scholar]
- 33. Esch R. K., Wang Y., Errede B. (2006) Eukaryotic Cell 5, 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slessareva J. E., Routt S. M., Temple B., Bankaitis V. A., Dohlman H. G. (2006) Cell 126, 191–203 [DOI] [PubMed] [Google Scholar]
- 35. Li E., Cismowski M. J., Stone D. E. (1998) Mol. Gen. Genet. 258, 608–618 [DOI] [PubMed] [Google Scholar]
- 36. Siekhaus D. E., Drubin D. G. (2003) Nat. Cell Biol. 5, 231–235 [DOI] [PubMed] [Google Scholar]
- 37. Poritz M. A., Malmstrom S., Kim M. K., Rossmeissl P. J., Kamb A. (2001) Yeast 18, 1331–1338 [DOI] [PubMed] [Google Scholar]
- 38. Muratani M., Kung C., Shokat K. M., Tansey W. P. (2005) Cell 120, 887–899 [DOI] [PubMed] [Google Scholar]
- 39. Hao N., Nayak S., Behar M., Shanks R. H., Nagiec M. J., Errede B., Hasty J., Elston T. C., Dohlman H. G. (2008) Mol. Cell 30, 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Whiteway M., Hougan L., Thomas D. Y. (1990) Mol. Cell Biol. 10, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cole G. M., Reed S. I. (1991) Cell 64, 703–716 [DOI] [PubMed] [Google Scholar]
- 42. Laine A., Ronai Z. (2005) Sci. STKE 2005, re5. [DOI] [PubMed] [Google Scholar]
- 43. Torres M. P., Lee M. J., Ding F., Purbeck C., Kuhlman B., Dokholyan N. V., Dohlman H. G. (2009) J. Biol. Chem. 284, 8940–8950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dunn R., Hicke L. (2001) J. Biol. Chem. 276, 25974–25981 [DOI] [PubMed] [Google Scholar]
- 45. Shcherbik N., Kee Y., Lyon N., Huibregtse J. M., Haines D. S. (2004) J. Biol. Chem. 279, 53892–53898 [DOI] [PubMed] [Google Scholar]
- 46. Radivojac P., Vacic V., Haynes C., Cocklin R. R., Mohan A., Heyen J. W., Goebl M. G., Iakoucheva L. M. (2010) Proteins 78, 365–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen R. E., Thorner J. (2007) Biochim. Biophys. Acta 1773, 1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sabbagh W., Jr., Flatauer L. J., Bardwell A. J., Bardwell L. (2001) Mol. Cell 8, 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Y., Abu Irqeba A., Ayalew M., Suntay K. (2009) PLoS ONE 4, e7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoffman G. A., Garrison T. R., Dohlman H. G. (2002) Methods Enzymol. 344, 617–631 [DOI] [PubMed] [Google Scholar]
- 51. Lan K. L., Sarvazyan N. A., Taussig R., Mackenzie R. G., DiBello P. R., Dohlman H. G., Neubig R. R. (1998) J. Biol. Chem. 273, 12794–12797 [DOI] [PubMed] [Google Scholar]
- 52. Sun L., Chen Z. J. (2004) Curr. Opin. Cell Biol. 16, 119–126 [DOI] [PubMed] [Google Scholar]
- 53. Sasaki A. T., Carracedo A., Locasale J. W., Anastasiou D., Takeuchi K., Kahoud E. R., Haviv S., Asara J. M., Pandolfi P. P., Cantley L. C. (2011) Sci. Signal 4, ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.