Abstract
We have shown that cytokine-like 1 (Cytl1) is a novel autocrine regulatory factor that regulates chondrogenesis of mouse mesenchymal cells (Kim, J. S., Ryoo, Z. Y., and Chun, J. S. (2007) J. Biol. Chem. 282, 29359–29367). In this previous work, we found that Cytl1 expression was very low in mesenchymal cells, increased dramatically during chondrogenesis, and decreased during hypertrophic maturation, both in vivo and in vitro. Moreover, exogenous addition or ectopic expression of Cytl1 caused chondrogenic differentiation of mouse limb bud mesenchymal cells. In the current study, we generated a Cytl1 knock-out (Cytl1−/−) mouse to investigate the in vivo role of Cytl1. Deletion of the Cytl1 gene did not affect chondrogenesis or cartilage development. Cytl1−/− mice also showed normal endochondral ossification and long bone development. Additionally, ultrastructural features of articular cartilage, such as matrix organization and chondrocyte morphology, were similar in wild-type and Cytl1−/− mice. However, Cytl1−/− mice were more sensitive to osteoarthritic (OA) cartilage destruction. Compared with wild-type littermates, Cytl1−/− mice showed more severe OA cartilage destruction upon destabilization of the medial meniscus of mouse knee joints. In addition, expression levels of Cytl1 were markedly decreased in OA cartilage of humans and experimental mice. Taken together, our results suggest that, rather than regulating cartilage and bone development, Cytl1 is required for the maintenance of cartilage homeostasis, and loss of Cytl1 function is associated with experimental OA cartilage destruction in mice.
Keywords: Bone, Extracellular Matrix, Gene Knockout, Gene Regulation, Gene Transcription, Cartilage Development, Cytokine-like 1 (CYTL1), Endochonral Ossification, Knockout Mice, Osteoarthritis
Introduction
Cartilage development is initiated by the differentiation of mesenchymal cells into chondrocytes (1–4). Differentiated chondrocytes synthesize cartilage-specific extracellular matrix (ECM)3 components, such as type II collagen and sulfated proteoglycan, to form cartilage tissue. Developed cartilage either remains as a permanent cartilage tissue, such as joint articular cartilage, or serves as a template for long bone development during endochondral ossification. Endochondral ossification requires hypertrophic maturation of chondrocytes. The sequential events that occur during chondrogenesis and cartilage and bone development are precisely regulated by various regulatory factors released from cartilage elements and the perichondrium (1–4). Although a number of regulatory factors have been identified, the precise regulatory mechanisms underlying chondrogenesis and cartilage and bone development remain to be elucidated.
In permanent cartilage tissue, cartilage homeostasis is maintained by chondrocytes, which are a cell type unique to cartilage tissue. Cartilage homeostasis is disrupted in osteoarthritis (OA), leading to eventual cartilage destruction. OA is a progressive and degenerative disorder of the joint primarily characterized by articular cartilage destruction. A variety of potential OA-causing mechanisms have been proposed (5–7). Biophysical and biochemical factors, such as mechanical stress and pro-inflammatory cytokines, respectively, are responsible for disruption of cartilage homeostasis and initiation of the catabolic pathway. This, in turn, activates intracellular pathways in chondrocytes that lead to the production of pro-inflammatory cytokines, inflammation, degradation of the ECM by matrix-degrading enzymes, and cessation of ECM synthesis via dedifferentiation and apoptosis of chondrocytes (5–7).
We have previously shown that cytokine-like 1 (Cytl1) is a novel autocrine regulatory factor involved in chondrogenesis of mouse mesenchymal cells in vitro (8). CYTL1 was originally cloned as a functionally unknown cytokine candidate from human bone marrow and cord blood mononuclear cells bearing the CD34 surface marker (9). The CYTL1 protein contains four α-helices and six conserved cysteine residues, which may form intra-disulfide bonds to yield a globular structure, a common structural characteristic of cytokines. CYTL1 also contains an N-terminal secretory signal peptide, and its secretion is associated with post-translational modifications, although the nature of these modifications remains to be established (8). The CYTL1 gene is found in vertebrates, including humans, chickens, and Fugu rubripes, but has not been detected in invertebrates, such as Drosophila or Caenorhabditis elegans. Thus, CYTL1 appears to have originated early in vertebrate evolution, suggesting a possible functions in cartilage and bone development. Indeed, CYTL1 expression has been found in cartilaginous tissues, such as mouse inner ear and human articular cartilage (10, 11). In our previous study (8), we showed that mouse Cytl1 expression was chondrocyte-specific, and further demonstrated that Cytl1 expression was very low in mesenchymal cells, dramatically increased during chondrogenesis, and decreased during hypertrophic maturation, both in vivo and in vitro. Exogenous addition of Cytl1 protein or lentivirus-mediated overexpression of Cytl1 caused chondrogenic differentiation of mouse limb bud mesenchymal cells during micromass culture. However, Cytl1 did not affect the hypertrophic maturation of chondrocytes. We have also demonstrated that mouse Cytl1 exerts its chondrogenic effect via stimulation of Sox9 transcriptional activity and induction of insulin-like growth factor (Igf) 1, which has the capacity to induce chondrogenesis (8).
The chondrocyte-specific expression pattern of Cytl1 and its chondrogenic effects observed in vitro suggest a possible role of Cytl1 in cartilage and bone development and/or maintenance of cartilage homeostasis. To investigate the in vivo role of Cytl1, we generated Cytl1 knock-out (Cytl1−/−) mice. We report here that deletion of the Cytl1 gene did not affect cartilage or bone development, despite the in vitro chondrogenic effects of Cytl1. However, Cytl1−/− mice were more sensitive to OA cartilage destruction, suggesting a role in the maintenance of cartilage homeostasis.
EXPERIMENTAL PROCEDURES
Animals
Male mice (C57BL/6, Cytl1−/−, STR/ort, and CBA/CaCrl) were used for cartilage and bone development and experimental OA studies. Cytl1−/− mice were generated as described below. STR/ort and CBA/CaCrl mice were obtained from Harlan Laboratories, Inc. (Borchen, Germany). Animals were maintained under pathogen-free conditions. All experiments were approved by the Gwangju Institute of Science and Technology Animal Care and Use Committee.
Generation of Cytl1−/− Mice
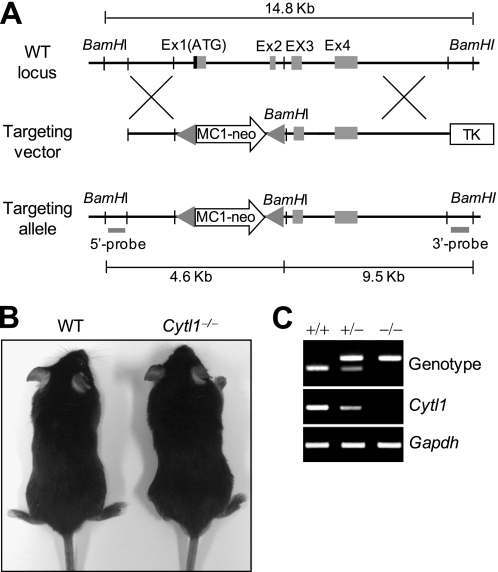
A targeting vector for deleting a segment containing a sequence of the Cytl1 gene from exon 1 to exon 2 (∼2.1 kb) was constructed using a 5′ short arm fragment (2.7 kb) and 3′ long arm fragment (7.3 kb) ligated into the pOsdupdel vector (Open Biosystems, Huntsville, AL). Short arm and long arm fragments were amplified by polymerase chain reaction (PCR) from genomic DNA obtained from 129/SvJ mouse J1 embryonic stem cells. The 2.7-kb NheI-XhoI fragment (5′ short arm) was amplified using forward (5′-gctagcaaaggagccacagaaatat-3′) and reverse (5′-ctcgagtgggacactgaattcttt-3′) primers containing NheI and XhoI sites, respectively (underlined). The 7.3-kb BamHI-NotI fragment (3′ long arm) was amplified using forward (5′-ggatccagctcagtctctggccctct-3′) and reverse (5′-gcggccgccaccagtgaaaagaattaa-3′) primers containing BamHI and NotI sites, respectively (underlined). The targeting vector was designed to replace the ∼2.1 kb genomic fragment with a positive selection marker containing a polyoma enhancer/herpes simplex virus thymidine kinase promoter and neomycin resistance gene. A negative selection marker containing an HSV-1 promoter derived from the thymidine kinase gene was appended to the construct to select against nonhomologous recombination. The targeting vector was linearized with NotI and electroporated into 129/SvJ mouse J1 embryonic stem cells. Clones resistant to G418 and ganciclovir were selected, and homologous recombination was confirmed by Southern blotting. The clones containing the targeted mutation were injected into C57BL/6 blastocysts, which were subsequently transferred into pseudopregnant foster mothers. The resulting male chimeric mice were bred to C57BL/6 females to obtain heterozygous Cytl1+/− mice. Heterozygous mice were crossed to generate homozygous Cytl1−/− mice. For Southern blot analysis, mouse genomic DNA obtained from 129/SvJ mouse J1 embryonic stem cells was digested with BamHI and hybridized with a 314-bp probe amplified from genomic DNA by PCR using the primers 5′-cctctacactctccacattcc-3′ (forward) and 5′-aagggcatcgaaggagtcctt-3′ (reverse). The probe detects 14.8-kb and 4.6-kb DNA fragment in WT and Cytl1−/− mice, respectively. Genotypes of WT and Cytl1−/− mice were determined by RT-PCR analysis from tail genomic DNA. PCR primers and conditions are summarized in Table 1.
TABLE 1.
PCR primers and conditions
| Gene | Strand | Primer sequences | Size (bp) | ATa | Origin | Detection |
|---|---|---|---|---|---|---|
| bp | °C | |||||
| Cytl1 | Sb | 5′-AGATCACCCGCGACTTCAAC-3′ | 302 | 60 | Human | RT-PCR |
| Asc | 5′-TTAGCGCTGACGATCTGGC-3′ | |||||
| S | 5′-CCACCTGCTACTCTCGGATG-3′ | 309 | 50 | Mouse | RT-PCR | |
| As | 5′-CCTCGGGAATTGGGTCTTC-3′ | |||||
| S | 5′-GATTCCTGTGTGAGGTAC-3′ | 342 | 60 | Mouse | Genotyping (WT) | |
| As | 5′-CAGAAGGAGTTCATGATG-3′ | |||||
| S | 5′-GAGGATCTCGTCGTGAC-3′ | 615 | 60 | Mouse | Genotyping (KO) | |
| As | 5′-CAGAAGGAGTTCATGATG-3′ | |||||
| Col2a1 | S | 5′-GGGTCTCCTGCCTCCTCCTGCTC-3′ | 204 | 62 | Mouse | RT-PCR |
| As | 5′-TCCTTTCTGCCCCTTTGGCCCTAATTTTCGGG-3′ | |||||
| Acan | S | 5′-GAAGACGACATCACCATCCAG-3′ | 581 | 56 | Mouse | RT-PCR |
| As | 5′-CTGTCTTTGTCACCCACACATG-3′ | |||||
| Lect1 | S | 5′-ACAGTGACCAAGCAGAGCATC-3′ | 504 | 62 | Mouse | RT-PCR |
| As | 5′-GCGGCATCCTTGGTAATTG-3′ | |||||
| Sox9 | S | 5′-GCGCGTGCAGCACAAGAAGGACCACCCGGATTACAAGTAC-3′ | 385 | 50 | Mouse | RT-PCR |
| As | 5′-CCTCGGGAATTGGGTCTTC-3′ | |||||
| Gapdh | S | 5′-CGTCTTCACCACCATGGAGA-3v | 300 | 62 | Human | RT-PCR |
| As | 5′-CGGCCATCACGCCACAGTTT-3′ | |||||
| S | 5′-TCACTGCCACCCAGAAGAC-3′ | 450 | 60 | Mouse | RT-PCR | |
| As | 5′-TGTAGGCCATGAGGTCCAC-3′ |
a AT, annealing temperature.
b S, sense primer.
c As, antisense primer.
Primary Culture of Chondrocytes and Mesenchymal Cells
Micromass cultures of mesenchymal cells were performed as described previously (8, 12). Briefly, mesenchymal cells obtained from E11.5 embryos of WT and Cytl1−/− mice were digested with 1% trypsin and 0.2% type II collagenase (381 units/mg solid; Sigma) for 10 min. A total of 2 × 107 cells were suspended in 1 ml of Dulbecco's modified Eagle's medium (DMEM)/F-12 medium (2:3; Invitrogen) containing 0.1% or 10% (v/v) fetal bovine serum. The cells were spotted as 15 μl drops on culture dishes and maintained for 5 days to induce chondrogenesis. Chondrogenesis was determined by examining the expression of type II collagen (Col2a1) by reverse transcription-PCR (RT-PCR) and by detecting accumulation of sulfate proteoglycans via Alcian blue staining. Rib chondrocytes were prepared from 2-day-old newborn wild-type (WT) and Cytl1−/− mice (8). The cartilage rib cages were preincubated for 1 h at 37 °C with 0.2% trypsin and 0.2% collagenase type II in DMEM, and rinsed with PBS. The tissues were digested with 0.2% type II collagenase in DMEM for 90 min. The cells were maintained in DMEM containing 10% (v/v) fetal bovine serum, 50 μg/ml streptomycin, and 50 units/ml penicillin, and were plated on culture dishes at a density of 5.0 × 104 cells/cm2.
Sox9 Reporter Gene Assay
Sox9 reporter gene activity in chondrifying mesenchymal cells was determined as described previously (8). Briefly, a reporter gene containing the 48 bp Sox9 binding site in the first intron of Col2a1 was inserted into the pGL3-promoter luciferase reporter vector (Promega, Madison, WI). pGL3-control or Sox9 reporter gene (1 μg) was transfected into isolated mesenchymal cells 6 h after spotting using the Lipofectamine 2000 reagent (Invitrogen). The cells were co-transfected with pCMV-β-galactosidase expression vector (0.2 μg) as an internal control to ensure transfection efficiency. The cells were maintained as micromass culture for 4 days in complete medium. Luciferase activity was normalized against β-galactosidase activity.
OA Cartilage and Experimental OA
Human normal cartilage was obtained from individuals undergoing arthroscopic meniscectomy or meniscal repair. Human OA knee joint cartilage was obtained from individuals (age 51–72 years) undergoing arthroplasty (13, 14). The Review Board of the Wonkwang University Hospital approved the use of these materials, and all individuals provided full written informed consent before the procedure. The International Cartilage Repair Society (ICRS) grading system was used to score cartilage destruction (14, 15). The classification ranges from healthy cartilage (grade 0) to the absence of cartilage with exposed subchondral bone (grade 4). The score criteria are the quantity and depth of lesions, either visually inspected by arthroscopy or non-invasively by magnetic resonance imaging (15). Spontaneous OA in STR/ort mice was examined at 28 weeks of age and compared with gender-matched CBA control mice (14, 16). Experimental OA was induced by destabilization of the medial meniscus (DMM) and a sham operation was used as a control, as described previously (17). Cartilage destruction was examined by Safranin-O staining and scored by Mankin's method (14, 18). This score is the sum of cartilage structure (0–6), cellularity (0–3), and Safranin-O staining (0–4). For histological analyses, human normal and OA cartilage were frozen, sectioned (6 μm thick), and fixed in paraformaldehyde. Sulfate proteoglycan was detected by Alcian blue staining. Control and OA cartilage tissues from mice were fixed in paraformaldehyde, decalcified, embedded in paraffin, sectioned (6 μm), and stained with Safranin-O, as described previously (14). Col2a1 and Prg4 (lubricin) proteins were detected by a standard immunohistochemical staining method using mouse anti-chicken Col2a1 antibody (Millipore) and rabbit anti-human Prg4 antibody (Abcam, Cambridge, UK).
Histological Analysis of Cartilage and Skeletons
Whole embryos and postnatal mice were skinned, eviscerated, and fixed with 95% ethanol for 4 days and treated with acetone for 3 days. Cartilage was stained with Alcian blue (1 volume of 0.3% Alcian blue 8GX in 70% ethanol, 1 volume of 100% acetic acid, and 18 volumes of 100% ethanol), as described previously (19). Skeletal staining was performed for 10 days with freshly prepared staining solution (1 volume of 0.1% Alizarin red S in 95% ethanol, 1 volume of 0.3% Alcian blue 8GX in 70% ethanol, 1 volume of 100% acetic acid, and 17 volumes of 100% ethanol). The samples were sequentially destained with 1% KOH for up to 48 h, and 20% glycerol containing 1% KOH for 7 days. Humerus, ulna, femur, and tibia longitudinal lengths were measured with the microscope image-analysis program, AxioVision (Carl Zeiss), using Alcian blue- and Alizarin red-stained samples. Histological analyses of cartilage and bones were performed as described previously (14, 19). Briefly, limbs of embryos and postnatal bones were fixed in 4% paraformaldehyde, and the tissues were embedded in paraffin. For postnatal tissue, bones were decalcified in 0.5 m EDTA (pH 7.4) for 7 days at 4 °C before paraffin embedding. The paraffin blocks were sectioned (5 μm), deparaffinized in xylene, and hydrated with a series of graded ethanol. The deparaffinized sections were stained with 1% Alcian blue in 0.1 n HCl for 10 min and then washed in 0.1 n HCl for 5 min. Serial sections were stained with 2% Alizarin red in water for 5 min, washed in acetone, and cleared in xylene. Von Kossa staining was performed by incubating sections with 5% silver nitrate for 1 h and then with 5% sodium thiosulfate for 3 min, followed by washing in water and clearing in xylene. For hematoxylin and eosin (H&E) staining, the sections were incubated in Harry's hematoxylin for 1 min, washed in running tap water, soaked in 95% ethanol for 1 min, and then in eosin for 10 s. After hydrating in graded ethanol, the samples were cleared with xylene (14).
Transmission Electron Microscopy of Mouse Articular Cartilage
For transmission electron microscopy (TEM) analyses, articular cartilage from 4-week-old WT and Cytl1−/− mice was gently removed using a scalpel and subjected to a double fixation procedure. Cartilage tissue was first fixed with Karnowsky fixative for 4 h at 4 °C, washed in phosphate buffer (pH 7.4), and post-fixed in 1% osmium tetroxide in the same buffer for 1 h. The specimens were dehydrated through a graded ethanol series, exchanged with propylene oxide, and embedded in a mixture of Epon 812 and Araldite (Polysciences Inc., Washington, PA). The embedded tissues were thin-sectioned on a Leica Em UC6 Ultramicrotome, stained with uranyl and lead citrate, and viewed in a FEL Tecnai G2 electron microscope (FEI, Eindhoven, Netherlands) operated at 80 kV.
Conventional RT-PCR and Quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from cartilage tissue and primary cultured chondrocytes. The RNA was reverse-transcribed, and the resulting cDNA was amplified by PCR using the primers and experimental conditions summarized in Table 1. Transcript levels were quantified by qRT-PCR as described previously (20, 21).
Phenotypic Analysis of Heart and Lung
Histological analyses of heart were performed as described previously (22). Briefly, hearts from 2-week-old mice were fixed in 4% paraformaldehyde and paraffin embedded. Sections (6 μm) were stained with H&E solution, as described above. Lung sections were obtained from 2-week-old mice, as described previously (23). After embedding in paraffin, lung paraffin blocks were sectioned (6 μm), deparaffinized in xylene, and stained with H&E.
Statistical Analysis
Statistical significance was evaluated using Student's t test. Significance was accepted at the 0.05 level of probability (p < 0.05).
RESULTS
Normal Chondrogenesis and Cartilage Development in Cytl1−/− Mice
We have previously shown that chondrocyte-specific Cytl1, as a novel autocrine factor, regulates in vitro chondrogenesis during micromass culture of mouse mesenchymal cells without directly affecting hypertrophic maturation of chondrocytes (8). To elucidate the in vivo function of Cytl1 in cartilage and bone development and maintenance of cartilage homeostasis, we generated Cytl1−/− mice by deleting a segment (∼2.1 kb) of the Cytl1 gene from exon 1 to exon 2, as described under “Experimental Procedures” (Fig. 1A). Cytl1−/− mice were viable and showed normal postnatal growth (Fig. 1B). Genotypes of offspring and knock-out of Cytl1 were confirmed by RT-PCR analysis (Fig. 1C).
FIGURE 1.
Generation of Cytl1−/− mice. A, vector construct used for the generation of Cytl1−/− mice. B, images of 10-week-old WT and Cytl1−/− mice. C, genotypes of WT (+/+), heterozygous (+/−), and homozygous (−/−) mice were determined by RT-PCR from tail genomic DNA. Cytl1 expression levels were determined by RT-PCR analysis from rib chondrocytes.
We first examined phenotypic changes in chondrogenesis of mesenchymal cells and cartilage development in Cytl1−/− mice. The role of Cytl1 in chondrogenesis was examined by micromass culture of mesenchymal cells isolated from WT and Cytl1−/− E11.5 embryos. Similar patterns of chondrogenesis were observed in WT and Cytl1−/− mesenchymal cells, as determined by Alcian blue staining of sulfate proteoglycan (Fig. 2A). Additionally, expression patterns of the chondrocyte markers Col2a1, aggrecan (Acan), chondromodulin 1 (Lect1), Sox9, and Igf1, and Sox9 transcriptional activity were similar between WT and Cytl1−/− cells (Fig. 2B). Thus, deletion of the Cytl1 gene did not affect chondrogenesis of mesenchymal cells in vitro. Chondrogenesis and cartilage development in vivo was examined by Alcian blue staining of limb buds and whole embryos. Patterns of cartilage development in limb buds of E12.5 and E13.5 embryos and whole embryo at E14.5 were similar in WT and Cytl1−/− mice (Fig. 2C). Collectively, these results indicate that the patterns of in vitro chondrogenesis and in vivo cartilage development are normal in Cytl1−/− mice.
FIGURE 2.
Normal chondrogenesis and cartilage and bone development in Cytl1−/− mice. A, mesenchymal cells isolated from the limb buds of wild-type (WT) and Cytl1−/− (KO) mice were maintained as micromass culture for 1–5 days to induce chondrogenesis. Left: accumulation of sulfate proteoglycan in the presence of 10% or 0.1% FBS at day 5 determined by Alcian blue staining. Right: Alcian blue-stained cells were extracted and quantified by measuring absorbance. B, left: expression levels of chondrocyte markers Col2a1, Acan, Lect1, Sox9, and Igf1 were determined by RT-PCR in chondrifying mesenchymal cells isolated from WT and Cytl1−/− (KO) mice. Right: Sox9 transcriptional activity was determined by the reporter gene assay in chondrifying mesenchymal cells isolated from WT and Cytl1−/− (KO) mice. Values represent means ± S.E. (n = 3). C, Alcian blue staining of limb buds of WT and Cytl1−/− (KO) E12.5 (left) and E13.5 (middle) embryos. Cartilage structure of WT and Cytl1−/− (KO) E14.5 whole embryos detected by Alcian blue staining (right). Scale bars: 400 μm (left and middle) and 3 mm (right).
Normal Endochondral Ossification and Bone Development in Cytl1−/− Mice
Because cartilage serves as a template for long bone development during endochondral ossification (2, 4), we next examined endochondral ossification and long bone developmental patterns in Cytl1−/− mice. At E18.5, the skeletal structure of whole embryos (Fig. 3A), autopod developmental patterns (Fig. 3B), and lengths of long bones (i.e. ulna, humerus, tibia, and femur; Fig. 3C) were similar in Cytl1−/− and WT littermates. The lengths of mineralized zones, determined by Alcian blue or von Kossa staining, of radius and ulna at E16.5, and lengths of hypertrophic zones were also similar in WT and Cytl1−/− mice (Fig. 3D). Additionally, the overall patterns of growth plate, zones of proliferating and hypertrophic chondrocytes, and secondary ossification center were also similar in 2-week-old WT and Cytl1−/− mice (Fig. 3E). Taken together, our results indicate that endochondral ossification and long bone development patterns are normal in Cytl1−/− mice.
FIGURE 3.
Normal endochondral ossification in Cytl1−/− mice. A, WT and Cytl1−/− (KO) E18.5 whole embryos stained with Alcian blue and Alizarin red to detect skeletal structure. Scale bar: 3 mm. B, Alizarin red and Alcian blue staining of autopods of WT and Cytl1−/− (KO) E18.5 embryos. Scale bar: 1 mm. C, left: humerus and ulna (forelimb) and femur and tibia (hind limb) from Cytl1−/− (KO) and WT E18.5 embryos stained with Alcian blue and Alizarin red. Right: bone length measured using a microscope image analysis program. Values represent means ± S.E. (n > 10). Scale bar: 1 mm. D, left: radius and ulna of WT and Cytl1−/− (KO) E16.5 embryos stained with Alcian blue or von Kossa to detect the mineralized zone. Scale bar: 150 μm. Right: enlarged images of the hypertrophic zone. Scale bar: 300 μm. E, left: Alcian blue and H&E staining of growth plate (GP) and secondary ossification center (OC) in the metatarsal bone of 2-week-old WT and Cytl1−/− (KO) mice. Right: enlarged images of the growth plate showing proliferating chondrocytes (PC), hypertrophic chondrocytes (HC), and trabecular bone (TB). Scale bar: 400 μm.
Enhanced OA Cartilage Destruction Caused by DMM Surgery in Cytl1−/− Mice
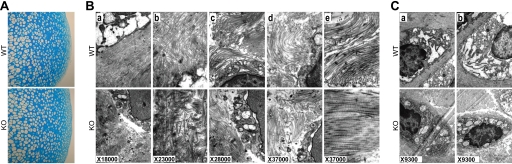
Next, we examined whether deletion of the Cytl1 gene affected ultrastructure and maintenance of articular cartilage. Alcian blue staining of articular cartilage from 4-week-old WT and Cytl1−/− mice showed no apparent differences in the overall structure of cartilage (Fig. 4A). Ultrastructural features of articular cartilage, such as ECM organization and morphology of chondrocytes, were examined by TEM using articular cartilage explants from 4-week-old WT and Cytl1−/− mice. The density of the matrix and the thickness of the collagen fibril network in articular cartilage were similar in WT and Cytl1−/− mice (Fig. 4B). Additionally, there were no evident differences in chondrocyte morphology between WT and Cytl1−/− mice (Fig. 4C).
FIGURE 4.
Ultrastructure of articular cartilage in WT and Cytl1−/− mice. A, Alcian blue staining of mouse articular cartilage from 4-week-old WT and Cytl1−/− (KO) mice. B and C, TEM images of cartilage matrix (B) and chondrocytes (C) in articular cartilage of 4-week-old WT and Cytl1−/− (KO) mice.
The role of Cytl1 in OA cartilage destruction was examined using an experimental OA model. We induced OA cartilage destruction in both WT and Cytl1−/− mice by DMM surgery. DMM surgical instability in the mice was caused by a 3 mm longitudinal incision over the distal patella to proximal tibial plateau. This model induced OA with a high reproducibility and slow progression of the disease (17). Mice were sacrificed 7 weeks after surgery, and cartilage destruction was examined by Safranin-O staining and scored using a Mankin's methods (18). DMM surgery caused OA cartilage destruction in both WT and Cytl1−/−, leading to the loss of Safranin-O stained proteoglycans in comparison to their respective sham controls. However, the degree of cartilage destruction was more remarkable in Cytl1−/− than in WT. We consistently observed a marked reduction in cartilage thickness and irregular cartilage surface in Cytl1−/−, both of which corresponded to the characteristics of more advanced OA (Fig. 5A). Indeed, scoring of cartilage destruction by Mankin's method indicated statistically significant augmentation of cartilage destruction in Cytl1−/− mice (Fig. 5B).
FIGURE 5.
Enhanced DMM-induced OA cartilage destruction in Cytl1−/− mice. A, WT and Cytl1−/− (KO) mice were subjected to a sham operation (control) or DMM surgery to induce OA cartilage destruction. Articular cartilage was stained with Safranin-O. The images at right are enlarged versions of the marked region in the left images. Scale bar: 400 μm. B, Mankin score in WT and Cytl1−/− (KO) mice after sham operations and DMM surgery. Values represent means ± S.E. (n = 16). C, immunohistochemical staining of Col2a1 and Prg4 in joint articular cartilage of WT and Cytl1−/− (KO) mice after sham operation and DMM surgery. Scale bar: 400 μm. D, Safranin-O staining (left) and Makin score (right) of 10-month-old WT and Cytl1−/− (KO) mice joint articular cartilage. Scale bar: 400 μm. Values represent means ± S.E. (n = 6). *, p < 0.05.
In addition, immunostaining of Col2a1 and Prg4, whose down-regulations are associated with OA pathogenesis, indicated that DMM caused reduction in the expression levels of Col2a1 and Prg4 in degenerating cartilage, and this effect was more evident in Cytl1−/− mice (Fig. 5C). Finally, we examined whether Cytl1−/− mice exhibit spontaneous OA with age. Safranin-O staining and scoring by Mankin's method indicated enhanced cartilage destruction in 10-month-old Cytl1−/− mice compared with corresponding WT mice (Fig. 5D). However, the degree of spontaneous cartilage destruction was relatively weak at this age when compared with OA cartilage destruction by DMM surgery (Fig. 5, B and D).
To confirm the association between Cytl1 expression and OA cartilage destruction, we examined CYTL1 mRNA levels in OA cartilage. CYTL1 mRNA levels in OA cartilage in DMM model animals were markedly decreased compared with those in sham-operated controls (Fig. 6A). To extend these observations, we also examined Cytl1 expression in the cartilage of STR/ort mice, which are genetically predisposed to develop OA-like lesions in the medial tibial cartilage. The majority (>85%) of male STR/ort mice have been shown to display signs of cartilage destruction at 6 months of age (16). Compared with gender-matched CBA control mice, STR/ort mice showed severe cartilage destruction and down-regulation of Cytl1 expression (Fig. 6B). Finally, we examined CYTL1 expression in normal human cartilage obtained from individuals experiencing meniscus problems during arthroscopic meniscectomy or meniscal repair and OA-affected human cartilage obtained from individuals undergoing arthroplasty. CYTL1 mRNA levels were markedly reduced in human OA cartilage compared with those in undamaged normal cartilage (Fig. 6C). Cartilage damage in OA-affected individuals was confirmed by scoring of Alcian blue-stained samples according to the ICRS grading system (14, 15). Thus, all examined OA cartilage showed marked down-regulation of Cytl1, and deletion of Cytl1 enhanced OA cartilage destruction, suggesting the involvement of Cytl1 in the maintenance of cartilage homeostasis. Finally, we examined Cytl1 mRNA levels in OA cartilage at different time points after DMM surgery. Notably, decrease in Cytl1 expression levels, as determined by qRT-PCR analysis, preceded OA cartilage destruction, which was determined by Mankin score. The decrease in Cytl1 expression was detected as early as 1 day after DMM surgery, whereas cartilage destruction occurred 2 weeks after DMM surgery (Fig. 6D), suggesting a possible protective role of Cytl1 in OA cartilage destruction.
FIGURE 6.
Down-regulation of Cytl1 expression in OA cartilage. A, WT mice were subjected to a sham operation (left knee joint) or DMM surgery (right knee joint) to induce OA cartilage destruction. Expression levels of Cytl1 were quantified by qRT-PCR 7 weeks after surgery. Values represent means ± S.E. (n = 6). **, p < 0.005. B, cartilage sections from STR/ort and CBA control mice. Left: Safranin-O staining; middle: Mankin score (n = 5); right: Cytl1 mRNA levels quantified by qRT-PCR. Values represent means ± S.E. n = 6). **, p < 0.005. Scale bar: 400 μm. C, undamaged and damaged human cartilage section. Left: Alcian blue staining of human cartilage sections from an OA-affected damaged region and an undamaged region. Middle: ICRS grade of normal cartilage and OA cartilage (n = 6). Right: Cytl1 mRNA levels in normal and OA cartilage quantified by qRT-PCR. Values represent means ± S.E. (n = 6). **, p < 0.005. Scale bar: 400 μm. D, WT mice were subjected to a sham operation (left knee joint) or DMM surgery (right knee joint) to induce OA cartilage destruction. At indicated time points, expression levels of Cytl1 and Mankin scores were determined. Cytl1 mRNA levels are relative mean values in right knee joints cartilage (DMM surgery) against sham operation (left knee joints cartilage). Values represent means ± S.E. (n = 7). *, p < 0.05 and **, p < 0.005 against sham operation in each indicated time point.
Normal Development of Lung and Heart in Cytl1−/− Mice
We have previously shown that Cytl1 is also expressed in lung and heart, although at much lower levels than in cartilage (8). We therefore examined possible phenotypic changes in the lungs and hearts of Cytl1−/− mice (Fig. 7). The size and weight of lungs in 2-week- and 12-week-old mice were essentially the same in Cytl1−/− mice and WT littermates. Additionally, H&E staining of lung sections from Cytl1−/− mice showed an alveolar structure similar to that of WT littermates (Fig. 7A). Moreover, there were no apparent differences in the size or weight of hearts or histology of heart sections between WT and Cytl1−/− mice (Fig. 7B), indicating that lung and heart developmental patterns in Cytl1−/− mice are normal.
FIGURE 7.
Normal lung and heart phenotypes in Cytl1−/− mice. A, left: lung images and weights for 2-week-old WT and Cytl1−/− mice. Values represent means ± S.E. (n = 5). Scale bar: 0.5 mm. Right: H&E-stained lung sections from WT and Cytl1−/− mice. Scale bar: 100 μm. B, left: Heart images and weights for 2-week-old WT and Cytl1−/− mice. Values represent means ± S.E. (n = 5). Scale bar: 0.5 mm. Right: H&E-stained lung sections from WT and Cytl1−/− mice. Scale bar: 100 μm.
DISCUSSION
We demonstrated in this study that deletion of the Cytl1 gene in mice does not affect cartilage development, endochondral ossification, or long bone development, despite the in vitro chondrogenic effects of Cytl1. However, Cytl1 appears to be required for the maintenance of cartilage homeostasis. This conclusion is suggested by the decreased expression of Cytl1 in OA cartilage and the more severe OA cartilage destruction in Cytl1−/− mice compared with WT mice.
An examination of Cytl1 expression in a number of mouse tissues, including lung, heart, trachea, testis, eye, brain, kidney, muscle, spleen, and liver, has shown that Cytl1 is predominantly expressed in cartilage. Additionally, Cytl1 expression is detected only in primary cultured chondrocytes but not in HTB-94 chondrosarcoma or ATDC5 chondroprogenitor cells (8). Indeed, Cytl1 expression is specific to differentiated chondrocytes; its expression is not detected in undifferentiated mesenchymal cells, hypertrophic chondrocytes, or de-differentiated chondrocytes generated by serial subculture as a monolayer. Cytl1 expression in vivo is also chondrocyte-specific; it is detected only in Col2a1-expressing chondrocytes but not in other cell types (e.g. hypertrophic chondrocytes) in developing cartilage and bone (8). Therefore, we initially hypothesized that Cytl1 regulates chondrogenesis, maintenance of the differentiated phenotype of chondrocytes, and/or hypertrophic maturation of chondrocytes. Indeed, in vitro experiments using micromass culture of mouse mesenchymal cells demonstrated that Cytl1 has the capacity to induce chondrogenesis of mesenchymal cells by activating Sox9 transcriptional activity (8). However, in this previous study, we were not able to assess the effects of Cytl1 silencing because we were inexplicably unable to knock down Cytl1 expression with any of the various siRNAs and shRNAs tested.
In the current study, we examined the in vivo function of Cytl1 by generating Cytl1−/− mice. Unexpectedly, and contrary to the in vitro chondrogenic effects of Cytl1, we found that Cytl1−/− mice exhibited normal cartilage development and endochondral ossification. We also observed that chondrogenesis of mesenchymal cells induced by micromass culture was normal in mesenchymal cells isolated from Cytl1−/− mice. Therefore, although exogenous Cytl1 or ectopic expression Cytl1 causes in vitro chondrogenesis, Cytl1 does not appear to be essential for the induction of chondrogenesis. However, this interpretation should be considered with care. Chondrogenesis of mesenchymal cells by micromass culture is normally induced in the presence of 10% fetal calf serum. In our previous study, the chondrogenic effects of exogenous Cytl1 or ectopically expressed Cytl1 were not observed in 10% serum, but were evident under low serum (0.1%) conditions (8). In the current study, we observed no differences in chondrogenesis of mesenchymal cells isolated from WT and Cytl1−/− mice in the presence of 10% serum, whereas chondrogenesis was not observed in 0.1% serum (Fig. 2A). Thus, it is possible that certain factors present in serum may overcome the effects of Cytl1 deletion. Nevertheless, the fact that cartilage development in Cytl1−/− mice is normal indicates that Cytl1 is not essential for chondrogenesis or subsequent cartilage development in vivo. Consequently, endochondral ossification and long bone development are also not affected by Cytl1 deletion in mice. Thus, our current results demonstrate that Cytl1 does not have a critical role in chondrogenesis, cartilage development, or endochondral ossification during long bone development in mice.
Although Cytl1 is not involved in cartilage or bone development, we demonstrated in this study that it is associated with the maintenance of articular cartilage homeostasis. Expression of Cytl1 was markedly decreased in degenerating OA cartilage of humans and mice. Additionally, deletion of the Cytl1 gene enhanced OA cartilage destruction caused by DMM surgery. Although the stimulatory effects of Cytl1 deletion in OA cartilage destruction were not dramatic, they were statistically significant. Therefore, it is likely that the loss of Cytl1 function facilitates OA progression. Indeed, the decrease in Cytl1 expression preceded OA cartilage destruction, suggesting that Cytl1 exerts protective effects against cartilage degeneration. The current study did not elucidate the stimulatory mechanisms underlying OA cartilage destruction in Cytl1−/− mice. In conditions that predispose toward OA, such as aging and genetic defects, OA cartilage destruction is caused by mechanical stimuli, which activate intracellular signaling pathways in chondrocytes that lead to a variety of effects, including the production of pro-inflammatory cytokines, inflammation, degradation of the ECM by matrix metalloproteinases and ADAMTS (a disintegrin and metalloproteinases with thrombospondin motifs), and cessation of ECM synthesis via dedifferentiation and apoptosis of chondrocytes (5–7). Because ultrastructural features of articular cartilage such as ECM organization and chondrocyte morphology are normal in Cytl1−/− mice, it is likely that other factors, such as biochemical pathways, are altered in Cytl1−/− mice during OA pathogenesis.
This work was supported by a grant from the Korea Healthcare Technology R&D project (A101793).
- ECM
- extracellular matrix
- Cytl1
- cytokine-like 1
- DMM
- destabilization of the medial meniscus
- H&E
- hematoxylin and eosin
- IL1β
- interleukin 1β
- KO mouse
- knock-out mouse
- OA
- osteoarthritis
- qRT-PCR
- quantitative RT-PCR
- RT-PCR
- reverse transcription-polymerase chain reaction
- TEM
- transmission electron microscope.
REFERENCES
- 1. Goldring M. B., Tsuchimochi K., Ijiri K. (2006) J. Cell. Biochem. 97, 33–44 [DOI] [PubMed] [Google Scholar]
- 2. Kronenberg H. M. (2003) Nature. 423, 332–336 [DOI] [PubMed] [Google Scholar]
- 3. Mariani F. V., Martin G. R. (2003) Nature. 423, 319–325 [DOI] [PubMed] [Google Scholar]
- 4. Provot S., Schipani E. (2005) Biochem. Biophys. Res. Commun. 328, 658–665 [DOI] [PubMed] [Google Scholar]
- 5. Goldring M. B., Goldring S. R. (2007) J. Cell. Physiol. 213, 626–634 [DOI] [PubMed] [Google Scholar]
- 6. Martel-Pelletier J., Boileau C., Pelletier J. P., Roughley P. J. (2008) Best Pract. Res. Clin. Rheumatol. 22, 351–384 [DOI] [PubMed] [Google Scholar]
- 7. Hashimoto M., Nakasa T., Hikata T., Asahara H. (2008) Med. Res. Rev. 28, 464–481 [DOI] [PubMed] [Google Scholar]
- 8. Kim J. S., Ryoo Z. Y., Chun J. S. (2007) J. Biol. Chem. 282, 29359–29367 [DOI] [PubMed] [Google Scholar]
- 9. Liu X., Rapp N., Deans R., Cheng L. (2000) Genomics 65, 283–292 [DOI] [PubMed] [Google Scholar]
- 10. Ficker M., Powles N., Warr N., Pirvola U., Maconochie M. (2004) Dev. Biol. 268, 7–23 [DOI] [PubMed] [Google Scholar]
- 11. Hermansson M., Sawaji Y., Bolton M., Alexander S., Wallace A., Begum S., Wait R., Saklatvala J. (2004) J. Biol. Chem. 279, 43514–43521 [DOI] [PubMed] [Google Scholar]
- 12. Oh C. D., Chun J. S. (2003) J. Biol. Chem. 278, 36563–36571 [DOI] [PubMed] [Google Scholar]
- 13. Hwang S. G., Ryu J. H., Kim I. C., Jho E. H., Jung H. C., Kim K., Kim S. J., Chun J. S. (2004) J. Biol. Chem. 279, 26597–26604 [DOI] [PubMed] [Google Scholar]
- 14. Yang S., Kim J., Ryu J. H., Oh H., Chun C. H., Kim B. J., Min B. H., Chun J. S. (2010) Nat. Med. 16, 687–693 [DOI] [PubMed] [Google Scholar]
- 15. Kleemann R. U., Krocker D., Cedraro A., Tuischer J., Duda G. N. (2005) Osteoarthritis Cartilage 13, 958–963 [DOI] [PubMed] [Google Scholar]
- 16. Mason R. M., Chambers M. G., Flannelly J., Gaffen J. D., Dudhia J., Bayliss M. T. (2001) Osteoarthritis Cartilage 9, 85–91 [DOI] [PubMed] [Google Scholar]
- 17. Glasson S. S., Blanchet T. J., Morris E. A. (2007) Osteoarthritis Cartilage 15, 1061–1069 [DOI] [PubMed] [Google Scholar]
- 18. Mankin H. J., Dorfman H., Lippiello L., Zarins A. (1971) J. Bone Joint Surg. Am. 53, 523–537 [PubMed] [Google Scholar]
- 19. Huh Y. H., Ryu J. H., Shin S., Lee D. U., Yang S., Oh K. S., Chun C. H., Choi J. K., Song W. K., Chun J. S. (2009) Gene 448, 7–15 [DOI] [PubMed] [Google Scholar]
- 20. Ryu J. H., Chun J. S. (2006) J. Biol. Chem. 281, 22039–22047 [DOI] [PubMed] [Google Scholar]
- 21. Huh Y. H., Ryu J. H., Chun J. S. (2007) J. Biol. Chem. 282, 17123–17131 [DOI] [PubMed] [Google Scholar]
- 22. Scott N. J., Ellmers L. J., Lainchbury J. G., Maeda N., Smithies O., Richards A. M., Cameron V. A. (2009) Biochim. Biophys. Acta 1792, 1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kurotani R., Tomita T., Yang Q., Carlson B. A., Chen C., Kimura S. (2008) Am. J. Resp. Crit. Care Med. 178, 389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]