Abstract
Controlling cell fate-determining gene expression is key to stem cell differentiation, tissue regeneration, and cancer therapy. To date, custom-built transcription factors recognize the information encoded in specific DNA sequences. Chromatin proteins undergo covalent modifications and form complexes that encode a second layer of information that determines proximal gene activity. Here, we employ a novel gene-targeting approach that exploits a specific chromatin modification to reactivate silenced loci in human cells. We used the human Polycomb chromatin protein and homologues from other species to construct modular synthetic transcription factors, called Pc-TFs, that recognize the repressive trimethyl-histone H3 lysine 27 (H3K27me3) signal and switch silenced genes to an active state. Pc-TF expression in U2OS osteosarcoma cells leads to increased transcription of the senescence locus CDKN2A (p16) and other loci in a chromodomain- and activation module-dependent manner, a switch to a senescence phenotype, and reduced cell proliferation. These results indicate that silenced developmental regulators can be reactivated by a synthetic transcription factor that interacts with chromatin rather than DNA, resulting in an altered cell state. As such, our work extends the flexibility of transcription factor engineering and is the first example of chromatin-mediated synthetic transcription factor targeting.
Keywords: Cell Differentiation, Chromatin, Gene Regulation, Histone Methylation, Transcription Regulation, Polycomb, Synthetic Transcription Factors
Introduction
Chromatin plays a pivotal role in regulating gene expression by establishing heritable cell states. Unlike transcription factors that bind specific DNA sequences, many chromatin-associated transcription regulators bind to the histone proteins that package DNA into nucleosomes. Post-translational modification of the residues along the unfolded N-terminal histone tails is a dynamic mechanism that allows a single genetic locus to assume different transcriptional states. Enzymes catalyze the covalent attachment and removal of methyl, acetyl, and other molecular groups. Effector proteins specifically recognize these marks and establish gene silencing or activation by altering the accessibility of the locus to RNA polymerase and other transcription factors (1, 2). One of the most well studied chromatin modification systems is Polycomb, an evolutionarily conserved set of proteins that was first characterized in Drosophila (3). Tumor suppressors and cell differentiation genes are subject to Polycomb-mediated regulation in mammals (4, 5).
The Polycomb silencing mechanism involves two core complexes that generate and interact with histone methylation. Polycomb repressive complex 2 methylates histone H3 lysine 27 (Fig. 1A). The trimethylated lysine (H3K27me3)2 is bound by the Polycomb chromodomain of CBX, a member of Polycomb repressive complex 1 (Fig. 1B) (3, 6), which may create condensed chromatin that impedes RNA polymerase activity (6). Transformations of tissue identity in developing animals upon misexpression of Polycomb proteins (7–9) underscore the importance of this gene regulation system in cell fate determination.
FIGURE 1.
Using the Polycomb system to engineer a chromatin-targeted transcription factor. A, model of H3K27 methylation by EZH1/2 of Polycomb repressive complex 2 (PRC2) (3, 6). B, gene repression by Polycomb repressive complex 1 (PRC1) (3, 6). Trimethylation of H3K27 acts as a docking site for the PRC1 complex. C, gene activation by Pc-TF. A conserved 62 amino acid Polycomb chromodomain from CBX8 of the PRC1 complex, fused to a transcription activation domain, binds H3K27me3. As a result, the silencing mark is translated into gene activation. A, EED, embryonic ectoderm development; EZH, Enhancer of zeste homologue; SU(Z)12, Suppressor of zeste 12; RBAP, retinoblastoma binding protein; BMI1, Polycomb ring finger oncogene; PCGF and MEL18, Polycomb group ring finger proteins; PHC, polyhomeotic homolog; RING1, Ring finger protein 1.
Our knowledge of the role of chromatin in development and disease has spurred progress in artificial manipulation of epigenetic states. Several studies have revealed Polycomb factor misregulation and abnormal accumulation of silenced chromatin in cancer cells (10–12), prompting the development of anti-cancer drugs that globally inhibit repressive chromatin modifications (13). To advance the technology for manipulating transcriptional states, we have developed a modular Polycomb-based artificial transcription factor (Pc-TF), consisting of the methyl-histone-binding Polycomb chromodomain and the VP64 activator (14), that targets and activates H3K27me3-associated Polycomb-repressed loci (Fig. 1C). Unlike small molecule drugs, synthetic transcription factors are modular, a characteristic that allows us to potentially tune Pc-TF specificity.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293 and U2OS Flp-in T-REx cells were grown in Dulbecco's modified Eagle's medium (DMEM) or McCoy's 5A medium (respectively) supplemented with 10% tetracycline-free fetal bovine serum and 1% penicillin and streptomycin. Cells were grown at 37 °C in a humidified CO2 incubator. Transient transfections of HEK293 cells were carried out in 24-well culture dishes using 1.8 μl of FuGENE 6 (Roche Applied Science) in 1 ml of medium. U2OS stable cell lines were generated by transfecting 5 × 105 cells with 2 μg of plasmid DNA plus 4 μl of Lipofectamine 2000 (Invitrogen) in antibiotic-free medium for 6 h. 2 days after transfection wash out, cells were grown under 1.0 μg/ml puromycin selection and screened for inducible mCherry (mCh) expression upon treatment with 1 μg/ml doxycycline (dox), and 2–3 clones were expanded and maintained as stable lines.
Plasmid Constructs
DNA fragments with universal cloning sites (EcoRI, NotI, XbaI, SpeI, and PstI) were generated by PCR and assembled according to a modified BioBrick DNA assembly method. 3Human codon optimized human, fly, and zebrafish Polycomb chromodomains were synthesized by GenScript. The Gal4 transcription activator and deletion variants were cloned as EcoRI/XbaI fragments into pcDNA3.1(+) (Invitrogen) for constitutive expression. Pc-TFs and deletion variants were cloned into pcDNA3.1(+) (Invitrogen) or ligated downstream of a CMV-TetO2 fragment (from pcDNA5/FRT/TO, Invitrogen) and cloned as EcoRI/PstI fragments into a pcDNA3.1(+)-based vector in which neomycin was replaced with puromycin resistance and the constitutive CMV promoter was deleted via SpeI digestion.
Luciferase Activity Assays
HEK293 23;4;9 cells (16) were treated with 1 μg/ml dox expression for 2, 4, 6, or 8 days to induce Gal4-EED expression. 4-day dox-treated or untreated cells were plated at ∼1 × 105 cells/well in 24-well culture dishes, transfected with 200 ng of plasmid DNA, grown without dox for 48 h, and harvested for luciferase assays (Bright-Glo kit, Promega). A Scepter digital counter (Millipore) was used for cell counting.
Microscopy
Transiently transfected HEK293 cells were imaged in 24-well plates at 200× using a Nikon fluorescence microscope (Model TE2000-E). U2OS cell lines were seeded at ∼5 × 104 cells/well on glass coverslips in 24-well culture dishes, treated with 1 μg/ml dox overnight, fixed with 4% paraformaldehyde/1× PBS for 10 min, and permeabilized with 0.5% Triton X-100/1× PBS for 10 min. Slips were inverted into vinol mounting medium (P8136 polyvinyl alcohol, Sigma; 33% glycerol; 0.1% sodium azide in 1× PBS) on glass slides and imaged at 600×. Microscopy of live colonies was carried out on cells plated at low density (∼600 cells/cm2) in 6-well plates and grown in the presence or absence of 1 μg/ml dox for 5 days.
Antibodies for Western Blot Analysis
Primary staining with rabbit anti-DsRed 632946 (Clontech) was followed by secondary staining with donkey anti-rabbit-HRP (Promega) and mouse anti-GAPDH-HRP conjugate G9295 (Sigma-Aldrich).
Reverse Transcription PCR
SuperScript III (Invitrogen) was used to generate cDNA from 2 μg of total RNA isolated with either TRIzol (Invitrogen) or the RNeasy kit (Qiagen). See supplemental Table S1 for primers. Real-time quantitative PCR reactions (15 μl each) contained SYBR Green master mix (Applied Biosystems), 2.25 pmol of primers, and 2 μl of a 1:10 cDNA dilution (1:1000 dilution for GAPDH and mCh). “Relative mRNA level” was calculated as 2[Ct GAPDH − Ct experimental gene].
Cross-linked Chromatin Immunoprecipitation
∼1 × 108 cells were treated with 1 μg/ml dox for 5 days, collected by scraping, and incubated with 25 ml of 1% formaldehyde/1× Dulbecco's PBS for 10 min at room temperature. Cross-linking was quenched with 25 mm glycine for 5 min. Cross-linked cells were washed once with 1× PBS and twice in 175 μg/ml PMSF/1× PBS.
Cell lysis and chromatin preps were carried out in buffers supplemented with 175 μg/ml PMSF and 12.5 mg/ml PLAAC protease inhibitor mixture. Cells were incubated for 10 min at 4 °C in 10 ml of lysis buffer (62.5 mm HEPES-KOH, pH 7.6; 146 mm NaCl; 1 mm EDTA; 10.4% glycerol; 0.5% Nonidet P-40; 0.26% Triton X-100), washed once in 8 ml of wash buffer (200 mm NaCl; 1 mm EDTA; 0.5 mm EGTA; 20 mm Tris, pH 8.0), and disrupted in 6 ml of sonication buffer (10 mm EDTA; 5 mm EGTA; 204 mm Tris, pH 8.0) using a Fisher Scientific Model 550 Sonic Dismembrator. Soluble chromatin was incubated with 5 mg/ml N-laurylsarcosine for 10 min and spun at ∼10,000 × g to remove impurities. DNA was purified from a chromatin sample by phenol/chloroform extraction and resolved via electrophoresis to confirm ∼500-bp fragments.
For immunoprecipitations, each 6-ml sample was supplemented with 1.2% Triton X-100, 0.12% sodium deoxycholate, 0.8× TE, pH 7.5, PLAAC, and PMSF. U2OS Flp-in T-REx chromatin was incubated with rabbit anti-H3K27me3 07-449 (Millipore) or rabbit IgG (sc-2027, Santa Cruz Biotechnology.) overnight and protein A-Sepharose beads for 3 h at 4 °C. Chromatin from Pc-TF- or TF-expressing cells was precipitated with mouse anti-Myc 3400 or mouse IgG 3420 bead conjugates (Cell Signaling Technology). Chromatin-antibody-bead complexes were washed essentially as described by Millipore (ChIP assay MCPROTO407). DNA-protein complexes were eluted in 0.5 ml of 1% SDS/TE. 100 μl of chromatin (input) was brought up to 500 μl with 1% SDS/TE. Samples were treated with Pronase for 1 h at 42 °C and then incubated at 65 °C for 48 h to reverse cross-linking. DNA was extracted twice with phenol:chloroform:isoamyl alcohol (25:24:1), once with chloroform, supplemented with 15 μg of GlycoBlue (Ambion) and 200 mm NaCl, and EtOH-precipitated. DNA was resuspended in 0.5 ml of 10 mm Tris, pH 8.0.
ChIP Real-time Quantitative PCR
Real-time quantitative PCR reactions (15 μl each) contained SYBR Green master mix, 2 μl of immunoprecipitated (IP), mock-IP, or input template DNA, and 2.25 pmol of primers. % IP DNA bound was calculated as 100 × 2[Ct input − Ct IP]. % mock-IP bound (100 × 2[Ct input − Ct mock-IP] was subtracted from % IP DNA bound to calculate “% IP DNA enrichment relative to mock-IP.”
Senescence-associated β-Galactosidase Assays
Optimization of the senescence associated β-galactosidase (β-gal) detection assay for U2OS cells was carried out as follows. Cells were plated in 6-well dishes (∼5 × 105 cells/well), stressed with 0.1–0.4 μg/ml rotenone or DMSO for 96 h, stained with 0.015–15 μg/ml C12FDG at 37 °C for 1 h, and then harvested and resuspended in 0.5 ml of 1× Dulbecco's PBS for flow cytometry. Pc-TF cells were induced with 1 μg/ml dox for 96 h, and U2OS Flp-in T-REx cells were treated with 0.2 μg/ml rotenone, DMSO, or dox for 96 h, stained with 1.5 μg/ml C12FDG, and collected for flow cytometry.
RESULTS
Pc-TF Reactivates a Polycomb-repressed Reporter Gene
To determine whether Pc-TF recognizes methylated histones and reactivates a silenced locus, we used an inducible Polycomb silencing reporter system in HEK293 cells (16). Polycomb chromatin formation at the GAL4TK-Luc transgene (luc) in HEK293 cells is mediated by Gal4-EED (silencer). dox treatment activates expression of the silencer, leading to luc repression and accumulation of H3K27me3 and Polycomb proteins at the promoter (16). Expression of a synthetic activator that consists of a Gal4 DNA-binding domain (Gal4DB), red fluorescent mCherry tag, VP64, and an SV40 nuclear localization signal leads to an ∼10-fold increase in luc expression over basal expression levels (supplemental Fig. S1A). Conversely, 4–8 days of dox-induced silencer expression lead to an ∼7-fold decrease of luc activity (supplemental Fig. S1B). Following luc silencing for 4 days, recruitment of VP64 directly upstream of the promoter leads to luc reactivation at levels ∼10-fold greater than initial basal expression levels (supplemental Fig. S1C). Deletion of the Gal4DB or the VP64 domain abolished luc reactivation, indicating that VP64 stimulates transcription when it is recruited near the promoter and that the Gal4DB alone is not sufficient to reactivate luc.
The synthetic transcription activator was reengineered to recognize H3K27me3 by replacing the Gal4DB with a Polycomb chromodomain (PCD). hPCD-TF includes amino acids 1–62 from human CBX8, a peptide that preferentially binds H3K27me3 in vitro (17). We also built Pc-TFs using the chromodomains from a distant vertebrate (zebrafish) and flies (Drosophila) to investigate whether variations in amino acids around the conserved hydrophobic residues (Fig. 2A) might alter Pc-TF activity. fshPCD-TF includes amino acids 2–63 of zebrafish Pc1, a CBX homologue that acts during embryonic development (18). flyPCD-TF includes amino acids 16–77 of Drosophila Polycomb (Pc), a peptide that shows preferential H3K27me3 binding (19, 20) and confers Polycomb-like genomic localization when the peptide is tethered to a heterologous protein (19, 21).
FIGURE 2.
Pc-TF-mediated activation of a Polycomb-repressed reporter gene in HEK293 cells. A, fusion protein gene constructs. A ClustalW alignments of the Polycomb chromodomains from human CBX8 (hPCD), zebrafish CBX2 (fshPCD), and Drosophila Pc (flyPCD) is shown. Boxes mark conserved aromatic residues within the H3K27me3-binding pocket (19, 20). Pc-TF fusion proteins (hPCD-TF, fshPCD-TF, and flyPCD-TF) contain a red fluorescent mCh protein, VP64 activation domain, SV40 nuclear localization signal (NLS), and Myc epitope tag. VP64 deletions (hPCD-RFP, fshPCD-RFP, and flyPCD-RFP) and a PCD deletion (TF) serve as negative controls for Pc-TF activity assays. B, nuclear localization of fusion proteins in transiently transfected HEK293 cells. Dashed lines mark the periphery of representative mCherry-positive cells. Scale bar = 20 μm. C, hPCD-TF-mediated GAL4TK-Luc (luciferase) reporter gene activation requires prior exposure to Polycomb silencing. The schematic illustration (left) shows the predicted behavior of the synthetic Pc-TF in the absence or presence of histone methylation, controlled by doxycycline-induced expression of Gal4-EED. Pc-TF fusion proteins and deletion controls were expressed in cells before and after dox treatment. Error bars represent S.D., n = 3 measurements. A one-tailed t test was used to compare the values under the horizontal bracket (*, p < 0.01). TK, thymidine kinase minimal promoter; EED, embryonic ectoderm development.
Human Pc-TF-mediated reactivation of the Polycomb-silenced luc reporter relies upon a previously established silenced state. Mitotically stable luc silencing (16) was generated by transient expression of the repressor (4 days of dox treatment), followed by dox wash-out and growth in dox-free medium for 10 days. Cells expressing the human Pc-TF show a modest significant increase in luc expression as compared with the unsilenced reporter (Fig. 2C). Reactivation by human Pc-TF requires previous Gal4-EED expression and the PCD module, suggesting that Pc-TF is recruited to the reporter gene through a PCD-H3K27me3 interaction.
Pc-TFs Activate Transcription at Methyl-histone-associated Loci
We identified Pc-TF targets by looking for genes that were up-regulated upon Pc-TF expression. Transgene constructs were placed under the control of a dox-inducible CMV promoter (supplemental Fig. S2A) and integrated into a U2OS cell line that expresses a tetracycline repressor (Flp-in T-REx, referred to herein as the “parent line”). Quantitative reverse transcription PCR (qRT-PCR) (supplemental Fig. S2B) and Western blots (supplemental Fig. S2C) confirmed dox-inducible expression. The mCh tag showed clear nuclear localization in all cell lines (Fig. 3A). To identify Pc-TF target genes, total RNA from cells grown with or without dox for 96 h was used for NanoString quantitative RNA analysis of 500 development- and differentiation-associated genes (22). We tested 14 positive hits by qRT-PCR of duplicate RNA samples (supplemental Fig. S3). CDKN2A (p16) senescence-associated gene, matrix metallopeptidase 12 (MMP12), natriuretic peptide (NPPA), and tumor necrosis factor (TNF) were up-regulated by one or more of the Pc-TF cell lines in a VP64- and PCD-dependent manner.
FIGURE 3.
Endogenous genes targeted by synthetic Pc-TFs in U2OS cells. A, expression of Pc-TFs and deletion variants in U2OS cells. Dotted lines mark the periphery of representative mCherry-positive cells. Scale bar = 20 μm. B, reverse transcription PCR of cDNA synthesized from total RNA of untreated or dox-treated cells. Relative expression levels are quantitative PCR values normalized to the untreated parental line (P.L., −dox). Error bars represent S.E., n = 2 RNA samples. C, ChIP analysis of Pc-TF target loci. IP DNA was amplified using primers 1–16 (supplemental Table S1). Probes n1 and n2, located at the constitutively active GAPDH promoter, are negative controls. Mean enrichments of IP DNA are shown as percentages of input minus background (nonspecific mock-IP DNA). Error bars represent S.D., n = 3 quantitative PCR reactions. A one-tailed t test was used to identify Pc-TF enrichments that were significantly higher (*, p < 0.01) than a randomly cross-linked protein that has no chromatin-binding domain (TF).
Cross-linked chromatin immunoprecipitation (ChIP) assays showed enrichment of histone methylation near the promoters of CDKN2A, MMP12, and NPPA (Fig. 3C, supplemental Fig. S4), but not at TNF (data not shown), suggesting that the first three genes are direct targets of Pc-TF. hPCD-TF was significantly enriched (p < 0.01) near the p16 promoter (probe 3) as compared with a non-targeted protein in which the PCD was deleted. All three Pc-TF homologues showed significant enrichment near the MMP12 promoter (probe 6). Thus, the ChIP data are consistent with the qRT-PCR data for p16 and MMP12, indicating that these genes are direct targets of the Pc-TFs. We did not detect enrichment of Pc-TFs at promoters lacking H3K27me3 (TNF, ATOH1, ACTC1, and CASP10, data not shown), indicating that H3K27me3 is required for Pc-TF binding at endogenous loci. Interestingly, the H3K27me3-enriched promoters of NPPA and eomesodermin (EOMES), an additional gene included in our analysis (Fig. 3C, supplemental Fig. S4), show no Pc-TF enrichment, demonstrating that Pc-TF may preferentially bind a subset of H3K27me3 sites.
Human Pc-TF Expression Is Accompanied by a Switch to Cellular Senescence
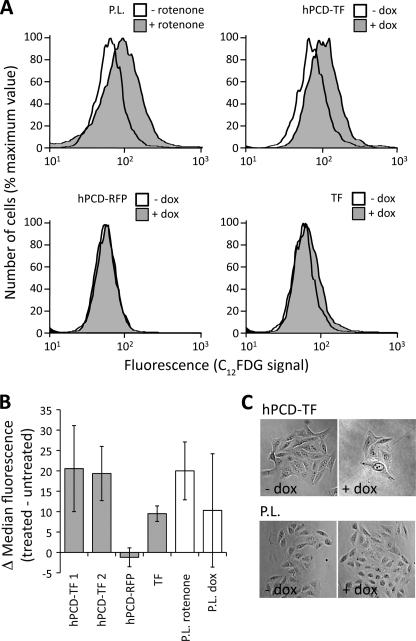
Consistent with p16 activation, hPCD-TF-expressing cells showed senescence-associated phenotypes. Hallmarks of senescence include increased levels of β-gal activity and decreased cell proliferation (23). Treatment with C12FDG followed by flow cytometry has been used to measure senescence-associated β-gal (SA-β-gal) in rotenone-stressed or aged senescent human fibroblasts (24). We optimized the assay for U2OS cells and found that 94 h of exposure to 0.2 μg/ml rotenone followed by staining with 1.5 μg/ml C12FDG was sufficient to detect an increase in SA-β-gal activity over background levels. We observed senescence-associated SA-β-gal activity in Pc-TF-expressing cells at levels comparable with cells stressed with rotenone. We repeated these results in two independent hPCD-TF-expressing cell lines and not in cells expressing VP64 or PCD deletion proteins or in parental cells (Fig. 4, A and B).
FIGURE 4.
Senescence-associated phenotypes in Pc-TF-expressing U2OS cells. A, histograms of senescence-associated β-gal activity detected by C12FDG staining and flow cytometry (24). Rotenone treatment of the parental cell line (P.L.) was used as a positive control. B, average differences in median fluorescence. Error bars represent S.D., n = 2 independent induction trials. C, proliferation of Pc-TF-expressing cells (hPCD-TF, +dox) as compared with untreated cells and the parental line.
To measure changes in cell proliferation, we plated cells at low density and observed growth of isolated colonies after 5 days of dox treatment. Colony size is diminished in hPCD-TF-expressing cells as compared with cells that do not express hPCD-TF (Fig. 4C). Our results demonstrate that the chromatin-targeted synthetic transcription activator induces a switch in cell state, from rapid proliferation to senescence, in the U2OS cancer cell line.
DISCUSSION
Synthetic biology presents opportunities to deepen our understanding of molecular biology while creating useful new tools (25). Using the PCD to target a transcription activator to methylated histones has revealed some complexities underlying chromatin interaction. The H3K27me3-associated luc reporter (Fig. 2C) and CDKN2A are targeted by the human Pc-TF but not by the zebrafish or fly homologues. Perhaps the human PCD participates in human protein-specific interactions with other chromatin-associated proteins in addition to H3K27me3. Interestingly, MMP12 is targeted by all three Pc-TFs in a PCD-dependent manner. Thus, at least one locus has a chromatin structure that permits general PCD binding. Finally, our data indicate that not all H3K27me3-associated regions are bound by Pc-TF. hPCD-TF is enriched at the region immediately upstream of the CDKN2A promoter, but not at the other H3K27me3-enriched regions. Two H3K27me3-associated promoters (NPPA and EOMES) showed no Pc-TF enrichment, indicating that certain regions may be more accessible to Pc-TF binding than others.
So far, we have confirmed p16 as a target of Pc-TF, which has implications for cancer therapy. Polycomb-mediated suppression of the senescence-associated p16 gene may augment cancer proliferation (23, 26). Recently, drugs that broadly disrupt chromatin-mediated gene silencing in cancer cells have entered clinical trials (13) but lack specificity at the target gene level and may cause off-target effects. Pc-TFs integrate the information from epigenetic modifications with transcriptional regulation, providing a way to target therapeutic genes in a disease state-specific manner.
Manipulating Polycomb-mediated gene regulation is also relevant for regenerative medicine. Human stem cells can be differentiated in vitro by depleting Polycomb factor levels and activating repressed genes (27). Using Pc-TFs to reverse epigenetic silencing at these loci has the possible advantage of enabling reconfiguration of the modular domains to adjust the range of activated target genes. Thus far, we have shown that changing the PCD module influences target gene specificity in the case of p16 and MMP12 (Fig. 3, B and C). Changing the TATA motif-dependent VP64 module (14) to a different activator, such as SP1 or p65 (28, 29), could expand the range of Pc-TF-activated genes. Ongoing identification of chromatin recognition domains (30) suggests great potential for synthetic transcription factors that interact with a variety of chromatin modifications. Our development of synthetic Polycomb-based transcription activators establishes a novel strategy that integrates the information encoded in chromatin with targeted gene regulation.
Supplementary Material
Acknowledgments
For the generous gifts of cell lines, DNA, and other resources, we thank K. Hansen (HEK293 23;4;9 cells), S. Blacklow (U2OS Flp-in T-REx cells), and A. Meissner and C. Gifford (NanoString assay).
This work was supported, in whole or in part, by National Institutes of Health Grants GM057476 and GM36373 (to P. A. S.) and 1F32GM087860 (to K. A. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
I. Phillips and P. A. Silver, personal communication.
- H3K27me3
- histone H3 trimethylated at lysine 27
- Pc-TF
- Polycomb-based artificial transcription factor
- Gal4DB
- Gal4 DNA-binding domain
- PCD
- Polycomb chromodomain
- mCh
- mCherry
- dox
- doxycycline
- TE
- Tris-EDTA
- DMSO
- dimethyl sulfoxide
- IP
- immunoprecipitated
- qRT-PCR
- quantitative reverse transcription PCR
- SA-β-gal
- senescence-associated β-gal
- FDG
- fluorescein di-β-d-galactopyranoside
- CBX
- chromobox homolog.
REFERENCES
- 1. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 2. Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sawarkar R., Paro R. (2010) Dev. Cell 19, 651–661 [DOI] [PubMed] [Google Scholar]
- 4. Bracken A. P., Helin K. (2009) Nat. Rev. Cancer 9, 773–784 [DOI] [PubMed] [Google Scholar]
- 5. Schuettengruber B., Cavalli G. (2009) Development 136, 3531–3542 [DOI] [PubMed] [Google Scholar]
- 6. Simon J. A., Kingston R. E. (2009) Nat. Rev. Mol. Cell Biol. 10, 697–708 [DOI] [PubMed] [Google Scholar]
- 7. Alkema M. J., van der Lugt N. M., Bobeldijk R. C., Berns A., van Lohuizen M. (1995) Nature 374, 724–727 [DOI] [PubMed] [Google Scholar]
- 8. Akasaka T., Kanno M., Balling R., Mieza M. A., Taniguchi M., Koseki H. (1996) Development 122, 1513–1522 [DOI] [PubMed] [Google Scholar]
- 9. Takihara Y., Tomotsune D., Shirai M., Katoh-Fukui Y., Nishii K., Motaleb M. A., Nomura M., Tsuchiya R., Fujita Y., Shibata Y., Higashinakagawa T., Shimada K. (1997) Development 124, 3673–3682 [DOI] [PubMed] [Google Scholar]
- 10. Djabali M., Selleri L., Parry P., Bower M., Young B. D., Evans G. A. (1992) Nat. Genet. 2, 113–118 [DOI] [PubMed] [Google Scholar]
- 11. Varambally S., Cao Q., Mani R. S., Shankar S., Wang X., Ateeq B., Laxman B., Cao X., Jing X., Ramnarayanan K., Brenner J. C., Yu J., Kim J. H., Han B., Tan P., Kumar-Sinha C., Lonigro R. J., Palanisamy N., Maher C. A., Chinnaiyan A. M. (2008) Science 322, 1695–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sneeringer C. J., Scott M. P., Kuntz K. W., Knutson S. K., Pollock R. M., Richon V. M., Copeland R. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 20980–20985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gravina G. L., Festuccia C., Marampon F., Popov V. M., Pestell R. G., Zani B. M., Tombolini V. (2010) Mol. Cancer. 9, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beerli R. R., Segal D. J., Dreier B., Barbas C. F., 3rd (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14628–14633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deleted in proof.
- 16. Hansen K. H., Bracken A. P., Pasini D., Dietrich N., Gehani S. S., Monrad A., Rappsilber J., Lerdrup M., Helin K. (2008) Nat. Cell Biol. 10, 1291–1300 [DOI] [PubMed] [Google Scholar]
- 17. Bernstein E., Duncan E. M., Masui O., Gil J., Heard E., Allis C. D. (2006) Mol. Cell Biol. 26, 2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kawamura A., Yokota S., Yamada K., Inoue H., Inohaya K., Yamazaki K., Yasumasu I., Higashinakagawa T. (2002) Biochem. Biophys. Res. Commun. 294, 456–463 [DOI] [PubMed] [Google Scholar]
- 19. Fischle W., Wang Y., Jacobs S. A., Kim Y., Allis C. D., Khorasanizadeh S. (2003) Genes Dev. 17, 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Min J., Zhang Y., Xu R. M. (2003) Genes Dev. 17, 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Platero J. S., Hartnett T., Eissenberg J. C. (1995) EMBO J. 14, 3977–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geiss G. K., Bumgarner R. E., Birditt B., Dahl T., Dowidar N., Dunaway D. L., Fell H. P., Ferree S., George R. D., Grogan T., James J. J., Maysuria M., Mitton J. D., Oliveri P., Osborn J. L., Peng T., Ratcliffe A. L., Webster P. J., Davidson E. H., Hood L., Dimitrov K. (2008) Nat. Biotechnol. 26, 317–325 [DOI] [PubMed] [Google Scholar]
- 23. Kuilman T., Michaloglou C., Mooi W. J., Peeper D. S. (2010) Genes Dev. 24, 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Noppe G., Dekker P., de Koning-Treurniet C., Blom J., van Heemst D., Dirks R. W., Tanke H. J., Westendorp R. G., Maier A. B. (2009) Cytometry A 75, 910–916 [DOI] [PubMed] [Google Scholar]
- 25. Haynes K. A., Silver P. A. (2009) J. Cell Biol. 187, 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sparmann A., van Lohuizen M. (2006) Nat. Rev. Cancer 6, 846–856 [DOI] [PubMed] [Google Scholar]
- 27. Bracken A. P., Dietrich N., Pasini D., Hansen K. H., Helin K. (2006) Genes Dev. 20, 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emami K. H., Navarre W. W., Smale S. T. (1995) Mol. Cell Biol. 15, 5906–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu P. Q., Rebar E. J., Zhang L., Liu Q., Jamieson A. C., Liang Y., Qi H., Li P. X., Chen B., Mendel M. C., Zhong X., Lee Y. L., Eisenberg S. P., Spratt S. K., Case C. C., Wolffe A. P. (2001) J. Biol. Chem. 276, 11323–11334 [DOI] [PubMed] [Google Scholar]
- 30. Vermeulen M., Eberl H. C., Matarese F., Marks H., Denissov S., Butter F., Lee K. K., Olsen J. V., Hyman A. A., Stunnenberg H. G., Mann M. (2010) Cell 142, 967–980 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.